Abstract
Background
Copy number variation (CNV) is a complex genomic rearrangement that has been linked to a large number of human diseases. Chromosome 15q13 microduplication is a rare form of CNV, which has been proved to be associated with multiple human disorders; however, the association between chromosome 15q13 microduplication and cardiac disorders has not been fully understood.
Case presentation
A fetus with fetal cardiac developmental defects was detected by Color Doppler ultrasound imaging; however, further chromosomal G-banding revealed no abnormal karyotype. Then, chromosomal microarray analysis (CMA) was performed and revealed a 1.8 Mb-duplication of the chromosome 15q13.2q13.3 region containing 7 genes (TRPM1, KLF13, OTUD7A, CHRNA7, FAN1, MIR211 and RAHGAP11A). Cardiac ultrasound follow-up displayed significant enlargement of the space-occupying lesion in the fetal heart with extension of the gestational age, and the space-occupying lesion was finally pathologically diagnosed as cardiac rhabdomyoma. Next-generation sequencing revealed no mutations in the TSC1 or TSC2 gene in the fetus, the mother or the father.
Conclusions
This is the first report to demonstrate the potential association between chromosome 15q13 microduplication and fetal cardiac rhabdomyoma. It is recommended that CMA be employed in fetuses with abnormal cardiac development diagnosed by routine cardiac color Doppler ultrasound imaging for early detection of congenital genetic abnormality, which may provide valuable information for prenatal diagnostic consultation and the decision on pregnancy termination.
Keywords: Chromosome 15q13 microduplication, Cardiac rhabdomyoma, Copy number variation, Chromosomal microarray analysis
Background
Copy number variation (CNV), a type of genomic structural variation, has been defined as a DNA segment of one kilobase (kb) or larger that is present at a variable copy number as compared to a reference genome [1]. As a complex genomic rearrangement [2], CNV has been linked to a large number of human diseases [3–5]. Chromosome 15q13 microduplication, a rare form of CNV, has been associated with cognitive impairment, autism, seizures, and attention-deficit hyperactivity disorder, and oral clefts [6–14], and the common recurrent BP4-BP5 duplication in the chromosomal region 15q13.3 was detected in a 14-year-old male with Tetralogy of Fallot and developmental delay [15]. However, the association between chromosome 15q13 microduplication and cardiac disorders has not been fully understood. Here, we report a fetus with cardiac rhabdomyoma who had a 1.8 Mb-duplication of the chromosome 15q13.2q13.3 region as detected by chromosomal microarray analysis (CMA).
Case presentation
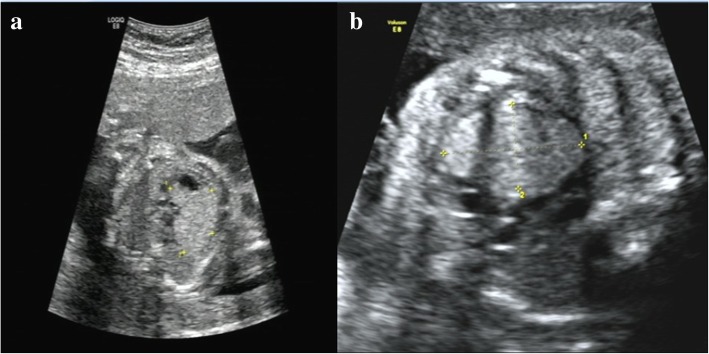
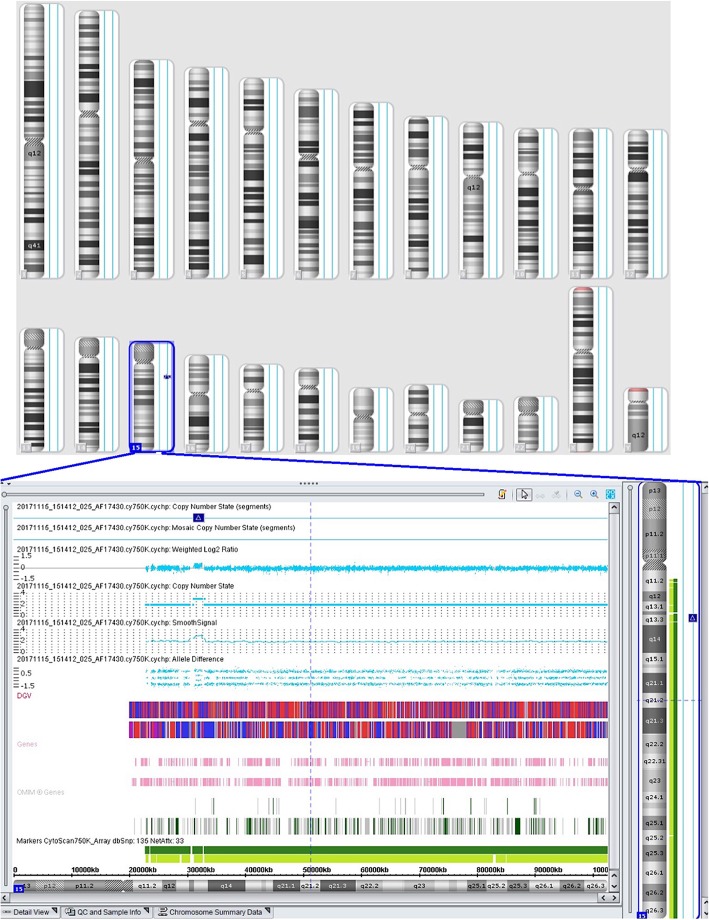
A 33-year-old pregnant woman, G1P0, at a gestational age of 23 + 4 weeks was referred to our hospital on November 3, 2017. Color Doppler ultrasound imaging showed a hyperechogenic mass in the fetal left ventricle, measuring 1.8 cm × 1.57 cm, broadening of the left lateral ventricle (1.11 cm) and a strong dot-like echo in the left ventricle (Fig. 1a), and cardiac rhabdomyoma was suspected. On November 7, 2017, approximately 30 mL of amniotic fluid was collected by ultrasound-guided transabdominal puncture for conventional chromosomal G-banding karyotype analysis and CMA with the BioChip Detection System of Affymetrix GeneChip following the manufacturer’s instructions (Zhejiang Biosan Biochemical Technologies Co., Ltd.; Hangzhou, China). G-banding analysis revealed a 46, XY karyotype, with no abnormal karyotype detected, and CMA detected a 1.8 Mb-duplication of the chromosome 15q13.2q13.3 region (arr [hg19] 15q13.2q13.3(31,073,668-32,920,694)× 3) containing 7 genes (TRPM1, KLF13, OTUD7A, CHRNA7, FAN1, MIR211 and RAHGAP11A), which occurred in the region between BP4-BP5 on chromosome 15q13.3 (Fig. 2).
Fig. 1.
Color Doppler ultrasound imaging. a on November 3, 2017 (23 + 4 weeks of gestation), color Doppler ultrasound imaging showed a hyperechogenic mass in the fetal left ventricle, measuring 1.8 cm × 1.57 cm, broadening of the left lateral ventricle (1.11 cm) and a strong dot-like echo in the left ventricle; b on December 7, 2017, color Doppler ultrasound reexaminations displayed multiple strong echoes in the fetal left ventricle (measuring 3.3 cm × 2.03 cm), compression of the left ventricular outflow tract, obvious enlargement of the tumor, and a 0.92 cm internal diameter of posterior horn of the left lateral ventricle
Fig. 2.
Chromosomal microarray analysis detects a 1.8 Mb-duplication of the chromosome 15q13.2q13.3 region containing 7 genes, which occurs in the region between BP4-BP5 on chromosome 15q13.3
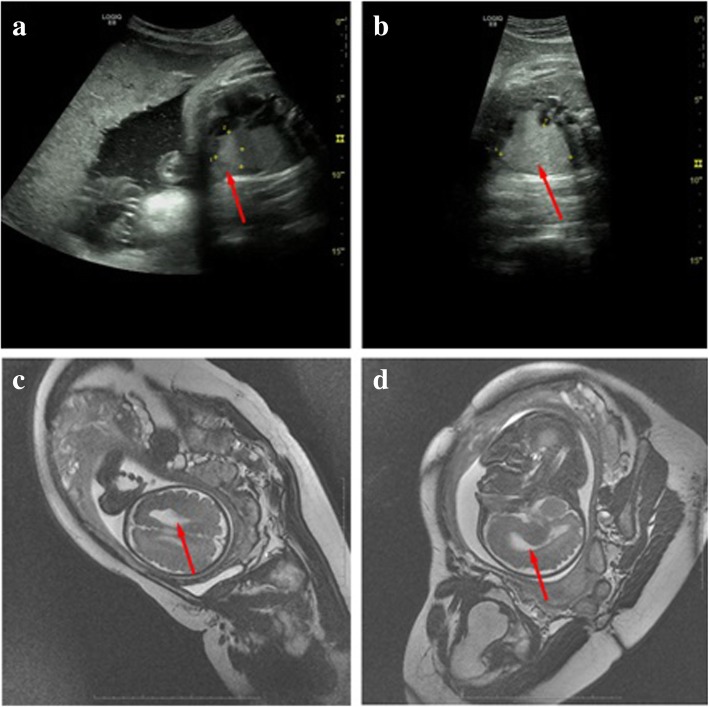
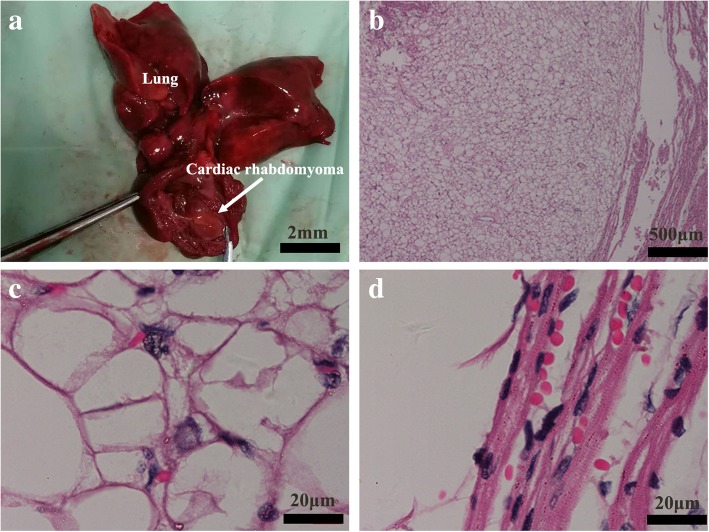
At a gestational age of 28 + 3 weeks (December 7, 2017), color Doppler ultrasound reexaminations displayed multiple strong echoes in the fetal left ventricle (measuring 3.3 cm × 2.03 cm), compression of the left ventricular outflow tract, obvious enlargement of the tumor, and a 0.92 cm internal diameter of posterior horn of the left lateral ventricle (Fig. 1b). On January 15, 2018 (34 weeks of gestation), fetal brain magnetic resonance imaging (MRI) in Fujian Provincial Maternity and Children’s Hospital (Fuzhou, China) revealed abnormal morphology of the left frontal lobe with irregular extension of anterior horn of the left ventricle (Fig. 3c and d). Lissencephaly in the left frontal lobe and cortical dysplasia were therefore diagnosed. On January 25, 2018 (35 + 2 weeks of gestation), color Doppler ultrasound reexaminations of the fetal heart displayed multiple hyperechogenic masses (measuring 4.35 cm × 3.13 cm) in the fetal left ventricle, abnormal morphology of anterior horn of the left lateral ventricle with broadening of the internal diameter (2.02 cm) (Fig. 3a and b), indicating a high likelihood of cardiac rhabdomyoma. After careful consideration, the pregnant woman and her family decided to terminate pregnancy. Labor was induced and a 3 kg male infant was delivered. Anatomical findings showed a solid space-occupying lesion in the fetal left ventricle (approximately 4 cm × 3 cm) (Fig. 4a). Histology of the fetal heart specimen revealed typical spider-shaped cells, which were consisted of abundant glycogen-rich cytoplasm and cytofilaments that were extended and radiated to pericellular regions, and pathologic examinations confirmed that the solid space-occupying lesion in the fetal left ventricle was cardiac rhabdomyoma (Fig. 4b, c and d). Next-generation sequencing revealed no mutations in the tuberous sclerosis complex 1 (TSC1) gene or TSC2 gene in the fetus. On August 12, 2018, peripheral blood was sampled from the fetus’ parents, and subjected to CMA. Then, a 2.5 Mb-duplication of the chromosome 15q13.2q13.3 region (arr [hg19] 15q13.2q13.3(30,386,398-32,915,089)× 3) was detected in the woman, and no mutations were identified in the TSC1 or TSC2 gene. These testing suggested that the fetal chromosome with the microduplication was inherited from his mother.
Fig. 3.
Color Doppler ultrasound and fetal brain magnetic resonance imaging examinations. a and b On January 25, 2018 (35 + 2 weeks of gestation), color Doppler ultrasound reexaminations of the fetal heart display multiple hyperechogenic masses (measuring 4.35 cm × 3.13 cm) in the fetal left ventricle, abnormal morphology of anterior horn of the left lateral ventricle with broadening of the internal diameter (2.02 cm); c and d on January 15, 2018 (34 weeks of gestation), fetal brain magnetic resonance imaging reveals abnormal morphology of the left frontal lobe with irregular extension of anterior horn of the left ventricle
Fig. 4.
Anatomical and histological findings. a anatomical findings show a solid space-occupying lesion in the fetal left ventricle (approximately 4 cm × 3 cm); b, c and d histology of the fetal heart specimen reveals typical spider-shaped cells, which are consisted of abundant glycogen-rich cytoplasm and cytofilaments that are extended and radiated to pericellular regions, and pathologic examinations confirm that the solid space-occupying lesion in the fetal left ventricle is cardiac rhabdomyoma
The pregnant woman reported a regular menstrual cycle, a natural pregnancy, and smooth pregnancy, and she had no special discomfort, no smoking or alcohol consumption history, no family history of fetal anomaly, and no history of viral infections. In addition, she denied oral administration of special medications during the pregnancy. During the pregnancy, non-invasive prenatal testing (NIPT) revealed a low risk, and color Doppler ultrasound imaging of the woman heart displayed no remarkable abnormality of cardiac structure or functions.
This study was approved by the Ethics Review Committee of Fuzhou Municipal First Hospital Affiliated to Fujian Medical University (approval no. FZSY-201700132). Written informed consent was obtained from the fetus’s parents following a detailed description of the study purpose. All experimental procedures described in this study were in accordance with international and national laws, regulations and guidelines. The pregnant woman and her husband involved in this study agreed to publish related demographic and clinical features.
Discussion
CMA, also known as array comparative genomic hybridization or comparative genomic hybridization, is a high-resolutiongenome-wide screening testing to detect a majority of chromosomal imbalance and CNV (microdeletion and microduplication) [16–18]. As compared to the conventional karyotype analysis, CMA presents a higher resolution, which may detect 50 to 100 bp microdeletions and microduplications, and the CNV neighboring the rearranged breakpoints that were seemingly balanced in karyotyping [19–21]. It is therefore considered that CMA is replacing karyotyping as a first-line test in the evaluation of pregnancies in the era of omics [22–24].
In this study, routine color Doppler ultrasound imaging displayed fetal cardiac developmental defects, and further chromosomal G-banding revealed no abnormal karyotype; however, CMA detected a 1.8 Mb-duplication of the chromosome 15q13.2q13.3 region containing 7 genes (TRPM1, KLF13, OTUD7A, CHRNA7, FAN1, MIR211 and RAHGAP11A), which occurred in the region between BP4-BP5 on chromosome 15q13.3. Neuronal acetylcholine receptor subunit alpha-7, also known as nAChRα7, a protein that in humans is encoded by the CHRNA7 gene that contains 479 amino acids, is a major component of the neuronal nicotinic receptor, which is widely distributed in brain and spinal cord [25]. It has been reported that subjects with chromosome 15q13 microduplications may develop cognitive impairment, autism, seizures, and attention-deficit hyperactivity disorder, and oral clefts [6–14], and these disorders may be strongly associated with 15q13.3 microduplications involving CHRNA7 [7, 9, 10, 14, 26, 27]. Scan statistic-based analysis of exome sequencing data showed that FAN1 is a key driver in the 15q13.3 locus for the associated psychiatric and neurodevelopmental phenotypes, and FAN1 at 15q13.3 has been identified as a susceptibility gene for schizophrenia and autism [28], while MIR211 was identified as a putative attenuator of cholinergic-mediated seizures by intersecting forebrain microRNA profiles and dynamic miR-211 decreases were found to induce hypersynchronization and nonconvulsive and convulsive seizures, accompanied by expression changes in cholinergic and TGFBR2 pathways [29]. In addition, the OTUD7A gene has been identified as a critical gene regulating neurodevelopmental phenotypes [30], and the TRPM1 gene is closely associated with fetal intelligence development [31–33]. In this study, the fetus harboring the 1.8 Mb-duplication of the chromosome 15q13.2q13.3 region containing the CHRNA7, TRPM1 and OTUD7A genes, suggesting that a high possibility of developmental neuropsychiatric disorders in the fetus.
In a previous report, a recurrent duplication 15q13.3 (BP4-BP5) was detected in 150 patients with congenital heart defects, suggesting the possible link between chromosome 15q13 duplication and heart disorders [15]; however, the association between chromosome 15q13 microduplication and cardiac developmental defects has not fully undefined until now. In this study, the 1.8 Mb-duplication of the chromosome 15q13.2q13.3 region contained the KLF13 gene. KLF13, an evolutionally highly-conserved gene, is a member of the Krüppel-like family that encodes zinc-finger proteins, which mainly functions as a regulator of cardiac gene expression and heart development by interacting with GATA4 [34, 35]. Immunohistochemical staining showed high KLF13 protein expression in myocardial and endocardial tissues [35]. In addition, knockdown of KLF13 in Xenopus embryos was found to cause abnormal cardiac development, including ventricular trabecular formation, atrioventricular septal defects, delayed atrioventricular cushion formation and delayed heart valve maturation [36]. In this study, cardiac ultrasound follow-up revealed significant enlargement of the space-occupying lesion in the fetal heart with the extension of the gestational age, and the space-occupying lesion was finally pathologically diagnosed as cardiac rhabdomyoma. In addition, no mutations were identified in the TSC1 or TSC2 gene in the fetus, the mother or the father by next-generation sequencing, which may rule out tuberous sclerosis complex [37, 38]. Taken these findings together, it is hypothesized that the development of fetal cardiac rhabdomyoma may be attributed to the aberrant KLF13 expression caused by chromosome 15q13 microduplication, which is in agreement with previous studies [15]; however, more clinical evidence is required to examine the association between chromosome 15q13 microduplication and cardiac rhabdomyoma.
Conclusions
The results of the present study demonstrate, for the first time, the potential association between chromosome 15q13 microduplication and fetal cardiac rhabdomyoma. Since CMA is effective to detect hereditary diseases that G-banding karyotyping fails to do [19–21], it is therefore recommended that CMA be employed in fetuses with abnormal cardiac development diagnosed by routine cardiac color Doppler ultrasound imaging for early detection of congenital genetic abnormality, which may provide valuable information for prenatal diagnostic consultation and the decision on pregnancy termination.
Acknowledgements
Not applicable.
Abbreviations
- CMA
Chromosomal microarray analysis
- CNV
Copy number variation
- NIPT
Non-invasive prenatal testing
Authors’ contributions
BRQ conceived and designed the study. CZL, JSH, YDH and XQH performed the study. CZL collected and analyzed the data. CZL prepared the first version of the manuscript. BRQ revised and finalized the manuscript. All authors read and approved the final version of the manuscript.
Funding
This study was supported by the grant from the Middle-aged and Young Scientific Research Project of Fuzhou Municipal Health and Family Planning Commission (grant no. 2018-S-wq3) and the Clinical Medicine Center Construction Program of Fuzhou City (grant no. 2018080301).
Availability of data and materials
The data reported in this study are available upon request by contact with the corresponding author.
Ethics approval and consent to participate
This study was approved by the Ethics Review Committee of Fuzhou Municipal First Hospital Affiliated to Fujian Medical University (approval no. FZSY-201700132). Written informed consent was obtained from the fetus’s parents following a detailed description of the study purpose. All experimental procedures described in this study were in accordance with international and national laws, regulations and guidelines.
Consent for publication
The pregnant woman and her husband involved in this study agreed to publish related demographic and clinical features.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chen-Zhao Lin, Email: 6456969@sina.com.
Bi-Ru Qi, Phone: +86-591-88301752, Email: applewang814@163.com.
Jian-Su Hu, Email: 812148446@qq.com.
Yu-Dian Huang, Email: hydfz0591@163.com.
Xiu-Qiong Huang, Email: 1564471138@qq.com.
References
- 1.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, González JR, Gratacòs M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macé A, Kutalik Z, Valsesia A. Copy number variation. Methods Mol Biol. 2018;1793:231–258. doi: 10.1007/978-1-4939-7868-7_14. [DOI] [PubMed] [Google Scholar]
- 3.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu L, Yao X, Huang H, Guo Z, Cheng X, Xu Y, Shen Y, Xu B, Li D. Clinical significance of germline copy number variation in susceptibility of human diseases. J Genet Genomics. 2018;45:3–12. doi: 10.1016/j.jgg.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 5.McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat Genet. 2007;39:S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 6.Zhou D, Gochman P, Broadnax DD, Rapoport JL, Ahn K. 15q13.3 duplication in two patients with childhood-onset schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2016;171:777–783. doi: 10.1002/ajmg.b.32439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szafranski P, Schaaf CP, Person RE, Gibson IB, Xia Z, Mahadevan S, Wiszniewska J, Bacino CA, Lalani S, Potocki L, Kang SH, Patel A, Cheung SW, Probst FJ, Graham BH, Shinawi M, Beaudet AL, Stankiewicz P. Structures and molecular mechanisms for common 15q13.3 microduplications involving CHRNA7: benign or pathological? Hum Mutat. 2010;31:840–850. doi: 10.1002/humu.21284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beal JC. Neuronal migration disorder associated with chromosome 15q13.3 duplication in a boy with autism and seizures. J Child Neurol. 2014;29:NP186–NP188. doi: 10.1177/0883073813510356. [DOI] [PubMed] [Google Scholar]
- 9.Bacchelli E, Battaglia A, Cameli C, Lomartire S, Tancredi R, Thomson S, Sutcliffe JS, Maestrini E. Analysis of CHRNA7 rare variants in autism spectrum disorder susceptibility. Am J Med Genet A. 2015;167A:715–723. doi: 10.1002/ajmg.a.36847. [DOI] [PubMed] [Google Scholar]
- 10.Soler-Alfonso C, Carvalho CM, Ge J, Roney EK, Bader PI, Kolodziejska KE, Miller RM, Lupski JR, Stankiewicz P, Cheung SW, Bi W, Schaaf CP. CHRNA7 triplication associated with cognitive impairment and neuropsychiatric phenotypes in a three-generation pedigree. Eur J Hum Genet. 2014;22:1071–1076. doi: 10.1038/ejhg.2013.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koochek M, Harvard C, Hildebrand MJ, Van Allen M, Wingert H, Mickelson E, Holden JJ, Rajcan-Separovic E, Lewis ME. 15q duplication associated with autism in a multiplex family with a familial cryptic translocation t(14;15)(q11.2;q13.3) detected using array-CGH. Clin Genet. 2006;69:124–134. doi: 10.1111/j.1399-0004.2005.00560.x. [DOI] [PubMed] [Google Scholar]
- 12.Melchior L, Bertelsen B, Debes NM, Groth C, Skov L, Mikkelsen JD, Brøndum-Nielsen K, Tümer Z. Microduplication of 15q13.3 and Xq21.31 in a family with Tourette syndrome and comorbidities. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:825–831. doi: 10.1002/ajmg.b.32186. [DOI] [PubMed] [Google Scholar]
- 13.Williams NM, Franke B, Mick E, Anney RJ, Freitag CM, Gill M, Thapar A, O'Donovan MC, Owen MJ, Holmans P, Kent L, Middleton F, Zhang-James Y, Liu L, Meyer J, Nguyen TT, Romanos J, Romanos M, Seitz C, Renner TJ, Walitza S, Warnke A, Palmason H, Buitelaar J, Rommelse N, Vasquez AA, Hawi Z, Langley K, Sergeant J, Steinhausen HC, Roeyers H, Biederman J, Zaharieva I, Hakonarson H, Elia J, Lionel AC, Crosbie J, Marshall CR, Schachar R, Scherer SW, Todorov A, Smalley SL, Loo S, Nelson S, Shtir C, Asherson P, Reif A, Lesch KP, Faraone SV. Genome-wide analysis of copy number variants in attention deficit hyperactivity disorder: the role of rare variants and duplications at 15q13.3. Am J Psychiatry. 2012;169:195–204. doi: 10.1176/appi.ajp.2011.11060822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie Y. Is chromosome 15q13.3 duplication involving CHRNA7 associated with oral clefts? Child Neurol Open. 2015;2:2329048X15618918. doi: 10.1177/2329048X15618918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derwińska K, Bartnik M, Wiśniowiecka-Kowalnik B, Jagła M, Rudziński A, Pietrzyk JJ, Kawalec W, Ziółkowska L, Kutkowska-Kaźmierczak A, Gambin T, Sykulski M, Shaw CA, Gambin A, Mazurczak T, Obersztyn E, Bocian E, Stankiewicz P. Assessment of the role of copy-number variants in 150 patients with congenital heart defects. Med Wieku Rozwoj. 2012;16:175–182. [PubMed] [Google Scholar]
- 16.Friedman JM. High-resolution array genomic hybridization in prenatal diagnosis. Prenat Diagn. 2009;29:20–28. doi: 10.1002/pd.2129. [DOI] [PubMed] [Google Scholar]
- 17.Roa BB, Pulliam J, Eng CM, Cheung SW. Evolution of prenatal genetics: from point mutation testing to chromosomal microarray analysis. Expert Rev Mol Diagn. 2005;5:883–892. doi: 10.1586/14737159.5.6.883. [DOI] [PubMed] [Google Scholar]
- 18.Levy B, Wapner R. Prenatal diagnosis by chromosomal microarray analysis. Fertil Steril. 2018;109:201–212. doi: 10.1016/j.fertnstert.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wapner RJ, Martin CL, Levy B, Ballif BC, Eng CM, Zachary JM, Savage M, Platt LD, Saltzman D, Grobman WA, Klugman S, Scholl T, Simpson JL, McCall K, Aggarwal VS, Bunke B, Nahum O, Patel A, Lamb AN, Thom EA, Beaudet AL, Ledbetter DH, Shaffer LG, Jackson L. Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med. 2012;367:2175–2184. doi: 10.1056/NEJMoa1203382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hillman SC, McMullan DJ, Hall G, Togneri FS, James N, Maher EJ, Meller CH, Williams D, Wapner RJ, Maher ER, Kilby MD. Use of prenatal chromosomal microarray: prospective cohort study and systematic review and meta-analysis. Ultrasound Obstet Gynecol. 2013;41:610–620. doi: 10.1002/uog.12464. [DOI] [PubMed] [Google Scholar]
- 21.Society for Maternal-Fetal Medicine (SMFM) The use of chromosomal microarray for prenatal diagnosis. Am J Obstet Gynecol. 2016;215:B2–B9. [Google Scholar]
- 22.Bi W, Borgan C, Pursley AN, Hixson P, Shaw CA, Bacino CA, Lalani SR, Patel A, Stankiewicz P, Lupski JR, Beaudet AL, Cheung SW. Comparison of chromosome analysis and chromosomal microarray analysis: what is the value of chromosome analysis in today's genomic array era? Genet Med. 2013;15:450–457. doi: 10.1038/gim.2012.152. [DOI] [PubMed] [Google Scholar]
- 23.Fiorentino F, Napoletano S, Caiazzo F, Sessa M, Bono S, Spizzichino L, Gordon A, Nuccitelli A, Rizzo G, Baldi M. Chromosomal microarray analysis as a first-line test in pregnancies with a priori low risk for the detection of submicroscopic chromosomal abnormalities. Eur J Hum Genet. 2013;21:725–730. doi: 10.1038/ejhg.2012.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breman A, Pursley AN, Hixson P, Bi W, Ward P, Bacino CA, Shaw C, Lupski JR, Beaudet A, Patel A, Cheung SW, Van den Veyver I. Prenatal chromosomal microarray analysis in a diagnostic laboratory; experience with >1000 cases and review of the literature. Prenat Diagn. 2012;32:351–361. doi: 10.1002/pd.3861. [DOI] [PubMed] [Google Scholar]
- 25.Chini B, Raimond E, Elgoyhen AB, Moralli D, Balzaretti M, Heinemann S. Molecular cloning and chromosomal localization of the human alpha 7-nicotinic receptor subunit gene (CHRNA7) Genomics. 1994;19:379–381. doi: 10.1006/geno.1994.1075. [DOI] [PubMed] [Google Scholar]
- 26.Gillentine MA, Berry LN, Goin-Kochel RP, Ali MA, Ge J, Guffey D, Rosenfeld JA, Hannig V, Bader P, Proud M, Shinawi M, Graham BH, Lin A, Lalani SR, Reynolds J, Chen M, Grebe T, Minard CG, Stankiewicz P, Beaudet AL, Schaaf CP. The cognitive and behavioral phenotypes of individuals with CHRNA7 duplications. J Autism Dev Disord. 2017;47:549–562. doi: 10.1007/s10803-016-2961-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gillentine MA, Schaaf CP. The human clinical phenotypes of altered CHRNA7 copy number. Biochem Pharmacol. 2015;97:352–362. doi: 10.1016/j.bcp.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ionita-Laza I, Xu B, Makarov V, Buxbaum JD, Roos JL, Gogos JA, Karayiorgou M. Scan statistic-based analysis of exome sequencing data identifies FAN1 at 15q13.3 as a susceptibility gene for schizophrenia and autism. Proc Natl Acad Sci U S A. 2014;111:343–348. doi: 10.1073/pnas.1309475110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bekenstein U, Mishra N, Milikovsky DZ, Hanin G, Zelig D, Sheintuch L, Berson A, Greenberg DS, Friedman A, Soreq H. Dynamic changes in murine forebrain miR-211 expression associate with cholinergic imbalances and epileptiform activity. Proc Natl Acad Sci U S A. 2017;114:E4996–E5005. doi: 10.1073/pnas.1701201114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uddin M, Unda BK, Kwan V, Holzapfel NT, White SH, Chalil L, Woodbury-Smith M, Ho KS, Harward E, Murtaza N, Dave B, Pellecchia G, D'Abate L, Nalpathamkalam T, Lamoureux S, Wei J, Speevak M, Stavropoulos J, Hope KJ, Doble BW, Nielsen J, Wassman ER, Scherer SW, Singh KK. OTUD7A regulates neurodevelopmental phenotypes in the 15q13.3 microdeletion syndrome. Am J Hum Genet. 2018;102:278–295. doi: 10.1016/j.ajhg.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irie S, Furukawa T. TRPM1. Handb Exp Pharmacol. 2014;222:387–402. doi: 10.1007/978-3-642-54215-2_15. [DOI] [PubMed] [Google Scholar]
- 32.Oancea E, Wicks NL. TRPM1: new trends for an old TRP. Adv Exp Med Biol. 2011;704:135–145. doi: 10.1007/978-94-007-0265-3_7. [DOI] [PubMed] [Google Scholar]
- 33.Prasun P, Hankerd M, Kristofice M, Scussel L, Sivaswamy L, Ebrahim S. Compound heterozygous microdeletion of chromosome 15q13.3 region in a child with hypotonia, impaired vision, and global developmental delay. Am J Med Genet A. 2014;164A:1815–1820. doi: 10.1002/ajmg.a.36535. [DOI] [PubMed] [Google Scholar]
- 34.McConnell BB, Yang VW. Mammalian Krüppel-like factors in health and diseases. Physiol Rev. 2010;90:1337–1381. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavallée G, Andelfinger G, Nadeau M, Lefebvre C, Nemer G, Horb ME, Nemer M. The Kruppel-like transcription factor KLF13 is a novel regulator of heart development. EMBO J. 2006;25:5201–5213. doi: 10.1038/sj.emboj.7601379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nemer M, Horb ME. The KLF family of transcriptional regulators in cardiomyocyte proliferation and differentiation. Cell Cycle. 2007;6:117–121. doi: 10.4161/cc.6.2.3718. [DOI] [PubMed] [Google Scholar]
- 37.Jang MA, Hong SB, Lee JH, Lee MH, Chung MP, Shin HJ, Kim JW, Ki CS. Identification of TSC1 and TSC2 mutations in Korean patients with tuberous sclerosis complex. Pediatr Neurol. 2012;46:222–224. doi: 10.1016/j.pediatrneurol.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 38.Aicher LD, Campbell JS, Yeung RS. Tuberin phosphorylation regulates its interaction with hamartin. Two proteins involved in tuberous sclerosis. J Biol Chem. 2001;276:21017–21021. doi: 10.1074/jbc.C100136200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data reported in this study are available upon request by contact with the corresponding author.