Short abstract
Neuropathic pain is a type of chronic pain induced by either central or peripheral nerve injury. MicroRNAs have been recently linked to many diseases, including neuropathic pain. However, the role of miR-7a in neuropathic pain still remains elusive. Thus, we aim to investigate the effects of miR-7a on neuropathic pain based on the spinal nerve ligation rat model. After establishment of spinal nerve ligation rat models, rats were infected with adeno-associated virus-neurofilament light polypeptide, adeno-associated virus-miR-7a or treated with metformin. The paw withdrawal threshold and paw withdrawal latency were assessed afterward, and the expression of miR-7a and neurofilament light polypeptide as well as their interaction was determined. Subsequently, miR-7a was overexpressed or silenced in dorsal root ganglion cells to investigate the role of miR-7a in neuropathic pain. Furthermore, the regulatory effect of neurofilament light polypeptide on neuropathic pain was detected using plasmid overexpressing neurofilament light polypeptide. Spinal nerve ligation rat model exhibited upregulation of neurofilament light polypeptide but downregulation of miR-7a. In addition, neurofilament light polypeptide accumulation or miR-7a inhibition decreased paw withdrawal threshold and paw withdrawal latency. Then, neurofilament light polypeptide accumulation or miR-7a inhibition was observed to increase the phosphorylation level of signal transducer and activator of transcription. miR-7a was found to directly target neurofilament light polypeptide and downregulate neurofilament light polypeptide. In addition, inhibiting the signal transducer and activator of transcription signaling pathway was also revealed to increase paw withdrawal threshold and paw withdrawal latency. Collectively, our study demonstrated that miR-7a ameliorated neuropathic pain via blocking the signal transducer and activator of transcription signaling pathway by repressing neurofilament light polypeptide. These findings, if taken further, can be of important clinical significance in treating patients with neuropathic pain.
Keywords: Neuropathic pain, microRNA-7a, neurofilament light polypeptide, signal transducer and activator of transcription signaling pathway
Introduction
Neuropathic pain is associated with genetic variations in the dorsal root ganglion (DRG).1 Besides, neuropathic pain can also be triggered by dysfunction of the somatosensory system, which is often found in a particular area of the body.2 Neuropathic pain affects approximately 7% to 10% of the general population, and this number increases due to enhanced tumor survival after chemotherapy, elevated diabetes mellitus occurrence rate, and aged tendency of population worldwide.3 Current treatment approaches for neuropathic pain include a combination of surgical, interventional, medical, auxiliary, and traditional therapies.4 However, owing to the complexity in the pathogenesis of neuropathic pain, current clinical outcomes of current therapeutic options remain unsatisfactory.5 This therefore warrants other potential therapeutic approaches to help treat patients with neuropathic pain.
Interestingly, microRNAs (miRs) have been found to serve as key switches of chronic pain maintenance and progression as well as main regulators of neuronal processes.6 The causal roles of miRNAs and therapeutic potential in chronic pain, such as intractable neuropathic pain, have been recently reported.7,8 The aberrant expression of miRNAs has been found to be induced in the DRG; among those miR-1 and miR-206 were found to be downregulated in the DRG in mouse model of sciatic nerve partial ligation.9 Another miR, miR-146a-5p is dysregulated in spinal astrocytes in neuropathic pain condition while its restoration has been revealed to alleviate spinal nerve ligation (SNL)-induced neuropathic pain in the spinal cord.10 Interestingly, miR-7a, abundantly expressed in the hypothalamus, has been revealed to regulate hypothalamic neurons post-transcriptionally.11 Furthermore, miR-7a has been reported to modulate neuronal excitability to reduce the maintenance of neuropathic pain.12 Moreover, the expression of miR-7a has been found to be downregulated in ipsilateral lumbar 5 DRGs.13 However, the changes of miR-7a in DRG in the SNL model and molecular mechanism how it functions in neuropathic pain currently remain unclear. miR-7a has been predicted to target neurofilament light polypeptide (NEFL) based on an analysis from the online prediction website microRNA.org. NEFL plays a role in the nerve conduction velocity and axonal transport by encoding a neuronal protein that is vital for neurofilament formation.14 Mutations in the NEFL gene have been associated with motor neuron diseases in mice and Charcot-Marie-Tooth disease in human.15 The mutation of NEFL could cause a painful predominantly sensory neuropathy.16 Signal transducer and activator of transcription (STAT3) has been proved to be involved in cell differentiation of the nervous system.17 The STAT3 signaling pathway has been identified as a critical participant in neuroinflammation, and its activation has been found in the spinal dorsal horn of both spinal cord injury and neuropathic pain models.18 The current study has shown that miR-7a could affect neuropathic pain development via the STAT3 signaling pathway by regulating NEFL based on the rat model of SNL, which offers a better understanding for the underlying molecular mechanisms in neuropathic pain progression.
Materials and methods
Ethics statement
The study protocol was approved by the Experimental Animal Ethics Committee of Xiangya Hospital, Central South University. Animal experiments were carried out in accordance with the principles of the International Association for Study of pain which aims to minimize the pain inflicted on animals during the experiment.
Animal model establishment
A total of 300 Sprague-Dawley male rats weighing approximately 200 g to 240 g were purchased from the Experimental Animal Center of Nantong University. SNL models were established according to operation procedures reported by Kim and Chung.19 The 180 rats were then anesthetized by 3% pentobarbital sodium and fixed on a specific shelf in a prone position. A 5-cm incision was cut approximately 1 cm along the left side of the back line, with the middle point of incision at ligature of the superior ilium on both sides. The fascia was cut open to carefully detach the muscles, and a crown opener was used to stretch surrounding muscles to expose the sixth lumbar transverse process. The surrounding muscles were bluntly separated, and the sixth lumbar transverse process was removed. A bipolar coagulation was performed to stop bleeding. Following that, the lumbar 5 (L5) and L5 spinal nerve were carefully detached, and 4-0 silk threads were slightly introduced to ligate the L5 and L6 nerves together. The muscles were put back into place, and fascia and skin were sutured. Spinal nerve was exposed but not ligated in 120 rats in the sham group. After the operation, rats were fed in a rearing cage with constant body temperature. Throughout the experiment, rats were fed in a specific pathogen-free experimental animal center, with constant room temperature and humidity, 12-h cycles of light/darkness, and free access to food and water.20
Transfection of adenovirus vectors
Adenovirus vectors were purchased from Shanghai Genechem Co., Ltd. (Shanghai, China), and metformin was purchased from Sigma (St. Louis, MO, USA). Rats were anesthetized by pentobarbital sodium and fixed on a specific shelf in a prone position. Adenovirus vectors were diluted into 1 × 1014 vector genomes (vg)/mL with phosphate buffer saline (PBS). Next, 5 μL adenovirus vectors were slowly injected into the L4-L5 dorsal spinal cord using a 27-gauge needle microsyringe.21 In details, after rats were anesthetized, adeno-associated virus (AAV)-NEFL or AAV-miR-7a alone was injected into the L4-L5 segments of the dorsal spinal cord as described in the previously reported method.21 For the combined treatment of both AAV-NEFL and AAV-miR-7a, the rats injected with AAV-NEFL were left to recover in cages. After 12 h and recovery, the rats were anesthetized and then injected with AAV-miR-7a into the dorsal spinal cord and left to recuperate again in their cages. On the seventh day after injection, the L4-L5 gap of dorsal spinal cord was injected with an equal amount of metformin or saline (control) at a dose of 200 mg/kg22 using a microsyringe for seven consecutive days.23 During the establishment of SNL models, 26 rats died during the process; and 15 rats in each group were utilized for pain-related behavior assessment. Five rats in the sham and SNL groups were used for subsequent tissue experiments on the 3rd, 7th, and 14th day, while 15 rats from the remaining groups were used for experiments on the 14th day.
Animal grouping
Rats in the sham group were untreated or treated with either or two of the following viruses or drugs: AAV-negative control (NC), AAV expressing NEFL (AAV-NEFL), AAV expressing miR-7a (AAV-miR-7a), AAV-NEFL and AAV-miR-7a, metformin, or saline. SNL-treated rats were untreated or treated with either or two of the following viruses or drugs: AAV-NC, AAV-NEFL, AAV-miR-7a, AAV-NEFL and AAV-miR-7a, AAV-NC and saline, AAV-NC and metformin, AAV-NEFL and saline, or AAV-NEFL and metformin.
Pain-related behavior assessment
Pain-related behaviors such as the paw withdrawal threshold (PWT) and paw withdrawal latency (PWL) were assessed on 15 SNL-treated rats from each group on the 0, 3rd, 7th, 10th, 12th, and 14th day after SNL model establishment. PWT was assessed according to the following procedures: von Frey filaments (Stoelting Co., Ltd., Wood Dale, IL, USA) was used to stimulate the rat paw to assess mechanical hypersensitivity. The instrument was placed in a quiet room, and rats were placed inside transparent organic glass cages (30 cm × 30 cm × 30 cm) with a metal mesh-constituted bottom that was fixed on a shelf approximately 50 cm high. After 15-min adaption in the cage, the test began. Von Frey filaments, starting from 0.008 g and increasing in size, were used to stimulate the planta pedis of rat hind paws. After stimulation, the acupuncture intensity (g) of rat paw withdrawal was recorded. Each filament was used to stimulate rat paws five times with every stimulation lasting no more than 1 min during the whole process. Next, PWL was measured when rats were placed in an organic glass boxes with a radiative heat source. The radiative heat source was concentrated on the surface of the planta pedis of rat hind paws. When rats showed pain sensations on the planta pedis and withdrew their paws, the PWL was recorded. Every rat was assessed five times, with 10-min intervals between each time. Heat radiation lasted for 10 s,24 and a cut-off time of 30 s was set to avoid unwanted tissue damage.25
Immunofluorescence staining
Rats were deeply anesthetized through injection of 100 mg/kg pentobarbital sodium (P3761, Sigma, St. Louis, MO, USA) and fixed in a prone position. An incision was made at the middle of the back. After separation of the muscles on both sides of the spine, the transverse and spinous processes were exposed, after which the spinous process was excised, and then transverse processes on the vertebral plate and both sides were excised. After the spinal cord was lifted with forceps, the spinal nerve can be seen at the spinal canal and the intervertebral foramen was exposed. Spinal cord L4-L5 DRG tissues were collected. L4-L5 DRG tissues of rats in the sham and SNL groups were fixed with 4% paraformaldehyde overnight and continuously dehydrated by 20% and 30% cane sugar. DRG tissues were sectioned into 8-μm-thick serial slices and treated with 10% donkey serum, 3% bovine serum albumin, and 0.3% Triton X-100 at room temperature for 2 h. Slices were then incubated with primary mouse-anti NEFL antibody (sc71678, 1: 500, Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA), a C-fiber neuron marker, isolectin B4 (IB4; ALX-650-001-M001, Enzo Life Sciences, Farmingdale, NY, USA), and an A-fiber neuron marker, neurofilament 200 (NF200; ab82259, Abcam Inc., Cambridge, MA, USA) at 4°C overnight. On the following day, after rinsing in PBS three times, the slices were incubated with fluorescein isothiocyanate-labeled goat anti-mouse secondary antibody (ab6785, Abcam Inc., Cambridge, MA, USA) for 1 h, followed by PBS rinse six times. The slices were then photographed and analyzed under a fluorescence microscope (Leica Biosystems, Shanghai, China).26
Rat DRG treatment
DRGs of rats were purchased from BeNa Culture Collection (BNCC338571, Beijing, China) and cultured according to the instructions. After the cells were cultured in a T-25 m2 culture bottle at 5% CO2 and 37°C for 4 to 6 h, the culture solution was replaced with high-glucose Dulbecco’s modified Eagle medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum. The culture medium was replaced every two to three days. Plasmids were constructed by Invitrogen (Carlsbad, CA, USA) based on the known sequences of miR-7a and NEFL from the national center of biotechnology information. After reaching the desired confluency, cells were inoculated into six-well plates and transfected 12 h later, according to the instructions provided by the Lipofectamine 2000 kit (Invitrogen, Carlsbad, CA, USA). In brief, cells were harvested after trypsinization and seeded in a 60-mm tissue culture dish at a density of 4 × 105 to 8 × 105 cells/cm2 in complete medium. An appropriate size of the culture dish was selected according to the experiment purpose to allow an approximately 70% to 90% of cells to adhere to the culture dish. Next, cells were incubated at 37°C with 5% CO2 for 8 to 24 h, and cells were allowed to fully adhere to the wall before transfection. Prior to transfection, 2 mL pre-warmed serum-free medium was added to the cells. Lipofectamine 2000-DNA mixture was prepared with 420 µL as an example of the total reaction volume for a 60-mm tissue culture dish. Next, 2 to 8 µg plasmid DNA was diluted with 240 µL Hepes-buffered saline (HBS) in a 1.5 mL centrifuge tube and then mixed. In another tube, 100 µM Lipofectamine 2000 storage solution was diluted to 10 µM with HBS, and then mixed. Afterwards, 180 µL diluted Lipofectamine 2000 solution (10 µM) was then fully mixed with the HBS containing DNA at room temperature for 20 to 30 min. Finally, a total of 420 µL polyethylenimine-DNA mixture was added in a dropwise manner to the monolayer cell culture medium and gently shaken and then incubated at 37°C with 5% to 7% CO2. The above transfection was performed with the following plasmids: miR-7a-mimic (upregulating miR-7a expression), miR-7a-inhibitor (inhibiting miR-7a expression), NEFL overexpression plasmid (oe-NEFL), and their corresponding NCs (miR-7a mimic NC, miR-7a inhibitor NC, and oe-NC). The cells were harvested for further experiments after 48 h.
Dual luciferase reporter assay
Target gene analysis of miR-7a was performed using the biological prediction website microRNA.org. A dual luciferase reporter assay was carried out to determine whether NEFL was a direct target gene of miR-7a. The synthesized wild-type (WT) NEFL 3′UTR fragment and a mutant type (MUT) sequence in which the putative binding sites of miR-7a were mutated were separately ligated into the pMIR-reporter (Beijing, Huayueyang Biotechnology, Beijing, China). The reporter plasmids WT and MUT were then co-transfected with miR-7a mimic or miR-7a NC, respectively, into HEK-293T cells (Shanghai Beinuo Biotechnology Co., Ltd., Shanghai, China). The cells were lysed after 48 h. Luciferase activity was detected using a Glomax20/20 luminometer (Promega Corp., Madison, Wisconsin, USA) according to instructions provided by the luciferase assay kits (K801-200, BioVision Inc., Milpitas, CA, USA). The experiment was repeated three times independently.
Reverse transcription quantitative polymerase chain reaction
Total RNA was extracted from the rat dorsal spinal cord L4-L5 DRG tissues or cells using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Reverse transcription was conducted using random primers or stem-loop methods according to the instructions of reverse transcription kit (Takara, Tokyo, Japan). The reverse primer sequence used in the stem-loop method was miR-7a stem loop with a sequence: 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACAACAAA-3′.27 Quantitative polymerase chain reaction (qPCR) reaction was conducted according to the instructions of SYBR Green PCR Master Mix kit (Roche Diagnostics GmbH, Mannheim, Germany) on a real-time fluorescence quantitative PCR instrument (ABI Company, Oyster Bay, NY, USA) with the following reaction conditions: pre-denaturation at 95°C for 5 min, 40 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 20 s, extension at 72°C for 20 s, and final extension at 78°C for 20 s. Primers for miR-7a, NEFL, U6, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were designed and synthesized by Invitrogen (Carlsbad, CA, USA) (Table 1). U6 and GAPDH were used as references, and the gene expression ratio of the experimental group to the control group was expressed as 2ΔΔCt: ΔΔCT = ΔCtexperimental group – ΔCtcontrol group, ΔCt = CT(target gene) – CT(internal control). Ct refers to the number of amplification cycles when the real-time fluorescence intensity of reaction reaches the set threshold, during which the amplification reaches logarithmic growth. The experiment was repeated three times independently.
Table 1.
The primer sequence for reverse transcription quantitative polymerase chain reaction.
| Gene | Primer sequence (5′–3′) |
|---|---|
| miR-7a | F: GTGTCTCCAGTGTATCGGCG |
| R: TACTGGCACCACTGGAAACC | |
| NEFL | F: AGGACGAGGTGTCGGAAAG |
| R: TTCTCCAGTTTGTTGATTGTGTC | |
| U6 | F: CTCGCTTCGGCAGCACA |
| R: AACGCTTCACGAATTTGCGT | |
| GAPDH | F: CAAGGTCATCCATGACAACTTTG |
| R: GTCCACCACCCTGTTGCTGTAG |
F: forward; R: reverse; miR-7a: microRNA-7a; NEFL: neurofilament light polypeptide; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
Western blot analysis
Dorsal spinal cord L4-L5 DRG tissues of 15 rats in each group and DRG cells (at least five independent samples in each group) were washed by PBS twice, lysed with lysis buffer, and centrifuged at 25,764 × g at 4°C for 30 min. The supernatant was collected, and the concentration of total protein was determined using a bicinchoninic acid kit. Next, 50 μg protein was first separated with 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene fluoride membrane by a wet transfer method. After that, the membrane was blocked with 5% skimmed milk powder at room temperature for 1 h and incubated with the following mouse-anti primary antibodies: NEFL (sc71678, 1: 500), STAT3 (sc8019, 1: 500), p-STAT3 (sc56747, 1: 500), and GAPDH (sc47724). The above antibodies were all purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA). After three washes with Tris-buffered saline Tween 20, the membrane was incubated with goat anti-mouse immunoglobulin G antibody (IgG, ab6789, 1: 5000, Abcam Inc., Cambridge, MA, USA) at room temperature for 1 h. After being washed and developed with enhanced chemiluminescence solution (Invitrogen, Carlsbad, CA, USA), the membrane was photographed under a Bio-Rad image analysis system (Bio-Rad Inc., Hercules, CA, USA) and analyzed using Quantity One v4.6.2 software. The relative protein levels were expressed as the ratio of the gray value of related protein band to that of GAPDH band. The experiment was repeated three times independently.
Statistical analysis
All data analysis was conducted using SPSS 21.0 software (IBM Corp., Armonk, NY, USA). The measurement data were presented as the mean ± standard deviation. All data were initially examined for normal distribution and homogeneity of variance. If the data were consistent with normal distribution or homogeneity of variance, comparisons between two groups were analyzed by an un-paired t-test, while comparisons among multiple groups were analyzed using one-way analysis of variance or repeated measures analysis of variance with Tukey’s post hoc tests for multiple comparisons. Values of p < 0.05 were considered to be statistically significant.
Results
NEFL is upregulated in the SNL rat model
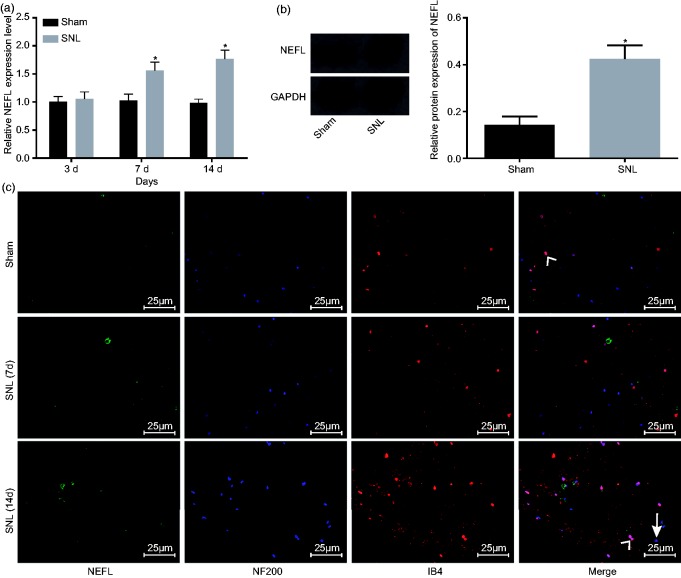
In order to detect the expression patterns of NEFL in SNL models, the mRNA levels of NEFL in the dorsal spinal cord L4-L5 DRG tissues of rats that received SNL or sham-operated rats were determined by reverse transcription (RT)-qPCR on the 3rd, 7th, and 14th day after operation. Protein levels of NEFL were determined by western blot analysis on the 14th day after operation. It was found that both mRNA and protein levels of NEFL were significantly higher in the L4-L5 dorsal spinal cord DRG tissues of rats that received SNL compared to the sham-operated rats on the 14th day after operation (p < 0.05) (Figure 1(a) and (b)). Immunofluorescence staining indicated that the expression levels of NEFL, IB4, and NF200 were obviously higher in the DRG tissues of rats that received SNL on the 7th and 14th day after operation (Figure 1(c)). The above results demonstrated that NEFL levels were upregulated in SNL rat model.
Figure 1.
The SNL rats exhibit upregulation of NEFL. (a) mRNA expression of NEFL in rat dorsal spinal cord L4-L5 DRG tissues in the sham-operated or SNL-treated rats on the 3rd, 7th, and 14th day after operation (n = 15, five rats at each time point); (b) protein level of NEFL in rat dorsal spinal cord L4-L5 DRG tissues in the sham-operated or SNL-treated rats on the 14th day after operation; *p < 0.05 versus the sham-operated rats; (c) expression of NEFL, IB4, and NF200 in rat dorsal spinal cord L4-L5 DRG tissues in rats of the SNL model on the 7th and 14th day after operation measured by immunofluorescence; white arrow represents double staining of NEFL with IB4, and white arrow head represents double staining of NEFL with NF200, scale bar = 25 µm; all data were measurement data, expressed as mean ± standard deviation, and analyzed by un-paired t-test; and the experiment was repeated three times independently.
NEFL: neurofilament light polypeptide; SNL: spinal nerve ligation; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
Overexpression of NEFL triggers neuropathic pain
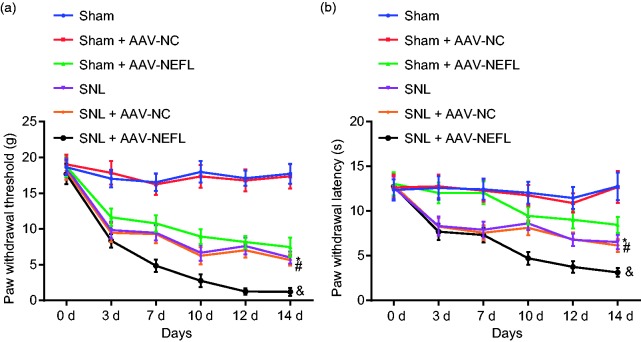
Neuropathic pain after model establishment was assessed by PWT and PWL testing. SNL-treated rats exhibited a lower PWT and PWL compared to their sham-operated rat counterparts (p < 0.05). Rats that received SNL or sham-operated rats were infected with AAV-NC and AAV-NEFL, respectively, to determine the relationship between NEFL and neuropathic pain. Compared with the rats infected with AAV-NC in the sham-operated rats, the rats infected with AAV-NEFL exhibited a lower score of PWT and PWL (p < 0.05). Rats that received SNL by AAV-NEFL treatment also had lower PWT and PWL scores than those treated with AAV-NC (Figure 2(a) and (b)). These results suggested that neuropathic pain could be aggravated by NEFL.
Figure 2.
Overexpression of NEFL triggers neuropathic pain. (a) The effect of NEFL accumulation on PWT on the day 0, 3, 7, 10, 12, and 14 after operation; (b) the effect of NEFL accumulation on PWL on the day 0, 3, 7, 10, 12, and 14 after operation; *p < 0.05 versus the sham group; #p < 0.05 versus the sham + AAV-NC group; &p < 0.05 versus the SNL + AAV-NC group; all data were measurement data, expressed as mean ± standard deviation, and analyzed by repeated measures ANOVA, followed by Tukey’s post hoc tests for multiple comparisons (n = 15).
NEFL: neurofilament light polypeptide; SNL: spinal nerve ligation; NC: negative control; AAV: adeno-associated virus.
Inactivation of the STAT3 signaling pathway attenuates neuropathic pain triggered by NEFL
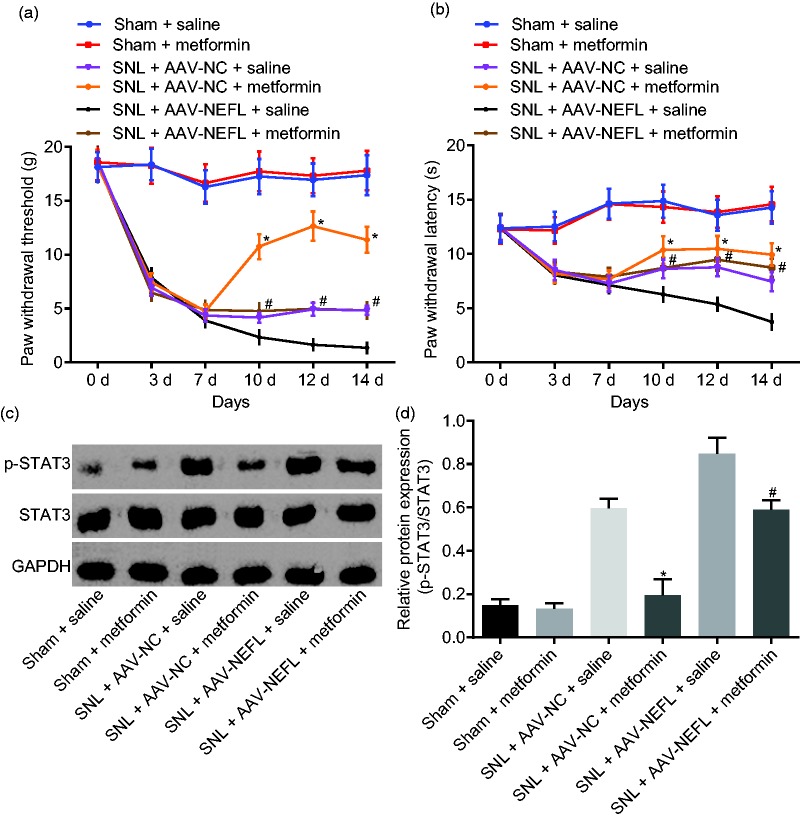
In order to investigate the relationship between NEFL and the STAT3 signaling pathway in neuropathic pain, rats that received SNL were treated with AAV-NEFL, or metformin which could inhibit the STAT3 signaling pathway.18 It was shown that among the rats that received SNL, compared with rats treated with AAV-NC and saline, rats treated with AAV-NC and metformin exhibited elevated PWT and PWL and reduced ratios of STAT3 phosphorylation to STAT3 protein in the dorsal spinal cord L4-L5 DRG tissues (p < 0.05). This suggests that inhibition of STAT3 signaling pathway by metformin could alleviate neuropathic pain. The PWT and PWL scores were higher, and the ratio of the extent of STAT3 phosphorylation to STAT3 protein was lower in the L4-L5 dorsal spinal cord DRG tissues in SNL rats treated with AAV-NEFL and injected with metformin compared to those infected with AAV-NEFL and saline (p < 0.05; Figure 3(a) to (c)). These results demonstrated that blocking STAT3 signaling pathway by metformin could alleviate neuropathic pain triggered by NEFL.
Figure 3.
Inactivation of STAT3 signaling pathway attenuates neuropathic pain induced by NEFL. (a) PWT on the day 0, 3, 7, 10, 12, and 14 after operation after alteration of NEFL and the STAT3 signaling pathway; (b) PWL on the day 0, 3, 7, 10, 12, and 14 after operation after alteration of NEFL and the STAT3 signaling pathway; (c-d) extent of STAT3 phosphorylation and protein levels of STAT3 in dorsal spinal cord L4-L5 DRG tissues on the 14th day after operation, the 7th day after infection with AAV-NEFL, and the 7th day after administrated with metformin; *p < 0.05 versus the SNL + AAV-NC + saline group; #p < 0.05 versus the SNL + AAV-NEFL + saline group; all data were measurement data and expressed as mean ± standard deviation; the data in panel a and b were analyzed using repeated measures ANOVA, followed by Tukey’s post hoc tests for multiple comparisons while the data in panel c were analyzed using one-way ANOVA, followed by Tukey’s post hoc tests for multiple comparisons (n = 15). NEFL: neurofilament light polypeptide; STAT3: signal transducer and activator of transcription; SNL: spinal nerve ligation; NC: negative control; AAV: adeno-associated virus; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
miR-7a directly targets NEFL and downregulates NEFL expression
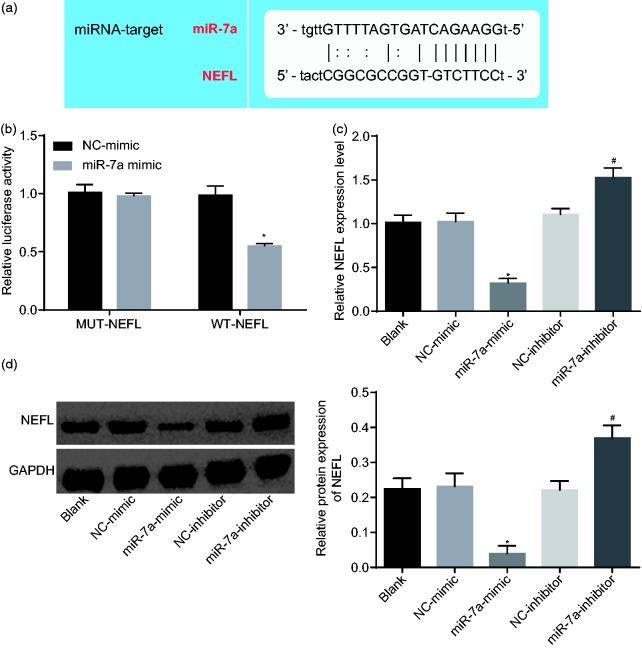
A binding site between miR-7a and NEFL was predicted using the online biological prediction website (Figure 4(a)), and dual luciferase reporter gene assay was subsequently conducted to verify whether NEFL was a target gene of miR-7a. As shown in Figure 4(b), the luciferase activity of WT-NEFL was remarkably decreased when co-transfected with miR-7a mimic compared with mimic NC (p < 0.05), while the luciferase activity of MUT-NEFL had no significant change when co-transfected with mimic-NC or miR-7a mimic (p > 0.05). RT-qPCR and western blot analysis were conducted to detect the expression of NEFL after alteration of miR-7a. The expression of NEFL was significantly reduced at both mRNA and protein levels after the transfection of miR-7a mimic, while markedly increased after the transfection of miR-7a inhibitor (p < 0.05, Figure 4(c) and (d)). All these results suggested that NEFL was a target gene of miR-7a, and miR-7a inhibited NEFL expression.
Figure 4.
NEFL is a target gene of miR-7a. (a) The binding site between miR-7a and NEFL predicted by online prediction software; (b) the binding of miR-7a to NEFL verified by dual luciferase reporter assay; *p < 0.05 versus co-transfection of NC mimic and WT-NEFL; (c) NEFL expression at mRNA level with overexpression or silencing of miR-7a; (d) expression of NEFL at protein level with overexpression or silencing of miR-7a; *p < 0.05 versus the NC-mimic group; #p < 0.05 versus the NC-inhibitor group; all data were measurement data and expressed as mean ± standard deviation and analyzed by un-paired t-test; the experiment was repeated three times independently.
miR-7a: microRNA-7a; NEFL: neurofilament light polypeptide; NC: negative control; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; MUT: mutant type; WT: wild type.
miR-7a inhibits NEFL to inactivate the STAT3 signaling pathway
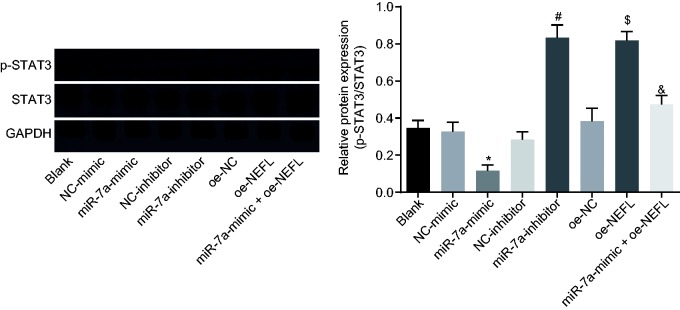
The effect of miR-7a on the STAT3 signaling pathway was further explored in DRG cells that were infected with miR-7a mimic, inhibitor, and plasmids overexpressing NEFL. The total level and phosphorylation level of STAT3 were determined by western blot analysis. Results showed that the ratio of the extent of STAT3 phosphorylation to STAT3 protein was significantly lower after the treatment with miR-7a mimic, but increased following treatment with miR-7a inhibitor (p < 0.05). In addition, DRG cells overexpressing NEFL produced significantly higher ratios of STAT3 phosphorylation to STAT3 protein (p < 0.05). On the other hand, we found that the level of phosphorylated STAT3 was significantly decreased in DRG cells transfected with miR-7a mimic and NEFL when compared with those transfected with NEFL only (p < 0.05, Figure 5). Taken together, miR-7a could inactivate the STAT3 signaling pathway by negatively regulating NEFL.
Figure 5.
miR-7a blocks the STAT3 signaling pathway by downregulating NEFL. The total protein level of STAT3 and the extent of STAT3 phosphorylation in DRG cells after transfected with miR-7a mimic or inhibitor or NEFL detected by western blot analysis; *p < 0.05 versus the NC-mimic group; #p < 0.05 versus the NC-inhibitor group; $p < 0.05 versus the oe-NC group; &p < 0.05 versus the oe-NEFL group; all data were measurement data, expressed as mean ± standard deviation, and analyzed by one-way ANOVA, followed by Tukey’s post hoc tests for multiple comparisons; the experiment was repeated three times independently.
miR-7a: microRNA-7a; NEFL: neurofilament light polypeptide: STAT3: signal transducer and activator of transcription; NC: negative control; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
miR-7a inhibits NEFL and inactivates the STAT3 signaling pathway to alleviate neuropathic pain
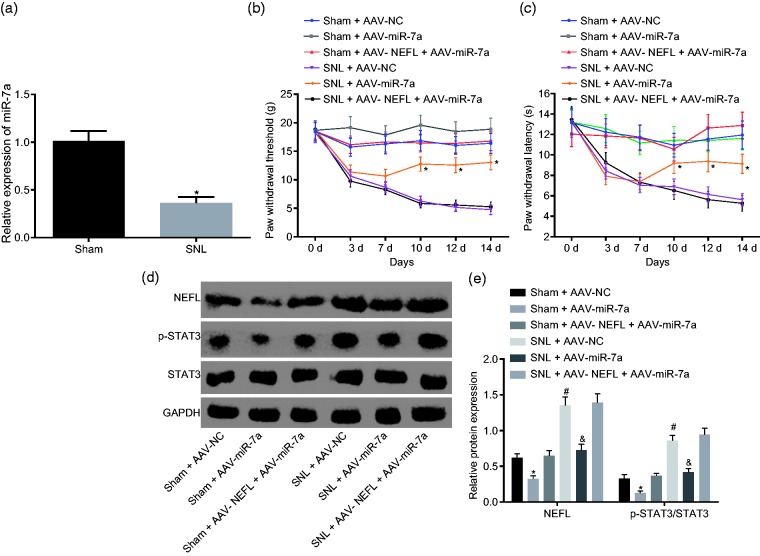
Then the underlying mechanism of miR-7a, NEFL and STAT3 signaling pathway involved in neuropathic pain was further explored. RT-qPCR results showed that miR-7a expression was decreased significantly in the rats that received SNL as compared to the sham-operated rats (p < 0.05; Figure 6(a)). The measurement of PWT and PWL was carried out after this. As suggested in Figure 6(b) and (c), SNL-treated rats infected with AAV-miR-7a displayed increased PWT and PWL compared to SNL-treated rats infected with AAV-NC (p < 0.05). Western blot analysis was performed to determine the protein levels of NEFL and STAT3 as well as the extent of STAT3 phosphorylation, which indicated that NEFL expression and the ratio of phosphorylated to total STAT3 protein were higher in the SNL-treated rats infected with AAV-NC than in rats in the sham-operated rats infected with AVV-NC (p < 0.05). However, SNL-treated rats infected with AAV-miR-7a displayed decreased NEFL expression as well as reduced ratio of phosphorylated to total STAT3 protein compared to the SNL-treated rats infected with AAV-NC (p < 0.05). Compared to the sham-operated rats infected with AAV-NEFL and AAV-miR-7a, SNL-treated rats infected with AAV-NEFL and AAV-miR-7a presented with significantly higher NEFL expression and higher ratio of phosphorylated to total STAT3 protein (p < 0.05, Figure 6(d) and (e)). Hence, miR-7a alleviated neuropathic pain through inactivation of the STAT3 signaling pathway.
Figure 6.
miR-7a attenuates neuropathic pain by suppressing NEFL expression and the STAT3 signaling pathway. (a) miR-7a expression in the sham-operated or SNL-treated rats; *p < 0.05 versus the sham group; (b) PWT of sham-operated or SNL-treated rats after the infection of AAV-miR-7a on the day 0, 3, 7, 10, 12, and 14 after operation; (c) PWL level of sham-operated or SNL-treated rats in response to the infection of AAV-miR-7a on the day 0, 3, 7, 10, 12, and 14 after operation; (d) and (e) protein bands and levels of NEFL and STAT3 as well as the extent of STAT3 phosphorylation in SNL-treated rats in response to infection of AAV-miR-7a at the 14th day after operation; *p < 0.05 versus the sham + AAV-NC group; #p < 0.05 versus the SNL + AAV-NC group; &p < 0.05 versus the sham + AAV-NEFL + AAV-miR-7a group; all data were measurement data and expressed as mean ± standard deviation; the data in panel A were analyzed using t-test, and data in panel B and C were analyzed using repeated measures ANOVA, followed by Tukey’s post hoc tests for multiple comparisons; while the data in panel E were analyzed using ANOVA, followed by Tukey’s post hoc tests for multiple comparisons (n = 15).
miR-7a: microRNA-7a; NEFL: neurofilament light polypeptide; STAT3: signal transducer and activator of transcription; SNL: spinal nerve ligation; AAV: adeno-associated virus; NC: negative control; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
Discussion
Neuropathic pain is a difficult disease to cure due to the lack of new forms of treatments and the highly intricate syndromes.28 Presently, miRNAs have been reported to play a role in many human diseases including neuropathic pain through regulating various genes and pathways.8 Thus, our study is focused on studying the role of miR-7a in the development of neuropathic pain. Our results demonstrated that miR-7a was able to alleviate neuropathic pain through blocking the STAT3 signaling pathway by downregulating NEFL.
We found that NEFL was expressed at high levels in the dorsal spinal cord L4-L5 DRG tissues of SNL rat. The overexpression of NEFL exaggerated neuropathic pain, which was reflected by lower PWT and PWL values. Sainio et al.29 have found that NEFL acts as a transcript and is highly expressed in neurons. Increasing evidence has revealed that neurofilament has been found to be expressed in neurons and that NEFL expression is related to neurological diseases.30 In addition, neurofilament proteins have been frequently found to be abnormally aggregated in neurodegenerative diseases, such as NEFL mutants-induced subtypes of the peripheral sensory-motor neuropathies.31 A recent study has also reported that NEFL expression is correlated with the development of neuroblastoma and that high levels of NEFL could contribute to enhanced survival rates of neuroblastomas.32
According to the online prediction software and dual luciferase reporter assay, NEFL was found to be a target gene of miR-7a. In line with our findings, both miR-183 and miR-25 have been previously reported to be involved in accelerating glioma cell invasion and proliferation by directly binding to NEFL.33,34 Besides, miR-7a has been found to dominate dopaminergic neurons spatial derivation in forebrain by targeting Pax6.35 A study showed that NEFL knockout enhances STAT3-stathmin interaction, which stabilizes the microtubules in pmn mutant motoneurons, consequently blocking axonal maintenance.36 In this study, overexpression of NEFL was observed to increase the phosphorylated-total STAT3 protein ratio. STAT3 phosphorylation is essential for its activation. Hence, higher phosphorylated levels of STAT3 by NEFL overexpression suggested that NEFL activates the STAT3 signaling pathway. We further determined that upregulation of miR-7a reversed NEFL-induced increase in phosphorylated–total STAT3 protein ratio. This suggests that miR-7a inactivates the STAT3 signaling pathway by negatively regulating NEFL. Consistent with our findings, miR-124 has been shown to promote immune clearance mediated by T-cells in gliomas by blocking of the STAT3 signaling pathway.37 Moreover, miR-17-5p and STAT3 play vital roles in the treatment of sciatic nerve conditioning injury through accelerating repair of damaged dorsal column in the primary sensory neurons from DRGs.38 Interestingly, our study mainly suggested the functions of miR-7a/NEFL/STAT3 axis in affecting neuropathic pain in dorsal spinal cord L4-L5 DRG tissues.
The most significant finding of our study was that miR-7a can downregulate NEFL, thereby attenuating neuropathic pain via blocking the activation of STAT3 signaling pathway. This was suggested by the increased PWT and PWL. Both PWT and PWL are known to be standard hallmarks and indicators of neuropathic pain, and they correlate with thermal hyperalgesia and mechanical allodynia.39 Neuropathic pain leads to tolerance of opioid receptor by means of activation of the endogenous kappa opioid system in spinal cords of mouse.40 Consistently, miR-7a has been suggested to attenuate neuropathic pain by modulating neuronal excitability which is indicative of its role as a potential biomarker in the treatment of chronic neuropathic pain.12 Similarly, miR-30b alleviates neuropathic pain in SNL rat models by mediating voltage-gated sodium channel nav1.3.41 MiR-146a-5p was previously demonstrated to restrain the TRAF6 signaling pathway to ameliorate neuropathic pain, suggesting that miR-146a-5p may act as a therapeutic target for treating chronic neuropathic pain.10 As mentioned above, the overexpression of NEFL exaggerated neuropathic pain. However, the upregulation of miR-7a alleviated NEFL-induced neuropathic pain, suggesting that miR-7a attenuates neuropathic pain through downregulation of NEFL. In addition, spinal cord microglia with peripheral nerve damage exhibited activated JAK-STAT3 signaling pathways. When the JAK-STAT3 signaling pathway is activated, an alleviation of the mechanical allodynia can be observed in the SNL models.42 It has been found that the JAK-STAT3 signaling pathway exerts a regulatory impact on neuropathic pain maintenance through interleukin 6 in the red nucleus.43 Moreover, miR-93 serves as a suppressor in neuroinflammation and neuropathic pain progression through blocking STAT3 in rats with chronic constriction sciatic nerve injury.44 MiR-544 has also been found to be associated with ameliorated neuropathic pain through repressing STAT3 in a rat model.45 Our study further demonstrated that inhibition of the STAT3 signaling pathway by metformin contributed to lower PWT and PWL values, alleviating neuropathic pain. Those findings supported the conclusion that miR-7a is highly able to attenuate neuropathic pain through repressing the activation of the STAT3 signaling pathway.
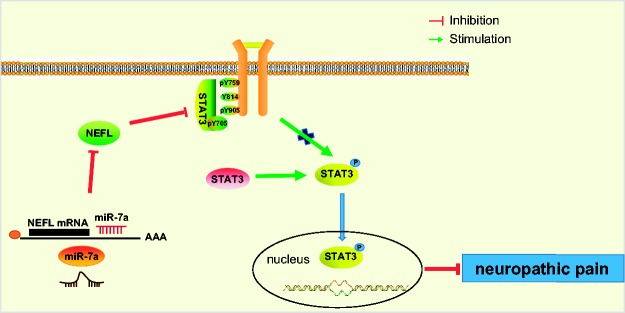
In conclusion, the current study revealed that miR-7a repressed NEFL expression by directly targeting NEFL, thereby inhibiting the activation of the STAT3 signaling pathway and ultimately relieving neuropathic pain in SNL rat models (Figure 7). Although miR-based therapeutics is still in their early developmental stages, our findings on the role of miR-7a in neuropathic pain have provided a basis for a potential treatment target of neuropathic pain.
Figure 7.
Molecular mechanism underlying how miR-7a alleviates neuropathic pain. miR-7a could specifically target NEFL and inhibit NEFL expression to inactivate the STAT3 signaling pathway, thus ameliorating neuropathic pain.
NEFL: neurofilament light polypeptide; STAT3: signal transducer and activator of transcription.
Author contributions
FRY and QLG designed the study. HY and LYP were involved in data collection. JC, HY, and XLH performed the statistical analysis and preparation of figures. FRY, JC, and QLG drafted the paper. All authors read and approved the final manuscript.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by National Natural Science Foundation of China (No. 81300971), Natural Science Foundation of Hunan Province (No. 2016JJ4079), and Foundation of Hunan Provincial Health and Family Planning Commission (No. B2016129).
References
- 1.Yuan J, Wen J, Wu S, Mao Y, Mo K, Li Z, Su S, Gu H, Ai Y, Bekker A, Zhang W, Tao YX. Contribution of dorsal root ganglion octamer transcription factor 1 to neuropathic pain after peripheral nerve injury. Pain 2018; 160: 375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allegri M, Baron R, Hans G, Correa-Illanes G, Mayoral Rojals V, Mick G, Serpell M. A pharmacological treatment algorithm for localized neuropathic pain. Curr Med Res Opin 2016; 32: 377–384. [DOI] [PubMed] [Google Scholar]
- 3.Colloca L, Ludman T, Bouhassira D, Baron R, Dickenson AH, Yarnitsky D, Freeman R, Truini A, Attal N, Finnerup NB, Eccleston C, Kalso E, Bennett DL, Dworkin RH, Raja SN. Neuropathic pain. Nat Rev Dis Primers 2017; 3: 17002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones RC, 3rd, Lawson E, Backonja M. Managing neuropathic pain. Med Clin North Am 2016; 100: 151–167. [DOI] [PubMed] [Google Scholar]
- 5.Han G, Li L, Meng LX. Effects of hyperbaric oxygen on pain-related behaviors and nitric oxide synthase in a rat model of neuropathic pain. Pain Res Manag 2013; 18: 137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leinders M, Uceyler N, Pritchard RA, Sommer C, Sorkin LS. Increased miR-132-3p expression is associated with chronic neuropathic pain. Exp Neurol 2016; 283: 276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakai A, Suzuki H. microRNA and pain. Adv Exp Med Biol 2015; 888: 17–39. [DOI] [PubMed] [Google Scholar]
- 8.Jiangpan P, Qingsheng M, Zhiwen Y, Tao Z. Emerging role of microRNA in neuropathic pain. Curr Drug Metab 2016; 17: 336–344. [DOI] [PubMed] [Google Scholar]
- 9.Kusuda R, Cadetti F, Ravanelli MI, Sousa TA, Zanon S, De Lucca FL, Lucas G. Differential expression of microRNAs in mouse pain models. Mol Pain 2011; 7: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Y, Cao DL, Jiang BC, Yang T, Gao YJ. MicroRNA-146a-5p attenuates neuropathic pain via suppressing TRAF6 signaling in the spinal cord. Brain Behav Immun 2015; 49: 119–129. [DOI] [PubMed] [Google Scholar]
- 11.Herzer S, Silahtaroglu A, Meister B. Locked nucleic acid-based in situ hybridisation reveals miR-7a as a hypothalamus-enriched microRNA with a distinct expression pattern. J Neuroendocrinol 2012; 24: 1492–1504. [DOI] [PubMed] [Google Scholar]
- 12.Sakai A, Saitow F, Miyake N, Miyake K, Shimada T, Suzuki H. miR-7a alleviates the maintenance of neuropathic pain through regulation of neuronal excitability. Brain 2013; 136: 2738–2750. [DOI] [PubMed] [Google Scholar]
- 13.Bali KK, Kuner R. Noncoding RNAs: key molecules in understanding and treating pain. Trends Mol Med 2014; 20: 437–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Gu LQ, Burnette WB, Li J. N98S mutation in NEFL gene is dominantly inherited with a phenotype of polyneuropathy and cerebellar atrophy. J Neurol Sci 2016; 365: 46–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agrawal PB, Joshi M, Marinakis NS, Schmitz-Abe K, Ciarlini PD, Sargent JC, Markianos K, De Girolami U, Chad DA, Beggs AH. Expanding the phenotype associated with the NEFL mutation: neuromuscular disease in a family with overlapping myopathic and neurogenic findings. JAMA Neurol 2014; 71: 1413–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doppler K, Kunstmann E, Kruger S, Sommer C. Painful Charcot-Marie-Tooth neuropathy type 2E/1F due to a novel NEFL mutation. Muscle Nerve 2017; 55: 752–755. [DOI] [PubMed] [Google Scholar]
- 17.Miura M, Miyatsuka T, Katahira T, Sasaki S, Suzuki L, Himuro M, Nishida Y, Fujitani Y, Matsuoka TA, Watada H. Suppression of STAT3 signaling promotes cellular reprogramming into insulin-producing cells induced by defined transcription factors. EBioMedicine 2018; 36: 358–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge A, Wang S, Miao B, Yan M. Effects of metformin on the expression of AMPK and STAT3 in the spinal dorsal horn of rats with neuropathic pain. Mol Med Rep 2018; 17: 5229–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SH, Chung JM. An experimental model for peripheral neuropathy produced by segmental spinal nerve ligation in the rat. Pain 1992; 50: 355–363. [DOI] [PubMed] [Google Scholar]
- 20.Shin J, Yin Y, Park H, Park S, Triantafillu UL, Kim Y, Kim SR, Lee SY, Kim DK, Hong J, Kim DW. p38 siRNA-encapsulated PLGA nanoparticles alleviate neuropathic pain behavior in rats by inhibiting microglia activation. Nanomedicine (Lond) 2018; 13: 1607–1621. [DOI] [PubMed] [Google Scholar]
- 21.Mestre C, Pelissier T, Fialip J, Wilcox G, Eschalier A. A method to perform direct transcutaneous intrathecal injection in rats. J Pharmacol Toxicol Methods 1994; 32: 197–200. [DOI] [PubMed] [Google Scholar]
- 22.Melemedjian OK, Asiedu MN, Tillu DV, Sanoja R, Yan J, Lark A, Khoutorsky A, Johnson J, Peebles KA, Lepow T, Sonenberg N, Dussor G, Price TJ. Targeting adenosine monophosphate-activated protein kinase (AMPK) in preclinical models reveals a potential mechanism for the treatment of neuropathic pain. Mol Pain 2011; 7: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreira MWL, Rodrigues J, Kumar N, Al-Muhtadi J, Korotaev V. Nature-inspired algorithm for training multilayer perceptron networks in e-health environments for high-risk pregnancy care. J Med Syst 2018; 42: 51. [DOI] [PubMed] [Google Scholar]
- 24.Lai A, Moon A, Purmessur D, Skovrlj B, Winkelstein BA, Cho SK, Hecht AC, Iatridis JC. Assessment of functional and behavioral changes sensitive to painful disc degeneration. J Orthop Res 2015; 33: 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Su Z, Liu HL, Li L, Wei M, Ge DJ, Zhang ZJ. Effects of miR-26a-5p on neuropathic pain development by targeting MAPK6 in in CCI rat models. Biomed Pharmacother 2018; 107: 644–649. [DOI] [PubMed] [Google Scholar]
- 26.Wang B, Liu S, Fan B, Xu X, Chen Y, Lu R, Xu Z, Liu X. PKM2 is involved in neuropathic pain by regulating ERK and STAT3 activation in rat spinal cord. J Headache Pain 2018; 19: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Cui H, Huang X, Zhu B, Guan S, Cheng W, Lai Y, Zhang X, Hua ZC. MiR-7a is an important mediator in Fas-associated protein with death domain (FADD)-regulated expression of focal adhesion kinase (FAK). Oncotarget 2016; 7: 51393–51407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouhassira D, Attal N. Translational neuropathic pain research: a clinical perspective. Neuroscience 2016; 338: 27–35. [DOI] [PubMed] [Google Scholar]
- 29.Sainio MT, Ylikallio E, Maenpaa L, Lahtela J, Mattila P, Auranen M, Palmio J, Tyynismaa H. Absence of NEFL in patient-specific neurons in early-onset Charcot-Marie-Tooth neuropathy. Neurol Genet 2018; 4: e244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Z, Zhuo Y, Shen Z, Wang Y, Wang L, Li H, Chen J, Chen W. The role of NEFL in cell growth and invasion in head and neck squamous cell carcinoma cell lines. J Oral Pathol Med 2014; 43: 191–198. [DOI] [PubMed] [Google Scholar]
- 31.Gentil BJ, Mushynski WE, Durham HD. Heterogeneity in the properties of NEFL mutants causing Charcot-Marie-Tooth disease results in differential effects on neurofilament assembly and susceptibility to intervention by the chaperone-inducer, celastrol. Int J Biochem Cell Biol 2013; 45: 1499–1508. [DOI] [PubMed] [Google Scholar]
- 32.Capasso M, Diskin S, Cimmino F, Acierno G, Totaro F, Petrosino G, Pezone L, Diamond M, McDaniel L, Hakonarson H, Iolascon A, Devoto M, Maris JM. Common genetic variants in NEFL influence gene expression and neuroblastoma risk. Cancer Res 2014; 74: 6913–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang ZY, Xiong J, Zhang SS, Wang JJ, Gong ZJ, Dai MH. Up-regulation of microrna-183 promotes cell proliferation and invasion in glioma by directly targeting NEFL. Cell Mol Neurobiol 2016; 36: 1303–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng G, Yuan X, Yuan J, Liu Q, Dai M, Shen C, Ma J, Liao Y, Jiang W. miR-25 promotes glioblastoma cell proliferation and invasion by directly targeting NEFL. Mol Cell Biochem 2015; 409: 103–111. [DOI] [PubMed] [Google Scholar]
- 35.de Chevigny A, Core N, Follert P, Gaudin M, Barbry P, Beclin C, Cremer H. miR-7a regulation of Pax6 controls spatial origin of forebrain dopaminergic neurons. Nat Neurosci 2012; 15: 1120–1126. [DOI] [PubMed] [Google Scholar]
- 36.Yadav P, Selvaraj BT, Bender FL, Behringer M, Moradi M, Sivadasan R, Dombert B, Blum R, Asan E, Sauer M, Julien JP, Sendtner M. Neurofilament depletion improves microtubule dynamics via modulation of Stat3/stathmin signaling. Acta Neuropathol 2016; 132: 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei J, Wang F, Kong LY, Xu S, Doucette T, Ferguson SD, Yang Y, McEnery K, Jethwa K, Gjyshi O, Qiao W, Levine NB, Lang FF, Rao G, Fuller GN, Calin GA, Heimberger AB. miR-124 inhibits STAT3 signaling to enhance T cell-mediated immune clearance of glioma. Cancer Res 2013; 73: 3913–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z, Yuan W, Li B, Chen X, Zhang Y, Chen C, Yu M, Xiu Y, Li W, Cao J, Wang X, Tao W, Guo X, Feng S, Wang T. PEITC promotes neurite growth in primary sensory neurons via the miR-17-5p/STAT3/GAP-43 axis. J Drug Target 2019; 27: 82–93. [DOI] [PubMed] [Google Scholar]
- 39.Wu FX, Bian JJ, Miao XR, Huang SD, Xu XW, Gong DJ, Sun YM, Lu ZJ, Yu WF. Intrathecal siRNA against Toll-like receptor 4 reduces nociception in a rat model of neuropathic pain. Int J Med Sci 2010; 7: 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu M, Petraschka M, McLaughlin JP, Westenbroek RE, Caron MG, Lefkowitz RJ, Czyzyk TA, Pintar JE, Terman GW, Chavkin C. Neuropathic pain activates the endogenous kappa opioid system in mouse spinal cord and induces opioid receptor tolerance. J Neurosci 2004; 24: 4576–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su S, Shao J, Zhao Q, Ren X, Cai W, Li L, Bai Q, Chen X, Xu B, Wang J, Cao J, Zang W. MiR-30b attenuates neuropathic pain by regulating voltage-gated sodium channel Nav1.3 in rats. Front Mol Neurosci 2017; 10: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dominguez E, Rivat C, Pommier B, Mauborgne A, Pohl M. JAK/STAT3 pathway is activated in spinal cord microglia after peripheral nerve injury and contributes to neuropathic pain development in rat. J Neurochem 2008; 107: 50–60. [DOI] [PubMed] [Google Scholar]
- 43.Ding CP, Guo YJ, Li HN, Wang JY, Zeng XY. Red nucleus interleukin-6 participates in the maintenance of neuropathic pain through JAK/STAT3 and ERK signaling pathways. Exp Neurol 2018; 300: 212–221. [DOI] [PubMed] [Google Scholar]
- 44.Yan XT, Ji LJ, Wang Z, Wu X, Wang Q, Sun S, Lu JM, Zhang Y. MicroRNA-93 alleviates neuropathic pain through targeting signal transducer and activator of transcription 3. Int Immunopharmacol 2017; 46: 156–162. [DOI] [PubMed] [Google Scholar]
- 45.Jin H, Du XJ, Zhao Y, Xia DL. XIST/miR-544 axis induces neuropathic pain by activating STAT3 in a rat model. J Cell Physiol 2018; 233: 5847–5855. [DOI] [PubMed] [Google Scholar]