Abstract
Background
We have previously identified 11 promising candidate genes for milk composition traits by resequencing the whole genomes of 8 Holstein bulls with extremely high and low estimated breeding values for milk protein and fat percentages (high and low groups), including FCGR2B, CENPE, RETSAT, ACSBG2, NFKB2, TBC1D1, NLK, MAP3K1, SLC30A2, ANGPT1 and UGDH those contained 25 indels between high and low groups. In this study, the purpose was to further examine whether these candidates have significant genetic effects on milk protein and fat traits.
Results
With PCR product sequencing, 13 indels identified by whole genome resequencing were successfully genotyped. With association analysis in 769 Chinese Holstein cows, we found that the indel in FCGR2B was significantly associated with milk yield, protein yield and protein percentage (P = 0.0041 to 0.0297); five indels in CENPE and one indel in MAP3K1 were markedly relevant to milk yield, fat yield and protein yield (P < 0.0001 to 0.0073); polymorphism in RETSAT was evidently associated with milk yield, fat yield, protein yield and protein percentage (P = 0.0001 to 0.0237); variant in ACSBG2 affected fat yield and protein percentage (P = 0.0088 and 0.0052); one indel in TBC1D1 was with respect to fat percentage and protein percentage (P = 0.0224 and 0.0209). Significant associations were shown between indels in NLK and protein yield and protein percentage (P = 0.0012 to 0.0257); variant in UGDH was related to the milk yield (P = 0.0312). The two exonic indels in FCGR2B and CENPE were predicted to change the mRNA and protein secondary structures, and resulted in the corresponding protein dysfunction.
Conclusion
Our findings presented here provide the first evidence for the associations of eight functional genes with milk yield and composition traits in dairy cattle.
Electronic supplementary material
The online version of this article (10.1186/s12863-019-0751-y) contains supplementary material, which is available to authorized users.
Keywords: Indel, Candidate gene, Genetic effect, Milk composition traits, Dairy cattle
Backgroud
In dairy cattle, milk yield and milk composition traits are the most important economic traits, which are controlled by numerous environmental factors and genes [1–4]. Over the past decades, unraveling the major genes and causal mutations with large effect on milk yield and composition traits is one of the important research fields for researchers. Quantitative trait locus (QTL) mapping and genome-wide association study (GWAS) have been widely applied to identify the QTLs, candidate genes and mutations affecting milk production traits in dairy cattle [5–8], and a large number of QTLs and genetic associations have been detected using such two approaches so far (http://www.animalgenome.org/cgi-bin/QTLdb/index). In recent years, short insertion and deletion (indel), as the second main form of genomic variation, has been increasingly paid more attention and has made great contribution to investigations on genetic and phenotypic diversities in human, chicken, pig and dairy cattle [9–13]. A previous study found that 2–18 base pairs (bp) indel located upstream of TAL bHLH transcription factor 1 (TAL1) was responsible for the T-cell acute lymphoblastic leukemia (T-ALL) [14]. In chicken, the 9–15 bp indel of premelanosome protein (PMEL17) gene was confirmed to be the causative mutation for the plumage color (Dominant white, Dun and Smoky) [10]. In pig, an intronic inserted retrotransposon of sperm flagellar 2 (SPEF2) led to the immotile short-tail sperm defect [15]. In Belgian blue cattle, a 11-bp indel in myostatin (MSTN) gene resulted in double-muscled phenotype [9], and an exonic 15-bp insertion in coagulation factor XI (F11) gene caused the factor XI deficiency in Japanese black cattle [11]. However, up to now, limited research of indel polymorphisms associated with milk production traits in dairy cattle has been reported [16].
With the rapidly emergence of next-generation sequencing (NGS), whole genome resequencing has been an important tool in the efforts to detect polymorphsims which were contributed to the complex traits or economic traits in human and domestic animals [17–20]. In our previous whole genome resequencing study, we identified over 0.9 million short indels and 3625 common differential indels with the same allelic distribution directions based on the 8 Holstein bulls with extremely high or low estimated breeding values (EBVs) of milk protein and fat percentages (high and low groups) [21]. Based on this, 11 genes were identified as the promising candidates affecting milk compositions traits in dairy cattle, including FCGR2B, CENPE, RETSAT, ACSBG2, NFKB2, TBC1D1, NLK, MAP3K1, SLC30A2, ANGPT1 and UGDH, which contained 25 differential indels [21]. Thus, the aim of this study was to further validate whether these identified indels in the 11 genes significantly impact on milk yield and compositions traits in Chinese Holstein population.
Results
Indel verification and genotyping
Based on two DNA pools from 40 Holstein sires, with PCR product sequencing, 22 of 25 indels identified by whole genome resequencing [21] were confirmed as true ones (Additional file 2), among them, four indels were identified for the first time (Table 1). Subsequently, 13 indels in 8 genes were successfully genotyped and performed for association analysis. Of the 13 indels, two indels, including rs381714237 in FCGR2B, ss2137349053 in CENPE were located in the exons, whilst, the remaining 11 indels were located in the intronic regions. Chi-squared test showed that all the 13 indels were in Hardy-Weinberg equilibrium (P > 0.05). The genotype frequencies and allele frequencies of the 13 indels were summarized in Table 2.
Table 1.
Detailed information of 24 indels of 11 genes identified in Chinese Holstein cattle
| Indel | Gene | Location | GenBank no. | Position in UMD_3.1 | Indel Sequence | Confirmed? |
|---|---|---|---|---|---|---|
| 1 N ins | FCGR2B | exon7 | rs381714237 | chr3:7930047 | G | 1 |
| 3 N ins | CENPE | exon58 | ss2137349053 | chr6:23018080 | AGA | 1 |
| 3 N del | CENPE | exon68 | – | chr6:23026632–23026634 | TAG | 3 |
| 3 N ins | CENPE | intron13 | rs385060942 | chr6:22983076 | GTT | 1 |
| 1 N ins | CENPE | intron13 | ss2137349051 | chr6:22983397 | T | 1 |
| 1 N del | CENPE | intron18 | rs384082187 | chr6:22989805 | A | 2 |
| 21 N ins | CENPE | intron24 | rs377812754 | chr6:22996564–22996573 | ACTTAAGTATATAACCTTAAC | 2 |
| 2 N del | CENPE | intron41 | rs453960300 | chr6:23018994–23018995 | CC | 1 |
| 1 N del | CENPE | intron49 | rs378415122 | chr6:23036105 | C | 1 |
| 4 N ins | CENPE | intron51 | ss2137349056 | chr6:23040582 | ACAC | 4 |
| 2 N del | RETSAT | 3’UTR | rs136527375 | chr11:49489416–49489417 | AA | 2 |
| 9 N ins | RETSAT | intron6 | rs134985825 | chr11:49485899 | ATTCTGGGG | 1 |
| 1 N ins | ACSBG2 | intron7 | rs377943075 | chr7:19476990 | G | 1 |
| 2 N ins | NFKB2 | 5′ regulatory region | – | chr26:22891203 | GG | 3 |
| 1 N del | TBC1D1 | intron1 | rs136639319 | chr6:58898979 | T | 1 |
| 1 N ins | NLK | intron1 | rs137724510 | chr19:20180649 | T | 2 |
| 2 N del | NLK | intron1 | rs379188781 | chr19:20189055–20189056 | AT | 1 |
| 1 N del | NLK | intron3 | rs135129224 | chr19:20264835 | A | 2 |
| 4 N del | NLK | intron3 | rs134444531 | chr19:20276109–20276112 | AAAA | 1 |
| 5 N del | MAP3K1 | intron16 | ss2137349058 | chr20:22365627–22365631 | CATTT | 1 |
| 6 N del | SLC30A2 | intron2 | ss2137349049 | chr2:127640012–127640017 | TTTTTG | 2 |
| 2 N ins | ANGPT1 | intron1 | ss2137349057 | chr14:59305051 | AT | 2 |
| 1 N ins | UGDH | intron7 | rs383327605 | chr6:60236955 | T | 2 |
| 1 N ins | UGDH | intron2 | ss2019489562 | chr6:60252782 | G | 1 |
Note: 1 indels were genotyped successfully; 2 indels were failed to genotype using MALDI-TOF MS; 3 indels were not polymorphic in current population; 4 primers of indel were failed to design
Table 2.
The genotypic and allelic frequencies of 13 indels of 8 genes
| Locus | Gene | Genotype | Genotype frequencies | Allele | Allele frequencies |
|---|---|---|---|---|---|
| rs381714237 | FCGR2B | del/del | 0.115 | del | 0.369 |
| del/ins | 0.508 | ins | 0.631 | ||
| ins/ins | 0.377 | ||||
| ss2137349053 | CENPE | del/del | 0.208 | del | 0.462 |
| del/ins | 0.507 | ins | 0.538 | ||
| ins/ins | 0.284 | ||||
| rs385060942 | CENPE | del/del | 0.206 | del | 0.462 |
| del/ins | 0.512 | ins | 0.538 | ||
| ins/ins | 0.282 | ||||
| ss2137349051 | CENPE | del/del | 0.277 | del | 0.534 |
| del/ins | 0.513 | ins | 0.466 | ||
| ins/ins | 0.210 | ||||
| rs453960300 | CENPE | ins/ins | 0.212 | ins | 0.467 |
| ins/del | 0.511 | del | 0.533 | ||
| del/del | 0.277 | ||||
| rs378415122 | CENPE | ins/ins | 0.210 | ins | 0.467 |
| ins/del | 0.514 | del | 0.533 | ||
| del/del | 0.276 | ||||
| rs134985825 | RETSAT | del/del | 0.276 | del | 0.524 |
| del/ins | 0.497 | ins | 0.476 | ||
| ins/ins | 0.227 | ||||
| rs377943075 | ACSBG2 | del/del | 0.082 | del | 0.276 |
| del/ins | 0.388 | ins | 0.724 | ||
| ins/ins | 0.530 | ||||
| rs136639319 | TBC1D1 | ins/ins | 0.025 | ins | 0.186 |
| ins/del | 0.321 | del | 0.814 | ||
| del/del | 0.654 | ||||
| rs379188781 | NLK | ins/ins | 0.374 | ins | 0.608 |
| ins/del | 0.468 | del | 0.392 | ||
| del/del | 0.158 | ||||
| rs134444531 | NLK | ins/ins | 0.265 | ins | 0.532 |
| ins/del | 0.532 | del | 0.468 | ||
| del/del | 0.202 | ||||
| ss2137349058 | MAP3K1 | ins/ins | 0.207 | ins | 0.466 |
| ins/del | 0.518 | del | 0.534 | ||
| del/del | 0.275 | ||||
| ss2019489562 | UGDH | del/del | 0.220 | del | 0.466 |
| del/ins | 0.491 | ins | 0.534 | ||
| ins/ins | 0.289 |
Associations between indels and five milk production traits
The results of associations between the 13 indels and five milk production traits were shown in Table 3. It was observed that all these indels were significantly associated with at least one of the milk traits (P < 0.0001 to P = 0.0312) as described below.
Table 3.
Association results of the thirteen indels in eight genes on the five milk production traits (least squares mean ± SE)
| Locus | Gene | Genotype (No.) | MY | FY | FP | PY | PP |
|---|---|---|---|---|---|---|---|
| rs381714237 | FCGR2B | del/del(86) | 10,891 ± 99.80 | 385.05 ± 4.21 | 3.59 ± 0.040 | 319.32 ± 3.10ab | 2.95 ± 0.014a |
| ins/del(380) | 10,702 ± 64.80 | 383.94 ± 2.89 | 3.66 ± 0.027 | 315.86 ± 2.11Aa | 2.98 ± 0.009ab | ||
| ins/ins(282) | 10,834 ± 68.20 | 385.61 ± 3.03 | 3.64 ± 0.028 | 321.83 ± 2.20Bb | 2.99 ± 0.009b | ||
| P value | 0.0297* | 0.7926 | 0.1738 | 0.0041** | 0.0198* | ||
| ss2137349053 | CENPE | del/del(153) | 10,403 ± 55.66A | 374.36 ± 2.82Aa | 3.61 ± 0.034 | 308.40 ± 2.65A | 2.97 ± 0.024 |
| del/ins(373) | 10,661 ± 47.88B | 378.61 ± 2.43Aa | 3.57 ± 0.027 | 315.70 ± 2.14Ba | 2.97 ± 0.017 | ||
| ins/ins(209) | 10,852 ± 51.03C | 385.99 ± 2.59B | 3.58 ± 0.031 | 321.05 ± 2.37Bb | 2.97 ± 0.020 | ||
| P value | < 0.0001** | < 0.0001** | 0.3733 | < 0.0001** | 0.9882 | ||
| rs385060942 | CENPE | del/del(152) | 10,494 ± 84.00Aa | 376.49 ± 3.60Aa | 3.60 ± 0.034 | 310.45 ± 2.62B | 2.96 ± 0.012 |
| del/ins(377) | 10,717 ± 65.77Aa | 381.44 ± 2.93ab | 3.57 ± 0.027 | 318.18 ± 2.13Aa | 2.97 ± 0.009 | ||
| ins/ins(208) | 10,871 ± 74.83B | 387.82 ± 3.26Bb | 3.59 ± 0.031 | 322.24 ± 2.38Aa | 2.97 ± 0.010 | ||
| P value | < 0.0001** | 0.0045** | 0.6063 | < 0.0001** | 0.6166 | ||
| ss2137349051 | CENPE | ins/ins(206) | 10,817 ± 75.26A | 386.81 ± 3.28A | 3.60 ± 0.031 | 321.11 ± 2.39A | 2.97 ± 0.010 |
| del/ins(382) | 10,586 ± 65.52Ba | 378.41 ± 2.92Ba | 3.59 ± 0.027 | 314.42 ± 2.12Ba | 2.98 ± 0.009 | ||
| del/del(156) | 10,397 ± 83.94Bb | 374.49 ± 3.61Ba | 3.61 ± 0.034 | 308.61 ± 2.63Bb | 2.97 ± 0.012 | ||
| P value | < 0.0001** | 0.0010** | 0.7289 | < 0.0001** | 0.9571 | ||
| rs453960300 | CENPE | ins/ins(158) | 10,380 ± 83.33Aa | 375.15 ± 3.58 | 3.63 ± 0.034 | 307.63 ± 2.61Aa | 2.98 ± 0.012 |
| ins/del(381) | 10,586 ± 66.10b | 380 ± 2.95 | 3.61 ± 0.027 | 313.34 ± 2.15b | 2.98 ± 0.009 | ||
| del/del(207) | 10,764 ± 74.83Bc | 385.96 ± 3.26 | 3.61 ± 0.031 | 318.18 ± 2.38Bc | 2.97 ± 0.010 | ||
| P value | < 0.0001** | 0.0073** | 0.7589 | 0.0002** | 0.9461 | ||
| rs378415122 | CENPE | ins/ins(158) | 10,373 ± 83.79Aa | 373.03 ± 3.60Aa | 3.62 ± 0.034 | 306.81 ± 2.62Aa | 2.97 ± 0.012 |
| ins/del(386) | 10,547 ± 65.79Aa | 377.75 ± 2.93a | 3.60 ± 0.027 | 312.15 ± 2.13Aa | 2.97 ± 0.009 | ||
| del/del(207) | 10,785 ± 74.23B | 386.21 ± 3.23Bb | 3.60 ± 0.030 | 319.19 ± 2.36B | 2.97 ± 0.010 | ||
| P value | < 0.0001** | 0.0004** | 0.8913 | < 0.0001** | 0.9535 | ||
| rs134985825 | RETSAT | del/del(205) | 10,774 ± 75.77Aa | 383.94 ± 3.30ab | 3.58 ± 0.031 | 318.23 ± 2.40ab | 2.96 ± 0.010a |
| del/ins(370) | 10,563 ± 66.51B | 378.10 ± 2.95Aa | 3.60 ± 0.027 | 314.44 ± 2.15Aa | 2.98 ± 0.009ab | ||
| ins/ins(169) | 10,823 ± 79.45Aa | 388.92 ± 3.43Bb | 3.61 ± 0.032 | 323.7 ± 2.50Bb | 2.99 ± 0.011b | ||
| P value | 0.0003** | 0.0009** | 0.6500 | 0.0001** | 0.0237* | ||
| rs377943075 | ACSBG2 | del/del(62) | 10,746 ± 111.22 | 378.40 ± 4.68Aa | 3.56 ± 0.045 | 318.61 ± 3.42 | 3.00 ± 0.016ab |
| del/ins(294) | 10,776 ± 69.04 | 387.18 ± 3.03ab | 3.61 ± 0.028 | 319.22 ± 2.19 | 2.99 ± 0.009Ab | ||
| ins/ins(401) | 10,884 ± 63.81 | 391.00 ± 2.84Bb | 3.62 ± 0.026 | 319.31 ± 2.05 | 2.97 ± 0.008Ba | ||
| P value | 0.1334 | 0.0088** | 0.4393 | 0.9749 | 0.0052** | ||
| rs136639319 | TBC1D1 | ins/ins(19) | 10,752 ± 182.68 | 391.92 ± 7.47 | 3.65 ± 0.073ab | 320.14 ± 5.45 | 2.98 ± 0.026ab |
| del/ins(242) | 10,564 ± 72.44 | 383.75 ± 3.18 | 3.65 ± 0.030a | 314.38 ± 2.32 | 2.99 ± 0.010a | ||
| del/del(493) | 10,659 ± 64.49 | 380.06 ± 2.88 | 3.58 ± 0.027b | 314.42 ± 2.1 | 2.96 ± 0.009b | ||
| P value | 0.2493 | 0.1198 | 0.0224* | 0.5384 | 0.0209* | ||
| rs379188781 | NLK | ins/ins(277) | 10,676 ± 70.45 | 377.09 ± 3.11 | 3.56 ± 0.029 | 318.23 ± 2.26 | 2.99 ± 0.009Aa |
| del/ins(347) | 10,627 ± 67.41 | 381.09 ± 3.00 | 3.60 ± 0.028 | 314.48 ± 2.18 | 2.97 ± 0.009b | ||
| del/del(117) | 10,749 ± 90.28 | 384.24 ± 3.83 | 3.57 ± 0.037 | 315.92 ± 2.79 | 2.95 ± 0.013Bb | ||
| P value | 0.3389 | 0.1020 | 0.2615 | 0.1279 | 0.0047** | ||
| rs134444531 | NLK | ins/ins(197) | 10,748 ± 78.14 | 384.18 ± 3.40 | 3.58 ± 0.032 | 322.04 ± 2.47Aa | 2.99 ± 0.011a |
| del/ins(395) | 10,723 ± 65.31 | 386.23 ± 2.91 | 3.61 ± 0.027 | 317.93 ± 2.12a | 2.97 ± 0.009ab | ||
| del/del(150) | 10,563 ± 85.88 | 381.58 ± 3.68 | 3.63 ± 0.035 | 312.32 ± 2.68Bb | 2.96 ± 0.012b | ||
| P value | 0.0733 | 0.3026 | 0.4078 | 0.0012** | 0.0257* | ||
| ss2137349058 | MAP3K1 | ins/ins(152) | 10,482 ± 82.27Aa | 372.63 ± 3.54Aa | 3.58 ± 0.034 | 310.01 ± 2.58Aa | 2.97 ± 0.011 |
| del/ins(380) | 10,613 ± 66.30Aa | 376.47 ± 2.94Aa | 3.56 ± 0.027 | 314.12 ± 2.14Aa | 2.97 ± 0.009 | ||
| del/del(202) | 10,909 ± 77.27Bb | 390.47 ± 3.35Bb | 3.59 ± 0.032 | 323.13 ± 2.44Bb | 2.97 ± 0.011 | ||
| P value | < 0.0001** | < 0.0001** | 0.4863 | < 0.0001** | 0.8495 | ||
| ss2019489562 | UGDH | del/del(405) | 10,860 ± 64.35a | 387.55 ± 2.89 | 3.64 ± 0.027 | 318.31 ± 2.11 | 2.99 ± 0.009 |
| del/ins(4) | 9902 ± 384.75b | 376.58 ± 15.49 | 3.81 ± 0.153 | 299.74 ± 11.30 | 3.02 ± 0.056 | ||
| ins/ins(342) | 10,798 ± 66.31ab | 388.28 ± 2.95 | 3.64 ± 0.027 | 315.05 ± 2.13 | 2.97 ± 0.009 | ||
| P value | 0.0312* | 0.7267 | 0.5077 | 0.0532 | 0.0567 |
Note:*significant association at the significance level of 0.05; **significant association at the significance level of 0.01
The different superscripts (A,B within the same column with different superscripts indicate P < 0.01; a,b indicate P < 0.05) adjusted after correction for multiple testing indicate significant differences among the genotypes
MY milk yield, FY fat yield, FP fat percentage, PY protein yield, PP protein percentage
Exonic indels
The exonic indel rs381714237 in FCGR2B was associated with milk yield (P = 0.0297), protein yield (P = 0.0041) and protein percentage (P = 0.0198). The other exonic indel ss2137349053 in CENPE, was strongly associated with milk yield (P < 0.0001), fat yield (P < 0.0001) and protein yield (P < 0.0001).
Intronic indels
The four intronic indels (rs385060942, ss2137349051, rs453960300 and rs378415122) in CENPE were significantly associated with milk yield (P < 0.0001), fat yield (P = 0.0004 to 0.0073) and protein yield (P < 0.0001 to 0.0002). Additionally, the five indels (four intronic indels and one exonic indel above) of CENPE gene were found to be highly linked (r2 > 0.98), and one haplotype block was inferred as presented in Fig. 1. Haplotype-based association analysis showed that the haplotype combination was evidently associated with milk yield, fat yield and protein yield as well (P < 0.0001 to P = 0.0076) (Table 4).
Fig. 1.
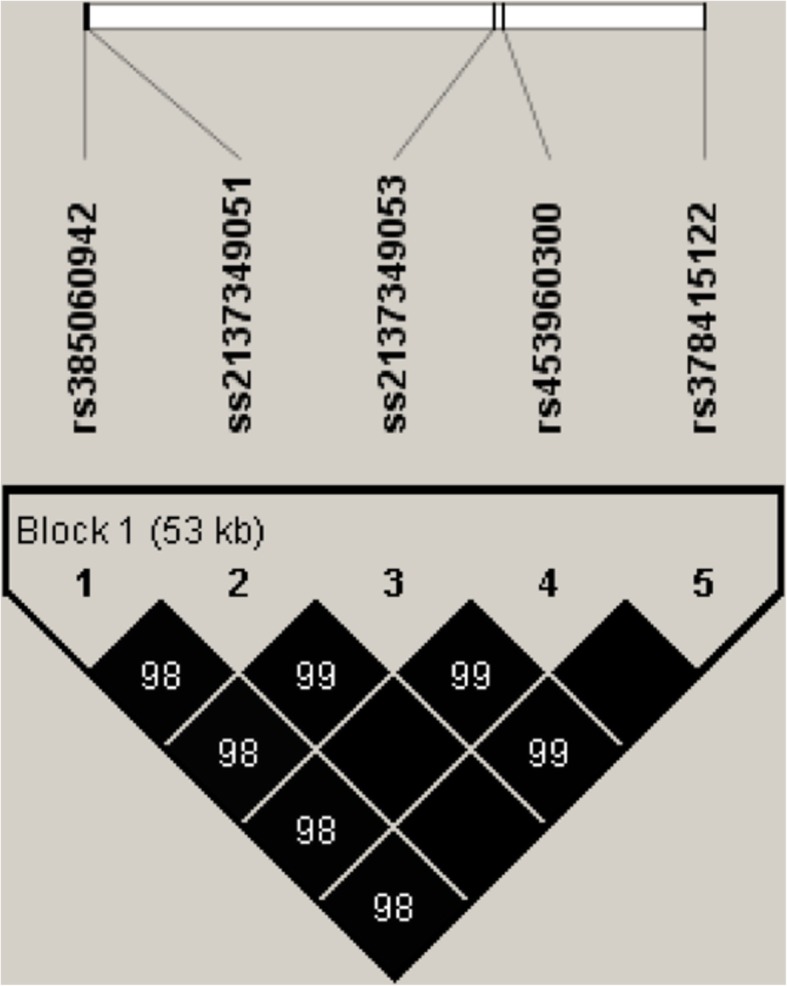
Linkage disequilibrium estimated of the five indels in CENPE gene. The values in boxes are pair-wise indel correlations (r2)
Table 4.
Haplotype analysis of CENPE gene (least squares mean ± SE)
| Haplotype (No.) | MY | FY | FP | PY | PP |
|---|---|---|---|---|---|
| H1H1 (198) | 10,874 ± 77.45Aa | 389.53 ± 3.38Aa | 3.60 ± 0.031 | 321.43 ± 2.46Aa | 2.97 ± 0.011 |
| HIH2 (356) | 10,734 ± 68.09Aa | 382.37 ± 3.02b | 3.58 ± 0.028 | 316.68 ± 2.20Ab | 2.96 ± 0.009 |
| H2H2 (140) | 10,498 ± 88.34Bb | 378.32 ± 3.78Bb | 3.62 ± 0.036 | 309.40 ± 2.75B | 2.95 ± 0.012 |
| P value | 0.0003** | 0.0076** | 0.4045 | < 0.0001** | 0.7803 |
Note:*significant association at the significance level of 0.05; **significant association at the significance level of 0.01
The different superscripts (A,B within the same column with different superscripts indicate P < 0.01; a,b indicate P < 0.05) adjusted after correction for multiple testing indicate significant differences among the genotypes
MY milk yield, FY fat yield, FP fat percentage, PY protein yield, PP protein percentage
Indel rs134985825 in the intron 6 of RETSAT showed remarkable effects on milk yield, protein yield, fat yield and protein percentage (P = 0.0001 to 0.0237). For ACSBG2, indel rs377943075 in the intron 7 was significantly associated with fat yield (P = 0.0088) and protein percentage (P = 0.0052). Variant rs136639319 in the intron 3 of TBC1D1was significantly associated with fat percentage (P = 0.0224) and protein percentage (P = 0.0209).
For the indel rs379188781 in the intron 1 of NLK, it was found to be associated with protein percentage (P = 0.0047), the other indel rs134444531 in the intron 3 was associated with protein yield (P = 0.0012) and protein percentage (P = 0.0257). While, no LD was observed between such two indels (r2 = 0.14).
For MAP3K1, indel ss2137349058 in the intron 16 was markedly associated with milk yield, fat yield and protein yield (P < 0.0001).
For the intronic indel of UGDH gene, the indel ss2019489562 located in intron 2 was significantly associated with milk yield (P = 0.0312).
Additionally, the significant additive, dominant and allele substitution effects of the 13 indels on the five milk traits were observed as well (Table 5).
Table 5.
Genetic effects of thirteen indels in eight genes on five milk production traits
| Locus | Gene | Gene effects | MY | FY | FP | PY | PP |
|---|---|---|---|---|---|---|---|
| rs381714237 | FCGR2B | Additive effect(a) | 66.13* | 0.83 | −0.0072 | 2.99** | 0.0051 |
| Dominant effect(d) | 123.27 | 0.27 | − 0.0618 | 0.47 | −0.0348** | ||
| Substitution effect(α) | 30.92 | 0.76 | 0.0105 | 2.85* | 0.0150* | ||
| ss2137349053 | CENPE | Additive effect(a) | 224.66** | 5.81** | −0.0174 | 6.32** | − 0.0002 |
| Dominant effect(d) | 33.43 | −1.57 | −0.0246 | 0.98 | 0.0028 | ||
| Substitution effect(α) | 227.29** | 5.69** | −0.0193 | 6.4** | 0.0001 | ||
| rs385060942 | CENPE | Additive effect(a) | −188.63** | −5.66** | 0.007 | −5.89** | −0.0042 |
| Dominant effect(d) | 34.37 | −0.71 | −0.0218 | 1.84 | 0.0065 | ||
| Substitution effect(α) | −191.51** | −5.60** | 0.0089 | −6.05** | −0.0048 | ||
| ss2137349051 | CENPE | Additive effect(a) | −209.61** | − 6.16** | 0.0093 | − 6.25** | −0.0009 |
| Dominant effect(d) | −20.82 | − 2.24 | −0.014 | −0.44 | 0.0022 | ||
| Substitution effect(α) | −208.06** | −6.00** | 0.0103 | −6.22** | −0.0011 | ||
| rs453960300 | CENPE | Additive effect(a) | − 192.46** | −5.40** | 0.0105 | − 5.27** | 0.0015 |
| Dominant effect(d) | 13.90 | −0.55 | −0.0106 | 0.43 | 0.0017 | ||
| Substitution effect(α) | −193.46** | −5.36** | 0.0113 | −5.31** | 0.0014 | ||
| rs378415122 | CENPE | Additive effect(a) | −205.95** | −6.59** | 0.0069 | −6.19** | −0.0011 |
| Dominant effect(d) | −32.03 | −1.87 | −0.0068 | − 0.85 | 0.0022 | ||
| Substitution effect(α) | −203.68** | −6.46** | 0.0073 | −6.13** | −0.0012 | ||
| rs134985825 | RETSAT | Additive effect(a) | 24.57 | 2.49 | 0.0156 | 2.74* | 0.0163** |
| Dominant effect(d) | − 235.31** | −8.32** | 0.0001 | −6.52** | 0.0036 | ||
| Substitution effect(α) | 29.53 | 2.67 | 0.0156 | 2.88* | 0.0162** | ||
| rs377943075 | ACSBG2 | Additive effect(a) | −69.28 | −6.30** | −0.0272 | −0.35 | 0.0154* |
| Dominant effect(d) | −39.32 | 2.48 | 0.0178 | 0.26 | 0.0108 | ||
| Substitution effect(α) | −56.34 | −7.12* | −0.033 | −0.44 | 0.0118 | ||
| rs136639319 | TBC1D1 | Additive effect(a) | 47.54 | −1.85 | −0.0342** | 0.02 | −0.0127** |
| Dominant effect(d) | 140.15 | 10.02 | 0.0346 | 5.74 | 0.0103 | ||
| Substitution effect(α) | 98.36 | 1.79 | −0.0216 | 2.11 | −0.0089 | ||
| rs379188781 | NLK | Additive effect(a) | −36.45 | −3.58 | −0.0057 | 1.16 | 0.0194** |
| Dominant effect(d) | −85.74 | 0.43 | 0.0343 | −2.59 | −0.0039 | ||
| Substitution effect(α) | −55.71 | −3.48 | 0.002 | 0.58 | 0.0185* | ||
| rs134444531 | NLK | Additive effect(a) | 92.55 | 1.30 | −0.0226 | 4.86** | 0.0169* |
| Dominant effect(d) | 67.33 | 3.36 | 0.0084 | 0.75 | −0.0055 | ||
| Substitution effect(α) | 97.88 | 1.57 | −0.0219 | 4.92** | 0.0164* | ||
| ss2137349058 | MAP3K1 | Additive effect(a) | −213.49** | −8.92** | −0.0046 | −6.56** | 0.0013 |
| Dominant effect(d) | −82.74 | −5.08* | −0.0264 | −2.45 | 0.0043 | ||
| Substitution effect(α) | −208.37** | −8.60** | −0.0029 | −6.41** | 0.0011 | ||
| ss2019489562 | UGDH | Additive effect(a) | 31.00 | −0.36 | − 0.0032 | 1.63 | 0.0097* |
| Dominant effect(d) | −925.95* | −11.33 | 0.1715 | −16.94 | 0.0462 | ||
| Substitution effect(α) | −42.62 | −1.27 | 0.0104 | 0.28 | 0.0134* |
Note: (a), (d), (α) means Additive, Dominant and Substitution effect, respectively
* means the additive, dominant or allele substitution effect of the locus indicated differ at P < 0.05 and ** means the additive, dominant or allele substitution effect of the locus indicated differ at P < 0.01
MY milk yield, FY fat yield, FP fat percentage, PY protein yield, PP protein percentage
Prediction the mRNA and protein structures
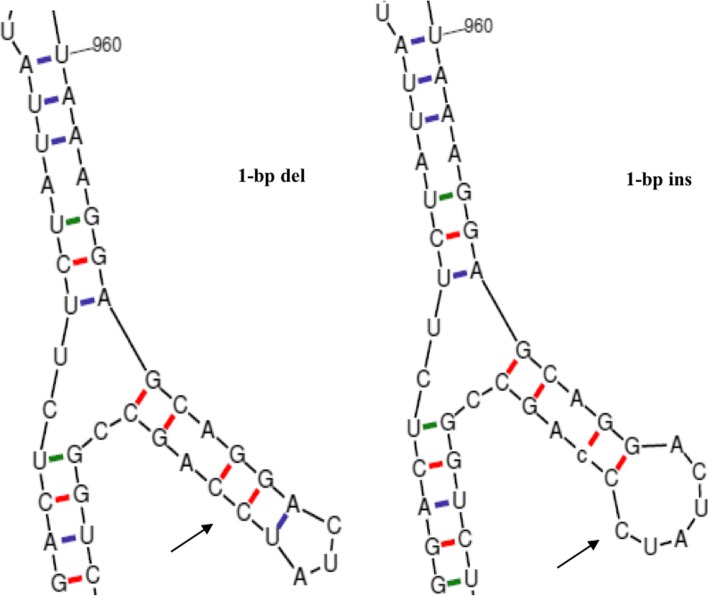
Using a statistical folding algorithm, the alteration of the most stable mRNA secondary structures caused by the two exonic indels for FCGR2B and CENPE were observed for both the ins/ins and del/del genotypes. As illustrated in Fig. 2, obvious structural differences spanning the position 971–980 between the ins/ins and del/del genotypes of the indel rs381714237 in FCGR2B gene were observed. The free energy (∆G) of the ins allele was predicted to be higher (∆G = − 468.70 kcal/mol) than the del allele (∆G = − 470.30 kcal/mol). Correspondingly, the ins allele was deduced to form one larger single loop structure, which potentially decreasing the stability of mRNA (∆∆G = + 1.6 kcal/mol). It is worth mentioning that previous studies have evidenced that the ∆∆G ranged from − 3.9 kcal/mol to + 4.0 kcal/mol could affect the mRNA stability [22–28]. In addition, indel rs381714237 of FCGR2B was predicted to decrease the number of amino acid by 38, which might change protein structure and function. As a result, differences of the protein secondary structures were predicted between the FCGR2B proteins corresponding to alleles del and ins with regard to alpha helix (21.64% vs. 16.45%), extended strand (23.10% vs. 24.67%), beta turn (7.02% vs. 7.89%) and random coil (48.25% vs. 50.99%) using the SOPMA program.
Fig. 2.
The predicted mRNA secondary structures corresponding to the exonic indel of FCGR2B gene
For the non-frameshiting indel, subtle change of mRNA secondary structures between the two homozygous genotypes of indel ss2137349053 in CENPE was occurred (data not shown). The free energy was altered from − 1816.10 kcal/mol for the del allele to − 1818.90 kcal/mol for the ins allele. While, slight difference was predicted for the CENPE protein in accordance between the del/del and ins/ins genotypes, alpha helix (72.57% vs. 72.69%), and random coil (13.94% vs. 13.82%). There was no change of extended strand and beta turn for CENPE protein.
Discussion
In the present work, we confirmed that 13 indels belonging to 8 candidate genes (FCGR2B, CENPE, RETSAT, ACSBG2, TBC1D1, NLK, MAP3K1 and UGDH) for milk compositions identified by our previous whole genome resequencing study [21] showed significant genetic effects on at least one of milk traits in dairy cattle. As far as our knowledge, this is the first report to connect these genes to milk production traits of dairy cattle.
Among the total 25 differential indels with the same allelic distribution directions between the bulls in high and low groups identified by our previous whole genome resequencing study [21], indel rs383700527 (3 N ins) located upstream of ACSBG2 gene was found to contribute to milk fat in a cis-regulatory manner (unpublished data). Thus, we investigated another 24 indels in the present study. Among them, one intronic indel (4 N ins in CENPE) was failed to be verified by Sanger sequencing due to the special characteristic of the flanking sequence with lower GC% and repetitive DNA sequences. Two indels (3 N del in CENPE and 2 N ins in NFKB2) didn’t show polymorphic in this study. Eight indels (1 N del and 21 N ins in CENPE, 2N del in RETSAT, 1 N ins and 1 N del in NLK, 6 N del in SLC30A2, 2 N ins in ANGPT1 and 1N ins in UGDH) were failed to be genotyped by using MALDI-TOF MS. The possible reason may be that MALDI-TOF MS for multiplex genotyping was relied on multiplex-PCR primers and extended primers to genotype multiple loci [29], simultaneously, the primer design was depended on sequence composition, molecular weight, annealing temperature and reaction efficiencies of each locus [29]. Hence, a total of 13 polymorphic indels were successfully genotyped and performed for association analysis.
Significant associations between candidate genes and milk production traits
Six indels in FCGR2B and CENPE
For indel rs381714237 in FCGR2B, we demonstrated that ins/ins genotype had higher protein percentage. As a regulator, FCGR2B was contributed to immune response [30]. Additionally, bovine mammary gland is a product of the innate immune system and active during lactation. Thus, these evidences indicated that FCGR2B might affect milk protein percentage through impacting the cows on immune response during lactation.
For the five indels in CENPE, the association results revealed that ins/ins genotypes were dominant compared with del/del genotypes for milk yield, fat yield and protein yield. Previous report has found that CENPE acted as a monitor protein and was necessary for cell cycle [31]. Thus, it appeared that the CENPE might affect these traits through modulating bovine mammary gland development.
Seven indels located in six genes
Our association analysis confirmed that the ins/ins genotype of the indel rs134985825 in RETSAT gene increased milk yield, fat yield, and protein yield. RETSAT was considered as a regulator for liver metabolism, and was critical for lipid accumulation and adipogenesis promotion [32]. Previous research has investigated that the polymorphisms of RETSAT gene were associated with premium cut yields and backfat thickness in pig [33]. Taken together, we speculated that RETSAT might affect milk traits through influencing the lipid metabolism.
Herein, we found that individuals with ins/ins genotype of indel rs377943075 in ACSBG2 showed higher fat yield than those with del/del genotype. The ACSBG2 gene encodes the protein that belongs to a member of the acyl-CoA synthetase family and participated in PPAR signaling pathway and involved in lipid metabolism and lipid droplet formation [34, 35]. Previous researchers have found that polymorphisms of ACSBG2 showed positive effects on yolk development and abdominal fat weight [36].
In current study, our results also showed a significant relationship between the indel rs136639319 in TBC1D1 and fat percentage as well as protein percentage. It was worth mentioning that TBC1D1, as a member of Rab GTPase-activating proteins (GAPs), was involved in translocation of GLUT4 to the plasma membrane. Polymorphisms in TBC1D1 have been observed to show significant effects on severe obesity or carcass in human [37] and chicken [38], respectively, suggesting exhibiting functions related to lipid and energy homeostasis as reported by Hargett et al. [39, 40].
Two intronic indel (rs379188781 and rs134444531) in NLK showed strong associations with protein yield and protein percentage. Interestingly, Cole et al. reported that one single nucleotide polymorphism (SNP) (ARS-BFGL-NGS-106227) significantly associated with protein percentage (P = 5.59 × 10− 8) was merely 90 kb away from the NLK gene [41]. Furthermore, NLK, as a member of MAPK subfamily, had an essential role in mediating the mTORC1 signaling pathway which was involved in milk protein synthesis [42, 43]. Together, these data suggested that significant variation of protein yield and protein percentage might be regulated by NLK.
As for MAP3K1, individuals with del/del genotype of indel polymorphism ss2137349058 had higher milk yield, fat yield and protein yield. MAP3K1 was known to be involved in the MAPK signaling pathway, and was considered to be a metabolic stimuli inducing cell proliferation [44, 45]. Meanwhile, it functioned as a candidate gene for type 2 diabetes (T2D) by interacting with insulin signaling pathway [46]. Thus, we concluded that MAP3K1 might regulate milk composition traits by modulating bovine mammary gland development.
The intronic indel ss2019489562 in UGDH showed significant effect on milk yield. UGDH encodes the protein that was implicated with biosynthesis of glycosaminoglycans, hyaluronan, chondroitin sulfate, and heparan sulfate. Previously, Xu et al. have demonstrated that two exonic SNPs in UGDH showed significant associations with milk production traits in Chinese Holstein population [47]. In particular, UGDH was close to the peak location of two reported QTLs for fat yield, fat percentage and protein yield [48–51]. Further, two previously reported significant SNPs for fat yield, protein yield, fat percentage and protein percentage [41] were near to the UGDH gene. Moreover, expression pattern in InnateDB showed that UGDH have the highest expression in liver which plays an indispensable role in metabolism of carbohydrates, fats and proteins in dairy cattle. Hence, these data demonstrated that UGDH gene might be a vital regulator for milk traits by affecting liver metabolism.
Conclusion
In the present study, we performed association analysis for the 13 short indels within 8 candidate genes for milk compositions identified by our previous whole genome resequencing study, including FCGR2B, CENPE, RETSAT, ACSBG2, TBC1D1, NLK, MAP3K1 and UGDH. As a result, the 13 indels were shown to have significant genetic effects on at least one of milk yield and composition traits. These results not only validated the candidate genes and indels from the previous whole genome resequencing work, but also provided novel molecular information for genetic improvement program of dairy cattle.
Methods
Ethics statement
All the procedures for sample collections and phenotypic observations of experimental individuals were carried out along with regular quarantine inspection of the farms and in strict accordance with the protocol reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at China Agricultural University, and the permit number is DK996.
Animals
The animals used for association analysis included a total of 769 Chinese Holstein cows those were daugters of 40 sire families. These daughters were collected from 22 herds of Beijing Sanyuanlvhe Dairy Farming Center, a leading dairy company in China. Phenotypic data of the five milk production traits including 305-day milk yield (MY), fat yield (FY), protein yield (PY), fat percentage (FP) and protein percentage (PP) those were calculated based on at least 6 test-day records in each lactation using a multiple trait random regression test-day model by the Dairy Data Center of China, Dairy Association of China (http://www.holstein.org.cn/).
Genomic DNA was isolated from whole blood of cows and frozen semen of sires as previously described by Yang et al. [16].
Indels selection, PCR amplification, sequencing and genotyping
Of the 25 short indels that identified by our previous whole-genome resequencing study, 24 indels were investigated the associations with the five milk production traits except for a three-nucleotide insertion (3 N ins) in ACSBG2 gene.
A total of 23 pairs of PCR primers were designed with Primer Premier 5.0 and Oligo 7.0 softwares based on the genomic sequences of the 11 candidate genes in Bos_taurus_UMD3.1 assembly (Additional file 1). To identify the twenty-four potential indel polymorphisms, two DNA pools for the above 40 sires were constructed with equal concentration of 50 ng/μl of each bull (20 individuals/pool). PCR products basd on the pooled DNA were purified with an EasyPure PCR Purification Kit (TransGen Biotech, Beijing, China) and then bi-directionally sequenced using ABI3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA).
To further confirm the position and sequence of the insertions and deletions, the purified PCR products were cloned into the pClone007 vector with a pClone 007 Vector Kit (TsingKe Biological Technology, Beijing, China). Positive clones including target indels were sequenced to search potential indels. The BLAST software (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and Chromas 2.0 (Technelysium, Australia) were applied for sequence alignment to the reference sequence of the corresponding gene referring to Bos_taurus_UMD_3.1 assembly. Finally, genotyping for the identified indels in 769 chinese cows was performed by using the Sequenom MassArray matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS).
Bioinformatics analysis
To further explore the potential impact of the exonic indels in FCGR2B and CENPE on the mRNA secondary structures as well as the second structures of corresponding proteins, the online RNA FOLDING FORM (version 2.3) software [52] and SPOMA program (http://npsa-pbil.ibcp.fr/) [53] were used, respectively.
Association analysis
Allele frequencies and genotype frequencies between the insertion and deletion genotypes, as well as the Hardy-Weinberg equilibrium were determined through a chi-square test. Associations between the 13 investigated indels and the five milk production traits were carried out by applying the mixed procedure in SAS 9.2 [54] based on the following linear mixed regression model:
Where Y is the phenotypic record for the analyzed trait of the cows, μ is the overall mean of the phenotypic record for each trait, hys is a fixed effect of herd, year and season, b is linear regression coefficient on calving month (M), M is effect of calving month, G is a fixed effect of indel genotype or haplotype, a is a random polygenic effect account for all known pedigree relationships, and e is a random residual.
Also, we estimated the additive (a), dominance (d) and allele substitution (α) effects using the equation of Falconer & Mackay [55]: where AA, BB and AB were the least square means of the phenotypic values for corresponding genotypes, and p and q indicates the allele frequencies of the corresponding alleles. Multiple t-tests with Bonferroni correction were used to compare the effects of the genotypes on each indel.
The linkage disequilibrium (LD) extent among the genotyped indels (five indels in CENPE gene and two indels in NLK gene) and haplotype blocks was estimated using Haploview 4.2 (Broad Institute of MIT and Harvard, Cambridge, MA, USA).
Additional files
Table S1. Primers used for pooled DNA sequencing for the 24 indels. (XLSX 42 kb)
Results of sanger and clone sequencing of the thirteen indels. (DOCX 332 kb)
Acknowledgements
We appreciate Beijing Dairy Cattle Center for providing the phenotypic data of milk production traits.
Abbreviations
- ACSBG2
Acyl-CoA synthetase bubblegum family member 2
- bp
Base pair
- CENPE
Centromere protein E
- EBV
Estimated breeding value
- F11
Coagulation factor XI
- FCGR2B
Fc fragment of IgG receptor IIb
- FP
Fat percentage
- FY
Fat yield
- GWAS
Genome-wide association study
- indel
Insertion and deletion
- MALDI-TOFMS
Matrix-assisted laser desorption/ionization time of flight mass spectrometry
- MAP3K1
Mitogen-activated protein kinase kinase kinase 1
- MSTN
Myostatin
- MY
Milk yield
- NGS
Next-generation sequencing
- NLK
Nemo like kinase
- PMEL17
Premelanosome protein
- PP
Protein percentage
- PY
Protein yield
- QTL
Quantitative trait locus
- RETSAT
Retinol saturase
- SNP
Single nucleotide polymorphism
- SPEF2
Sperm flagellar 2
- T2D
Type 2 diabetes
- TAL1
TAL bHLH transcription factor 1
- T-ALL
T-cell acute lymphoblastic leukemia
- TBC1D1
TBC1 domain family member 1
- UGDH
UDP-glucose 6-dehy- drogenase
Authors’ contributions
DS conceived and designed the experiments, JJ and GY analyzed the data, JJ, SL and LW prepared the DNA samples for SNP identification and genotyping, and the manuscript was prepared by JJ and DS. LY and LL provided the samples and participated in the result interpretation. All authors read and approved the final manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (31872330, 31802041), Beijing Dairy Industry Innovation Team (BAIC06–2018/2019), Beijing Science and Technology Program (D171100002417001), National Science and Technology Programs of China (2013AA102504), earmarked fund for Modern Agro-industry Technology Research System (CARS-36), and the Program for Changjiang Scholar and Innovation Research Team in University (IRT_15R62). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
All relevant data are available within the article and its additional files.
Ethics approval and consent to participate
All protocols for collection of the samples of experimental individuals and phenotypic observations were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at China Agricultural University. Samples were collected specifically for this study following standard procedures with the full agreement of the Beijing Sanyuanlvhe Dairy Farming Center who owned the Holstein cows and bulls, respectively.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jianping Jiang, Email: jiangjianping818@126.com.
Lin Liu, Email: liulin@bdcc.com.cn.
Yahui Gao, Email: 812956123@qq.com.
Lijun Shi, Email: 408529347@qq.com.
Yanhua Li, Email: yhli1976@163.com.
Weijun Liang, Email: westkcw@163.com.
Dongxiao Sun, Email: sundx@cau.edu.cn.
References
- 1.Blott S, Kim JJ, Moisio S, Schmidt-Kuntzel A, Cornet A, Berzi P, Cambisano N, Ford C, Grisart B, Johnson D, et al. Molecular dissection of a quantitative trait locus: a phenylalanine-to-tyrosine substitution in the transmembrane domain of the bovine growth hormone receptor is associated with a major effect on milk yield and composition. Genetics. 2003;163(1):253–266. doi: 10.1093/genetics/163.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grisart B, Farnir F, Karim L, Cambisano N, Kim JJ, Kvasz A, Mni M, Simon P, Frere JM, Coppieters W, et al. Genetic and functional confirmation of the causality of the DGAT1 K232A quantitative trait nucleotide in affecting milk yield and composition. Proc Natl Acad Sci U S A. 2004;101(8):2398–2403. doi: 10.1073/pnas.0308518100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen-Zinder M, Seroussi E, Larkin DM, Loor JJ, Everts-van der Wind A, Lee JH, Drackley JK, Band MR, Hernandez AG, Shani M, et al. Identification of a missense mutation in the bovine ABCG2 gene with a major effect on the QTL on chromosome 6 affecting milk yield and composition in Holstein cattle. Genome Res. 2005;15(7):936–944. doi: 10.1101/gr.3806705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang SH, Bi XJ, Xie Y, Li C, Zhang SL, Zhang Q, Sun DX. Validation of PDE9A gene identified in GWAS showing strong association with Milk production traits in Chinese Holstein. Int J Mol Sci. 2015;16(11):26530–26542. doi: 10.3390/ijms161125976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Georges M, Nielsen D, Mackinnon M, Mishra A, Okimoto R, Pasquino AT, Sargeant LS, Sorensen A, Steele MR, Zhao X, et al. Mapping quantitative trait loci controlling milk production in dairy cattle by exploiting progeny testing. Genetics. 1995;139(2):907–920. doi: 10.1093/genetics/139.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersson L. Genome-wide association analysis in domestic animals: a powerful approach for genetic dissection of trait loci. Genetica. 2009;136(2):341–349. doi: 10.1007/s10709-008-9312-4. [DOI] [PubMed] [Google Scholar]
- 7.Schennink A, Bovenhuis H, Leon-Kloosterziel KM, van Arendonk JA, Visker MH. Effect of polymorphisms in the FASN, OLR1, PPARGC1A, PRL and STAT5A genes on bovine milk-fat composition. Anim Genet. 2009;40(6):909–916. doi: 10.1111/j.1365-2052.2009.01940.x. [DOI] [PubMed] [Google Scholar]
- 8.Li C, Sun D, Zhang S, Wang S, Wu X, Zhang Q, Liu L, Li Y, Qiao L. Genome wide association study identifies 20 novel promising genes associated with milk fatty acid traits in Chinese Holstein. PLoS One. 2014;9(5):e96186. doi: 10.1371/journal.pone.0096186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grobet L, Martin LJ, Poncelet D, Pirottin D, Brouwers B, Riquet J, Schoeberlein A, Dunner S, Menissier F, Massabanda J, et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet. 1997;17(1):71–74. doi: 10.1038/ng0997-71. [DOI] [PubMed] [Google Scholar]
- 10.Kerje S, Sharma P, Gunnarsson U, Kim H, Bagchi S, Fredriksson R, Schutz K, Jensen P, von Heijne G, Okimoto R, et al. The dominant white, dun and smoky color variants in chicken are associated with insertion/deletion polymorphisms in the PMEL17 gene. Genetics. 2004;168(3):1507–1518. doi: 10.1534/genetics.104.027995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunieda M, Tsuji T, Abbasi AR, Khalaj M, Ikeda M, Miyadera K, Ogawa H, Kunieda T. An insertion mutation of the bovine F11 gene is responsible for factor XI deficiency in Japanese black cattle. Mamm Genome. 2005;16(5):383–389. doi: 10.1007/s00335-004-2462-5. [DOI] [PubMed] [Google Scholar]
- 12.Montgomery SB, Goode DL, Kvikstad E, Albers CA, Zhang ZD, Mu XJ, Ananda G, Howie B, Karczewski KJ, Smith KS, et al. The origin, evolution, and functional impact of short insertion-deletion variants identified in 179 human genomes. Genome Res. 2013;23(5):749–761. doi: 10.1101/gr.148718.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catalan A, Glaser-Schmitt A, Argyridou E, Duchen P, Parsch J. An Indel polymorphism in the MtnA 3' untranslated region is associated with gene expression variation and local adaptation in Drosophila melanogaster. PLoS Genet. 2016;12(4):e1005987. doi: 10.1371/journal.pgen.1005987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, Etchin J, Lawton L, Sallan SE, Silverman LB, et al. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346(6215):1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sironen A, Thomsen B, Andersson M, Ahola V, Vilkki J. An intronic insertion in KPL2 results in aberrant splicing and causes the immotile short-tail sperm defect in the pig. Proc Natl Acad Sci U S A. 2006;103(13):5006–5011. doi: 10.1073/pnas.0506318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang S, Li C, Xie Y, Cui X, Li X, Wei J, Zhang Y, Yu Y, Wang Y, Zhang S, et al. Detection of functional polymorphisms influencing the promoter activity of the SAA2 gene and their association with milk production traits in Chinese Holstein cows. Anim Genet. 2015;46(6):591–598. doi: 10.1111/age.12332. [DOI] [PubMed] [Google Scholar]
- 17.Xia Q, Guo Y, Zhang Z, Li D, Xuan Z, Li Z, Dai F, Li Y, Cheng D, Li R, et al. Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx) Science. 2009;326(5951):433–436. doi: 10.1126/science.1176620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER. The next-generation sequencing revolution and its impact on genomics. Cell. 2013;155(1):27–38. doi: 10.1016/j.cell.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daetwyler HD, Capitan A, Pausch H, Stothard P, van Binsbergen R, Brondum RF, Liao X, Djari A, Rodriguez SC, Grohs C, et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat Genet. 2014;46(8):858–865. doi: 10.1038/ng.3034. [DOI] [PubMed] [Google Scholar]
- 20.Li MZ, Tian SL, Yeung CKL, Meng XH, Tang QZ, Niu LL, Wang X, Jin L, Ma JD, Long KR, et al. Whole-genome sequencing of Berkshire (European native pig) provides insights into its origin and domestication. Sci Rep. 2014;4. [DOI] [PMC free article] [PubMed]
- 21.Jiang JP, Gao YH, Hou YL, Li WH, Zhang SL, Zhang Q, Sun DX. Whole-genome resequencing of Holstein bulls for Indel discovery and identification of genes associated with Milk composition traits in dairy cattle. PLoS One. 2016;11(12):e0168946. doi: 10.1371/journal.pone.0168946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin JG, Yuan ZQ, Fordyce K, Sreeramoju P, Kent TS, Kim J, Wang V, Schneyer D, Weber TK. A del T poly T (8) mutation in the 3 ' untranslated region (UTR) of the CDK2-AP1 gene is functionally significant causing decreased mRNA stability resulting in decreased CDK2-AP1 expression in human microsatellite unstable (MSI) colorectal cancer (CRC) Surgery. 2007;142(2):222–227. doi: 10.1016/j.surg.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Chatterjee R, Batra J, Ghosh B. A common Exonic variant of Interleukin21 confers susceptibility to atopic asthma. Int Arch Allergy Imm. 2009;148(2):137–146. doi: 10.1159/000155744. [DOI] [PubMed] [Google Scholar]
- 24.Lechner J, Bae HA, Guduric-Fuchs J, Rice A, Govindarajan G, Siddiqui S, Farraj LA, Yip SP, Yap M, Das M, et al. Mutational analysis of MIR184 in sporadic keratoconus and myopia. Invest Ophth Vis Sci. 2013;54(8):5266–5272. doi: 10.1167/iovs.13-12035. [DOI] [PubMed] [Google Scholar]
- 25.Pinsonneault JK, Frater JT, Kompa B, Mascarenhas R, Wang DX, Sadee W. Intronic SNP in ESR1 encoding human estrogen receptor alpha is associated with brain ESR1 mRNA isoform expression and behavioral traits. PLoS One. 2017;12(6):e0179020. doi: 10.1371/journal.pone.0179020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun WY, Lan J, Chen L, Qiu JJ, Luo ZG, Li MZ, Wang JY, Zhao JG, Zhang TH, Long X, et al. A mutation in porcine pre-miR-15b alters the biogenesis of MiR-15b\16-1 cluster and strand selection of MiR-15b. PLoS One. 2017;12(5):e0178045. doi: 10.1371/journal.pone.0178045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li C, Grove ML, Yu B, Jones BC, Morrison A, Boerwinkle E, Liu XM. Genetic variants in microRNA genes and targets associated with cardiovascular disease risk factors in the African-American population. Hum Genet. 2018;137(1):85–94. doi: 10.1007/s00439-017-1858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moya L, Lai J, Hoffman A, Srinivasan S, Panchadsaram J, Chambers S, Clements JA, Batra J, BioResour APC. Association analysis of a microsatellite repeat in the TRIB1 gene with prostate Cancer risk, aggressiveness and survival. Front Genet. 2018;9. [DOI] [PMC free article] [PubMed]
- 29.Ross P, Hall L, Smirnov I, Haff L. High level multiplex genotyping by MALDI-TOF mass spectrometry. Nat Biotechnol. 1998;16(13):1347–1351. doi: 10.1038/4328. [DOI] [PubMed] [Google Scholar]
- 30.Jonsson S, Sveinbjornsson G, de Lapuente Portilla AL, Swaminathan B, Plomp R, Dekkers G, Ajore R, Ali M, Bentlage AEH, Elmer E, et al. Identification of sequence variants influencing immunoglobulin levels. Nat Genet. 2017;49(8):1182–1191. doi: 10.1038/ng.3897. [DOI] [PubMed] [Google Scholar]
- 31.Orozco-Lucero E, Dufort I, Robert C, Sirard MA. Rapidly cleaving bovine two-cell embryos have better developmental potential and a distinctive mRNA pattern. Mol Reprod Dev. 2014;81(1):31–41. doi: 10.1002/mrd.22278. [DOI] [PubMed] [Google Scholar]
- 32.Schupp M, Lefterova MI, Janke J, Leitner K, Cristancho AG, Mullican SE, Qatanani M, Szwergold N, Steger DJ, Curtin JC, et al. Retinol saturase promotes adipogenesis and is downregulated in obesity. P Natl Acad Sci USA. 2009;106(4):1105–1110. doi: 10.1073/pnas.0812065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Montes AM, Fernandez A, Perez-Montarelo D, Alves E, Benitez RM, Nunez Y, Ovilo C, Ibanez-Escriche N, Folch JM, Fernandez AI. Using RNA-Seq SNP data to reveal potential causal mutations related to pig production traits and RNA editing. Anim Genet. 2017;48(2):151–165. doi: 10.1111/age.12507. [DOI] [PubMed] [Google Scholar]
- 34.Marszalek JR, Kitidis C, Dararutana A, Lodish HF. Acyl-CoA synthetase 2 overexpression enhances fatty acid internalization and neurite outgrowth. J Biol Chem. 2004;279(23):23882–23891. doi: 10.1074/jbc.M313460200. [DOI] [PubMed] [Google Scholar]
- 35.Fujimoto Y, Onoduka J, Homma KJ, Yamaguchi S, Mori M, Higashi Y, Makita M, Kinoshita T, Noda J, Itabe H, et al. Long-chain fatty acids induce lipid droplet formation in a cultured human hepatocyte in a manner dependent of acyl-CoA synthetase. Biol Pharm Bull. 2006;29(11):2174–2180. doi: 10.1248/bpb.29.2174. [DOI] [PubMed] [Google Scholar]
- 36.Sun CJ, Lu J, Yi GQ, Yuan JW, Duan ZY, Qu LJ, Xu GY, Wang KH, Yang N. Promising loci and genes for yolk and ovary weight in chickens revealed by a genome-wide association study. PLoS One. 2015;10(9):e0137145. doi: 10.1371/journal.pone.0137145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L, Chen QL, Xie BX, Quan C, Sheng Y, Zhu SS, Rong P, Zhou SL, Sakamoto K, MacKintosh C, et al. Disruption of the AMPK-TBC1D1 nexus increases lipogenic gene expression and causes obesity in mice via promoting IGF1 secretion. Proc Natl Acad Sci U S A. 2016;113(26):7219–7224. doi: 10.1073/pnas.1600581113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Xu HY, Gilbert ER, Peng X, Zhao XL, Liu YP, Zhu Q. Detection of SNPs in the TBC1D1 gene and their association with carcass traits in chicken. Gene. 2014;547(2):288–294. doi: 10.1016/j.gene.2014.06.061. [DOI] [PubMed] [Google Scholar]
- 39.Hargett SR, Walker NN, Hussain SS, Hoehn KL, Keller SR. Deletion of the Rab GAP Tbc1d1 modifies glucose, lipid, and energy homeostasis in mice. Am J Physiol-Endocrinol Metab. 2015;309(3):E233–E245. doi: 10.1152/ajpendo.00007.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hargett SR, Walker NN, Keller SR. Rab GAPs AS160 and Tbc1d1 play nonredundant roles in the regulation of glucose and energy homeostasis in mice. Am J Physiol-Endocrinol Metab. 2016;310(4):E276–E288. doi: 10.1152/ajpendo.00342.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cole John B, Wiggans George R, Ma Li, Sonstegard Tad S, Lawlor Thomas J, Crooker Brian A, Van Tassell Curtis P, Yang Jing, Wang Shengwen, Matukumalli Lakshmi K, Da Yang. Genome-wide association analysis of thirty one production, health, reproduction and body conformation traits in contemporary U.S. Holstein cows. BMC Genomics. 2011;12(1):408. doi: 10.1186/1471-2164-12-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bionaz M, Loor JJ. Gene networks driving bovine mammary protein synthesis during the lactation cycle. Bioinform Biol Insights. 2011;5:83–98. doi: 10.4137/BBI.S7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan HX, Wang Z, Yu FX, Li F, Russell RC, Jewell JL, Guan KL. NLK phosphorylates raptor to mediate stress-induced mTORC1 inhibition. Genes Dev. 2015;29(22):2362–2376. doi: 10.1101/gad.265116.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glubb DM, Maranian MJ, Michailidou K, Pooley KA, Meyer KB, Kar S, Carlebur S, O'Reilly M, Betts JA, Hillman KM, et al. Fine-scale mapping of the 5q11.2 breast Cancer locus reveals at least three independent risk variants regulating MAP3K1. Am J Hum Genet. 2015;96(1):5–20. doi: 10.1016/j.ajhg.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuo SH, Yang SY, You SL, Lien HC, Lin CH, Lin PH, Huang CS. Polymorphisms of ESR1, UGT1A1, HCN1, MAP3K1 and CYP2B6 are associated with the prognosis of hormone receptor-positive early breast cancer. Oncotarget. 2017;8(13):20925–20938. doi: 10.18632/oncotarget.14995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tabassum R, Chauhan G, Dwivedi OP, Mahajan A, Jaiswal A, Kaur I, Bandesh K, Singh T, Mathai BJ, Pandey Y, et al. Genome-wide association study for type 2 diabetes in Indians identifies a new susceptibility locus at 2q21. Diabetes. 2013;62(3):977–986. doi: 10.2337/db12-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Q, Mei G, Sun DX, Zhang Q, Zhang Y, Yin CC, Chen HY, Ding XD, Liu JF. Detection of genetic association and functional polymorphisms of UGDH affecting milk production trait in Chinese Holstein cattle. BMC Genomics. 2012;13:590. doi: 10.1186/1471-2164-13-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freyer G, Kuhn C, Weikard R, Zhang Q, Mayer M, Hoeschele I. Multiple QTL on chromosome six in dairy cattle affecting yield and content traits (vol 119, pg 60, 2002) J Anim Breed Genet. 2002;119(3):200. doi: 10.1046/j.1439-0388.2002.00350.x. [DOI] [Google Scholar]
- 49.Szyda J, Liu Z, Reinhardt F, Reents R. Estimation of quantitative trait loci parameters for milk production traits in German Holstein dairy cattle population. J Dairy Sci. 2005;88(1):356–367. doi: 10.3168/jds.S0022-0302(05)72695-3. [DOI] [PubMed] [Google Scholar]
- 50.Chen HY, Zhang Q, Yin CC, Wang CK, Gong WJ, Mei G. Detection of quantitative trait loci affecting milk production traits on bovine chromosome 6 in a Chinese Holstein population by the daughter design. J Dairy Sci. 2006;89(2):782–790. doi: 10.3168/jds.S0022-0302(06)72140-3. [DOI] [PubMed] [Google Scholar]
- 51.Kucerova J, Lund MS, Sorensen P, Sahana G, Guldbrandtsen B, Nielsen VH, Thomsen B, Bendixen C. Multitrait quantitative trait loci mapping for milk production traits in danish Holstein cattle. J Dairy Sci. 2006;89(6):2245–2256. doi: 10.3168/jds.S0022-0302(06)72296-2. [DOI] [PubMed] [Google Scholar]
- 52.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Geourjon C, Deleage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci. 1995;11(6):681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 54.Kolbehdari D, Wang Z, Grant JR, Murdoch B, Prasad A, Xiu Z, Marques E, Stothard P, Moore SS. A whole genome scan to map QTL for milk production traits and somatic cell score in Canadian Holstein bulls. J Anim Breed Genet. 2009;126(3):216–227. doi: 10.1111/j.1439-0388.2008.00793.x. [DOI] [PubMed] [Google Scholar]
- 55.Falconer DS, Mackay TFC: Introduction to quantitative genetics: 4th edn. Longman scientific and technical, New York.; 1996.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primers used for pooled DNA sequencing for the 24 indels. (XLSX 42 kb)
Results of sanger and clone sequencing of the thirteen indels. (DOCX 332 kb)
Data Availability Statement
All relevant data are available within the article and its additional files.