Abstract
Background
Radioresistance is one of the main obstacle limiting the therapeutic efficacy and prognosis of patients, the molecular mechanisms of radioresistance is still unclear. The purpose of this study was to identify the key genes and miRNAs and to explore their potential molecular mechanisms in radioresistant nasopharyngeal carcinoma.
Methods
In this study, we analysis the differentially expressed genes and microRNA based on the database of GSE48501 and GSE48502, and then employed bioinformatics to analyze the pathways and GO terms in which DEGs and DEMS target genes are involved. Moreover, Construction of protein-protein interaction network and identification of hub genes. Finally, analyzed the biological networks for validated target gene of hub miRNAs.
Results
A total of 373 differentially expressed genes (DEGs) and 14 differentially expressed microRNAs (DEMs) were screened out. The up-regulated gene JUN was overlap both in DEGs and publicly available studies, which was potentially targeted by three miRNAs, including hsa-miR-203, hsa-miR-24 and hsa-miR-31. Moreover, Pathway analysis showed that both up-regulated gene and DEMs target genes were enriched in TGF-beta signaling pathway, Hepatitis B, Pathways in cancer and p53 signaling pathway. Finally, we further constructed protein-protein interaction network (PPI) of DEGs and analyzed the biological networks for above mentioned common miRNAs, the result indicated that JUN was a core hub gene in PPI network, hsa-miR-24 and its target gene were significantly enriched in P53 signaling pathway.
Conclusions
These results might provide new clues to improve the radiosensitivity of Nasopharyngeal Carcinoma.
Electronic supplementary material
The online version of this article (10.1186/s12920-019-0507-6) contains supplementary material, which is available to authorized users.
Keywords: Nasopharyngeal carcinoma; microRNA, gene expression omnibus differentially expressed genes, bioinformatics analysis
Background
Radiotherapy is a mainly treatment for nasopharyngeal carcinoma (NPC). However, radioresistance is one of the major factors to affect the therapeutic efficacy and prognosis of patients [1–3]. Accordingly, identifying potential biomarkers and studying the molecular mechanisms associated with radioresistant nasopharyngeal carcinoma has become a hot topic both in basic and clinical research.
Microarrays are considered to be an important method for identifying potential biomarkers in many diseases at the molecular level with more effective and detailed insights [4]. Several microRNAs and mRNAs have been discovered to be involved in radioresistant NPC, whereas traditional methods have failed to elucidate the interaction of mRNAs and microRNAs and the molecular mechanisms of NPC due to the limitations on the comparative analysis [5–7]. Therefore, systematically investigating the interaction between microRNA and mRNA, and elucidating the molecular mechanism of radioresistant NPC is of great significance. With the development of bioinformatics, we can apply global analysis to process the data generated by microarray technology and find the interaction between DEGs and DEM, especially in the pathway interaction network, to summarize their potential mechanisms in diseases [8–10]. Based on above mentioned reasons, the present study aims to identify the key genes and miRNAs and to explore their potential molecular mechanisms in radioresistant nasopharyngeal carcinoma.
In this study, we analysis the differentially expressed genes and microRNA between radioresistant NPC CNE2-R cells and radiosensitive CNE2 cells based on the database of GSE48501 and GSE48502, and then employed bioinformatics to analyze the pathways and GO terms in which DEGs and DEMS target genes are involved. Moreover, Construction of protein-protein interaction network and identification of hub genes. Finally, analyzed the biological networks for validated target gene of hub miRNAs. Our data may provide an important contribution to identified biological markers and clarify the mechanisms of NPC radioresistance.
Results
DEGs and DEMs in radioresistant NPC cells compared with radioresistant NPC cells
GEO2R analyzed result shown that a total of 373 DEGs were identified in radioresistant NPC cells, including 291 mRNAs were up-regulated and 82 mRNAs were down-regulated (Table 1). The DEMs results indicated that there were 277 miRNAs were detected, 14 of which were differentially expressed with≥1.5 fold-change (t-test, P < 0.05), including 4 up-regulated miRNAs and 10 down-regulated miRNAs (Table 2). Moreover, DigSee software were used to identify the radioresistant related genes for publicly available studies, 37 related genes were retrieved. In addition, Venn diagram analyses revealed that JUN and SOD2 were common both in the DEGs and the DigSee (Fig. 1a). Furthermore, we identified JUN related microRNA by mirDIP software and analyzed the common microRNAs between the JUN-related microRNAs and DEMs by Venn diagram software. 35 JUN-related microRNA were retrieved, 3 down-regulated microRNAs were detected which were joint in JUN-related microRNAs and DEMs, including hsa-miR-203, hsa-miR-24 and hsa-miR-31 (Table 3 and Fig. 1b).
Table 1.
Differential mRNA expression profile of radioresistant nasopharyngeal carcinoma CNE2R versus CNE-2 cells (The Table 1 show the top 20 differential expression genes)
| Gene Symbol | Description | Fold Change |
|---|---|---|
| LXN | latexin | 22.53 |
| IGFBP3 | insulin-like growth factor binding protein 3 | 18.88 |
| ABCG1 | ATP-binding cassette, sub-family G (WHITE), member 1 | 16.82 |
| CP | ceruloplasmin (ferroxidase) | 14.76 |
| TRIM31 | tripartite motif-containing 31 | 12.30 |
| NNMT | nicotinamide N-methyltransferase | 10.96 |
| GDF15 | growth differentiation factor 15 | 10.15 |
| INHBE | inhibin, beta E | 9.59 |
| EGR1 | early growth response 1 | 7.95 |
| IL8 | interleukin 8 | 7.49 |
| METTL7A | methyltransferase like 7A | 7.31 |
| LOC387763 | hypothetical LOC387763 | 7.24 |
| LCN2 | lipocalin 2 | 6.82 |
| EDN2 | endothelin 2 | 6.57 |
| BMP2 | bone morphogenetic protein 2 | 6.56 |
| C8orf4 | chromosome 8 open reading frame 4 | 6.42 |
| ASNS | asparagine synthetase | 6.12 |
| SLC16A6 | solute carrier family 16, member 6 (monocarboxylic acid transporter 7) | 5.55 |
| PCK2 | phosphoenolpyruvate carboxykinase 2 (mitochondrial) | 5.44 |
| STEAP4 | STEAP family member 4 | 5.32 |
Table 2.
Differentially expressed miRNAs in GSE48502
| miRNA | Fold change | P-value |
|---|---|---|
| Up-regulated miRNA | ||
| hsa-miR-762 | 2.510 | 0.00337 |
| hsa-miR-1202 | 2.292 | 0.0008 |
| hsa-miR-193b | 1.530 | 0.00986 |
| hsa-let-7e | 1.521 | 0.00054 |
| Down-regulated miRNA | ||
| hsa-miR-203 | 3.337 | 0.01698 |
| hsa-miR-545 | 1.980 | 0.04888 |
| hsa-miR-4291 | 1.722 | 0.00271 |
| hsa-miR-183 | 1.677 | 0.03486 |
| hsa-miR-24 | 1.667 | 0.00032 |
| hsa-miR-130a | 1.598 | 0.01252 |
| hsa-miR-660 | 1.578 | 0.01531 |
| hsa-miR-31 | 1.535 | 0.00208 |
| hsa-miR-23a | 1.527 | 0.03552 |
| hsa-miR-30a | 1.526 | 0.0274 |
Fig. 1.
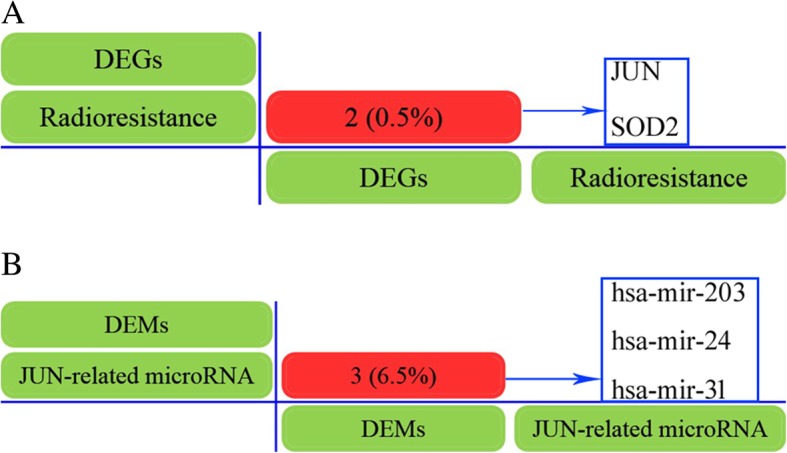
Screening common genes or miRNAs by Venn diagram software. a Identification common genes between the DEGs and the publicly available studies by Venn diagram. b Analyzed the common microRNAs between the JUN-related microRNAs and DEMs by Venn diagram software
Table 3.
Identification of JUN related microRNA by mirDIP software. Prediction analysis was performed by mirDIP online software. In this table, asterisk represents common microRNA in DEMs and JUN-related microRNAs by Veen analysis
| Gene Symbol | MicroRNA | Integrated Score | Score Class |
|---|---|---|---|
| JUN | hsa-miR-200b-3p | 0.8428 | Excellent |
| JUN | hsa-miR-139-5p | 0.7769 | Excellent |
| JUN | hsa-miR-200c-3p | 0.7693 | Excellent |
| JUN | hsa-miR-429 | 0.7576 | Excellent |
| JUN | hsa-miR-495-3p | 0.7162 | Excellent |
| JUN | hsa-miR-32-5p | 0.6837 | Excellent |
| JUN | hsa-miR-92a-3p | 0.6745 | Excellent |
| JUN | hsa-miR-216b-5p | 0.6528 | Excellent |
| JUN | hsa-miR-522-3p | 0.6392 | Excellent |
| JUN | hsa-miR-501-5p | 0.60082 | Excellent |
| JUN | hsa-miR-200a-3p | 0.5751 | Excellent |
| JUN | hsa-miR-524–5p | 0.5637 | Excellent |
| JUN | hsa-miR-520d-5p | 0.5365 | Excellent |
| JUN | hsa-miR-141–3p | 0.5211 | Excellent |
| JUN | hsa-miR-203* | 0.5019 | Excellent |
| JUN | hsa-miR-580-3p | 0.4817 | Excellent |
| JUN | hsa-miR-940 | 0.4770 | Excellent |
| JUN | hsa-miR-1299 | 0.4628 | Excellent |
| JUN | hsa-miR-9-5p | 0.4390 | Excellent |
| JUN | hsa-miR-612 | 0.4313 | Excellent |
| JUN | hsa-miR-583 | 0.4260 | Excellent |
| JUN | hsa-miR-455-3p | 0.4018 | Excellent |
| JUN | hsa-miR-637 | 0.3870 | Excellent |
| JUN | hsa-miR-92b-3p | 0.3700 | Excellent |
| JUN | hsa-miR-758-3p | 0.3659 | Excellent |
| JUN | hsa-miR-25-3p | 0.3602 | Excellent |
| JUN | hsa-miR-24* | 0.3585 | Excellent |
| JUN | hsa-miR-31* | 0.3585 | Excellent |
| JUN | hsa-miR-493-5p | 0.3318 | Excellent |
| JUN | hsa-miR-127-5p | 0.3255 | Excellent |
| JUN | hsa-miR-633 | 0.3227 | Excellent |
| JUN | hsa-miR-766-3p | 0.3199 | Excellent |
| JUN | hsa-miR-224-3p | 0.3097 | Excellent |
| JUN | hsa-miR-494-3p | 0.3081 | Excellent |
| JUN | hsa-miR-1285-3p | 0.3039 | Excellent |
Gene ontology analysis of DEMs target genes and DEGs
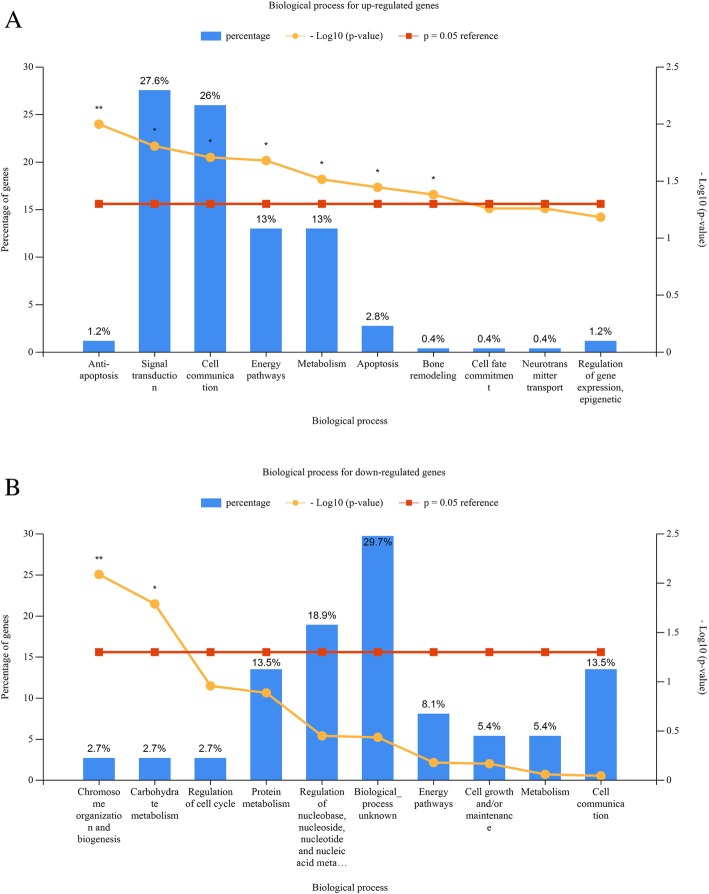
We performed gene ontology (GO) analysis of DEGs and DEMs target genes. Our result indicated that the significantly enriched GO terms of up-regulated and down-regulated microRNAs target genes were mainly involved in mitotic cell cycle; RNA binding; nucleoplasm; cytosol; biosynthetic process; gene expression; cellular nitrogen compound metabolic process; ion binding (Table 4). As shown in Fig. 2, the most significantly enriched GO terms corresponded to up-regulated DEGs were “Anti-apoptosis” (Ontology Biological Process), the most significant biological process for the down-regulated genes are Chromosome organization and biogenesis.
Table 4.
GO functional annotation of DEMs (Top 10)
| GO Category | Gene Target | miRNAs | P-value |
|---|---|---|---|
| Up-regulated miRNAs | |||
| mitotic cell cycle | 158 | 4 | 0 |
| protein binding transcription factor activity | 143 | 4 | 0 |
| RNA binding | 473 | 4 | 0 |
| nucleoplasm | 369 | 4 | 0 |
| cytosol | 664 | 4 | 0 |
| biosynthetic process | 909 | 4 | 0 |
| gene expression | 234 | 4 | 0 |
| viral process | 177 | 4 | 0 |
| cellular nitrogen compound metabolic process | 1112 | 4 | 0 |
| ion binding | 1145 | 4 | 0 |
| Down-regulated miRNAs | |||
| nucleoplasm | 612 | 18 | 0 |
| biosynthetic process | 1636 | 18 | 0 |
| gene expression | 389 | 18 | 0 |
| cellular nitrogen compound metabolic process | 2032 | 18 | 0 |
| organelle | 3947 | 18 | 0 |
| ion binding | 2198 | 16 | 0 |
| mitotic cell cycle | 221 | 15 | 0 |
| RNA binding | 812 | 15 | 0 |
| cellular protein modification process | 985 | 15 | 0 |
| cytosol | 1170 | 15 | 0 |
Fig. 2.
GO functional annotation of DEGs. a The top 20 significant biology process of up-regulated genes. b The significant biology process of down-regulated genes
Pathway enrichment analyses
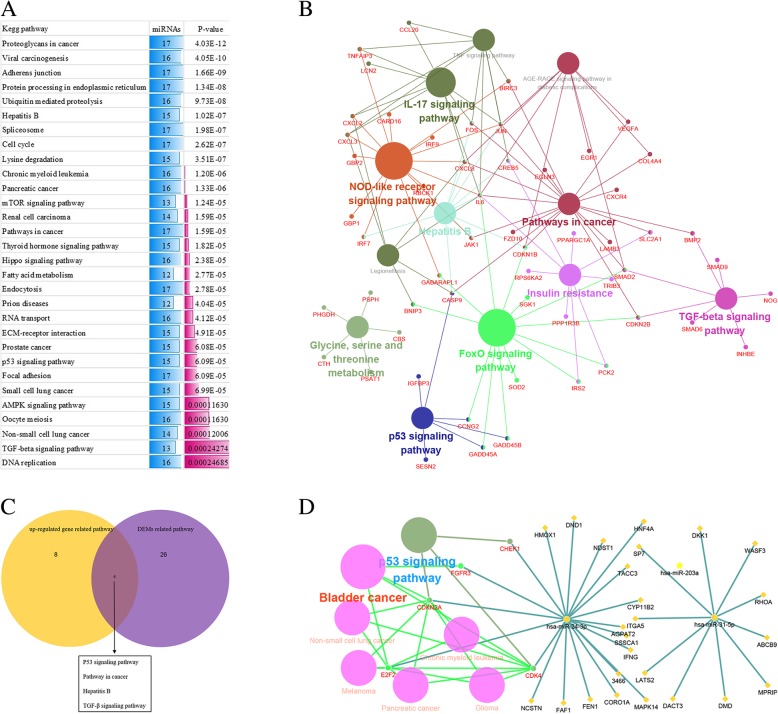
We performed pathways enrichment analysis of DEMs target genes and DEGs using DIANA miRPATH tool and Clue go software respectively. For the pathway analysis, the top of 30 significant pathways were selected in the DEMs target genes (Fig. 3a). Moreover, the upregulated DEGs were enriched in 12 kegg pathways (Fig. 3b). We further identified 4 significant pathways both in up-regulated DEGs and DEMs, including TGF-beta signaling pathway, Hepatitis B, Pathways in cancer and p53 signaling pathway which were considered as crucial pathways (Fig. 3c). Finally, we analyzed the above mentioned microRNAs (hsa-miR-203, hsa-miR-24 and hsa-miR-31) and their corresponding pathways using Clue go and Clue pedia software. The results indicated that hsa-miR-24 and 20 target genes were associated with 7 pathways, and the P53 signaling pathway is the most significant pathway (Fig. 3d). Our result indicated that P53 signaling pathway may be related to nasopharyngeal carcinoma radioresistance.
Fig. 3.
Pathway enrichment analysis of DEMs target genes and DEGs. a The top 30 enriched kegg pathway for DEMs target genes. b Significant pathways in up-regulated genes. c Venn diagrams show the common pathway between upregulated genes and DEMs target genes. d Biomolecular network about 5 validated genes (in red) targeted by the common microRNAs and corresponding pathways were analyzed by Clue Go and Clue Pedia. The yellow diamond nodes represent target gene, the violet circle and red circle nodes represent miRNA and their related pathway respectively
Protein-protein interaction network and subnetwork of DEGs
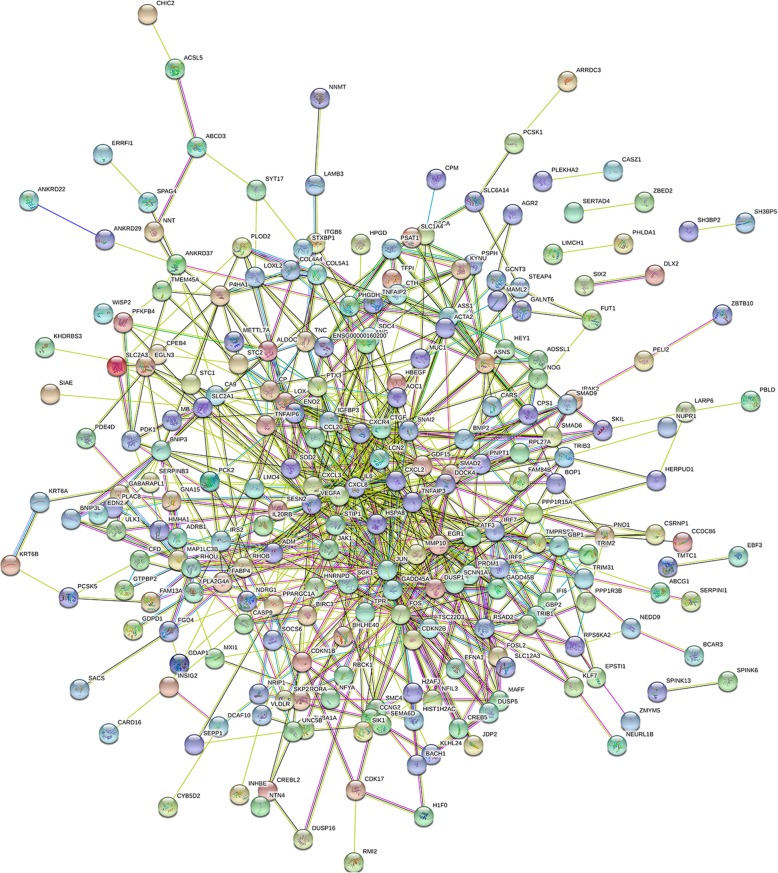
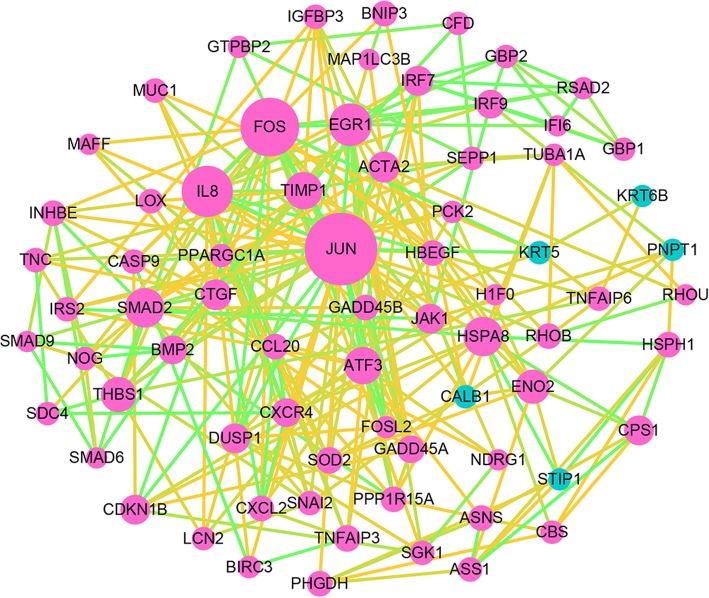
The PPI network was consisted of 339 nodes and 714 edges which were mapped by STRING software (Fig. 4). As shown in Fig. 5, the PPI network of DEGs was composed of 72 nodes that were interacted with each other. The connectivity degree of each node was calculated in this PPI network and the top 5 nodes with degree more than 20 were JUN, FOS, IL8, EGR, HSPA8. Among these genes, JUN with highest degree (54) in the Protein-Protein interaction network was considered as the hub node which have closely interacted with other genes. Therefore, we can infer that the up-regulated JUN may be a key node related with radioresistant nasopharyngeal carcinoma.
Fig. 4.
Constructed PPI network of DEGs by STRING software. Using the STRING software, proteins are represented with nodes and the interactions with continuous lines to represent direct interactions (physical), while indirect ones (functional) are presented by interrupted lines. Line thickness indicates the strength of data support
Fig. 5.
Significant subnetwork of DEGs. Red nodes represent up-regulated genes, while Green nodes denote down-regulated genes. The size of the nodes is positively correlated with the count of genes. The color of line is determined by the combined score provided by STRING
Discussion
Nasopharyngeal carcinoma is a geographically distributed disease, especially in southern china and southeast Asia. Although radiotherapy is considered to be the primary treatment for NPC, radioresistance-induced locoregional recurrence and distant metastases remains mainly obstacle to successful treatment [11, 12]. Therefore, in-depth study biomarkers and mechanisms of radioresistance in NPC is of great significance for improving the radiosensitivity of NPC and provide a new ideas for design of good therapy.
Previous studies have identified that radioresistance-associated molecules (mRNAs, microRNAs, and proteins) regulate radioresistance through different biological process, such as DNA repair, apoptosis, cell cycle, and protective autophagy [9]. However, the molecular mechanisms underlying NPC radioresistnce remain elusive.
In order to understand the mechanisms underlying in radioresistant NPC, we performed global analysis of key genes and microRNAs in radioresistant Nasopharyngeal Carcinoma by bioinformatics analysis. Our result demonstrated that a total of 373 DEGs and 14 DEMs between radioresistant NPC CNE2-IR cells and radiosensitive CNE2 cells were identified. Our study revealed that JUN was significantly up-regulated in radioresistant NPC CNE2-IR cell and which was overlap between the DEGs and the DigSee. Meanwhile, in Protein-Protein interaction network analysis, JUN was a core hub gene in PPI network. Recent research has shown that JUN is an important components of the activator protein-1 (AP-1) transcription factor and is closely related to cell proliferation, apoptosis and malignant transformation [13]. JUN could promote tumor growth and progression. Over-expression of c-jun was found to result in abnormal cell proliferation and loss of apoptosis. Some researchers have noted that inhibition the expression of c-jun can enhances radiosensitivity, induces cell cycle arrest and apoptosis [14]. Our previous study shown that the expression of c-jun was significantly up-regulated in CNE-2R cells, which may be associated with the radioresistance of NPC [15]. The kegg pathway analysis data indicated that JUN is involved in a variety of pathways. Such as TNF signaling pathway, Epstein-Barr virus infection, MAPK signaling pathway, Pathways in cancer. MAPK signaling activity confers inherent radioresistance to KRAS-mutant colorectal carcinoma cells by rapidly upregulating of heterogeneous nuclear ribonucleoprotein K (hnRNPK) [16]. Our result showed that JUN was involved in Hepatitis B, NOD-like receptor signaling pathway, AGE-RAGE signaling pathway in diabetic complications, Pathways in cancer, IL-17 signaling pathway, TNF signaling pathway (Fig. 3b). We speculated that JUN may be participated in the regulation of nasopharyngeal carcinoma radiosensitivity through these pathways, the internal mechanism may need to be further tested in the future experiments. Based on the above, we conclude that JUN is involved in NPC radioresistance, which may provide new clues to improve radiosensitivity of Nasopharyngeal Carcinoma.
microRNAs are single stranded, endogenous, 19–25 nucleotide (nt), which are thought to be modulated tumor radiosensitivity [17]. Therefore, identification of radioresistance associated miRNAs which may contribute to more effective treatments for NPC patients. In this study, we screened out 14 differentially expressed miRNAs in the radioresistant CNE2-R cells, including the up-regulated mirRNA-762, mirRNA-1202, mirRNA-193b, mirRNA-let-7e and down-regulated mirRNA-203, mirRNA-545, mirRNA-4291, mirRNA-183, mirRNA-24, mirRNA-130a, miRNA-660, miRNA-31, miRNA-30a, miRNA-23a, suggesting that the regulation of these miRNAs might be participate in the NPC radioresistance. Most of them have been shown to be associated with radioresistance of tumor [2, 18, 19]. mirRNA-let-7e and miRNA-31 have been recently discovered to involving in the acquisition of cancer cell radioresistance [1, 19]. Recent reports indicate that miRNA-23a is downregulated in the radioresistant NPC tissues, and is an independent predictor of poor prognosis in patients with nasopharyngeal carcinoma. up-regulated miRNA-23a improves NPC cell radiosensitivity in vivo and vitro. Downregulated miRNA-23a increases NPC radioresistance through activating IL-8/Stat3 signaling. Targeting miR-23a/IL-8/Stat3 signaling might be an effective approach to improving radiosensitivity of NPC [18]. It has been reported that miR-24 is frequently downregulated in NPC cell lines, and the consumption of miR-24 inhibited NPC cell growth and proliferation, while improving the radiosensitivity of NPC both in vitro and in vivo. In addition, it is reported that SP1 was verified as a target for miR-24, miR-24/SP pathway should help us understand the radiosensitivity mechanisms of human NPC, which may be a potential therapeutic target [20]. Our study detected that miRNA-24 is down-regulated in radioresistant NPC cells, and which not only was common in DEMs and JUN-related microRNAs, but also had significantly enriched in P53 signaling pathway. In conclusion, we can infer that the above mention microRNAs, especially miRNA-24 may be a key factor to affect the radiosensitivity of NPC, which may be helpful to predict radiosensitivity in NPC.
To completely understand the function of miRNAs and mRNAs in radioresistant NPC, we performed pathway enrichment analysis of DEGs and DEMs, the result demonstrated that 4 pathways are considered as the key pathways for the radiosensitivity of NPC, including TGF-beta signaling pathway, Hepatitis B, Pathways in cancer and p53 signaling pathway, which were regulated by miRNA and mRNA together. Moreover, hsa-miR-24 and its target gene were found have significantly enriched in P53 signaling pathway. P53 can predict cancer response to IR and chemotherapy [21]. Improving the radiosensitivity of non-small cell lung cancer cells by inhibition of TGF-β1 signaling [22]. It is reported that p53 signaling pathway correlates with the radioresponse of non-small cell lung cancer. Differentially expressed genes in the p53 signaling pathway related to DNA damage repair, apoptosis, cycle regulation, metastasis, deterioration and radioresistance [23]. Previous study has shown that p53 signaling pathway mediate inhibition and apoptosis induced by 12C6+ heavy ion beam irradiation on HepG2 cancer cells [24]. Accordingly, we can infer that hsa-miR-24 and p53 signaling pathway should provide an important contribution to understand the mechanisms of radiosensitivity in human NPC and that it may represent a potential therapeutic target.
Conclusion
In conclusion, this study demonstrates that the upregulated gene JUN was overlap both in DEGs and publicly available studies, was a core hub gene in PPI network, which was potentially targeted by three miRNAs, including hsa-miR-203, hsa-miR-24 and hsa-miR-31. Pathway analysis showed that both up-regulated gene and DEMs were enriched in TGF-beta signaling pathway, Hepatitis B, Pathways in cancer and p53 signaling pathway. Finally, we analyzed the biological networks for validated target gene of common miRNAs, the result indicated that miR-24 is frequently down-regulated in radioresistant NPC cell lines and significantly enriched in P53 signaling pathway. Based on these reasons, our study indicated that the JUN, miR-24 and P53 signaling pathway may be associated with radioresistance in Nasopharyngeal Carcinoma, and which may provide new clues for improving radiosensitivity in Nasopharyngeal Carcinoma. However, these results are only speculated by the combination of databases and bioinformatics methods, and still needs to be confirmed. In the following study, we explored the association between JUN expression and radioresistance in vitro. To further determine the clinical predictive value of JUN in NPC, we performed immunohistochemistry assays to examine the protein expression pattern of JUN in NPC specimens and normal nasopharyngeal epithelium specimens. And then, we performed a log-rank test analysis the Overall survival (OS) of patients with NPC based on JUN expression. Finally, Study the correlation between JUN expression and distant metastasis in patients with NPC.
Methods
Microarray data
The mRNA expression profile of GSE48501 and microRNA profile of GSE48502 were downloaded from the GEO database (Gene expression omnibus, http://www.ncbi.nlm.nih.gov/geo/) [7] (Additional file 1). As previousy described, the raw data was preprocessed by the application of bioconductor package ‘affy’ [6]. The sample of GSE48501 included 2 radioresistant NPC samples and 2 radiosensitive NPC in total, whereas the GSE48502 miRNA expression profiles included 3 radioresistant NPC samples and 3 radiosensitive NPC samples in total.
Identification of DEGs and DEMs
GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/) is a web tool that can analyze almost GEO series. DEGs and DEMs were screened with GEO2R [25] (Additional file 1). The differentially expressed mRNAs were selected using adjusted P -values< 0.05 and |logFC| ≥ 1. P < 0.05 and fold-change≥1.5 were set as the cut-off criterion in the DEMs. Furthermore, we applied to the online tool Morpheus (https://software.broadinstitute.org/morpheus/) to generate a heat map of DEGs [26] (Additional file 1).
Screening common genes between the DEGs and the publicly available studies by Venn diagram
We applied to disease gene search engine with evidence sentence (http://210.107.182.61/geneSearch/) to identify the radioresistant related genes for publicly available studies using the following keywords: “radioresistance” [9]. Then, the overlapping genes between the DEGs and the DigSee were screened by Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/) [27] (Additional file 1).
Gene ontology and pathway analysis of DEGs and DEMs target genes
Gene ontology (GO) biological process terms and pathway enrichment analysis of differentially expressed genes was performed using FunRich software (www.funrich.org/) and Clue Go software respectively [10, 28] (Additional file 1). The P-value < 0.05 was considered significant GO and pathway term. DIANA miRPATH tool (http://www.funrich.org/) was used to analyze gene ontology and pathway analysis of DEMs target genes [29] (Additional file 1). P-value< 0.05 was set as the cut-off criterion in the significant GO terms and kegg pathway.
PPI network construction and subnetwork mining
The STRING database (http://string-db.org/) is an online tool to construct protein-protein interaction network [30] (Additional file 1). The Cytoscape software is a tool for the visual exploration of interaction networks composed of protein, gene, and other types of interactions [31] (Additional file 1). In present study, the protein-protein interaction network (PPI) of DEGs was mapped by STRING and then visualized using Cytoscape. Combined score ≥ 0.4 was set as the cut-off criterion. The proteins with high degrees were considered as the hub nodes. In addition, we further constructed subnetwork mining in the PPI network based on CentiScape with centrality value is high/equals threshold 5.
Hub genes related microRNAs were predicted using mirDIP online software
mirDIP online software (http://ophid.utoronto.ca/mirDIP/index.jsp) integrates twelve microRNA prediction datasets from six different microRNA prediction databases [32] (Additional file 1). In present study, hub genes corresponding microRNAs were predicted by mirDIP software. Top 5% was named high, Score class = high was set as the cut-off criterion.
Analysis of biological networks for common miRNAs
Firstly, we used venn diagram to screen common microRNAs between JUN related microRNAs and DEMs. Then we established a regulatory network for common miRNAs, their target genes and pathways by Clue Go and Clue Pedia [1]. P-value≤0.05, cluster ≥3 and min genes ≥4% was named significant biological networks (Additional file 1).
Additional file
Related data in this article. (PDF 77 kb)
Acknowledgements
The authors will thank Ya Li Wang for her great help in bioinformatics. Thanks Xiao Z, Li X, Qu J for providing analyzable data for this study, including GSE48501and GSE48502 which can be downloaded from the GEO database.
Funding
The publication cost of this article was funded by Shaanxi Provincial Natural Science Foundation [grant numbers 2017JQ8057].
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Abbreviations
- DEGs
Differentially expressed genes
- DEMs
Differentially expressed microRNAs
- GEO
Gene expression omnibus
- GO
gene ontology
- NPC
nasopharyngeal carcinoma
- PPI
Protein-protein interaction network
Authors’ contributions
G Y wrote the main manuscript text and designed the experiment. M Y N and H Y prepared Figs. 1, 2 and 3. Z Y and Z S J prepared Figs. 4 and 5. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ya Guo, Phone: 0086-18629261930, Email: gy8569851@163.com.
Yang Zhang, Email: panduola_001@126.com.
Shu Juan Zhang, Email: 843677894@qq.com.
Yi Nan Ma, Email: yinanxue@126.com.
Yun He, Email: 583170597@qq.com.
References
- 1.Li G, Liu Y, Liu C, Su Z, Ren S, Wang Y, Deng T, Huang D, Tian Y, Qiu Y. Genome-wide analyses of long noncoding RNA expression profiles correlated with radioresistance in nasopharyngeal carcinoma via next-generation deep sequencing. BMC Cancer. 2016;16:719. doi: 10.1186/s12885-016-2755-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li G, Qiu Y, Su Z, Ren S, Liu C, Tian Y, Liu Y. Genome-wide analyses of radioresistance-associated miRNA expression profile in nasopharyngeal carcinoma using next generation deep sequencing. PLoS One. 2013;8(12):e84486. doi: 10.1371/journal.pone.0084486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin H, Chen Z, Zhu X, Li L, Qu S, Wei Z, Su F, Wei J, Liang Z, Mo Q, et al. Serum CD166: a novel biomarker for predicting nasopharyngeal carcinoma response to radiotherapy. Oncotarget. 2017;8(38):62858–62867. doi: 10.18632/oncotarget.16399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan L, Yu X, Huang Z, Zheng S, Zhou Y, Lv H, Zeng Y, Xu J, Zhu X, Yi X. Analysis of microarray-identified genes and MicroRNAs associated with idiopathic pulmonary fibrosis. Mediat Inflamm. 2017;2017:1804240. doi: 10.1155/2017/1804240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett T, Wilhite S, Ledoux P, Evangelista C, Kim I, Tomashevsky M, Marshall K, Phillippy K, Sherman P, Holko M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mou T, Zhu D, Wei X, Li T, Zheng D, Pu J, Guo Z, Wu Z. Identification and interaction analysis of key genes and microRNAs in hepatocellular carcinoma by bioinformatics analysis. World J Surg Oncol. 2017;15(1):63. doi: 10.1186/s12957-017-1127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Qu J, Yi H, Zhang P, Yi H, Wan X, He Q, Ye X, Yuan L, Zhu J, et al. Integrated analysis of differential miRNA and mRNA expression profiles in human radioresistant and radiosensitive nasopharyngeal carcinoma cells. PLoS One. 2014;9(1):e87767. doi: 10.1371/journal.pone.0087767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z, Zhuan B, Yan Y, Jiang S, Wang T. Identification of gene markers in the development of smoking-induced lung cancer. Gene. 2016;576(1):451–457. doi: 10.1016/j.gene.2015.10.060. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Yin W, Zhu X. Blocked autophagy enhances radiosensitivity of nasopharyngeal carcinoma cell line CNE-2 in vitro. Acta Otolaryngol. 2014;134(1):105–110. doi: 10.3109/00016489.2013.844365. [DOI] [PubMed] [Google Scholar]
- 10.Gao H, Wang H, Yang W. Identification of key genes and construction of microRNA-mRNA regulatory networks in multiple myeloma by integrated multiple GEO datasets using bioinformatics analysis. Int J Hematol. 2017;106(1):99–107. doi: 10.1007/s12185-017-2216-2. [DOI] [PubMed] [Google Scholar]
- 11.Chen W, Hu G. Biomarkers for enhancing the radiosensitivity of nasopharyngeal carcinoma. Cancer Biol Med. 2015;12(1):23–32. doi: 10.7497/j.issn.2095-3941.2014.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S, Liu T, Mo W, Hou Q, Zhou Y, Liu M, He Z, Liu Z, Chen Q, Wang H, et al. Prognostic value of phosphorylated Raf kinase inhibitory protein at serine 153 and its predictive effect on the clinical response to radiotherapy in nasopharyngeal carcinoma. Radiat Oncol. 2016;11(1):121. doi: 10.1186/s13014-016-0696-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaulian E. AP-1--the Jun proteins: oncogenes or tumor suppressors in disguise? Cell Signal. 2010;22(6):894–899. doi: 10.1016/j.cellsig.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Qing H, Gong W, Che Y, Wang X, Peng L, Liang Y, Wang W, Deng Q, Zhang H, Jiang B. PAK1-dependent MAPK pathway activation is required for colorectal cancer cell proliferation. Tumour Biol. 2012;33(4):985–994. doi: 10.1007/s13277-012-0327-1. [DOI] [PubMed] [Google Scholar]
- 15.Guo Y, Zhu X, Qu S, Li L, Su F, Li Y, Huang S, Li D. Identification of genes involved in radioresistance of nasopharyngeal carcinoma by integrating gene ontology and protein-protein interaction networks. Int J Oncol. 2012;40(1):85–92. doi: 10.3892/ijo.2011.1172. [DOI] [PubMed] [Google Scholar]
- 16.Eder S, Arndt A, Lamkowski A, Daskalaki W, Rump A, Priller M, Genze F, Wardelmann E, Port M, Steinestel K. Baseline MAPK signaling activity confers intrinsic radioresistance to KRAS-mutant colorectal carcinoma cells by rapid upregulation of heterogeneous nuclear ribonucleoprotein K (hnRNP K) Cancer Lett. 2017;385:160–167. doi: 10.1016/j.canlet.2016.10.027. [DOI] [PubMed] [Google Scholar]
- 17.Song Y, Zuo Y, Qian X, Chen Z, Wang S, Song L, Peng L. Inhibition of MicroRNA-21-5p promotes the radiation sensitivity of non-small cell lung Cancer through HMSH2. Cell Physiol Biochem. 2017;43(3):1258–1272. doi: 10.1159/000481839. [DOI] [PubMed] [Google Scholar]
- 18.Qu J, Yi H, Ye X, Li L, Zhu J, Xiao T, Yuan L, Li J, Wang Y, Feng J, et al. MiR-23a sensitizes nasopharyngeal carcinoma to irradiation by targeting IL-8/Stat3 pathway. Oncotarget. 2015;6(29):28341–28356. doi: 10.18632/oncotarget.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lynam-Lennon N, Reynolds J, Marignol L, Sheils O, Pidgeon G, Maher S. MicroRNA-31 modulates tumour sensitivity to radiation in oesophageal adenocarcinoma. J Mol Med. 2012;90(12):1449–1458. doi: 10.1007/s00109-012-0924-x. [DOI] [PubMed] [Google Scholar]
- 20.Kang M, Xiao J, Wang J, Zhou P, Wei T, Zhao T, Wang R. MiR-24 enhances radiosensitivity in nasopharyngeal carcinoma by targeting SP1. Cancer Med. 2016;5(6):1163–1173. doi: 10.1002/cam4.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rashi-Elkeles S, Elkon R, Shavit S, Lerenthal Y, Linhart C, Kupershtein A, Amariglio N, Rechavi G, Shamir R, Shiloh Y. Transcriptional modulation induced by ionizing radiation: p53 remains a central player. Mol Oncol. 2011;5(4):336–348. doi: 10.1016/j.molonc.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y, Wang L, Huang Q, Jiang Y, Wang J, Zhang L, Tian Y, Yang H. Radiosensitization of non-small cell lung Cancer cells by inhibition of TGF-β1 signaling with SB431542 is dependent on p53 status. Oncol Res. 2016;24(1):1–7. doi: 10.3727/096504016X14570992647087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung I, Kang H, Kim K, Kim I. PTEN/pAkt/p53 signaling pathway correlates with the radioresponse of non-small cell lung cancer. Int J Mol Med. 2010;25(4):517–523. doi: 10.3892/ijmm_00000372. [DOI] [PubMed] [Google Scholar]
- 24.Liu K, Zhao X, Gu J, Wu J, Zhang H, Li Y. Effects of 12C6+ heavy ion beam irradiation on the p53 signaling pathway in HepG2 liver cancer cells. Acta Biochim Biophys Sin Shanghai. 2017;49(11):989–998. doi: 10.1093/abbs/gmx096. [DOI] [PubMed] [Google Scholar]
- 25.Qi Y, Chen X, Wu N, Ma C, Cui X, Liu Z. Identification of risk factors for sepsis-associated mortality by gene expression profiling analysis. Mol Med Rep. 2018. [DOI] [PubMed]
- 26.Mi B, Liu G, Zhou W, Lv H, Liu Y, Liu J. Identification of genes and pathways in the synovia of women with osteoarthritis by bioinformatics analysis. Mol Med Rep. 2018. [DOI] [PMC free article] [PubMed]
- 27.Niu Yuqing, Hu Bei, Li Xiaoquan, Chen Houbin, Takáč Tomáš, Šamaj Jozef, Xu Chunxiang. Comparative Digital Gene Expression Analysis of Tissue-Cultured Plantlets of Highly Resistant and Susceptible Banana Cultivars in Response to Fusarium oxysporum. International Journal of Molecular Sciences. 2018;19(2):350. doi: 10.3390/ijms19020350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Z, Wu H, Wang G, Feng Y. Identification of potential candidate genes for hypertensive nephropathy based on gene expression profile. BMC Nephrol. 2016;17(1):149. doi: 10.1186/s12882-016-0366-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Aiming, Lou Lixia, Zhai Jianying, Zhang Dongmei, Chai Limin, Nie Bo, Zhu Haiyan, Gao Yonghong, Shang Hongcai, Zhao Mingjing. miRNA Expression Profile and Effect of Wenxin Granule in Rats with Ligation-Induced Myocardial Infarction. International Journal of Genomics. 2017;2017:1–10. doi: 10.1155/2017/2175871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou K, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Zhao Q, Lan N, Wang S. Identification of methylated genes and miRNA signatures in nasopharyngeal carcinoma by bioinformatics analysis. Mol Med Rep. 2018. [DOI] [PMC free article] [PubMed]
- 32.Tokar T, Pastrello C, Rossos A, Abovsky M, Hauschild A, Tsay M, Lu R, Jurisica I. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018;46(D1):D360–D370. doi: 10.1093/nar/gkx1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related data in this article. (PDF 77 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its supplementary information files].