Abstract
Background
Brassinosteroids (BRs) play crucial roles in drought tolerance, but the underlying molecular mechanisms remain unclear in the important oilseed and fiber crop, cotton (Gossypium hirsutum L.).
Results
To elucidate how BRs mediate drought tolerance in cotton, a cotton brassinosteroid (BR)-deficient mutant, pag1 (pagoda1), was employed for analysis. Importantly, the pag1 mutant showed increased sensitivity to drought stress, with shorter primary roots and fewer lateral roots. The number of stomata was significantly increased in the mutant, and the stomata aperture was much wider than that of the control plants. These mutant plants therefore showed an increased water loss rate. Furthermore, the abscisic acid (ABA) content, photosynthetic efficiency and starch content of the mutant were significantly lower than those of the wild type. The overall performance of the mutant plants was worse than that of the wild-type control under both normal and drought conditions. Moreover, Proteomic analysis revealed reduced levels of stress-related proteins in pag1 plants.
Conclusions
These results suggest that BRs may modulate the drought tolerance of cotton by regulating much genes that related to drought stress and multiple organ responses to drought, including root growth, stomata development, the stomata aperture and photosynthesis. This study provides an important basis for understanding drought resistance regulated by BRs and cultivating drought-resistant cotton lines.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1832-9) contains supplementary material, which is available to authorized users.
Keywords: ABA, Brassinosteroid (BR), Cotton (Gossypium hirsutum L.), Drought stress, pag1, Proteomic
Background
Brassinosteroids (BRs), a group of plant-specific polyhydroxylated steroidal hormones were first isolated from B. napus pollens [1] and are regarded as essential in regulating multiple processes during normal plant growth and development, including cell division and elongation, vascular differentiation, root development, guard cell development, reproductive processes, and responses to various biotic and abiotic stresses [2–4]. The levels of bioactive endogenous BRs in plants are regulated by both biosynthesis and catabolism. CYP734A1/BAS1, a cytochrome P450 monooxygenase, catalyzes the C-26 hydroxylation of castasterone (CS) and brassinolide (BL), which are active BRs [5, 6]. PAG1 is the cotton homolog of CYP734A1/BAS1 and encodes a cytochrome P450 monooxygenase. The pag1 mutant shows reduced fiber elongation, in addition to the typical phenotypes of BR mutants, such as dwarfed stature and dark green leaves [7].
The function of BRs in stress tolerance has been frequently reported. A study by Kagale showed that 24-epi-BL (24-epibrassinolide) treatment could enhance drought tolerance in both Arabidopsis thaliana and B. napus seedlings [8]. In maize, BL application improves drought tolerance by modulating enzymatic antioxidants and the leaf gas exchange system [9]. Another study demonstrated that BRs could enhance tolerance to oxidative stress induced by polyethylene glycol (PEG) treatment through BR-induced NO production and NO-activated ABA biosynthesis in maize leaves [10]. In cucumber, elevated H2O2 levels due to enhanced NADPH oxidase activity are involved in BR-induced tolerance to stresses, such as photo-oxidative and cold stress and Cucumber mosaic virus infection [11]. The Arabidopsis BR-deficient mutant det2–1 was found to be more sensitive to salt stress than wild-type plants [12]. Moreover, overexpression of the BR biosynthesis gene AtDWF4 in Arabidopsis increases the cold tolerance of transgenic seedlings [13]. One of the mechanisms through which BRs participate in cold tolerance might be regulation of pectin methylesterase (PME) activity [14]. In addition, overexpression of AtDWF4 in B. napus simultaneously increases seed yield and stress tolerance, and transcriptome analysis supported the integrated effects of BRs on growth and stress responses [15]. In tomato, the BR-deficient mutant Micro-Tom shows hypersensitivity to drought stress [16].
Drought is one of the most important abiotic factors that seriously threatens modern agriculture worldwide by limiting plant growth and ecosystem production [17]. As an important renewable resource, cotton ranks second among oilseed crops and is the world’s leading natural fiber crop; it is grown in more than seventy countries worldwide, and its production plays a vital role in the global economy. However, the productivity of cotton is adversely affected by a variety of biotic and abiotic stresses, including drought stress. Therefore, the importance things for enhancing cotton production are investigating molecular mechanisms of cotton stress tolerance and cultivating new drought-resistant cotton varieties [18].
BRs confer tolerance to a wide range of abiotic stresses. However, most studies on the effects of BRs on plant stress responses have been conducted using exogenous BRs, and the mechanisms underlying the functions of BRs in abiotic stresses are still unclear. Thus, additional genetic evidence is required to determine the function of BRs in stress tolerance and reveal their functional mechanisms in abiotic stresses. Here, we performed a detailed evaluation of the drought tolerance of pag1 to determine the role of BRs in the response of cotton to drought. We found that pag1 exhibits increased sensitivity to drought stress. In addition, compared with CCRI24 control plants, pag1 plants have lower CS content, increased stomata number and aperture, increased water loss rate, lower ABA content, reduced primary root length and lateral root numbers, lower expression of GhPIN2 and GhLAX3 (which encode polar auxin transport carriers), lower photosynthetic efficiency and starch content, and reduced agronomic traits under both normal and drought conditions. Proteomics analysis of pag1 and CCRI24 (Chinese cotton research institution 24) plants after PEG6000 treatment indicated that a number of stress-related proteins were down-regulated in the pag1 mutant. Our study is one of the first to reveal the function of BRs in stress tolerance using a BR-deficient cotton mutant, and our data suggest that BRs play important roles in the cotton drought stress response. These findings provide an important basis for fully understanding the mechanism of drought resistance regulated by BRs and for developing BR-based breeding programs for crop improvement.
Results
The pag1 mutant is specifically defective in CS, an active BR
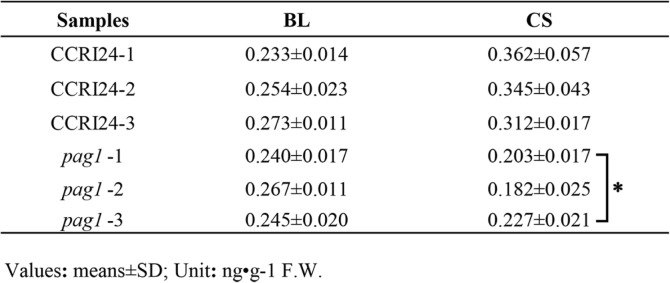
In Arabidopsis, BAS1 (CYP734A1) inactivates CS and BL which all belongs to BRs via C-26 hydroxylation [6], and PAG1 is closely related to BAS1 [7], suggesting that PAG1 might function similarly to BAS1 in the inactivation of CS and BL. However, the exact function of PAG1 on CS and BL and whether pag1 is truly a BR-deficient mutant or not require further analysis. To confirm the role of PAG1 in regulating CS and BL levels, the CS and BL contents of pag1 and wild-type CCRI24 plants were analyzed. The results showed that the average content of CS was 0.340 ng/g in CCRI24 plants and 0.204 ng/g in pag1 plants (Table 1); moreover, the test of significant differences showed that the contents of CS were significantly lower in pag1 plants than in CCRI24 plants (Table 1). Although the content of BL was slightly lower in pag1 plants than in CCRI24 plants, there was no significant difference in BL contents between the two lines (Table 1). These results suggest that pag1 is a BR-deficient mutant and that PAG1 primarily functions in the inactivation of CS.
Table 1.
Brassinosteroids (BRs) levels in the pag1 mutant and CCRI24
Student’s t test: *, P < 0.05
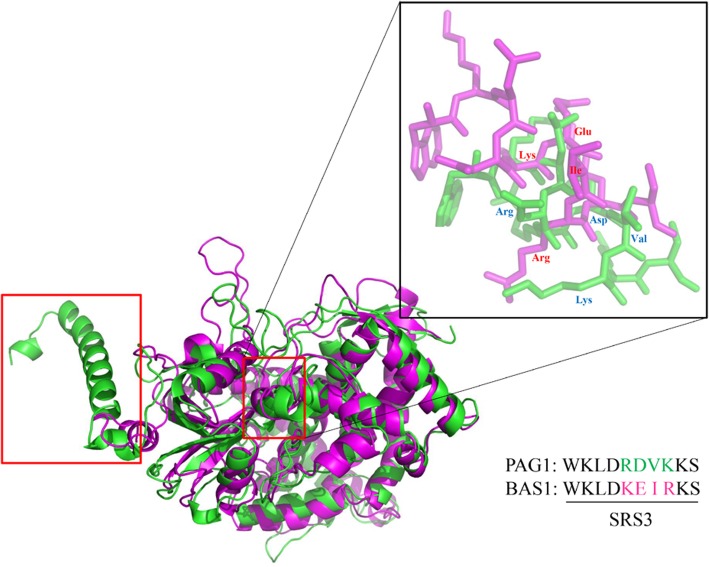
To analyze the specific function of PAG1 in cotton, the well-modeled PAG1 and BAS1 structures all built with Cytochrome P450 4B1 template (PDB ID: 6c93) were applied using the Swiss-Model server [19], then structural alignment of PAG1 with BAS1 was done and shown in Fig. 1. The results of structural analysis showed that there was an extra α-helix at the N-terminus of PAG1, and the third substrate recognition sites (SRS3) of PAG1 and BAS1 did not match perfectly (Fig. 1). The structural differences between PAG1 and BAS1 might be the reason that PAG1 mainly inactivated CS in cotton.
Fig. 1.
Structural alignment of PAG1 (green) with BAS1 (magenta). The red box indicates the areas of differences in the structures of PAG1 and BAS1. The inset shows the difference in the third substrate recognition site (SRS3) between PAG1 and BAS1
The pag1 mutant is sensitive to drought stress
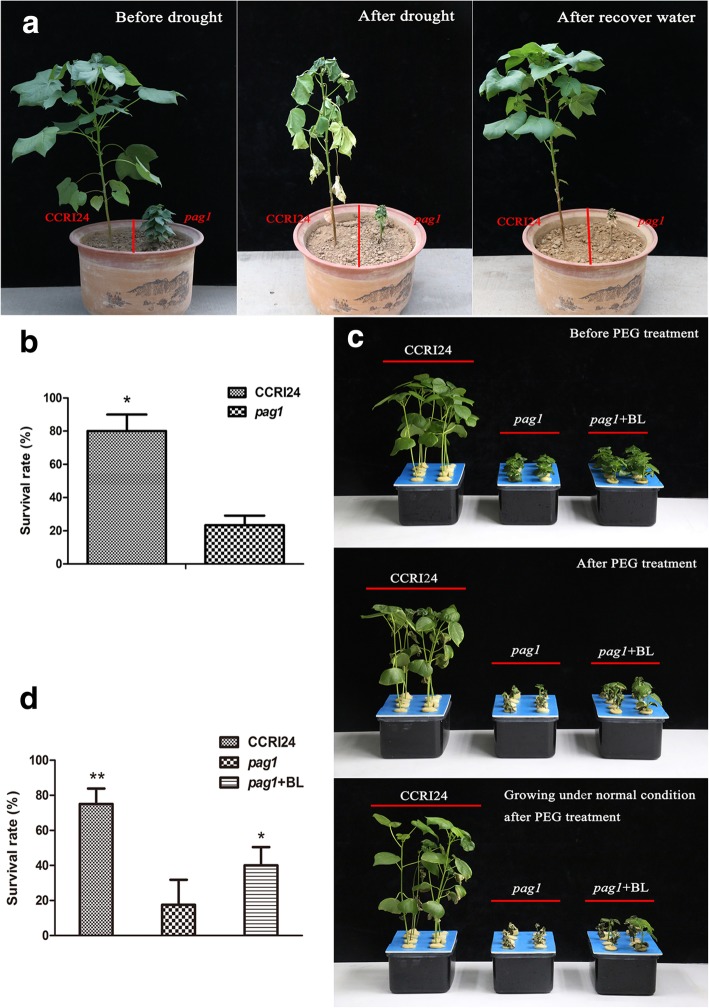
To evaluate the function of BRs in drought tolerance in cotton, the pag1 mutant and CCRI24 controls were grown for 50 days in pots, and then watering was stopped. After 25 days, all the leaves of pag1 plants were wilted, but majority of the CCRI24 leaves remained vigorous (Fig. 2a). After one week of water recovery, the CCRI24 plants resumed normal growth, but the pag1 plants had died (Fig. 2a), and the statistical result showed that CCRI24 plants had a significantly higher survival rate than pag1 plants (Fig. 2b).
Fig. 2.
Response of pag1 and CCRI24 to drought stress. a Phenotypes of pag1 and CCRI24 before drought stress, after drought stress and after water recovery while growing in soil. b Analysis of the survival rate after water recovery of pag1 and CCRI24, as shown in a. c Phenotype of pag1 and CCRI24 growing in cotton nutrient solution before PEG treatment, after PEG treatment and growing under normal conditions after PEG treatment. d Analysis of the survival rate pag1 and CCRI24 growing under normal conditions after PEG treatment as shown in c. (t test): *, P < 0.05; **, P < 0.01
To determine whether BRs could restore the ability of pag1 to resist drought stress, CCRI24 plants, pag1 plants, and pag1 plants treated with 24-epi-BL were grown in hydroculture, and drought stress was simulated by treating three-week-old seedlings with 6% PEG6000. After one week of treatment, a majority of the pag1 plants were dead, and only 13.3% survived (Fig. 2c, d). The CCRI24 line exhibited a significantly higher survival ratio of 73.3%, and the 24-epi-BL-treated pag1 plants also exhibited a higher survival ratio of 26.7% (Fig. 2d). These results demonstrated that PAG1 regulates cotton drought tolerance partially by affecting BR levels.
Increased leaf stomata density and wider stomata aperture in pag1
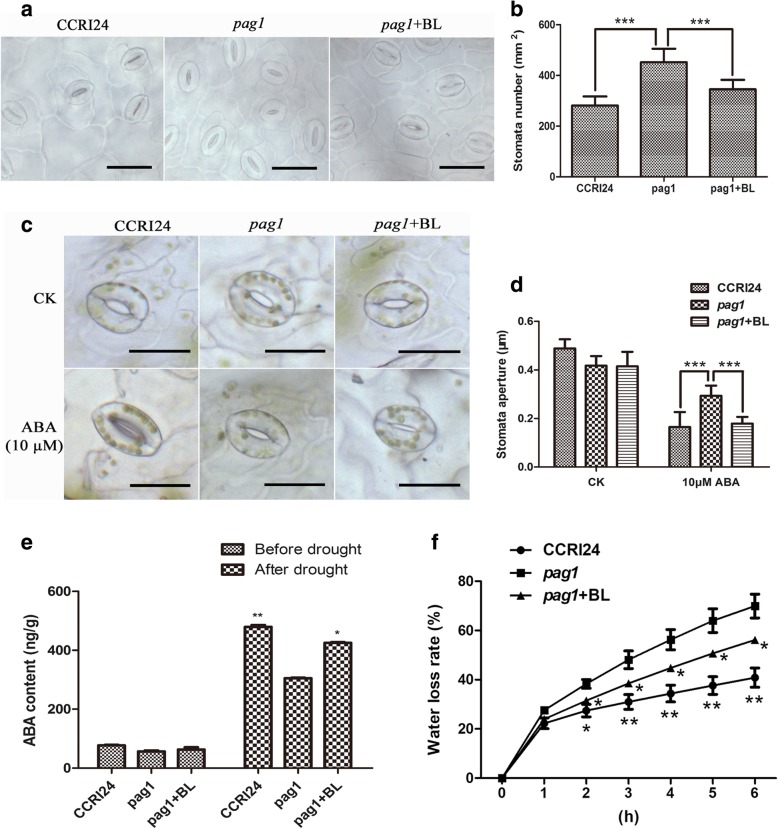
Stomata density and stomata movement (opening and closing) can regulate transpirational water loss, which is a key determinant of drought tolerance [20, 21]. To examine why pag1 is sensitive to drought stress, the stomata density of pag1 plants, 24-epi-BL-treated pag1 plants and CCRI24 plants was analyzed at the one-month-old seedling stage. The results showed that the average stomata density in the top fourth leaf of pag1 plants was 37.8% higher than in CCRI24 plants (Fig. 3a, b), and the stomata density of the 24-epi-BL-treated pag1 leaves was 23.7% lower than that of untreated pag1 plants (Fig. 3a, b).
Fig. 3.
Stomata analysis of pag1 and CCRI24. a Observations of the stomata of pag1, CCRI24 and 24-epi-BL-treated pag1 plants at the three-leaf stage. Bar = 100 μm. b Stomata number analysis of the pag1, CCRI24 and 24-epi-BL-treated pag1 plants shown in a. c Observations of stomata aperture of pag1, CCRI24 and 24-epi-BL-treated pag1 plants at the three-leaf stage under normal conditions and after ABA treatment. Bar = 50 μm. d Stomata aperture analysis of the pag1, CCRI24 and 24-epi-BL-treated pag1 plants shown in c. e ABA contents of pag1, CCRI24 and 24-epi-BL-treated pag1 plants under normal conditions and after drought treatment. f Water loss rate analysis of pag1, CCRI24 and 24-epi-BL-treated pag1 plants at the three-leaf stage. Student t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001
ABA modulates stomata closure by guard cells to avoid water loss during drought stress [22]. As shown in Fig. 3c and d, ABA treatment caused more pronounced stomata closure in CCRI24 and 24-epi-BL-treated pag1 leaves than in pag1 leaves. ABA content analysis showed that the ABA content in pag1 plants was not significantly different than in CCRI24 and 24-epi-BL-treated pag1 under normal conditions, but the ABA content was significantly lower in pag1 plants than in CCRI24 and 24-epi-BL-treated pag1 plants after drought treatment, even though ABA contents were increased in all three groups (Fig. 3e). These results demonstrate that BRs regulate the stomata aperture by regulating the ABA content in cotton.
Moreover, a water loss rate assay demonstrated that the water loss rate was significantly lower in both 24-epi-BL-treated pag1 plants and CCRI24 plants than in pag1 plants (Fig. 3f), and the water loss rate in the 24-epi-BL-treated pag1 plants was also higher than in the CCRI24 plants. These results were consistent with the results of the stomata density and stomata aperture analyses.
Altered root architecture resulting from lower expression levels of auxin polar transport genes in pag1
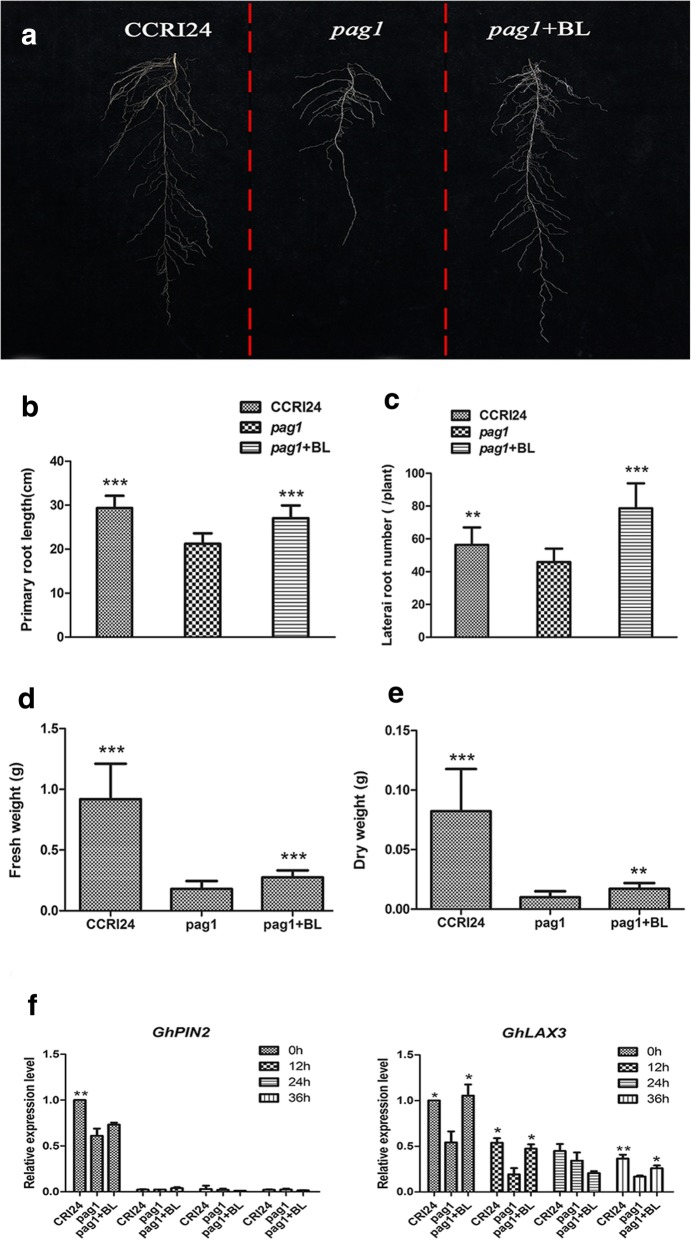
Deep root systems help to increase the water accessibility of plants. pag1 plants grown in hydroculture exhibited shorter primary root lengths and fewer lateral roots than CCRI24 plants at the trefoil stage; moreover, the primary root length and lateral root number were restored when pag1 plants were grown in a nutrient solution containing 10 nM 24-epi-BL (Fig. 4a, b, c). The root biomass of one-month-old pag1 seedlings was markedly decreased, whereas the root biomass of pag1 seedlings grown in a nutrient solution containing 10 nM 24-epi-BL was higher, but still lower than that of CCRI24 seedlings (Fig. 4d). Consequently, the root dry weight of pag1 seedlings was significantly lower than that of CCRI24 seedlings, and the root dry weight of 24-epi-BL-treated pag1 seedlings was considerably higher than that of untreated pag1 seedlings (Fig. 4e). The altered root architecture of pag1 plants might negatively contribute to drought tolerance, as shorter primary roots and fewer lateral roots could result in reduced water and nutrient uptake.
Fig. 4.
Root phenotype of pag1 and CCRI24. a Root phenotype of pag1 plants, CCRI24 plants and pag1 plants treated with 24-epi-BL at the three-leaf stage. b Primary root length analysis of the pag1, CCRI24 and 24-epi-BL-treated pag1 plants shown in a. c Lateral root number analysis of the pag1, CCRI24 and 24-epi-BL-treated pag1 plants shown in a. d Root fresh weight of pag1, CCRI24 and 24-epi-BL-treated pag1 plants at the three-leaf stage. e Root dry weight of pag1, CCRI24 and 24-epi-BL-treated pag1 plants at the three-leaf stage. f Expression levels of GhPIN2 and GhLAX3 in pag1, CCRI24 and 24-epi-BL-treated pag1 plants at the three-leaf stage under normal conditions and at 12 h, 24 h and 36 h after PEG treatment. (t test): *, P < 0.05; **, P < 0.01; ***, P < 0.001
Auxin is an important phytohormone that regulates root growth; thus, we speculated that BRs could promote the expression of auxin polar transport genes in plant roots [23]. Auxin polar transport requires auxin polar transport carriers, and qPCR was performed to analyze the gene expression levels of two auxin polar transport carrier genes, GhPIN2 and GhLAX3. The expression level of GhPIN2 was significantly lower in pag1 plants than in CCRI24 and 24-epi-BL-treated pag1 plants under normal conditions, but smaller differences were observed among pag1, CCRI24 and 24-epi-BL-treated pag1 plants after 12 h, 24 h, and 36 h of PEG6000 treatment (Fig. 4f). The expression of GhLAX3 was significantly lower in pag1 plants than in CCRI24 plants, both under normal conditions and after treatment with PEG6000; in addition, its expression was lower in pag1 plants than in 24-epi-BL-treated pag1 plants under the same conditions described above, except in plants treated with PEG6000 for 24 h (Fig. 4f). These results suggested that BRs regulate the expression of auxin polar transport genes to affect auxin polar transport and, in turn, regulate cotton root growth.
Reduced photosynthetic rate and starch content in the pag1 mutant
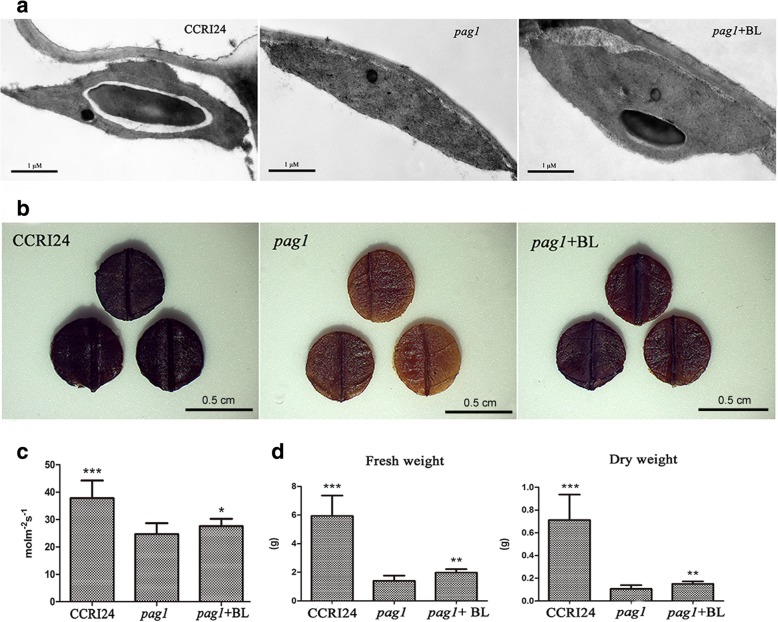
Studies have shown that BRs can promote the plant photosynthesis rate [24, 25]. During photosynthesis, fixed carbon is converted to starch in the leaves of most plant species; subsequently, starch accumulates within chloroplasts [26]. In our study, transmission electron microscopy images of sections of the top fourth leaves showed that CCRI24 plants, which contained large starch granules, exhibited a higher starch content than pag1 plants, which contained almost no starch in their chloroplasts (Fig. 5a). When pag1 leaves were treated with 24-epi-BL, starch deposits were observed in chloroplasts, but these deposits were smaller than those in CCRI24 leaves (Fig. 5a). Iodine staining was also used to detect starch reserves in the leaves of CCRI24, pag1 and 24-epi-BL-treated pag1 plants. Upon staining, CCRI24 leaves were found to harbor deep blue starch granules, while the staining in pag1 leaves was lighter. However, 24-epi-BL-treated pag1 leaves exhibited an intermediate blue shade after granule staining, which was deeper in color than that in pag1 leaves and slightly lighter than that in CCRI24 leaves (Fig. 5b). We also measured photosynthesis in pag1 and CCRI24 plants at the reproductive stage in the field and found that the photosynthetic rate was reduced by 34.7% in the pag1 plants compared with CCRI24 plants (Fig. 5c), while the photosynthetic rate of 24-epi-BL-treated pag1 plants was significantly higher than that of pag1 plants. The leaf photosynthesis rate is positively correlated with the total biomass of a plant [27]. Therefore, the seedling biomass was also examined. Although the fresh and dry weights of CCRI24 plants and 24-epi-BL-treated pag1 plants were significantly higher than those of pag1 plants, the biomass of 24-epi-BL-treated pag1 plants was still lower than that of CCRI24 plants (Fig. 5d).
Fig. 5.
Analysis of photosynthetic rates and starch contents in pag1 and CCRI24. a Observations of starch granules in the top fourth leaves of pag1, CCRI24 and 24-epi-BL-treated pag1 plants using transmission electron microscopy. Bar = 1 μm. b Observations of starch contents in the top fourth leaves of pag1, CCRI24 and 24-epi-BL-treated pag1 plants using the iodine staining method. Bar = 0.5 cm. c Photosynthetic rate analysis of pag1, CCRI24 and 24-epi-BL-treated pag1 plants. d Fresh weight and dry weight of pag1, CCRI24 and 24-epi-BL-treated pag1 plants at the three-leaf stage. (t test): *, P < 0.05; **, P < 0.01; ***, P < 0.001
Agronomic performance and cotton yield of the pag1 mutant in the field
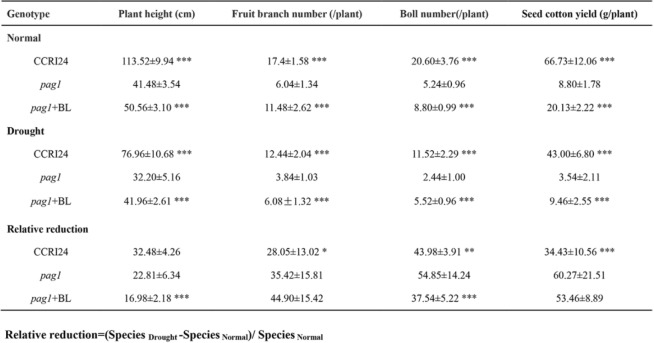
Yield is an important agronomic trait in analyzing stress tolerance of crops [28, 29]. To evaluate the agronomic traits of the pag1 mutant in the field, pag1, CCRI24 and 24-epi-BL-treated pag1 plants were grown in the field in Anyang, Henan Province, China. To evaluate field traits under drought conditions, pag1, 24-epi-BL-treated pag1 and CCRI24 plants were grown in an open green house in the same place to simulate normal field conditions, with the exception of rain. As shown in Table 2, under normal conditions, the pag1 plants showed significantly decreased height, fruit branch number, boll number per plant and seed cotton yield per plant compared with those of the CCRI24 and 24-epi-BL-treated pag1 plants. Under drought conditions, the seed cotton yield and other agronomic traits mentioned above were distinctly decreased in pag1 plants compared with CCRI24 and 24-epi-BL-treated pag1 plants (Table 2). These results suggest that BR deficiency has a negative effect on agronomic traits and cotton yield under both normal and drought stress conditions in the field. More importantly, under drought conditions, the relative reduction of boll number was much lower in CCRI24 and 24-epi-BL-treated pag1 plants than that in pag1 plants (Table 2). Besides, the relative reduction of seed cotton yield was much higher in pag1 plants than that in CCRI24 plants, and also high in pag1 plants than that in 24-epi-BL-treated pag1 plants though there was no significant difference between them (Table 2). These results suggest that BR deficiency results in easier reduction of yield traits of cotton under drought conditions.
Table 2.
Agronomic traits and cotton yields of pag1, CCRI24 and 24-epi-BL-treated pag1 under two below conditions
Student’s t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001
Proteomics reveals numerous proteins involved in the pag1 response to drought stress
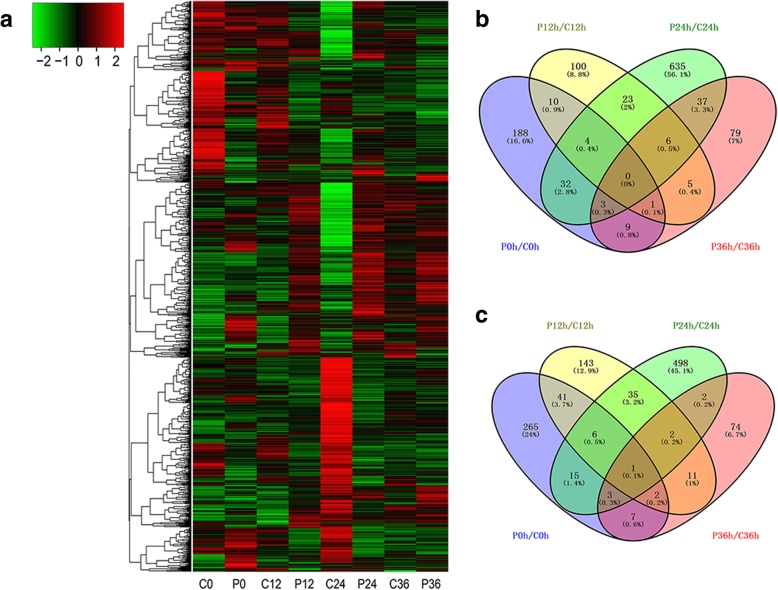
To explore the underlying molecular mechanisms of BR mediate drought tolerance in cotton, iTRAQ analysis was used to identify differentially abundant proteins in pag1 and CCRI24 roots, both under normal conditions and after treatment with 6% PEG6000. Under normal conditions, 587 proteins were up- or down-regulated by at least 1.2-fold in pag1 roots compared with CCRI24 roots. Moreover, at the three time points of PEG6000 treatment (12 h, 24 h and 36 h), 390, 1302 and 242 proteins with a greater than ±1.2-fold difference between pag1 and CCRI24 roots were identified. In addition, a cluster analysis was performed, and a heatmap was generated to visualize the expression patterns of all differentially abundant proteins in pag1 and CCRI24 plants (Fig. 6a). Venn diagram results indicated that only a few differentially abundant proteins overlapped between P0h/C0h (pag1-0 h/CCRI24-0 h), P12h/C12h (pag1-12 h/CCRI24-12 h), P24h/C24h (pag1-24 h/CCRI24-24 h) and P36h/C36h (pag1-36 h/CCRI24-36 h) (Fig. 6b, c). Among these 1245 down-regulated differentially abundant proteins, only 50 (4.5%), 25 (2.3%) and 13 (1.2%) proteins identified in P0h/C0h overlapped with those identified in P12h/C12h, P24h/C24h and P36h/C36h, respectively (Fig. 6c).
Fig. 6.
Overview of serial analysis of differentially abundant proteins between pag1 and CCRI24 after PEG treatment. a Hierarchical clustering analysis of all differentially abundant proteins based on iTRAQ data. b and c Venn diagrams of up- and down-regulated proteins in comparisons of P0/C0, P12/C12, P24/C24 and P36/C36. P, pag1; C, CCRI24; 0, 12, 24 and 36 represent the trefoil-stage roots of pag1 and CCRI24 after treatment with 6% PEG6000 for 0 h, 12 h, 24 h and 36 h, respectively
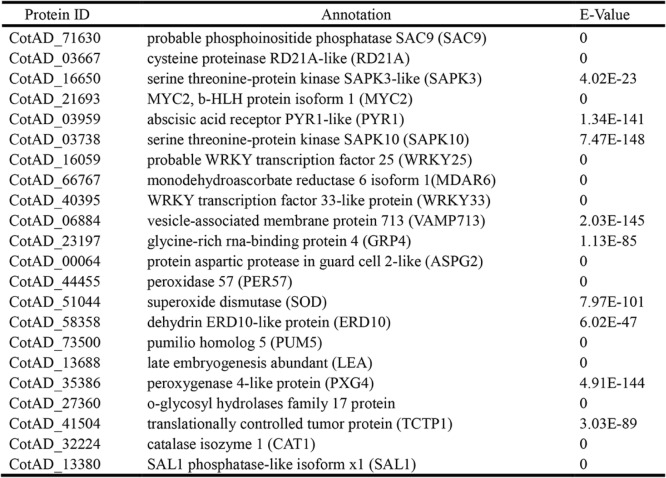
In addition to these differentially abundant proteins, many well-known stress-related genes were found to be down-regulated in pag1 roots (Table 3). Most of these proteins showed lower expression in pag1 roots than in CCRI24 roots both under normal conditions and after PEG6000 treatment. These results collectively suggest that the decrease of BRs in pag1 inhibited the protein abundant of stress-resistance genes under drought stress conditions, thus reducing pag1 drought tolerance. Among the proteins that were significantly inhibited in pag1 plants, quite a few were stress-responsive transcription factors, such as MYC2, WRKY25 and WRKY33, indicating that BRs may indirectly regulate the expression of numerous stress-responsive genes. To confirm the iTRAQ results, 10 stress-related genes were selected for qRT-PCR analysis using specific primers (Additional files 6: Table S5). The expression patterns of most of these genes were consistent with the iTRAQ results that the protein levels were lower in pag1 plants than in CCRI24 plants, with the exception of three genes for which larger differences were observed at different treatment time points (Additional file 1: Figure S1). These results suggest that the iTRAQ results are reliable and useful.
Table 3.
Down-regulated proteins related to stress tolerance in the pag1 mutant
Discussion
The PAG1 gene mainly affects CS content in cotton
The levels of bioactive endogenous BRs in plants are regulated by both biosynthesis and catabolism [30]. Mutation of CPD results in a typical BR-deficient phenotype, including an extremely dwarfed stature, dark green color, delayed flowering, and reduced male fertility [31, 32]. In vitro, biochemical analysis showed that CPD catalyzes the C-3 oxidation of early BR biosynthetic intermediates [33]. Ectopic overexpression of BAS1/CYP734A1, which encodes a BR-inactivating enzyme, in tobacco results in dwarf plants with dark-green leaves and short stems and petioles [5]. Moreover, the bas1-D/phyB-4 mutant exhibits low levels of BL and CS and accumulates 26-hydroxybrassinolide [5], primarily because the function of BAS1 is to catalyze the C-26 hydroxylation of CS and BL [5, 6]. Although a number of genes that regulate BR content have been identified in Arabidopsis, only a few genes have been reported to regulate BR levels in cotton. Here, we report an analysis of the PAG1 gene, which is a homolog of BAS1/CYP734A1 [7]. The cotton pag1 mutant, which exhibits high expression of the PAG1 gene, displays a typical BR-deficient phenotype, including severe dwarfism, dark green leaves and reduced fiber elongation [7]. These observations suggest that PAG1 functions similarly to BAS1 in catabolizing bioactive BRs in cotton. To confirm the characteristics of the PAG1 gene in cotton, BL and CS levels were determined in pag1. Our results showed that the CS level was significantly lower in the pag1 mutant than in CCRI24 plants; however, the BL level did not differ between pag1 and CCRI24 plants (Table 1). These results suggest that the main function of PAG1 might be to specifically catalyze the hydroxylation of CS and reduce total BR levels in cotton, and the reduction of an active BR could result in a typical BR-deficient phenotype in cotton. Structural analysis showed that PAG1 harbored an extra α-helix at N-terminus not found in BAS1 and that the third substrate recognition site (SRS3) of the two proteins exhibited a large difference (Fig. 1). Thus, the different functions of PAG1 and BAS1 might be a result of their protein structure differences and may indicate that PAG1 has a special function in inactivating CS in cotton, as homologous genes can have both similar and different functions in different plants.
BRs regulate stomata development and inhibit stomata closure induced by ABA
Recently, BRs have been found that it could activate the MAPK kinase kinase YDA to inhibit stomata formation in Arabidopsis [34–36]. In our study, we found that the stomata densities of CCRI24 and 24-epi-BL-treated pag1 plants were significantly lower than that of pag1 plants (Fig. 3a, b). This result indicates a similar function of BRs in inhibiting stomata formation in cotton.
ABA is an important phytohormone that influences stomata aperture movement (opening and closing) [37]. BR application has been shown to enhance the drought tolerance of Arabidopsis and B. napus seedlings, which is accompanied by ABA accumulation [8]. Our stomata aperture analysis indicated that the pag1 plants have larger stomata aperture than CCRI24 plants, while the stomata aperture of 24-epi-BL-treated pag1 plants was significantly reduced compared with that in untreated mutants (Fig. 3c, d). Moreover, the ABA content was significantly lower in pag1 plants than in CCRI24 plants and was remarkably elevated in 24-epi-BL-treated pag1 plants (Fig. 3e). These observations suggest that BRs regulate ABA-inducted stomata aperture movement, thereby regulating drought tolerance in cotton. In addition, ABA regulates plant responses by acting as a signaling molecule under normal or stress conditions [38, 39]. Thus, BRs might also affect stress-related genes that are controlled by ABA to regulate cotton drought tolerance (Additional file 2: Table S1).
Stomata density and stomata movement play key roles in regulating the plant leaf transpiration rate [20, 21]. In this study, we found that the water loss rate was significantly higher in pag1 plants than in CCRI24 and 24-epi-BL-treated pag1 plants (Fig. 3f), which was consistent with the results regarding stomata density and stomata aperture in pag1 plants (Fig. 3a-d). These observations suggest that BRs regulate stomata density and ABA-inducted stomata closure, which in turn helps regulate the water loss rate of cotton and, thus, controls drought tolerance.
BRs interact with auxin to regulate root development in cotton
A thick, deep root system enables plants to extract deep soil water and is considered important in determining plant drought tolerance [40]. It has been observed that a concentration of BL (1-100 nM) can promote root elongation in maize [41]. However, other studies indicated that a low concentration of BL (0.05–0.1 nM) can promote root elongation and 1–100 nM BL inhibits root elongation [23, 42]. In our study, we observed shorter primary root lengths in pag1 plants than in CCRI24 plants. The root length phenotype of pag1 mutants could be restored to that of CCRI24 plants after treatment with 24-epi-BL (Fig. 4a, b). These results suggest that a proper concentration of BRs can promote root elongation in plants and that BRs are required for root elongation. Moreover, it was found that wild-type plants exhibit more lateral roots than BR-deficient mutants [23]. Similarly, we observed fewer lateral roots in pag1 plants than in CCRI24 plants, although the phenotype could be restored to that of CCRI24 plants after treatment with 24-epi-BL (Fig. 4a, c). Our results provide further evidence that BRs regulate lateral root development in plants. Overall, it can be concluded that the function of BRs in root development is conservative.
Polar auxin transport, which results in distribution of auxin, is involved in almost all auxin-mediated plant growth processes, including root growth [43]. In addition, BRs enhance polar auxin transport and endogenous auxin distribution by up-regulating the expression of auxin efflux carriers, such as PIN2 [44]. Our study showed that the expression level of an auxin efflux carrier gene, GhPIN2, was significantly lower in pag1 plants than in CCRI24 plants and was elevated in 24-epi-BL-treated pag1 plants under normal conditions; furthermore, few differences in gene transcript levels were found in the three plant groups after PEG6000 treatment (Fig. 4f). The transcript level of GhLAX3, an auxin influx carrier gene, was significantly higher in CCRI24 and 24-epi-BL-treated pag1 plants than in pag1 plants under both normal and PEG6000 treatment conditions, except in samples of 24-epi-BL-treated pag1 plants that were treated with PEG6000 for 24 h, although the expression of GhLEA3 was down-regulated after PEG6000 treatment compared with that under normal conditions (Fig. 4f). These results suggest that BRs mediate root development by regulating the auxin distribution, which is affected by genes involved in polar auxin transport, and in turn play an important role in regulating drought tolerance in cotton.
BR deficiency inhibits the photosynthetic rate and reduces the yield of cotton
A number of studies have focused on the function of BRs in photosynthesis and determining yields. Xia et al. found that BRs could increase the plant photosynthesis rate and growth by positively regulating many photosynthetic enzymes [24]. Another study revealed that BRs could enhance CO2 assimilation, increase glucose pools in leaves, and increase the conversion of glucose to starch in seeds [25]. Furthermore, overexpression of AtDWF4, an important BR biosynthetic gene in B. napus was shown to increase the seed yield [15]. In our study, the pag1 mutant was found to exhibit a lower photosynthetic rate than CCRI24 and 24-epi-BL-treated pag1 plants (Fig. 5c). The biomass of pag1 plants was also lower than that of CCRI24 and 24-epi-BL-treated pag1 plants (Fig. 5d). In addition, the starch content was lower in pag1 plants than in the other two plant groups (Fig. 5a, b). Moreover, under both normal and drought conditions, the yield of pag1 plants was lower than that of CCRI24 plants and significantly increased following treatment with 24-epi-BL (Table 2). These results revealed that BR deficiency inhibits photosynthesis in cotton and reduces energy accumulation and cotton yield.
Photosynthesis plays a vital role in sucrose and starch synthesis, and sugars such as sucrose, glucose and fructose are involved in drought signaling pathways [45, 46]. The carbohydrate status in drought-stressed plants depends not only on the efficiency of the photosynthetic carbon reduction cycle and sucrose/starch synthesis but also on osmotic adjustment [47–49]. In our research, the pag1 mutant was found to be more sensitive to drought stress than control plants (Fig. 2a, b). Moreover, the relative reduction of yield traits (boll number and seed cotton yield) observed under drought conditions was much greater in pag1 plants than in CCRI24 plants and 24-epi-BL-treated pag1 plants, except for the seed cotton yield in pag1 plants treated with 24-epi-BL (Table 2). Thus, all the above results suggested that BR deficiency inhibits energy accumulation to support resistance to drought stress in cotton and increase the relative reduction of the cotton yield under drought conditions.
The profound impact of BRs on global protein abundance in pag1 is consistent with the observed phenotypes
Proteomics is a powerful tool for studying plant stress responses, as proteomic comparisons could facilitate the identification of key genes and regulatory mechanisms involved in drought tolerance. In this study, we analyzed differentially abundant proteins in pag1 and CCRI24 roots, both under normal conditions and after PEG6000 treatment. A total of 2100 differentially abundant proteins were found in pag1 and CCRI24 samples under both control and PEG6000-treated conditions, and 1105 proteins were revealed to exhibit low abundance in pag1 roots compared with CCRI24 roots (Fig. 6a, c). Moreover, many stress-related genes were down-regulated in the pag1 mutant (Table 3). These results suggest that BRs can regulate a large set of proteins, both under normal conditions and upon PEG6000 treatment. In addition, BR deficiency can reduce the protein expression of many well-known stress-responsive genes (Table 3), which may make cotton more sensitive to drought stress.
Through a detailed analysis of differentially abundant proteins between pag1 and CCRI24 under normal conditions and after PEG6000 treatment, we found that proteins involved in carbon metabolism, starch and sucrose metabolism, carbon fixation in photosynthetic organisms and photosynthesis showed significant reductions in pag1 plants (Additional file 3: Table S2), although we used the roots for proteomic analysis. These results suggest that BR effect on these proteins, which are mainly involved in photosynthesis, are far-reaching and extend to different organs. It has been reported that BRs can enhance the photosynthesis rate, and can increase seed yield in B. napus [15, 24]. Interestingly, our results showed a similar expression pattern of the above proteins in pag1 (Additional files 3: Table S2), which may partially explain the decreased photosynthetic rate and cotton yield observed in this BR-deficient mutant.
Moreover, many proteins involved in the ABA signal pathway, auxin signal pathway and root development were also found to be reduced in pag1 plants through proteomic analysis (Additional file 2: Table S1, Additional file 4: Table S3, Additional file 5: Table S4), and these proteomic results were consistent with the results of the present study (Figs. 3e, 4). Therefore, BR may function upstream of these factors and act as a master integrator of drought tolerance (Fig. 7). Thus, BR deficiency causes a variety of factors to induce cotton sensitivity to drought stress, and the genes involved in BR metabolism can serve as markers to identify plant drought resistance.
Fig. 7.
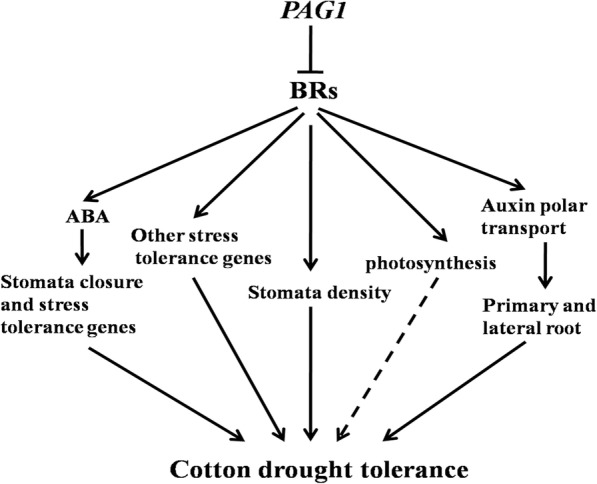
Hypothetical model of PAG1 and BR regulation of cotton drought tolerance. PAG1 inhibits endogenous BRs, which promotes ABA accumulation. ABA regulates stomata closure and stress tolerance genes to enhance drought tolerance. BRs regulate auxin polar transport to promote primary and lateral root development and, in turn, enhance drought tolerance. BRs also affect drought tolerance by regulating other stress tolerance genes (ABA independent genes), stomata density and photosynthesis
Conclusions
In the present study, we reported that BR deficiency results in cotton more sensitive to drought stress. Further study indicated many aspects such as root growth affected by auxin, stomata development, stomata aperture regulated by ABA, photosynthesis and starch accumulation, and with many stress-related genes were responsible for BR mutant, pag1, sensitivity to drought stress. This study demonstrates that BRs play important roles on drought tolerance of cotton and provides an important basis for understanding drought resistance mechanisms regulated by BRs. Overall, our study will provide a vital theoretical basis for genetic improvement and creation of excellent cotton germplasm resources related to drought resistance that regulated by BRs.
Methods
Plant materials and growth conditions
For our study, seeds of CCRI24 and pag1 were obtained from the Institute of Cotton Research of the Chinese Academy of Agricultural Sciences. To examine the resistance of pag1 to drought stress, seeds from the pag1 mutant and CCRI24 control plants were delinted, germinated and maintained in pot culture under natural conditions from May to July (2015) in Anyang, Henan Province, China (average daylight length: 14 h, average temperature: 27.7 °C day/17.7 °C nigh). When the cotton seedlings are fifty days old, we don’t water them until the difference in drought phenotypes occurs between pag1 and CCRI24 plants, then rewatered the cotton seedlings and the number of survival plants was recorded one week later for calculating the survival rate of pag1 and CCRI24 plants. Treated plants were photographed using a camera (Canon, EOS 70D) to document their phenotypes. Three biological replicates consisting of 10 plants were analyzed for each group.
For seedling hydroculture experiments, pag1 and CCRI24 seeds were germinated on wet filter paper placed vertically in 1000-ml beakers with 300 ml of water. The beakers were kept in an incubator at a constant temperature of 29 °C under a 16-h light/8-h dark photoperiod. After four days, the germinated cotton seedlings were transferred to cotton nutrient solution (Hoagland’s solution) for growth until the trefoil stage [50, 51]. In addition, approximately 200 pag1 seedlings were transplanted into cotton nutrient solution containing 24-epi-BL at a final concentration of 10 nM, and the shoots were sprayed twice a day with 10 μM 24-epi-BL. To further analyze the pag1 response to drought stress, pag1 seedlings, pag1 seedlings treated with 24-epi-BL and CCRI24 seedlings were removed from the nutrient solution at the trefoil stage and treated with 6% PEG6000 (drought simulation) for 5 days. Thereafter, the treated seedlings were transferred to normal nutrient solution for another 7 days to analyze the survival rate, and the phenotypes were photographed. Five biological replicates of 8 plants in each experimental group were included in this experiment. To calculate primary root length and lateral root numbers, the roots of 15 plants from each group at the trefoil stage cultivated in nutrient solution were used for analysis. To analyze the fresh root weight or whole-plant weight at the trefoil stage in pag1 plants, pag1 plants treated with 24-epi-BL and CCRI24 plants, their damp roots were dried with tissue paper to remove moisture, followed by weighing. Thereafter, the roots or whole plants were placed in a paper bag in an electric drying oven (WGLL-230BE) at 65 °C for one week, and the dry weights were then calculated for the roots or whole plants. In these experiments, 15 roots or whole plants were used for the analysis.
Measurement of stomata density and apertures
To measure stomata density, the top fourth leaves of one-month-old cotton plants were harvested, cut to similar-sized small pieces and placed into 50 ml centrifuge tubes containing 5 ml of reagent I (a mixture of anhydrous ethanol and glacial acetic acid; 9:1, v/v). The reagent was replaced with fresh reagent I after 3 h, and the tubes were incubated in the dark for total of 6 h at 22 °C. After reagent I was decanted, the tubes were rinsed five times each with 90 and 70% ethanol. Then, 5 ml of reagent II, containing glycerin, distilled water and anhydrous chloral (1 ml:1 ml:8 g), was added to the centrifuge tubes, and the samples were stored in the dark for 48 h at 22 °C. Finally, slides of the treated leaf samples were observed and photographed using an Olympus BX51 microscope. For statistical analysis of stomata density, three leaves were sampled from each plant from the pag1, pag1 treated with 24-epi-BL and CCRI24, and the stomata were counted under three random microscopic views for each leaf. A room temperature of 22 °C was which the experiment carried out.
Furthermore, to measure the stomata aperture of pag1 plants, 24-epi-BL-treated pag1 plants and CCRI24 plants, the top fourth leaves of one-month-old plants were detached and placed in a modified stomata-opening buffer (50 mM KCl, 10 mM CaCl2, 10 mM MES, pH 6.15) for 2 h exposed to light according to previous report [52], to open the stomata. Then, ABA was added to the stomata-opening buffer at a final concentration of 10 μM, and the samples were incubated for an additional 2 h while exposed to light [53]. For the 24-epi-BL-treated pag1 samples, 24-epi-BL was also added to the stomata-opening buffer at a final concentration of 1 μM. The lower leaf epidermis was removed using transparent scotch tape and transferred to slides. Finally, the stomata were observed and photographed with an Olympus BX51 microscope, and 30 stomata apertures were examined for each treatment. The entire experiment was carried out at a room temperature of 22 °C.
Hormone measurements
To measure the levels of BRs (BL and CS), approximately 5 g of leaf tissue was collected from one-month-old pag1 and CCRI24 plants grown under normal conditions in the open field. BRs were quantified by Wuhan Greensword Creation Technology Co., Ltd. as described previously [54]. Three biological replicates were analyzed for this experiment.
For ABA content analysis of the pag1, 24-epi-BL-treated pag1 and CCRI24 plants, pag1 and CCRI24 seeds were sown in pots containing a soil mixture (field soil: nutritional soil: vermiculite = 3:1:1). Approximately 5 g of leaf tissue was harvested from these plants at one month of age, after they had been grown under normal conditions or in the absence of water for one week. ABA levels were determined by Zoonbio Biotechnology Co., Ltd. as described by You et al. [55]. Three biological replicates from each plant group were analyzed.
Measurement of photosynthetic rates
The photosynthesis rate was measured using a portable photosynthesis system (Li-Cor LI-6400, USA) from 9 to 11 AM on the top fourth leaves of two-month-old cotton plants from the pag1, 24-epi-BL-treated pag1 and CCRI24 groups grown in the field. The photosynthetic measurements were taken at a constant photosynthetic photon flux density of 1000 mmol (photon) m− 2 s− 1. Twenty plants in each group were analyzed.
Starch staining experiment
To analyze starch accumulation in pag1, the top fourth leaves were harvested from two-month-old cotton plants of the pag1, 24-epi-BL-treated pag1 and CCRI24 groups, and 5-mm leaf pieces were cut out using a hole puncher and immersed in 95% ethanol in 50-ml centrifuge tubes for 24 h in the dark. The ethanol was then decanted, and the leaf pieces were stained with 2% iodine-potassium iodide (I2-KI) for half an hour [56]. Then, the leaf pieces were photographed using a stereomicroscope (Motic, Moticam Pro 285A). Three biological replicates from each group were used for this experiment.
Transmission electron microscopy (TEM) analysis
To observe starch accumulation in the chloroplasts of pag1, 24-epi-BL-treated pag1 and CCRI24 plants, the top fourth leaves were harvested from two-month-old cotton plants, and ultrathin tissue sections were obtained. Ultrathin sectioning was performed according to a previous report [57]. Finally, the sections were observed and photographed using a New Bio-TEM H-7500 transmission electron microscope (HITACHI, Japan). This experiment applied three biological replicates from each group.
RNA extraction and cDNA preparation
To extract total RNA, plant samples were quickly collected and frozen in liquid nitrogen, and RNA extraction and first-strand cDNA synthesis were performed as described previously [50]. The cDNA was used as a template in the subsequent real-time quantitative RT-PCR (qRT-PCR) analyses.
Quantitative RT-PCR
qRT-PCR was performed to assess the relative transcript levels of polar auxin transport carrier genes and stress-related genes expressed in the roots of the pag1, 24-epi-BL-treated pag1 and CCRI24 plants at the trefoil stage after treatment with 6% PEG6000 for different lengths of time (0, 12, 24 and 36 h). For qRT-PCR, the reactions were prepared and conducted as previously described [50]. Each amplification reaction was performed in triplicate, and three biological replicates were quantified. The G. hirsutum histone-3 gene (GhHIS3) was used as an internal control, and the results were calculated using the 2−ΔΔCT method.
Field trials
To evaluate the agronomic traits of pag1, 24-epi-BL-treated pag1 and CCRI24 plants, field trials were carried out from May to October (2015) in Anyang, Henan Province, China, under normal and drought conditions in the field. To evaluate the agronomic traits of plants under drought condition, pag1 and CCRI24 seeds were grown in the field under a water proof shed. This field trial involved three replicate plots of approximately 5 m2 each. Each plot comprised one row of CCRI24 plants and two rows of pag1 plants; one row of pag1 plants was sprayed with 10 μM 24-epi-BL twice a day beginning on the day the pag1 seedlings broke through the soil. Each plot included approximately 60 plants, with 30 cm inter-row spacing and 20 cm interplant spacing. The field was watered only once at sowing time, and during the remainder of the growing season, the cotton plants in each plot were grown under natural drought conditions. A duplicate set of plants was planted in another field with full irrigation to evaluate the agronomic traits of pag1, 24-epi-BL-treated pag1 and CCRI24 plants under normal conditions. Ten plants were randomly selected from each experimental group in each plot for agronomic trait and cotton yield analyses.
iTRAQ-based proteomic assay
To analyze the differentially abundant proteins in pag1 and CCRI24 plants, cotton seedlings were cultivated using the hydroculture method described above and treated with 6% PEG6000 for different lengths of time (0, 12, 24 and 36 h) at the trefoil stage. Next, 10 g root samples were collected, and proteomic analysis was performed by Shanghai Applied Protein Technology Co., Ltd. as described in a previous report [58]. Three biological replicates were analyzed for this experiment. Changes in protein abundance between pag1 and CCRI24 plants at four different treatment time points were determined to be significant if the abundance ratio was ≥1.2 and the P-value determined by Student’s t test was < 0.05. Gene ontology (GO) enrichment of the differentially abundant proteins was determined according to the methods used by Dong et al. [59]. A heatmap was generated using R software (version 3.4.1), and a Venn diagram was drawn using the Venny 2.1.0 [60].
Additional files
Figure S1. Expression patterns of ten genes determined from the proteomic results for pag1 and CCRI24 under normal conditions and at 12 h, 24 h and 36 h after PEG treatment. P, pag1; C, CCRI24; 0, 12, 24 and 36 represent the trefoil-stage roots of pag1 and CCRI24 after treatment with 6% PEG6000 for 0 h, 12 h, 24 h and 36 h. (DOCX 2427 kb)
Table S1. Down-regulated genes involved in the ABA signal pathway in the pag1 mutant. (DOCX 14 kb)
Table S2. Down-regulated genes involved in carbon metabolism, starch and sucrose metabolism, carbon fixation in photosynthetic organisms and photosynthesis in the pag1 mutant. (DOCX 14 kb)
Table S3. Down-regulated genes involved in the auxin signal pathway in the pag1 mutant. (DOCX 13 kb)
Table S4. Down-regulated genes involved in root development in the pag1 mutant. (DOCX 14 kb)
Table S5. Specific qRT-PCR primers used for analysis of the expression patterns of stress-related genes. (DOCX 13 kb)
Acknowledgements
We thank Dr. Feng Zhou for his help in protein structural alignment.
Funding
This work was supported by the Fund for Creative Research Groups (31621005). The funding agent did not involve in designing the study, the collection, analysis, and interpretation of data, and the drafting of the manuscript.
Availability of data and materials
The datasets used and analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- 24-epi-BL
24-epibrassinolide
- ABA
Abscisic acid
- BL
Brassinolide
- BR
Brassinosteroid
- BRs
Brassinosteroids
- CCRI24
Chinese cotton research institution 24
- CS
Castasterone
- GO
Gene ontology
- pag1
pagoda1
- PEG
Polyethylene glycol
- PME
Pectin methylesterase
- SRS3
The third substrate recognition sites
Authors’ contributions
EC, XZ and FL conceived and designed the research, EC, XZ and ZY performed the experiments, CZ, XW and XG carried out the data analysis, EC wrote and revised the manuscript. All authors have read and approved the current version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Eryong Chen, Email: lovelycheneryong@163.com.
Xueyan Zhang, Email: zhangxueyan_caas@126.com.
Zuoren Yang, Email: yangzuoren4012@163.com.
Chaojun Zhang, Email: zhangchaojun@caas.cn.
Xiaoqian Wang, Email: aywxiaoqian@163.com.
Xiaoyang Ge, Email: gexiaoyang613@163.com.
Fuguang Li, Phone: +86 372 2562204, Email: aylifug@163.com.
References
- 1.Grove MD, Spencer GF, Rohwedder WK, Mandava N, Worley JF, Warthen JD, Steffens GL, Flippen-Anderson JL, Cook JC. Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature. 1979;281:216–217. doi: 10.1038/281216a0. [DOI] [Google Scholar]
- 2.Clouse SD, Sasse JM. Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- 3.Zhu JY, Sae-Seaw J, Wang ZY. Brassinosteroid signalling. Development. 2013;140(8):1615–1620. doi: 10.1242/dev.060590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choudhary SP, Yu JQ, Yamaguchi-Shinozaki K, Shinozaki K, Tran LSP. Benefits of brassinosteroid crosstalk. Trends Plant Sci. 2012;17(10):594–605. doi: 10.1016/j.tplants.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 5.Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubuki M, Honda T, Takatsuto S, Yoshida S, et al. BAS1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci U S A. 1999;96(26):15316–15323. doi: 10.1073/pnas.96.26.15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Denzel MA, Torres QI, Neff MM. CYP72B1 inactivates brassinosteroid hormones: an intersection between photomorphogenesis and plant steroid signal transduction. Plant Physiol. 2003;133(4):1643–1653. doi: 10.1104/pp.103.030882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang ZR, Zhang CJ, Yang XJ, Liu K, Wu ZX, Zhang XY, Zheng W, Xun QQ, Liu CL, Lu LL, et al. PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytol. 2014;203(2):437–448. doi: 10.1111/nph.12824. [DOI] [PubMed] [Google Scholar]
- 8.Kagale S, Divi UK, Krochko JE, Keller WA, Krishna P. Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta. 2007;225(2):353–364. doi: 10.1007/s00425-006-0361-6. [DOI] [PubMed] [Google Scholar]
- 9.Anjum SA, Wang LC, Farooq M, Hussain M, Xue LL, Zou CM. Brassinolide application improves the drought tolerance in maize through modulation of enzymatic antioxidants and leaf gas exchange. J Agron Crop Sci. 2011;197(3):177–185. doi: 10.1111/j.1439-037X.2010.00459.x. [DOI] [Google Scholar]
- 10.Zhang A, Zhang J, Zhang J, Ye N, Zhang H, Tan M, Jiang M. Nitric oxide mediates brassinosteroid-induced ABA biosynthesis involved in oxidative stress tolerance in maize leaves. Plant Cell Physiol. 2011;52(1):181–192. doi: 10.1093/pcp/pcq187. [DOI] [PubMed] [Google Scholar]
- 11.Xia XJ, Wang YJ, Zhou YH, Tao Y, Mao WH, Shi K, Asami T, Chen Z, Yu JQ. Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber. Plant Physiol. 2009;150(2):801–814. doi: 10.1104/pp.109.138230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng H, Tang Q, Hua X. Arabidopsis brassinosteroid mutants det2-1 and bin2-1 display altered salt tolerance. J Plant Growth Regul. 2010;29(1):44–52. doi: 10.1007/s00344-009-9111-x. [DOI] [Google Scholar]
- 13.Divi UK, Krishna P. Overexpression of the brassinosteroid biosynthetic gene AtDWF4 in Arabidopsis seeds overcomes abscisic acid-induced inhibition of germination and increases cold tolerance in transgenic seedlings. J Plant Growth Regul. 2010;29(4):385–393. doi: 10.1007/s00344-010-9150-3. [DOI] [Google Scholar]
- 14.Qu T, Liu R, Wang W, An L, Chen T, Liu G, Zhao Z. Brassinosteroids regulate pectin methylesterase activity and AtPME41 expression in Arabidopsis under chilling stress. Cryobiology. 2011;63(2):111–117. doi: 10.1016/j.cryobiol.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Sahni S, Prasad BD, Liu Q, Grbic V, Sharpe A, Singh SP, Krishna P. Overexpression of the brassinosteroid biosynthetic gene DWF4 in Brassica napus simultaneously increases seed yield and stress tolerance. Sci Rep. 2016;6:28298. doi: 10.1038/srep28298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Shim D, Moon S, Kim H, Bae W, Kim K, Kim YH, Rhee SK, Hong CP, Hong SY, et al. Genome-wide transcriptomic analysis of BR-deficient micro-tom reveals correlations between drought stress tolerance and brassinosteroid signaling in tomato. Plant Physiol Bioch. 2018;127:553–560. doi: 10.1016/j.plaphy.2018.04.031. [DOI] [PubMed] [Google Scholar]
- 17.Boyer JS. Plant productivity and environment. Science. 1982;218(4571):443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- 18.Deeba F, Pandey AK, Ranjan S, Mishra A, Singh R, Sharma YK, Shirke PA, Pandey V. Physiological and proteomic responses of cotton (Gossypium herbaceum L.) to drought stress. Plant Physiol Bioch. 2012;53:6–18. doi: 10.1016/j.plaphy.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hetherington AM, Woodward FI. The role of stomata in sensing and driving environmental change. Nature. 2003;424(6951):901–908. doi: 10.1038/nature01843. [DOI] [PubMed] [Google Scholar]
- 21.Lake JA, Woodward FI. Response of stomatal numbers to CO2 and humidity: control by transpiration rate and abscisic acid. New Phytol. 2008;179(2):397–404. doi: 10.1111/j.1469-8137.2008.02485.x. [DOI] [PubMed] [Google Scholar]
- 22.Leung J, Giraudat J. Abscisic acid signal transduction. Annu Rev Plant Physiol. 1998;49:199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- 23.Bao F, Shen JJ, Brady SR, Muday GK, Asami T, Yang ZB. Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 2004;134(4):1624–1631. doi: 10.1104/pp.103.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia XJ, Huang LF, Zhou YH, Mao WH, Shi K, Wu JX, Asami T, Chen Z, Yu JQ. Brassinosteroids promote photosynthesis and growth by enhancing activation of rubisco and expression of photosynthetic genes in Cucumis sativus. Planta. 2009;230(6):1185–1196. doi: 10.1007/s00425-009-1016-1. [DOI] [PubMed] [Google Scholar]
- 25.Wu CY, Trieu A, Radhakrishnan P, Kwok SF, Harris S, Zhang K, Wang J, Wan J, Zhai H, Takatsuto S, et al. Brassinosteroids regulate grain filling in rice. Plant Cell. 2008;20(8):2130–2145. doi: 10.1105/tpc.107.055087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou R, Quebedeaux B. Changes in photosynthesis and carbohydrate metabolism in mature apple leaves in response to whole plant source-sink manipulation. J Am Soc Hortic Sci. 2003;128(1):113–119. doi: 10.21273/JASHS.128.1.0113. [DOI] [Google Scholar]
- 27.Peng S, Krieg DR, Girma FS. Leaf photosynthetic rate is correlated with biomass and grain production in grain sorghum lines. Photosynth Res. 1991;28(1):1–7. doi: 10.1007/BF00027171. [DOI] [PubMed] [Google Scholar]
- 28.Yu L, Wu SJ, Peng YS, Liu RN, Chen X, Zhao P, Xu P, Zhu JB, Jiao GL, Pei Y, et al. Arabidopsis EDT1/HDG11 improves drought and salt tolerance in cotton and poplar and increases cotton yield in the field. Plant Biotechnol J. 2016;14(1):72–84. doi: 10.1111/pbi.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu L, Chen X, Wang Z, Wang S, Wang Y, Zhu Q, Li S, Xiang C. Arabidopsis enhanced drought tolerance 1/HOMEODOMAIN GLABROUS 11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol. 2013;162(3):1378–1391. doi: 10.1104/pp.113.217596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao BL, Li J. Regulation of brassinosteroid biosynthesis and inactivation. J Integr Plant Biol. 2012;54(10):746–759. doi: 10.1111/j.1744-7909.2012.01168.x. [DOI] [PubMed] [Google Scholar]
- 31.Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T. Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 1996;9(5):701–713. doi: 10.1046/j.1365-313X.1996.9050701.x. [DOI] [Google Scholar]
- 32.Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85(2):171–182. doi: 10.1016/S0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 33.Ohnishi T, Godza B, Watanabe B, Fujioka S, Hategan L, Ide K, Shibata K, Yokota T, Szekeres M, Mizutani M. CYP90A1/CPD, a brassinosteroid biosynthetic cytochrome P450 of Arabidopsis, catalyzes C-3 oxidation. J Biol Chem. 2012;287(37):31551–31560. doi: 10.1074/jbc.M112.392720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim TW, Michniewicz M, Bergmann DC, Wang ZY. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature. 2012;482(7385):419–422. doi: 10.1038/nature10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell. 2007;19(1):63–73. doi: 10.1105/tpc.106.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lampard GR, MacAlister CA, Bergmann DC. Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science. 2008;322(5904):1113–1116. doi: 10.1126/science.1162263. [DOI] [PubMed] [Google Scholar]
- 37.Desikan R, Cheung MK, Bright J, Henson D, Hancock JT, Neill SJ. ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. J Exp Bot. 2004;55(395):205–212. doi: 10.1093/jxb/erh033. [DOI] [PubMed] [Google Scholar]
- 38.Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K. Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 2010;51(11):1821–1839. doi: 10.1093/pcp/pcq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hadiarto T, Tran LS. Progress studies of drought-responsive genes in rice. Plant Cell Rep. 2011;30(3):297–310. doi: 10.1007/s00299-010-0956-z. [DOI] [PubMed] [Google Scholar]
- 40.Kavar T, Maras M, Kidrič M, Šuštar-Vozlič J, Meglič V. Identification of genes involved in the response of leaves of Phaseolus vulgaris to drought stress. Mol Breeding. 2008;21(2):159–172. doi: 10.1007/s11032-007-9116-8. [DOI] [Google Scholar]
- 41.Kim SK, Chang SC, Lee EJ, Chung WS, Kim YS, Hwang S, Lee JS. Involvement of brassinosteroids in the gravitropic response of primary root of maize. Plant Physiol. 2000;123(3):997–1004. doi: 10.1104/pp.123.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Müssig C, Shin GH, Altmann T. Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 2003;133(3):1261–1271. doi: 10.1104/pp.103.028662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friml J. Auxin transport-shaping the plant. Curr Opin Plant Biol. 2003;6(1):7–12. doi: 10.1016/S1369526602000031. [DOI] [PubMed] [Google Scholar]
- 44.Li L, Xu J, Xu ZH, Xue HW. Brassinosteroids stimulate plant tropisms through modulation of polar auxin transport in Brassica and Arabidopsis. Plant Cell. 2005;17(10):2738–2753. doi: 10.1105/tpc.105.034397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinheiro C, Chaves MM. Photosynthesis and drought: can we make metabolic connections from available data? J Exp Bot. 2011;62(3):869–882. doi: 10.1093/jxb/erq340. [DOI] [PubMed] [Google Scholar]
- 46.Chaves MM, Flexas J, Pinheiro C. Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot. 2009;103(4):551–560. doi: 10.1093/aob/mcn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seki M, Umezawa T, Kim JM, Matsui A, To TK. Shinozaki K. Transcriptome analysis of plant drought and salt stress response. In: Jenks MA, Hasegawa PM, Jain SM, editors. Advances in molecular breeding toward drought and salt tolerant crops. Dordrecht: Springer Netherlands; 2007. pp. 261–283. [Google Scholar]
- 48.Langridge P, Paltridge N, Fincher G. Functional genomics of abiotic stress tolerance in cereals. Brief Funct Genomic Proteomic. 2006;4(4):343–354. doi: 10.1093/bfgp/eli005. [DOI] [PubMed] [Google Scholar]
- 49.Valliyodan B, Nguyen HT. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr Opin Plant Biol. 2006;9(2):189–195. doi: 10.1016/j.pbi.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 50.Chen E, Zhang X, Yang Z, Wang X, Yang Z, Zhang C, Wu Z, Kong D, Liu Z, Zhao G, et al. Genome-wide analysis of the HD-ZIP IV transcription factor family in Gossypium arboreum and GaHDG11 involved in osmotic tolerance in transgenic Arabidopsis. Mol Gen Genomics. 2017;292(3):593–609. doi: 10.1007/s00438-017-1293-5. [DOI] [PubMed] [Google Scholar]
- 51.Hoagland DR, Arnon DI. The water culture method for growing plants without soil. California Agricultural Experiment Station. 1950;347:1–32. [Google Scholar]
- 52.Li X, Ma X-G, He J. Stomatal bioassay in Arabidopsis leaves. Bio-protocol. 2013;3(19):e921. doi: 10.21769/BioProtoc.921. [DOI] [Google Scholar]
- 53.Desikan R, Griffiths R, Hancock J, Neill S. A new role for an old enzyme: nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2002;99(25):16314–16318. doi: 10.1073/pnas.252461999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding J, Mao LJ, Wang ST, Yuan BF, Feng YQ. Determination of endogenous brassinosteroids in plant tissues using solid-phase extraction with double layered cartridge followed by high-performance liquid chromatography-tandem mass spectrometry. Phytochem Analysis. 2013;24(4):386–394. doi: 10.1002/pca.2421. [DOI] [PubMed] [Google Scholar]
- 55.You C, Zhu H, Xu B, Huang W, Wang S, Ding Y, Liu Z, Li G, Chen L, Ding C, et al. Effect of removing superior spikelets on grain filling of inferior spikelets in rice. Front Plant Sci. 2016;7:1161. doi: 10.3389/fpls.2016.01161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J, Pang C, Wei H, Song M, Meng Y, Fan S, Yu S. Proteomic analysis of anthers from wild-type and photosensitive genetic male sterile mutant cotton (Gossypium hirsutum L.) BMC Plant Biol. 2014;14(1):390. doi: 10.1186/s12870-014-0390-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J, Pang C, Wei H, Song M, Meng Y, Ma J, Fan S, Yu S. iTRAQ-facilitated proteomic profiling of anthers from a photosensitive male sterile mutant and wild-type cotton (Gossypium hirsutum L.) J Proteome. 2015;126:68–81. doi: 10.1016/j.jprot.2015.05.031. [DOI] [PubMed] [Google Scholar]
- 58.Zeng Y, Du J, Wang L, Pan Z, Xu Q, Xiao S, Deng X. A comprehensive analysis of chromoplast differentiation reveals complex protein changes associated with plastoglobule biogenesis and remodeling of protein systems in orange flesh. Plant Physiol. 2015;168(4):1648–1665. doi: 10.1104/pp.15.00645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong M, Gu J, Zhang L, Chen P, Liu T, Deng J, Lu H, Han L, Zhao B. Comparative proteomics analysis of superior and inferior spikelets in hybrid rice during grain filling and response of inferior spikelets to drought stress using isobaric tags for relative and absolute quantification. J Proteome. 2014;109:382–399. doi: 10.1016/j.jprot.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 60.Oliveros JC. (2007-2015) Venny. An interactive tool for comparing lists with Venn’s diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Expression patterns of ten genes determined from the proteomic results for pag1 and CCRI24 under normal conditions and at 12 h, 24 h and 36 h after PEG treatment. P, pag1; C, CCRI24; 0, 12, 24 and 36 represent the trefoil-stage roots of pag1 and CCRI24 after treatment with 6% PEG6000 for 0 h, 12 h, 24 h and 36 h. (DOCX 2427 kb)
Table S1. Down-regulated genes involved in the ABA signal pathway in the pag1 mutant. (DOCX 14 kb)
Table S2. Down-regulated genes involved in carbon metabolism, starch and sucrose metabolism, carbon fixation in photosynthetic organisms and photosynthesis in the pag1 mutant. (DOCX 14 kb)
Table S3. Down-regulated genes involved in the auxin signal pathway in the pag1 mutant. (DOCX 13 kb)
Table S4. Down-regulated genes involved in root development in the pag1 mutant. (DOCX 14 kb)
Table S5. Specific qRT-PCR primers used for analysis of the expression patterns of stress-related genes. (DOCX 13 kb)
Data Availability Statement
The datasets used and analysed during the current study are available from the corresponding author on reasonable request.