Abstract
In several fields of research, molecular dynamics simulation techniques are exploited to evaluate the temporal motion of particles constituting water, ions, small molecules, macromolecules, or more complex systems over time. These techniques are considered difficult to setup, computationally demanding and require high specialization and scientific skills. Moreover, they need specialized computing infrastructures to run faster and make the simulation of big systems feasible. Here, we have simulated 3 systems of increasing sizes on scientific- and gaming-enabled graphic processing unit (GPU) cards with Amber, GROMACS, and NAMD and measured their performance accounting also for the market prices of the GPU cards where they were run on.
Keywords: molecular dynamics simulation, GPU computing, protein structure
Comment on: Biagini T, Chillemi G, Mazzoccoli G, et al. Molecular dynamics recipes for genome research. Brief Bioinform. 2018;19: 853-862. doi:10.1093/bib/bbx006. PubMed PMID: 28334084. https://www.ncbi.nlm.nih.gov/pubmed/28334084.
Introduction
Classical molecular dynamics (MD) is a computer simulation method that applies Newton’s laws to evaluate the motions of small and macromolecules or more complex systems, at the atomic level. With this method, the elementary interactions among atoms are parametrized, in accordance with the classical equations of motion, with the aim to simplify the simulation and making systems with high number of particles computationally tractable.
Results of an MD study strictly depend on the used software package, whose overall calculation time is bound, in turn, to the type of system under examination as well as to the available computing hardware. Usually, researchers need to achieve a broad compromise between these 2 needs. Speaking of computing hardware, graphic processing units (GPU) attracted increasing attention in the past few years because they are cheap, if compared to multi-core devices, and exhibit high performance. This is because GPUs have a highly parallel internal architecture. They consist of a high number of independent processing units (cores), in the order of several hundreds. This number is much higher than that normally available on the usual central processing units (CPUs). Each GPU core is designed to perform a large number of tasks, even thousands, called threads, which unlike CPUs, perform very simple operations. For these reasons, today, GPUs are successfully used as computational accelerators equally in the gaming and scientific worlds.
The leading company, NVIDIA Corporation, has set a reference standard hardware architecture for parallel processing, called compute unified device architecture (CUDA). In recent years, it was exploited in chemistry and computational biology, where we have seen a progressive adaptation of existing codes as well as the development of new software packages that were compliant with this type of hardware. MD appeared to be extremely suitable to be run on CUDA-enabled GPUs,1,2 as well as on other similar parallel hardware infrastructures. For this reason, NVIDIA introduced graphic cards in the market that were particularly designed for scientific computing.
Two important families of GPU cards are Quadro and Tesla. They have additional features compared with gaming-designed GPUs, like the GTX family. First, they were designed to bear intense and prolonged (in the order of several days) workloads. Moreover, the on-board memory is usually higher (24 GB) when compared with the others (6-11 GB). Another important feature that characterizes the former cards and not the latter is the support for error correcting codes (ECC). ECC is used to detect and possibly correct read errors from random access memory (RAM). This last feature requires extra hardware and thus increasing costs. This is why Quadro and Tesla GPU cards are not generally shipped with desktop computers. Nevertheless, the use of commercial graphics cards for gaming is today continuously increasing to the detriment of Tesla and Quadro cards because they provide stunning performance and extremely reasonable prices.2 It was demonstrated, in fact, that ECC events are rare over a Monte Carlo simulation and thus that the required time and memory overheads may outweigh the usage of this functionality.3 It should also be considered that the memory constraints for most real systems dropped dramatically over the years. A system of 60 000 atoms, for example, does not need more than half GB of RAM to be simulated and, in general, if there is no need to simulate systems with several millions atoms, few GB of memory are enough.
With these considerations, depending on the preferred MD code, the size and complexity of the system being examined, and the available budget, many research groups in the field of MD simulation and computing centers may feel the need to correctly setup their compute clusters. To support on this, in our previous work,4 we considered 3 leading molecular dynamics tools: AMBER,5 GROMACS,6 and NAMD.7 They were used to analyze 3 test cases, which mainly differed by size, ie, the atoms count and molecules types (protein, RNA, and lipids). The small system was made by ∼60 000 atoms. The medium system was made by ∼100 000 atoms, while the large system was made by around 1 million atoms. The aim was to evaluate whether a given hardware configuration was more advantageous than another and, eventually, if “commodity hardware” could be suitable for MD studies. In detail, each system was simulated by each MD tool on increasing numbers of AMD CPU cores (1-144), on increasing numbers of Intel CPU cores (1-8), and on increasing numbers of Tesla C2070 GPUs (1-4). Our results showed how the use of GPUs generally brought a great benefit in terms of performance, but depending on the considered tool. The best speedup obtained by Amber when using all available CPU cores was lower than the speedup obtained with just one GPU, for all 3 systems on all tested hardware infrastructures. This was valid for GROMACS and NAMD as well, but less markedly. Indeed, the speedup obtained with 4 GPUs working together matched that obtained with 48 CPUs on all 3 systems.
Thus, it was clear that MD takes advantage of high performance GPU cards and that the main benefit that these cards provide is due to their high number of GPU cores. In our previous article,4 we used 4 NVIDIA Tesla C2070 (Fermi) cards, which were equipped with 448 CUDA cores each. We here expand our considerations on modern NVIDIA cards and aim to further help researchers to identify the best hardware in terms of performance-to-price ratio. This study made use of Amber 18, NAMD version 2.13, and GROMACS 2018 for the production dynamics step, as they are the most used tools in the analysis of biomolecular systems.
Hardware
MD simulations were run on a dual Intel Xeon E5620 @2.40 GHz workstation (2 processors, 4 computing cores, and 16 threads, with hyper-threading enabled), equipped with 12 GB of RAM. This workstation had 1 Quadro P6000 (6999.99$), 2 GTX 1080Ti (829$), and 2 GTX 1070 (479$) onboard. These prices date back to March 2019. GPU cards were set up with default values of memory and core clock speeds and voltages; CUDA 9.2.88 was used to compile, where required; each process was set up to communicate by MPI and one MPI process was systematically mapped to one GPU. Each system, simulated for 50 ps by a time step of 2 fs (25 000 steps), was run 3 times. Nanoseconds elapsed per day (ns/day) varied by no more than 5% through each triplet. All benchmarks presented here refer to the best of the 3, in terms of ns/day.
Measures of performance
For each tested system and hardware configuration, we measured the production parameters in terms of simulated ns/day. Our measure of performance here is named price-nanosecond index (PR), which is calculated as the price of the most expensive GPU card on the price of the actual GPU card, multiplied by the number of elapsed ns/day in the simulation of a system.
Results and considerations
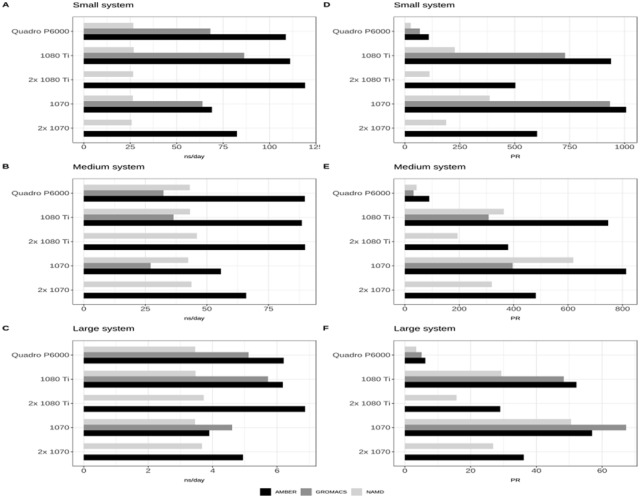
Figure 1 reports benchmark values for small (A), medium (B), and large (C) systems, when run on the considered GPU cards. Amber performed well with all 3 systems. Its best performance was achieved when using 2 GTX 1080Ti together. The speedup obtained with only one GPU outperformed systematically the best records of the other tools, when used in any configuration and on any system. It is worth mentioning that performance of the NVIDIA Quadro P6000 was close to that of a single GTX 1080Ti and that this was true for all 3 systems. NAMD showed similar performance when run on all hardware configurations and the use of 2 cards (a pair of GTX 1080Ti or a pair GTX 1070) led to only a small benefit with the medium and large systems. GROMACS achieved similar performance of AMBER for the large system when using a single GPU. The contribution of an additional GPU in GROMACS was not reported here because optimal performance with multiple GPUs typically requires tricky balancing configurations of simulation and launch parameters. However, the latest releases of GROMACS enabled the use of slower CPU in combination with faster GPU, encouraging the choice of multiple GPU setup.8 From these tests, GTX 1080Ti ran slightly better than Quadro P6000, systematically.
Figure 1.
Performance of Amber 18, NAMD version 2.13 and GROMACS 2018 measured as the number of ns/day required to simulate the small (A), medium (B) and large (C) systems, when run on the considered GPU cards. PR values for the same systems and GPUs are shown in C for the small system and in D and E for the medium and large systems, respectively.
In Figure 1, the performance/cost ratio was evaluated in terms of the PR index previously described. Amber achieved higher PR values for all hardware configurations and for all 3 systems and therefore it represents the most economical and convenient choice in terms of required hardware resources. The use of 2 GPUs was never advantageous in terms of costs, because simulations never scaled linearly with respect to the number of used GPU cards. It should be emphasized that the PR index takes only the price of GPUs into account and hence does not consider power consumption, required hardware components unrelated to the GPU, and licensing costs. AMBER, in fact, has a paid license while GROMACS and NAMD are free software. On the other hand, it should also be considered that the CPUs have no impact on the performance achieved with AMBER, because it is designed to delegate most computational loads to GPUs, leaving to underperforming and obsolete CPUs only secondary tasks. GROMACS and NAMD suffer with poorly performing CPUs.
Comparing game-enabled (GTX 1070 and GTX 1080Ti) versus scientific-enabled (Quadro P6000) cards in terms of the performance/cost ratio, it seems evident that Quadro P6000 obtains PR values that are considerably lower than those obtained with any other configuration equipped with GTX cards (Figure 1C to E). Hence, ignoring the eventual benefits brought by ECC, classical MD simulations appear to be advantaged by gaming GPUs, both in terms of costs and performance. The reader should acknowledge that, despite the good performance, high reliability, and reasonable prices, gaming cards were not designed for datacenter usage as stated in the NVIDIA drivers’ license agreement: “No Datacenter Deployment. The SOFTWARE is not licensed for datacenter deployment, except that blockchain processing in a datacenter is permitted” (https://www.nvidia.com/content/DriverDownload-March2009/licence.php?lang=us&type=TITAN).
Acknowledgments
We gratefully acknowledge NVIDIA Corporation and the Amber 18 Team for supporting this research.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the Italian Ministry of Health (Ricerca Corrente 2018 RC18DSCOMU).
Declaration of Conflicting Interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: TB and FP performed MD simulations. MT and AB performed data analysis of MD trajectories. RC provided technical assistence together with 3 of the 4 GPU cards used in this study, DC and SC critically read the paper and provided suggestions, MFT helped defining and modeling the systems being analyzed, MC supervised the biological side of the project. TM supervised the whole project and wrote the paper with TB.
ORCID iD: Tommaso Mazza  https://orcid.org/0000-0003-0434-8533
https://orcid.org/0000-0003-0434-8533
References
- 1. Lee TS, Cerutti DS, Mermelstein D, et al. GPU-accelerated molecular dynamics and free energy methods in Amber18: performance enhancements and new features. J Chem Inf Model. 2018;58:2043–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allec SI, Sun Y, Sun J, Chang CA, Wong BM. Heterogeneous CPU+GPU-enabled simulations for DFTB molecular dynamics of large chemical and biological systems [published online ahead of print March 27, 2019]. J Chem Theory Comput. doi: 10.1021/acs.jctc.8b01239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walker RC, Betz RM. An investigation of the effects of error correcting code on GPU-accelerated molecular dynamics simulations. In: Wilkins-Diehr N, ed. Proceedings of the Conference on Extreme Science and Engineering Discovery Environment: Gateway to Discovery San Diego, CA: ACM; 2013:1–3. [Google Scholar]
- 4. Biagini T, Chillemi G, Mazzoccoli G, et al. Molecular dynamics recipes for genome research. Brief Bioinform. 2018;19:853–862. [DOI] [PubMed] [Google Scholar]
- 5. Case DA, Cheatham TE, 3rd, Darden T, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abraham MJ, Murtola T, Schulz R, et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. [Google Scholar]
- 7. Phillips JC, Braun R, Wang W, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kutzner C, Páll S, Fechner M, Esztermann A, Groot BLD, Grubmüller H. More bang for your buck: improved use of GPU nodes for GROMACS 2018. https://arxiv.org/abs/1903.05918. [DOI] [PubMed]