Abstract
Background:
Elongation factor for RNA polymerase 2 (ELL2) and ELL associated factor 2 (EAF2) have been reported to have tumor suppressive properties in prostate epithelial cells.
Aims:
We investigated ELL2 expression in human prostate cancer specimens, and ELL2 protein stability and ubiquitination in prostate cancer cells.
Materials and Methods:
Immunostaining analysis of human prostate cancer specimens was used to determine ELL2 expression in tumor and normal tissues. ELL2 knockdown in prostate cancer cell lines LNCaP and C4-2 was used to compare proliferation and motility. Deletion and site-directed mutagenesis was used to identify amino acid residues in ELL2 that were important for degradation.
Results:
ELL2 protein was downregulated in prostate cancer specimens and was up-regulated by androgens in prostate cancer cell lines LNCaP and C4–2. ELL2 knockdown enhanced prostate cancer cell proliferation and motility. ELL2 protein has a short half-life and was stabilized by proteasome inhibitor MG132. Amino acid residues K584 and K599 in ELL2 were important for ELL2 degradation. EAF2 could stabilize ELL2 and inhibited its polyubiquitination.
Conclusion:
Our findings provide further evidence that ELL2 is a potential tumor suppressor frequently down-regulated in clinical prostate cancer specimens and provides new insights into regulation of ELL2 protein level by polyubiquitination and EAF2 binding.
Keywords: EAF2, ELL2, polyubiquitination, prostate cancer
1 ∣. INTRODUCTION
Prostate cancer is a major cancer world-wide, particularly in Western countries. In the United States, prostate cancer is the most frequently diagnosed cancer and the third leading cause of cancer death.1 Androgens play an important role in the development and progression of prostate cancer. However, the mechanisms by which androgens influence prostate carcinogenesis is incompletely understood. Elucidating the mechanisms by which androgen signaling modulates prostate carcinogenesis may lead to new approaches for the prevention and/or treatment of prostate cancer.
ELL2 (elongation factor), RNA polymerase 2 (previously called eleven-nineteen-lysine-rich leukemia 2) is encoded by an androgen-response gene.2-4 ELL2 was identified as a homolog of ELL,5 which was initially discovered based on its fusion to MLL (mixed-lineage leukemia), contributing to the pathogenesis of acute myeloid leukemia.6,7 ELL2, along with its other two homologs, ELL and ELL3, can function as an RNA polymerase 2 elongation factor through suppressing transient pulsing of RNA polymerase 2 activity.8 ELL family proteins are components of the super elongation complex (SEC) as well as the little elongation complex (LEC) (reviewed in ref. 9). ELL proteins are associated with EAF (ELL associated factor) family proteins EAF1 and EAF2. Both ELL and EAF family proteins can be present in the SEC and LEC complexes.
Our recent studies suggest that ELL2 plays an important role in prostate carcinogenesis. ELL2 mRNA expression was downregulated in high Gleason grade prostate cancer specimens.10 Knockdown of ELL2 by siRNA resulted in increased proliferation, migration, and invasion in prostate cancer cell lines.4,10 Furthermore, prostate-specific ELL2 knockout resulted in high-grade murine prostatic intraepithelial neoplasia in the mouse model.4 Considering the potential role of ELL2 in prostate carcinogenesis, it will be important to elucidate the mechanisms regulating ELL2 protein turnover. ELL2 has previously been shown to be polyubiquitinated by E3 ubiquitin ligase Siah1 in Hela cells.11 In the present study, we investigate ELL2 protein stability and ubiquitination in prostate cancer cells.
2 ∣. MATERIALS AND METHODS
2.1 ∣. Immunostaining of prostate cancer specimens
Prostate cancer tissues were collected from the Department of Urology at the Affiliated Cancer Hospital of Zhengzhou University. The study was reviewed and approved by institutional ethics committee. The normal prostate controls were collected from patients undergoing total cystectomy. For immunostaining, 5 μm sections of paraffin embedded tissues were de-paraffinized with xylene and endogenous peroxidase activity was quenched using 3% H2O2in methanol for 30 min in the dark. Tissue sections were dehydrated through graded alcohols and subjected to antigen retrieval using 10 mM sodium citrate. The slides were then washed with TBST (Tris Borate Saline Tween-20) and blocked with 5% BSA (Bovine Serum Albumin, Sigma-Aldrich Corp.,St. Louis, MO) for 1 h. Slides were incubated with primary ELL2 antibody (Catalog #A302-505A, Bethyl Laboratories, Inc., Montgomery, TX), 1:500 diluted in TBS. Slides were then washed for 5 min in TBST and incubated with HRP (Horse Radish Peroxidase) conjugated anti-mouse secondary antibody diluted with TBS for 1 hr. After washing, slides were incubated with DAB (3,3’-diaminobenzidine tetrahydrochloride) (Sigma-Aldrich) and immediately washed under tap water after color development. Slides were then counter stained with hematoxylin and observed under a light microscope (Carl Zeiss, Oberkochen, Germany).
All slides stained with ELL2 antibody were reviewed jointly by two of the authors (TY and ZZ, followed by a second, independent review by another pathologist (Jianbo Zhang, Affiliated Cancer Hospital of Zhengzhou University). ELL2 immunostaining in prostatic glandular cells was analyzed at a final magnification of 200X. The score for the IHC stained positive rate was scaled as (−) for <3%, (+) for 3-25%, (++) for 25-50%, and (+++) for >50% of tumor cells stained.
2.2. ∣. Isolation of total RNA and real-time RT-PCR
Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA). RNase-free DNase-treated total RNA was reverse-transcribed with a SuperScript III First-Strand Synthesis System (Promega, Madison, WI) following the manufacturer's instruction for quantitative real-time polymerase chain reaction (qPCR). Real-time RT-PCR was performed using SYBR Green method (Roche, Basel, Switzerland). Gene-specific primers are following: ELL2 forward 5′-CGC TGC CTC ATC TCC TCA GAA ACG-3′ and reverse 5′-TGC AGG GGG TGG TAG GAG GC-3′, GAPDH forward 5′-GTT GCT GAG TAT GTC GTG GA-3′ and reverse 5-CGG AGA TGA TGA CCC TTT TG-3′. PCR thermocycling parameters were 95°C for 10 min followed by 40 cycles (95°C for 15 s; 60°C for 5 s; 72°C for 20 s). Each sample was run in triplicate and was normalized to GAPDH RNA. Relative changes in gene expression were expressed as the fold change using 2-ΔΔCt method.12
2.3. ∣. Plasmids and reagents
The full-length ELL2 and ELL2 deletion mutants, ELL2(1-292), ELL2 (293-531), and ELL2(532-640), were generated using PCR and cloned into Flag expression vector (pCMV-3Tag-1A, Agilent Technologies, Santa Clara, CA). The ELL2 substitution mutants, K571R, K584R, and K599R, were generated using a QuickChange II site-directed mutagenesis kit (Catalog #200521, Agilent Technologies). All ELL2 mutant expression vectors were confirmed by DNA sequencing. Expression vectors for HA-ubiquitin plasmid was a kind gift from Dr Chunbing Zou from University of Pittsburgh, pCMV-myc-EAF1 and pCMV-myc-EAF2 were cloned as previously described..13 All vectors were sequence verified.
2.4. ∣. Cell culture and transfection
LNCaP, HEK 293, and 22Rv1 cell lines were purchased from American Type Culture Collection (ATCC) (Manassas, VA), and C4-2 was a gift from Dr Leland W.K. Chung. All the cell lines were maintained in RPMI-1640 medium, supplemented with 10% heat-inactivated fetal bovine serum (FBS). The LNCaP, C4-2 and 22Rv1 prostate cancer cell lines were authenticated in 2016 using DNA fingerprinting by examining microsatellite loci in a multiplex PCR reaction (AmpFlSTR® Identifiler® PCR Amplification Kit, Applied Biosystems, Foster City, CA) by the University of Pittsburgh Cell Culture and Cytogenetics Facility. The ATCC performed authentication for the HEK293 cell line using short tandem repeat profiling.
For knockdown experiments, cells were transfected with 100 pmol non-targeted siRNA (#D-001810-10-50, ON-TARGETplus Non-targeting Pool, Dharmacon) or siRNA against ELL2 (sequence 1-forward primer: 5′-ACA GAA UUC CUU AGG AGG UCC AUG CUA-3′ the reverse primer: 5′-GCA UGG ACC UCC UAA GGA AUU CUG T-3′; sequence 2- forward primer: 5′-AUU UAC AAU CUG AGG AGG AUG UGA GAU-3′ the reverse primer: 5′-CUC ACA UCC UCC UCA GAU UGU AAA T-3′; sequence 3- forward primer: 5′-AAG CAU UUC ACC UUG CAC AUU ACU GUU-3′, reverse primer: 5′-CAG UAA UGU GCA AGG UGA AAU GCU U-3′); or siRNA against EAF2 (sequence 1- forward primer: 5′-AAA CAG UUA CUG GUG GAG UUG AAC CUU-3′, reverse primer: 5′-AAG GUU CAA CUC CAC CAG UAA CUG UUU-3′, sequence 2- forward primer: 5′-CUG UUC ACC UUC ACC AAC CUC AAG GUA-3′, reverse primer: 5′-UAC CUU GAG GUU GGU GAA GGU GAA CAG-3′) using Lipofectamine 2000 (Invitrogen) in a six-well plate.
To determine androgen induction of ELL2 mRNA, LNCaP cells were cultured in the presence of increasing concentrations of synthetic androgen R1881 (0-10 nM) for 24 h prior to harvesting cells for RNA extraction. To determine androgen induction of ELL2 protein, LNCaP, or C4-2 prostate cancer cells were treated with 1 nM R1881 for 0, 24, 48, 72, or 96 h prior to preparation of cell lysates.
To determine the stability of ELL2 or ELL2 mutants, C4-2 cells were transiently transfected with various expression vectors for 24 h using PolyJet transfection reagent (Cat#SL100688, SignaGen Laboratories, Rockville, MD) transfection reagent, then treated with CHX (50 μg/mL) over a time course of 0 up to 24 h prior to preparation of cell lysates. MG132 (Z-leu-leu-leu-al, C2211) and Cycloheximide (CHX, C7698) were from Sigma-Aldrich.
After various treatment, cell lysates were prepared using RIPA buffer (50 mM Tris-HCl, pH 8.0,150 mM NaCl, 1 mM EDTA, 1% (v/v) NP-40, 0.1% SDS, 0.25% sodium deoxycholate, 1mM sodium orthovanadate, 1 mM PMSF, 1:100 dilution of protease inhibitor cocktail P8340 (Sigma-Aldrich) and protein concentration was determined by BCA Protein Assay (ThermoFisher Scientific, Pittsburgh, PA). The lysates were denatured by boiling in SDS sample buffer, separated on a NEXT GEL™10% gel (Amresco, Solon, OH) and transferred onto a nitrocellulose membrane. Blotted proteins were probed with primary antibodies: ELL2 (Catalog number: A302-505A, Bethyl Laboratories), b-actin (sc-47778, Santa Cruz Biotechnology, Dallas, TX, USA), Ub (sc-8017, Santa Cruz Biotechnology), flag (F1804, Sigma-Aldrich), b-tubulin (#2803, Cell Signaling Technology, Beverly, MA), Myc (MMS-150, Covance, Berkeley, CA, USA), HA-tag (MMS-101P, Covance), AR (N20, sc-816, Santa Cruz Biotechnology), EAF2,14 and GAPDH (sc-25778, Santa Cruz Biotechnology) followed by HRP (horseradish peroxidase) labeled secondary antibody (Santa Cruz Biotechnology). Signals were visualized using chemiluminescence (ECL™ Western Blotting Detection Reagents®, GE Healthcare, Pittsburgh, PA) and exposed to X-ray film (Fujifilm, Tokyo, Japan).
2.5. ∣. Cell proliferation assay
In brief, C4-2 cells were plated at a density of 3 × 103cells/well in 96-well microplates, incubated at 37°C and 5% CO2. After 24-h incubation, at approximately 70% confluence, cells were treated with two ELL2 siRNA and scramble oligonucleotides individually (Santa Cruz Biotechnology) following Lipofectamine 3000 Transfection Protocol (Invitrogen). Cell proliferation was measured after 48 h at 495 nm using a sulfonated tetrazolium salt, WST-1 (4-[3-[4-iodophenyl]-2-4(4-nitrophenyl)-2H-5-tetrazolio-1,3-benzene disulfonate], Catalog # 501594400 Roche).
2.6. ∣. Cell migration and invasion assay
Transwell migration and Matrigel invasion assays were performed using a transwell membrane (3422 and 35480, respectively, Corning Incorporated, Corning, NY) in a 24-well plate according to the manufacturer's instructions. Briefly 1 × 105 cells were seeded into the migration or Matrigel invasion chamber placed in a 24-well plate containing complete medium and incubated at 37°C and 5% CO2 for 24 h. To evaluate the effect of ELL2 knockdown, the cells were seeded in FBS free medium. After incubation, cells were washed and then fixed in 4% para-formaldehyde and stained with 0.2% crystal violet. The cells on the inner surface of the filter membrane were removed using a cotton swab. The membranes were moved from the chamber and fixed at a cover slide. Images of migrated or invaded cells where taken under a light microscope (Zeiss Lab.A1, Jena, Germany). Invasion and migration cells were counted in five random fields under a light microscope (200×).
2.7. ∣. Immunoprecipitation
HEK 293 cells were cultured in 10 cm plates to an optimal monolayer cell density of 80-90%. Then cells were transfected with plasmid using PolyJet transfection reagent (SignaGen Laboratories). After incubation for 18 h, medium was changed with/without 10 μM MG132 for another 10 h. Cells were then harvested and separated into two aliquots, one aliquot was lysed and the expression of transfected genes was confirmed by Western blot analysis of whole cell lysates. The second aliquot of cells was centrifuged and washed three times in PBS, then mixed with RIPA lysis buffer 500 μL (50 mM Tris-HCl PH 8.0,150 mM NaCL,1% NP-40,0.1% SDS, 0.5% Na-cl-Wxydilaic) containing PIC (protease inhibitors cocktail) 5μL, incubated on ice for 30 min. The mixture was then centrifuged at 12 000 G in 4°C for 10 min, then resuspended 1 mg into 600 μL lysis buffer. RIPA lysis buffer was used to wash 20 μL anti-Flag® M2 Affinity agarose beads (A2220-5ML, Sigma-Aldrich) followed by rotating for 10 min at 4°C, then beads were centrifuged at 60G for 2 min. Beads were washed in this fashion three times. Samples were added to the washed beads and rotated for 4 h at 4°C. Samples were then centrifuged at 12 000 G for 2 min at 4°C, discarding the supernatant. Samples were then washed with 600 μL RIPA lysis buffer with rotation for 10 min at 4°C followed by a wash with a buffer containing 250 mM NaCl, 20 mM Tris-HCl pH 7.5, 0.5% NP-40, 1.5 mM MgCl2, 15% Glycerol, and 2mM EDTA, in 4°C with rotating 10 min. After washing, samples were centrifuged and washed with buffer two times. Immunoprecipitates were boiled with SDS sample loading buffer for 5 min, then fractionated by SDS-polyacrylamide gel electrophoresis (PAGE), and transferred to nitrocellulose membranes for analysis by Western blotting.
3. ∣. RESULTS
3.1. ∣. ELL2 protein is downregulated in prostate cancer specimens.
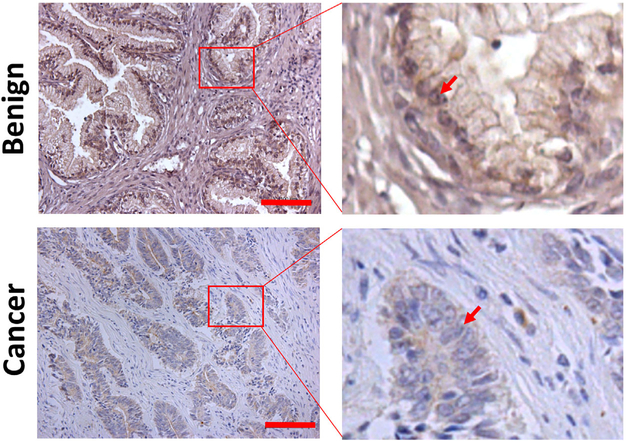
Our previous studies showed that the ELL2 mRNA was downregulated in prostate cancer specimens, particularly in high Gleason grade prostate cancer specimens.10 ELL2 mutation is not frequent in prostate cancer.4 However, the protein level of ELL2 was not evaluated previously in human prostate specimens. With a commercially available ELL2 antibody, we conducted immunostaining of ELL2 in a cohort of prostate cancer specimens, along with benign prostatic tissue controls to determine if ELL2 expression was altered at the protein level in prostate cancer (Figure 1). Of the 22 benign prostatic specimens, all were stained positive with ELL2 antibody, with 68% of them exhibiting strong staining (Table 1). ELL2 immunostaining intensity was reduced in prostate cancer specimens, with more dramatic downregulation in high-Gleason score prostate cancer specimens. ELL2 immunostaining were observed in all the low-Gleason score3-5 prostate cancer specimens. However, there was a shift toward reduced ELL2 staining in low-Gleason prostate specimens as compared to the benign prostate control (P = 0.0004). In Gleason 6–8 prostate specimens, no strong ELL2 staining was observed and 25% of the specimens exhibited no detectible ELL2 staining. We only had four high-Gleason score9-10 specimens and three of them had no ELL2 staining and one of them had only weak ELL2 staining. These data indicate that ELL2 protein expression was downregulated in prostate cancer progression.
FIGURE 1.
Immunostaining of ELL2 in clinical prostate cancer specimens. The red bars in the left panel of the images indicate 100 μm. The red arrows indicate the nuclei of luminal epithelial cells in the benign prostate tissue or the nuclei of cancer cells. [Color figure can be viewed at wileyonlinelibrary.com]
TABLE 1.
ELL2 immunostaining in human prostate specimens
| Grade of ELL2 expression |
||||||
|---|---|---|---|---|---|---|
| Group | − | + | ++ | +++ | Total | P |
| Benign Gleason score | 0 (0%) | 3 (14%) | 4 (18%) | 15 (68%) | 22 (100%) | |
| 3-5 | 0 (0%) | 3 (23%) | 4 (31%) | 6 (46%) | 13 (100%) | 0.0004 |
| 6-8 | 3 (25%) | 5 (42%) | 4 (33%) | 0 (0%) | 12 (100%) | |
| 9-10 | 3 (75%) | 1 (25%) | 0 (0%) | 0 (0%) | 4 (100%) | |
3.2. ∣. ELL2 expression is regulated by androgens in LNCaP and C4-2 prostate cancer cells.
Previous studies suggested that ELL2 expression is regulated by androgens in prostate cancer cells.2-4 We confirmed ELL2 induction by androgens in two different prostate cancer cell lines LNCaP and C4-2. Figure 2A showed ELL2 mRNA induction in C4-2 cells by synthetic androgen R1881 in a dose-dependent manner. Figures 2B and 2C showed R1881 induction of ELL2 protein over a time course (0-96 h) in Western blot analysis of LNCaP and C4-2 cells, respectively. The peak induction of ELL2 protein appeared to be at 48 h after R1881 treatment. These results provided further evidence for androgen induction of ELL2 expression in prostate cancer cells.
FIGURE 2.
A, Androgen induction of ELL2 mRNA in LNCaP prostate cancer cells. LNCaP cells in culture were treated with R1881 at the indicated dosage for 24 h. ELL2 mRNA expression was determined using real time RT-PCR. *P < 0.05 and **P < 0.01. B, Androgen induction of ELL2 protein in LNCaP cells. Cultured LNCaP cells were treated with 1 nM R1881 for indicated number of hours. The cell lysates were then isolated for western blot analysis using anti-ELL2 antibody. The Western membrane was re-probed using anti-GAPDH antibody as a loading control. C, Androgen induction of ELL2 protein in C4-2 prostate cancer cells. The experiment was conducted identically as described in panel B
The co-occurence of alterations in ELL2 and other androgen-responsive genes was examined using The Cancer Genome Atlas (TCGA), Provisional data set generated by the TCGA Research Network: http://cancergenome.nih.gov/. The TCGA, Provisional dataset contains data from 499 prostate tumor samples from 498 patients. Genes identified previously as androgen-responsive by Nelson et al2 and Bolton et al3 were queried to determine whether alterations in any of these genes co-occurred with alterations in ELL2 expression in prostate cancer specimens. Of these genes, 49 had a statistically significant tendency to co-occur in tumor specimens with ELL2 alteration, while two were mutually exclusive (Table 2). None of the identified AR response genes were identified previously as also being regulated by ELL24 or EAF2.15 Interestingly, only SPDEF and TM4SF1 were identified as having a statistically significant tendency to co-occur with EAF2 alteration in prostate cancer specimens (data not shown). These results suggest that several other androgen-responsive genes are altered similarly to ELL2 in prostate cancer, perhaps due in part to the dysregulation of AR activity.
TABLE 2.
ELL2 mutual exclusivity or co-occurrence with androgen responsive genes in prostate cancer specimens from TCGA
| Gene A | Gene B | Neither | A not B | B not A | Both | Log odds ratio | P-value | Tendency |
|---|---|---|---|---|---|---|---|---|
| ABHD18 | ELL2 | 427 | 28 | 35 | 9 | 1.366 | 0.003 | Co-occurrence |
| ABHD2 | ELL2 | 435 | 20 | 36 | 8 | 1.576 | 0.002 | Co-occurrence |
| ALDH1A3 | ELL2 | 445 | 10 | 36 | 8 | 2.291 | <0.001 | Co-occurrence |
| AP1G2 | ELL2 | 439 | 16 | 37 | 7 | 1.647 | 0.002 | Co-occurrence |
| AZGP1 | ELL2 | 432 | 23 | 35 | 9 | 1.575 | <0.001 | Co-occurrence |
| BIN1 | ELL2 | 433 | 22 | 31 | 13 | 2.111 | <0.001 | Co-occurrence |
| CDC14A | ELL2 | 436 | 19 | 37 | 7 | 1.468 | 0.005 | Co-occurrence |
| CERS6 | ELL2 | 430 | 25 | 36 | 8 | 1.341 | 0.005 | Co-occurrence |
| DBI | ELL2 | 425 | 30 | 29 | 15 | 1.992 | <0.001 | Co-occurrence |
| ELL2 | SELENOP | 444 | 35 | 11 | 9 | 2.34 | <0.001 | Co-occurrence |
| ELL2 | TMEM37 | 431 | 34 | 24 | 10 | 1.664 | <0.001 | Co-occurrence |
| ELL2 | CAMKK2 | 430 | 34 | 25 | 10 | 1.621 | <0.001 | Co-occurrence |
| ELL2 | IRF2BP2 | 423 | 37 | 32 | 7 | 0.917 | 0.045 | Co-occurrence |
| ELL2 | KRT8 | 437 | 39 | 18 | 5 | 1.135 | 0.043 | Co-occurrence |
| ELL2 | SLC30A1 | 439 | 39 | 16 | 5 | 1.258 | 0.03 | Co-occurrence |
| ELL2 | PNKD | 433 | 38 | 22 | 6 | 1.134 | 0.028 | Co-occurrence |
| ELL2 | TM4SF1 | 445 | 40 | 10 | 4 | 1.493 | 0.027 | Co-occurrence |
| ELL2 | MBL2 | 441 | 38 | 14 | 6 | 1.604 | 0.005 | Co-occurrence |
| ELL2 | RAB4A | 418 | 33 | 37 | 11 | 1.326 | 0.001 | Co-occurrence |
| EPB41L4B | ELL2 | 423 | 32 | 35 | 9 | 1.224 | 0.006 | Co-occurrence |
| FKBP5 | ELL2 | 430 | 25 | 38 | 6 | 0.999 | 0.045 | Co-occurrence |
| FST | ELL2 | 436 | 19 | 32 | 12 | 2.152 | <0.001 | Co-occurrence |
| GSR | ELL2 | 388 | 67 | 32 | 12 | 0.775 | 0.03 | Co-occurrence |
| HERC3 | ELL2 | 433 | 22 | 35 | 9 | 1.622 | <0.001 | Co-occurrence |
| INPP4B | ELL2 | 435 | 20 | 33 | 11 | 1.981 | <0.001 | Co-occurrence |
| IQGAP2 | ELL2 | 420 | 35 | 34 | 10 | 1.261 | 0.003 | Co-occurrence |
| KLK4 | ELL2 | 437 | 18 | 38 | 6 | 1.344 | 0.013 | Co-occurrence |
| KRT73 | ELL2 | 443 | 12 | 38 | 6 | 1.763 | 0.003 | Co-occurrence |
| LARP1B | ELL2 | 426 | 29 | 31 | 13 | 1.818 | <0.001 | Co-occurrence |
| MERTK | ELL2 | 433 | 22 | 38 | 6 | 1.134 | 0.028 | Co-occurrence |
| MME | ELL2 | 422 | 33 | 34 | 10 | 1.325 | 0.002 | Co-occurrence |
| MPST | ELL2 | 438 | 17 | 39 | 5 | 1.195 | 0.036 | Co-occurrence |
| NT5E | ELL2 | 392 | 63 | 20 | 24 | 2.01 | <0.001 | Co-occurrence |
| NUP88 | ELL2 | 390 | 65 | 42 | 2 | −1.253 | 0.046 | Mutual exclusivity |
| PGM3 | ELL2 | 405 | 50 | 19 | 25 | 2.366 | <0.001 | Co-occurrence |
| PLEKHA7 | ELL2 | 438 | 17 | 39 | 5 | 1.195 | 0.036 | Co-occurrence |
| PLK2 | ELL2 | 419 | 36 | 29 | 15 | 1.795 | <0.001 | Co-occurrence |
| PLPP1 | ELL2 | 420 | 35 | 31 | 13 | 1.616 | <0.001 | Co-occurrence |
| PTPN13 | ELL2 | 437 | 18 | 34 | 10 | 1.966 | <0.001 | Co-occurrence |
| SDC4 | ELL2 | 422 | 33 | 44 | 0 | <−3 | 0.043 | Mutual exclusivity |
| SMS | ELL2 | 437 | 18 | 30 | 14 | 2.427 | <0.001 | Co-occurrence |
| SORD | ELL2 | 436 | 19 | 38 | 6 | 1.287 | 0.016 | Co-occurrence |
| SPDEF | ELL2 | 437 | 18 | 38 | 6 | 1.344 | 0.013 | Co-occurrence |
| SRP19 | ELL2 | 424 | 31 | 27 | 17 | 2.153 | <0.001 | Co-occurrence |
| SSX2IP | ELL2 | 427 | 28 | 37 | 7 | 1.06 | 0.026 | Co-occurrence |
| STK39 | ELL2 | 425 | 30 | 34 | 10 | 1.427 | 0.001 | Co-occurrence |
| TNFAIP8 | ELL2 | 428 | 27 | 31 | 13 | 1.894 | <0.001 | Co-occurrence |
| UCK2 | ELL2 | 422 | 33 | 36 | 8 | 1.044 | 0.02 | Co-occurrence |
| VAPA | ELL2 | 411 | 44 | 35 | 9 | 0.876 | 0.032 | Co-occurrence |
| ZIC2 | ELL2 | 428 | 27 | 35 | 9 | 1.405 | 0.002 | Co-occurrence |
| ZMIZ1 | ELL2 | 428 | 27 | 33 | 11 | 1.665 | <0.001 | Co-occurrence |
3.3. ∣. ELL2 knockdown enhances prostate cancer cell proliferation and motility
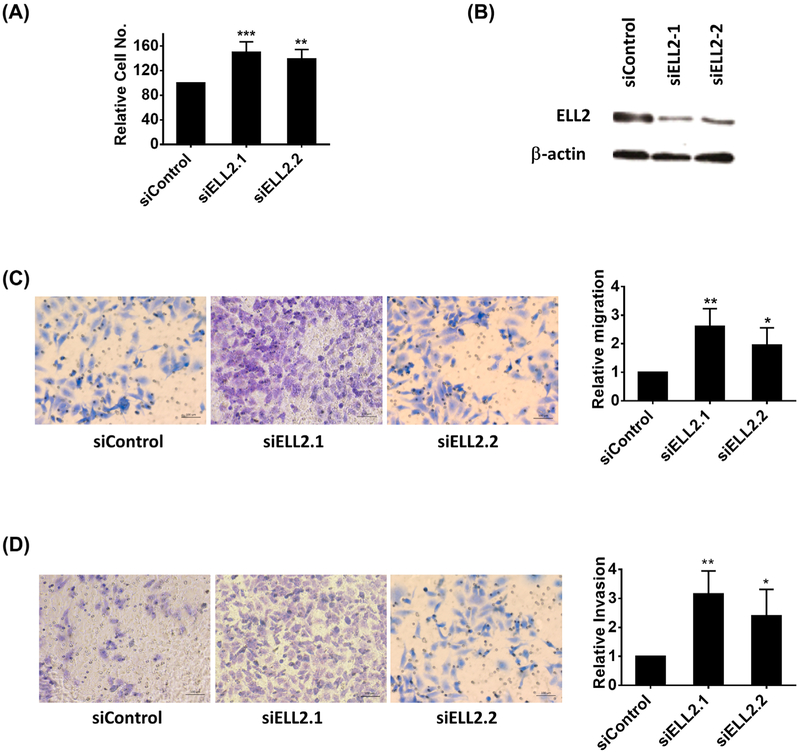
Previous studies suggested that siRNA knockdown of ELL2 in prostate cancer cells could increase proliferation in BrdU assay and cell migration in transwell assay.10 Furthermore, ELL2 knockdown in the murine prostate induced a significant increase in proliferation and in the development of neoplastic lesions.4 To confirm these findings, we used WST-1 assay to evaluate cell proliferation and transwell Matrigel invasion assay for cell motility. Figure 3 showed that knockdown of ELL2 in C4-2 cells using two independent siRNAs caused increased cell proliferation and invasion.
FIGURE 3.
A, The effect of ELL knockdown on proliferation of C4-2 cells. C4-2 cells cultured in 96-well plates were treated with ELL2 siRNA or scrambled siRNA control as described in section 2. The proliferation was measured using WST-1 assay. B, Western blot analysis of ELL2 knockdown by two independent siRNAs targeting ELL2. Western blot was conducted on cells 72 h after transfection with siRNA. C, Effect of ELL2 knockdown on C4-2 cell migration. After 24 h of transfection with siRNA, C4-2 cells were placed in transwell chambers and incubated at 37° for 24 h. The migrated cells on the lower surface of the transwell membrane were stained with crystal violet and quantified as described in the section 2. The quantitative data of relative migration is shown in the right panel. D, Effect of ELL2 knockdown on C4-2 cell invasion. C4-2 cells were treated with siRNA targeting ELL2 or control siRNA for 24 h, then trypsinized and suspended in serum-free medium. The cells were then added into each upper chamber in the Matrigel invasion assay. The transwell chambers were incubated at 37° for 48 h. The cells on the lower surface of the membrane were stained with crystal violet and quantified as described in section 2. The quantitative data was shown at the right panel. *P < 0.05, **P < 0.01, and ***P < 0.001. [Color figure can be viewed at wileyonlinelibrary.com]
3.4. ∣. ELL2 protein has a short half-life and is stabilized by proteasome inhibitor MG132
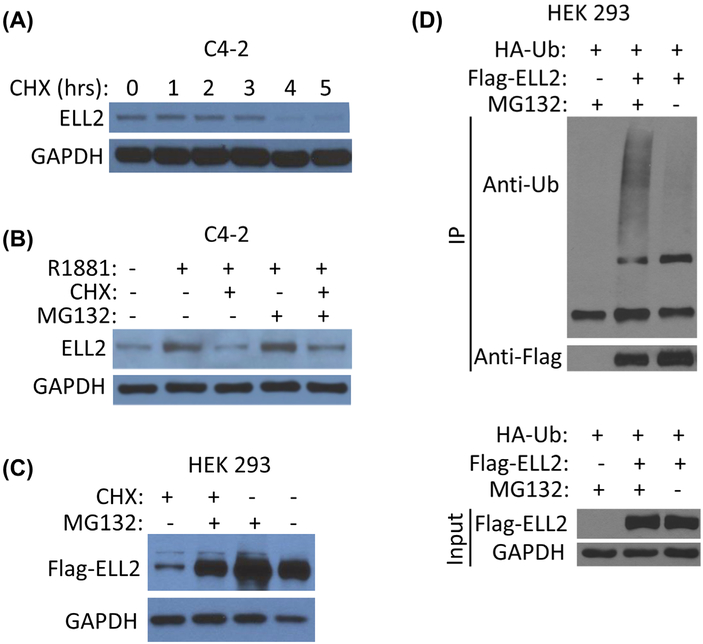
Since ELL2 plays an important role in regulating prostate cancer cell growth and motility, its protein level is likely to be regulated.Understanding the mechanism of regulating ELL2 protein turnover will enhance our understanding of mechanisms leading to ELL2 downregulation in prostate carcinogenesis. We first determined the half-life of ELL2 protein in the presence of protein synthesis inhibitor cycloheximide (CHX) in C4-2 cells. Figure 4A showed that ELL2 protein level were dramatically decreased between 3–4 h after CHX treatment, suggesting that the half-life of ELL2 in C4-2 cells should be less than 4 h. We next tested whether ELL2 turnover was mediated through the proteasome. In Figure 4B, as expected, CHX treatment caused a reduction in ELL2 protein level. In contrast, proteasome inhibitor MG132 enhanced ELL2 protein level. In C4-2 cells treated with R1881, ELL2 protein level in the presence of both CHX and MG132 was higher than its level in the presence of CHX, suggesting that MG132 can inhibit ELL2 degradation in the presence of CHX. This finding was consistent with the finding in Figure 4C using flag-tagged ELL2. The level of flag-ELL2 was enhanced by MG132 and reduced in the presence of cycloheximide, and MG132 can inhibit flag-ELL2 degradation in the presence of CHX. In Figure 4D, co-transfection of HA-tagged ubiquitin with flag-ELL2 showed that flag-ELL2 can be polyubiquitinated, suggesting that the ubiquitin-proteasome pathway was responsible for ELL2 degradation.
FIGURE 4.
The effect of cycloheximide (CHX) and/or MG132 on ELL2 protein level. A, Western blot analysis of ELL2 in C4-2 cells cultured in complete medium treated with CHX at the 50 μg/mL for indicated number of hours. GAPDH was probed as a loading control. B, Western blot analysis of ELL2 in C4-2 cells cultured in the presence or absence of 1 nM R1881 for 48 h and then cultured for an additional 16 h in the presence or absence of CHX at 50 μg/mL, with or without MG132 at 10 μM for 16 h. C, Western blot analysis of flag-ELL2 in HEK 293 cells. HEK 293 cells were transiently transfected with flag-ELL2 expression vector for 24 h and then treated with CHX (50 μg/mL) or vehicle control in the presence or absence of 10 μM MG132 for 16 h prior to cell lysate preparation. Anti-flag antibody was used for Western blot analysis. D, Analysis of flag-ELL2 polyubiquitination in HEK 293 cells. HEK 293 cells were co-transfected with flag-ELL2 and HA-ubiquitin. The cells were then cultured for 40 h after the transfection and treated for an additional 16 h with 0 or μM MG132. Flag-ELL2 protein was isolated from the denatured cell lysates using anti-FLAG antibody, followed by immunoblotting using both anti-FLAG and anti-ubiquitin antibodies. The whole cell lysate input was probed using anti-FLAG antibody to determine the expression of flag-ELL2. GAPDH was detected as loading control. Data shown are representative of 3 independent experiments. [Color figure can be viewed at wileyonlinelibrary.com]
3.5. ∣. Amino acid residues K584 and K599 in ELL2 are important for ELL2 degradation
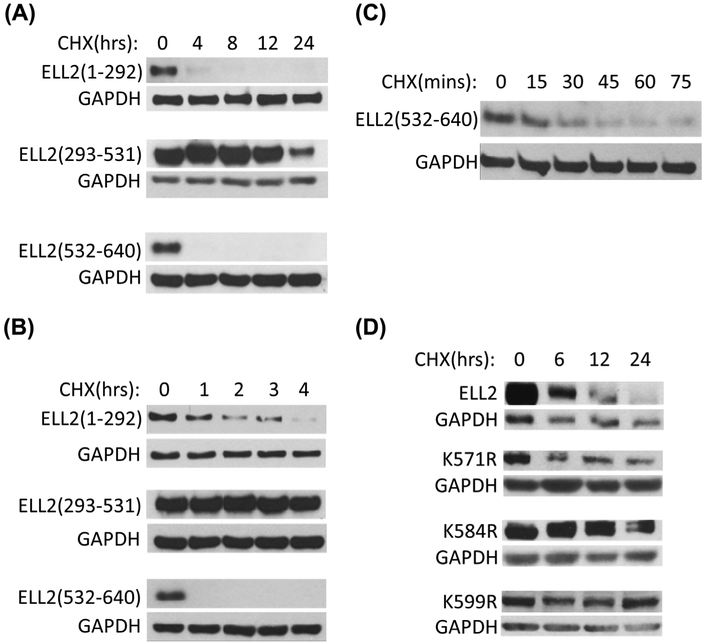
To identify amino acid residues in ELL2 important for its turnover, we have generated various deletion mutants of ELL2. Figure 5A showed the time courses of three ELL2 deletion mutant protein degradation in the presence of CHX. The half-life of ELL2 (293-531) appeared to be longer than 12 h. In contrast, ELL2 (1-292) and ELL2 (532-64D) had a half-life of less than 4 h. To further define the stability of these ELL2 deletion mutants, a short time course study for the stability of these ELL2 mutants in the presence of CHX was conducted (Figure 5B). The half-life of ELL2 (1-292) in the presence of CHX appeared to be around 1 h. In contrast, ELL2 (532-640) was less than one hour. Figure 5C showed an even shorter time course study to determine the stability of ELL2 (532-640) and the result indicated that ELL2 (532-640) had a half-life of about 15 min in the presence of CHX. This observation suggests that ELL2 (532-640) contains amino acid residues for polyubiquitination important for protein turnover. To identify potential ubiquitination sites, we utilized UbiPred to predict potential ubiquitylation sites for ELL2.16 We have generated three ELL2 mutants with point mutations at the K571, K584, and K599, generating K571R, K584R, and K599R. The ELL2 mutant, K571R exhibited a half-life less than 6 h, suggesting that K571 may not be an essential site for polyubiquitination. ELL2 mutant K584R was still expressed at high level 12 h after CHX treatment. Similarly, ELL2 mutant K599R was readily detectable even 24 h after CHX treatment. These findings suggest that K584 and K599 are important in mediating ELL2 polyubiquitination and degradation.
FIGURE 5.
Identification of amino acids residues important for ELL2 turnover. A, C4-2 cells were transfected with FLAG-tagged ELL2 deletion mutants, ELL2 (1-292), ELL2 (293-531), or ELL2 (532-640), for 24 h and then treated with CHX (50 μg/mL) for 0, 4, 8, 12, and 24 h. B, C4-2 cells were transfected with the ELL2 deletion mutants as described in A. and then treated with CHX at 50 μg/mL for 0, 1, 2, 3, and 4 h. C. C4-2 cells were transfected with ELL2 (532-640) deletion mutants and treated with CHX at 50 μg/mL for 0, 15, 30, 45, 60, and 75 min. D. C4-2 cells were transfected with wild-type ELL2 or ELL2 substitution mutants, K571R, K581R, or K599R for 24 h and then treated with CHX at the 50 μg/mL for 0, 6, 12, and 24 h. The whole cell lysates were analyzed by immunoblotting using anti-FLAG antibody. GAPDH was immunoblotted as a loading control.
3.6. ∣. EAF2 stabilizes ELL2 and inhibits its polyubiquitination
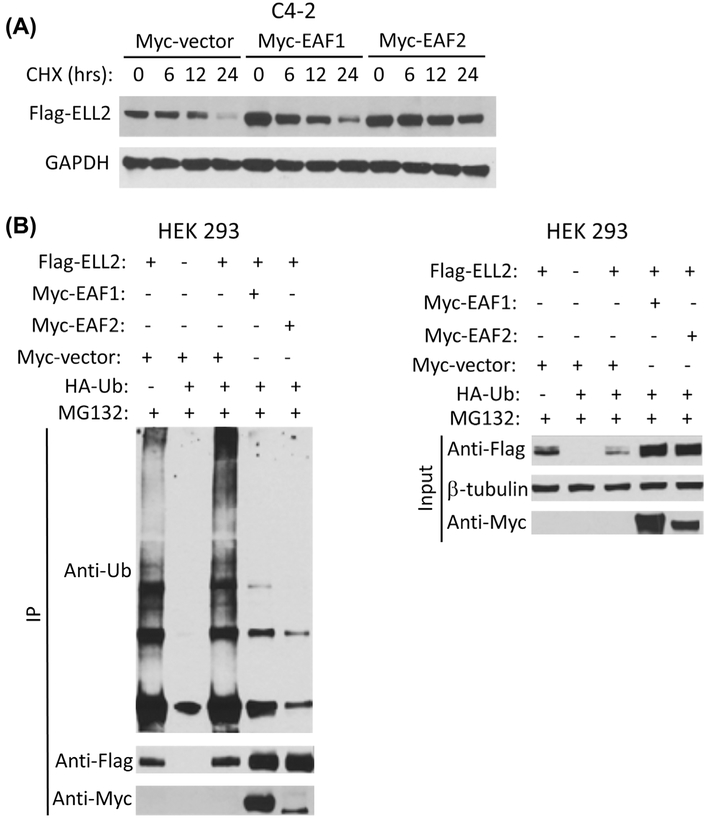
Since ELL2 is tightly associated with EAF family of proteins including both EAF1 and EAF2,8 we tested whether the binding of myc-tagged EAF1 or EAF2 could stabilize flag-tagged ELL2. Figure 6A showed that co-transfection of myc-EAF1 or myc-EAF2 could enhance the protein level of flag-ELL2. The level of flag-ELL2 was enhanced by either myc-EAF1 or myc-EAF2 co-transfection. The half-life of flag-ELL2 in the presence of CHX was slightly increased in the presence of myc-EAF1 and significantly increased in the presence of myc-EAF2 (Figure 6A). Furthermore, in Figure 6B, the presence of either myc-EAF1 or myc-EAF2 significantly inhibited polyubiquitination of flag-ELL2, with myc-EAF2 being more effective.
FIGURE 6.
The effect of EAF1 or EAF2 on ELL2 protein turnover and polyubiquitination. A, C4-2 cells were transiently transfected with FLAG-ELL2 expression vector together with empty Myc-vector, Myc-EAF1, or Myc-EAF2 expression vector for 24 h. The transfected cells were then treated with CHX at 50 μg/mL for 0, 6, 12, or 24 h. FLAG ELL2 expression level were determined by immunoblotting using anti-FLAG antibody. GAPDH was probed as a loading control. B, HEK 293 cells were co-transfected with FLAG-ELL2, Myc-EAF1, Myc-EAF2, empty Myc-vector, and/or HA-ubiquitin as indicated. The transfected cells were cultured for 18 h and then treated with 10 μM MG132 for an additional 16 h. The cell lysates were then prepared for immunoprecipitation using anti-FLAG antibody and analyzed by immunoblotting using indicated antibodies. Whole cell lysate input was also analyzed by Western blot using anti-FLAG and anti-Myc antibody. Beta-tubulin was probed as loading control. Data shown are representative of three independent experiments
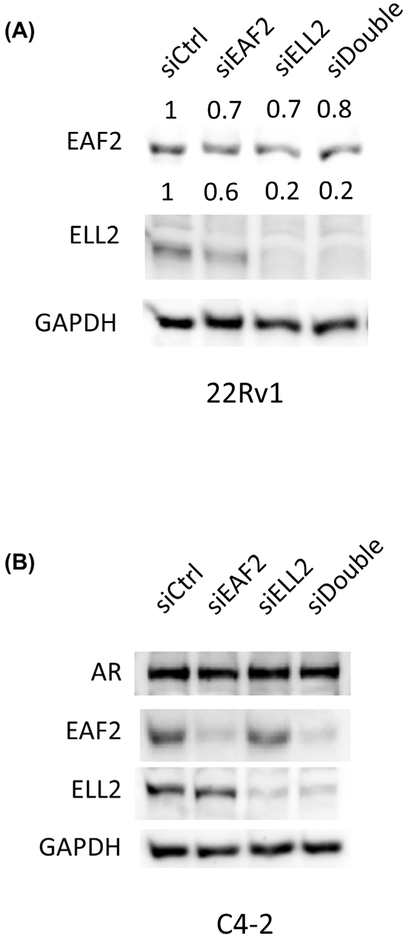
Knockdown of ELL2 resulted in a decrease in EAF2 protein level in 22Rv1 cells (Figure 7A). ELL2 knockdown in C4-2 and LNCaP cells showed variable results (data not shown). In C4-2 and LNCaP cells which also express EAF2,17 the expression of EAF2 protein was either decreased upon ELL2 knockdown or unchanged. The variability in EAF2 protein levels in response to ELL2 knockdown observed in C4-2 and LNCaP could be due in part to variability in cell cycle. ELL2 depletion has previously been associated with splicing in plasma cells.18 AR splice variants have been reported to be frequently observed in castration-resistant prostate cancer.19 We therefore investigated whether depletion of ELL2 could induce the expression of AR splice variants in C4-2 cells. Knockdown of ELL2 and/or EAF2 did not induced splicing in AR protein (Figure 7B).
FIGURE 7.
The effect of knockdown of EAF2 knockdown on ELL2 expression and AR splicing. A, Western immunoblotting analysis of EAF2 and ELL2 expression levels in 22Rv1 cells transfected with nontargeted control (siCtrl) siRNA; targeted to EAF2 (siEAF2), ELL2 (siELL2), and EAF2 and ELL2 (siDouble). B, The effect of EAF2 and/or ELL2 knockdown on AR splicing. Western immunoblotting analysis of C4-2 cells transfected with nontargeted control (siCtrl) siRNA; targeted to EAF2 (siEAF2), ELL2 (siELL2), and EAF2 and ELL2 (siDouble). GAPDH served as loading control. Data shown are representative of three independent experiments group
4. ∣. DISCUSSION
This study presents evidence for downregulation of an androgen-response gene ELL2 at the protein level in human prostate cancer specimens and further confirms a suppressive role for ELL2 in prostate cancer cell proliferation and motility. Using deletion and point mutagenesis coupled with protein stability assay in the presence of CHX, we have identified K584 and K599 being important amino acid resides in ELL2 that mediate proteasome degradation. Finally, we showed that binding of EAF family proteins, particularly EAF2 protein can significantly inhibit ELL2 polyubiquitination and enhance its stability.
Downregulation of ELL2 protein in prostate cancer specimens is consistent with its function as a potential tumor suppressor. Mice with conditional prostate-specific deletion of ELL2 exhibited prostatic defects including increased epithelial proliferation, vascularity, and PIN lesions.4 Microarray analysis of prostates from these mice identified several differentially expressed genes associated with proliferation, cellular motility, and epithelial and neural differentiation; and many of these genes were also down-regulated or deleted in prostate adenocarcinoma cases.4 The major signaling pathways altered in the mouse ELL2 knockout prostate are likely conserved and are also involved in ELL2 action in human prostate cancer cell lines. We have previously reported downregulation of ELL2 mRNA in high-Gleason score prostate cancer specimens.10 The downregulation of ELL2 protein in clinical prostate cancer specimens is consistent with its mRNA downregulation. Thus, one mechanism leading to ELL2 protein downregulation is likely mediated through downregulation of its mRNA. Knockdown of ELL2 in prostate cancer cell lines increased proliferation and cell migration. Cell proliferation was previously reported to be positively correlated with high Gleason score in patients.20 The downregulation of ELL2 in high Gleason score prostate tumor specimens and increased proliferation in prostate cancer cells treated with ELL2 knockdown suggests that ELL2 can modulate proliferation of prostate epithelial cells.
Our finding that EAF proteins can stabilize ELL2 through blocking its polyubiquitination suggests that loss of EAF proteins could cause downregulation of ELL2. EAF2 was reported as a potential tumor suppressor of prostate cancer and is downregulated frequently in prostate cancer specimens.17,21-24 Also, loss of EAF2 resulted in similar phenotype observed in ELL2 knockout mouse prostate.4 In C. elegans model, loss of EAF1 phenocopies the loss of ELL1.25 It appears that ELL and EAF family proteins require each other for their functions since ELL2 can be stabilized by EAF family proteins. Loss of EAF family proteins in prostate cancer cells can result in destabilization and subsequent functional loss of ELL2. In the prostate, EAF2 appears to play a more important role than EAF1 because EAF2 mRNA is expressed at levels higher than EAF1 mRNA.
Androgens appear to be able to regulate the expression of ELL2 at the both transcriptional and protein stability levels. According to the literature, AR can regulate ELL2 mRNA level in prostate cancer cells.2,3 The data from the present study showed that ELL2 protein stability can be enhanced by EAF2 protein, which is also encoded by an androgen-response gene. Co-induction of EAF2 and ELL2 by androgens can further enhance the ELL2 protein level. This mechanism can amplify the magnitude of androgen induction of ELL2 signaling.
A previous report showed that ELL2 polyubiquitination is mediated through Siah1 E3 ubiquitin ligase.11 It will be interesting to determine whether Siah1 is also responsible for ELL2 polyubiquitination in prostate cancer cells and whether the mechanism of ELL2 polyubiquitination inhibition by EAF family proteins involves blocking Siah1 access to ELL2.
Our previous studies showed that EAF2 can also undergo polyubiquitination, catalyzed by Siah2 E3 Ligase. Thus, both Siah1 and Siah2 could regulate ELL2 protein, with Siah1 being an ELL2 E3 Ligase directly modulates ELL2 protein turnover and Siah2 being an E3 Ligase for EAF2 and indirectly affects the stability of ELL2. These observations suggest that ELL2 expression can be controlled by multiple mechanisms, including AR regulation of ELL2 transcription, EAF family proteins binding, and Siah1-mediated polyubiquitination.
In summary, this study provided additional evidence for a tumor suppressive role of ELL2 in prostate cancer and suggested that ELL2 is downregulated at the protein level in advanced prostate cancer specimens. This study also identified K584 and K599 are potential ubiquitination sites important for regulating ELL2 stability. EAF family proteins, particularly EAF2, can stabilize ELL2 and block its polyubiquitination. These findings provide a foundation to further explore the mechanisms regulating ELL2 protein level in prostate cancer.
ACKNOWLEDGMENTS
We are grateful to Katherine O’Malley, Erica Parrinello, Aiyuan Zhang, and Jianhua Zhou for technical support. All slides stained with ELL2 antibody were reviewed by Dr Jianbo Zhang from Department of Urology, Affiliated Cancer Hospital of Zhengzhou University. This work was funded in part by NIH grants R01CA186780, 1R50CA211242, and P50 CA090386 and Science and Technology Project of Henan Province, China 162102310040. We thank B. Matija Peterlin from University of California, San Francisco for providing sequence 1 of siRNA against ELL2.
Funding information
Science and Technology Project of Henan Province (China), Grant number: 162102310040; National Cancer Institute, Grant numbers: P50CA090386,R01CA186780, R50CA211242
Footnotes
CONFLICT OF INTEREST
The authors have nothing to disclose.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Nelson PS, Clegg N, Arnold H, et al. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc Natl Acad Sci USA. 2002;99:11890–11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, Yamamoto KR. Cell-and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 2007;21:2005–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pascal LE, Masoodi KZ, Liu J, et al. Conditional deletion of ELL2 induces murine prostate intraepithelial neoplasia. J Endocrinol. 2017;235:123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shilatifard A, Duan DR, Haque D, et al. ELL2, a new member of an ELL family of RNA polymerase II elongation factors. Proc Natl Acad Sci USA. 1997;94:3639–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thirman MJ, Levitan DA, Kobayashi H, Simon MC, Rowley JD. Cloning of ELL, a gene that fuses to MLL in a t(11;19)(q23;p13.1) in acute myeloid leukemia. Proc Natl Acad Sci USA. 1994;91:12110–12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitani K, Kanda Y, Ogawa S, et al. Cloning of several species of MLL/MEN chimeric cDNAs in myeloid leukemia with t(11;19)(q23;p13.1) translocation. Blood 1995;85:2017–2024. [PubMed] [Google Scholar]
- 8.Kong SE, Banks CA, Shilatifard A, Conaway JW, Conaway RC. ELL-associated factors 1 and 2 are positive regulators of RNA polymerase II elongation factor ELL. Proc Natl Acad Sci USA. 2005;102:10094–10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo Z, Lin C, Shilatifard A. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol. 2012;13:543–547. [DOI] [PubMed] [Google Scholar]
- 10.Qiu X, Pascal LE, Song Q, et al. Physical and functional interactions between ELL2 and RB in the suppression of prostate cancer cell proliferation, migration, and invasion. Neoplasia 2017;19:207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu M, Hsu J, Chan C, Li Z, Zhou Q. The ubiquitin ligase Siah1 controls ELL2 stability and formation of super elongation complexes to modulate gene transcription. Mol Cell. 2012;46:325–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- 13.Jiang F, Ai J, Xiao W, Wang Z. FB1, an E2A fusion partner in childhood leukemia, interacts with U19/EAF2 and inhibits its transcriptional activity. Cancer Lett. 2007;253:265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pascal LE, Ai J, Masoodi KZ, et al. Development of a reactive stroma associated with prostatic intraepithelial neoplasia in EAF2 deficient mice. PLoS ONE 2013;8:e79542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su F, Correa BR, Luo J, Vencio RZ, Pascal LE, Wang Z. Gene expression profiling reveals regulation of ERK phosphorylation by androgen-induced tumor suppressor U19/EAF2 in the mouse prostate. Cancer Microenviron. 2013;6:247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tung CW, Ho SY. Computational identification of ubiquitylation sites from protein sequences. BMC Bioinformatics. 2008;9:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Pascal LE, Zhong M,et al. Combined loss of EAF2 and p53 induces prostate carcinogenesis in male mice. Endocrinology.2017;158:4189–4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benson MJ, Aijo T, Chang X, et al. Heterogeneous nuclear ribonucleoprotein L-like (hnRNPLL) and elongation factor, RNA polymerase II, 2 (ELL2) are regulators of mRNA processing in plasma cells. Proc Natl Acad Sci USA. 2012;109:16252–16257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.To SQ, Kwan EM, Fettke HC, et al. Expression of androgen receptor splice variant 7 or 9 in whole blood does not predict response to androgen-axis-targeting agents in metastatic castration-resistant prostate cancer. Eur Urol. 2018;73:818–821. [DOI] [PubMed] [Google Scholar]
- 20.Richardsen E, Andersen S, Al-Saad S, et al. Evaluation of the proliferation marker Ki-67 in a large prostatectomy cohort. PLoS ONE. 2017;12:e0186852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao W, Zhang Q, Habermacher G, et al. U19/Eaf2 knockout causeslung adenocarcinoma, B-cell lymphoma, hepatocellular carcinoma and prostatic intraepithelial neoplasia. Oncogene.2008;27:1536–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pascal LE, Vencio RZ, Page LS, et al. Gene expression relationship between prostate cancer cells of Gleason 3, 4 and normal epithelial cells as revealed by cell type-specific transcriptomes. BMC Cancer. 2009;9:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao W, Zhang Q, Jiang F, Pins M, Kozlowski JM, Wang Z. Suppression of prostate tumor growth by U19, a novel testosterone-regulated apoptosis inducer. Cancer Res. 2003;63:4698–4704. [PubMed] [Google Scholar]
- 24.Ai J, Pascal LE, O'Malley KJ, et al. Concomitant loss of EAF2/U19 and Pten synergistically promotes prostate carcinogenesis in the mouse model. Oncogene. 2014;33:2286–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai L, Phong BL, Fisher AL, Wang Z. Regulation of fertility, survival, and cuticle collagen function by the Caenorhabditis elegans eaf-1 and ell-1 genes. J Biol Chem. 2011;286:35915–35921. [DOI] [PMC free article] [PubMed] [Google Scholar]