Abstract
Members of the family Parvoviridae are small, resilient, non-enveloped viruses with linear, single-stranded DNA genomes of 4–6 kb. Viruses in two subfamilies, the Parvovirinae and Densovirinae, are distinguished primarily by their respective ability to infect vertebrates (including humans) versus invertebrates. Being genetically limited, most parvoviruses require actively dividing host cells and are host and/or tissue specific. Some cause diseases, which range from subclinical to lethal. A few require co-infection with helper viruses from other families. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the Parvoviridae, which is available at www.ictv.global/report/parvoviridae.
Keywords: Parvoviridae, Parvovirinae, Densovirinae, taxonomy, ICTV Report
Virion
Parvovirus virions are small, rugged, non-enveloped protein particles with T=1 icosahedral symmetry (Table 1 and Fig. 1). A single coat protein sequence is expressed as a nested set of virion proteins (VP) with a common C-terminal domain that forms the virion shell. VP1 N-termini may have phospholipase A2 (PLA2) activity [1, 2].
Table 1. Characteristics of the family Parvoviridae.
| Typical member: | human parvovirus B19-J35 G1 (AY386330), species Primate erythroparvovirus 1, genus Erythroparvovirus, subfamily Parvovirinae |
|---|---|
| Virion | Small, non-enveloped, T=1 icosahedra, 23–28 nm in diameter |
| Genome | Linear, single-stranded DNA of 4–6 kb with short terminal hairpins |
| Replication | Rolling hairpin replication, a linear adaptation of rolling circle replication. Dynamic hairpin telomeres prime complementary strand and duplex strand-displacement synthesis; high mutation and recombination rates |
| Translation | Capped mRNAs; co-linear ORFs accessed by alternative splicing, non-consensus initiation or leaky scanning |
| Host range | Parvovirinae: mammals, birds, reptiles. Densovirinae: insects, crustacea, echinoderms |
| Taxonomy | Two subfamilies, Parvovirinae and Densovirinae; 13 genera, >75 species |
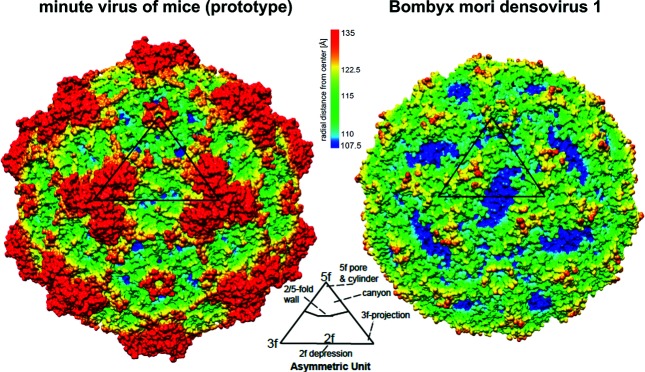
Fig. 1.
Virion morphology. Three-dimensional X-ray diffraction structures of exemplar parvoviruses at 3.1–3.4 Å resolution, obtained using PDBs 1MVM and 3P0S. Colour depicts distance from the virus centre; triangles outline one of 60 icosahedral units showing the 2-, 3- and 5-fold axes of symmetry.
Genome
Viruses package a single copy of a linear ssDNA molecule of 4–6 kb, which contains a long coding region bracketed by short (120–600 nt) dynamic hairpin termini that mediate DNA replication (Fig. 2). Packaged strands can be of negative or both senses. Two gene cassettes encode a replication initiator protein (NS1 or Rep) and a virion protein (VP), plus a few small genus-specific auxiliary proteins. Many parvoviruses are highly specialized for infecting particular host cells. Since host restrictions may be relaxed when cells undergo oncogenic transformation, viruses in some species, such as Rodent protoparvovirus 1, may be preferentially oncolytic [3]. In contrast, adeno-associated viruses co-opt helper viruses, such as adenoviruses or herpesviruses, to support their productive replication; the simplicity, durability, broad tissue specificity and lack of toxicity makes these viruses useful gene transfer vehicles for research and clinical studies [4].
Fig. 2.
Parvovirus genome organization shows genus-specific variations. Terminal hairpins are magnified relative to the coding region to show predicted secondary structures. ORFs are indicated by arrowed boxes. Angled arrows indicate transcriptional promoters and AAAAA indicates polyadenylation sites.
Replication
Parvoviruses bind glycosylated cell surface molecules and are internalized by receptor-mediated endocytosis. Virions are metastable [1]; in endosomes some undergo a conformational shift, exposing VP1 PLA2 domains required for lipid bilayer penetration. Intact virions enter the nucleus, where their genomic 3′-hairpin primes complementary strand synthesis by a host replication fork. This creates a duplex transcription template, allowing RNA polymerase II transcription to initiate gene expression (Fig. 2)
DNA replication proceeds via a ‘rolling hairpin’ mechanism, which relies on sequential unfolding and refolding of the hairpin termini. Unidirectional strand displacement synthesis generates continuous duplex intermediates, from which progeny single strands are excised by the endonuclease activity of NS1. Progeny genomes are packaged into preassembled viral particles by the NS1 helicase, via a portal at one of the icosahedral 5-fold axes. Progeny virions may be rapidly exported from living cells, or accumulate in the nucleus until liberated by cell lysis. Many viruses cause mild disease, whereas others, such as canine parvovirus (species Carnivore protoparvovirus 1) and most members of the subfamily Densovirinae, are highly pathogenic [2, 5].
Taxonomy
Members of the subfamily Parvovirinae infect vertebrates (mammals, birds and reptiles). This subfamily includes eight genera; Bocaparvovirus, Dependoparvovirus, Erythroparvovirus, Protoparvovirus, Tetraparvovirus, Amdoparvovirus, Aveparvovirus and Copiparvovirus, containing >55 species. Viruses that infect humans are present in seven species from the first five genera in this list. Members of subfamily Densovirinae infect invertebrates (insects, crustaceans and echinoderms). This subfamily contains one unassigned species and five genera; Ambidensovirus, Brevidensovirus, Hepandensovirus, Iteradensovirus and Penstyldensovirus, comprising >20 species.
Resources
Full ICTV Report on the family Parvoviridae: www.ictv.global/report/parvoviridae.
Funding information
Production of this summary, the online chapter and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA).
Acknowledgements
Members of the ICTV 10th Report Consortium are Elliot J. Lefkowitz, Andrew J. Davison, Stuart G. Siddell, Peter Simmonds, Sead Sabanadzovic, Donald B. Smith, Richard J. Orton and Balázs Harrach.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: NS1/Rep, replication initiator protein; PLA2, phospholipase A2; VP, virion protein.
References
- 1.Cotmore SF, Tattersall P. Parvoviruses: Small Does Not Mean Simple. Annu Rev Virol. 2014;1:517–537. doi: 10.1146/annurev-virology-031413-085444. [DOI] [PubMed] [Google Scholar]
- 2.Tijssen P, Pénzes JJ, Yu Q, Pham HT, Bergoin M. Diversity of small, single-stranded DNA viruses of invertebrates and their chaotic evolutionary past. J Invertebr Pathol. 2016;140:83–96. doi: 10.1016/j.jip.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Geletneky K, Nüesch JP, Angelova A, Kiprianova I, Rommelaere J. Double-faceted mechanism of parvoviral oncosuppression. Curr Opin Virol. 2015;13:17–24. doi: 10.1016/j.coviro.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Samulski RJ, Muzyczka N. AAV-mediated gene therapy for research and therapeutic purposes. Annu Rev Virol. 2014;1:427–451. doi: 10.1146/annurev-virology-031413-085355. [DOI] [PubMed] [Google Scholar]
- 5.Kailasan S, Agbandje-Mckenna M, Parrish CR. Parvovirus family conundrum: what makes a killer? Annu Rev Virol. 2015;2:425–450. doi: 10.1146/annurev-virology-100114-055150. [DOI] [PubMed] [Google Scholar]