Abstract
A metagenomics approach was used to detect novel and recognized RNA viruses in mosquitoes from the Yucatan Peninsula of Mexico. A total of 1359 mosquitoes of 7 species and 5 genera (Aedes, Anopheles, Culex, Mansonia and Psorophora) were sorted into 37 pools, homogenized and inoculated onto monolayers of Aedes albopictus (C6/36) cells. A second blind passage was performed and then total RNA was extracted and analysed by RNA-seq. Two novel viruses, designated Uxmal virus and Mayapan virus, were identified. Uxmal virus was isolated from three pools of Aedes (Ochlerotatus) taeniorhynchus and phylogenetic data indicate that it should be classified within the recently proposed taxon Negevirus. Mayapan virus was recovered from two pools of Psorophora ferox and is most closely related to unclassified Nodaviridae-like viruses. Two recognized viruses were also detected: Culex flavivirus (family Flaviviridae) and Houston virus (family Mesoniviridae), with one and two isolates being recovered, respectively. The in vitro host ranges of all four viruses were determined by assessing their replicative abilities in cell lines of avian, human, monkey, hamster, murine, lepidopteran and mosquito (Aedes, Anopheles and Culex) origin, revealing that all viruses possess vertebrate replication-incompetent phenotypes. In conclusion, we report the isolation of both novel and recognized RNA viruses from mosquitoes collected in Mexico, and add to the growing plethora of viruses discovered recently through the use of metagenomics.
Keywords: metagenomics, virus discovery, RNA-seq, mosquito, Negevirus, Nodaviridae, Culex flavivirus, Houston virus, Mexico
Introduction
The advent of next-generation sequencing (NGS) has revolutionized virus discovery [1–5]. This technology allows for the rapid and inexpensive detection of nucleic acids in diverse biological and environmental samples and offers distinct advantages over conventional virus detection techniques. Unfortunately, NGS is not always complemented by virus isolation experiments and therefore many newly discovered viruses are known solely from sequence data [6, 7]. Although many viruses cannot be easily propagated in cell culture, the importance of virus isolation cannot be understated because a newly discovered virus cannot be phenotypically characterized if an isolate is unavailable. Information on host range, transmissibility, pathogenesis, antigenicity and epidemiology are essential for the development and implementation of effective virus surveillance, diagnosis, control and prevention strategies.
A rapidly increasing number of novel RNA viruses have been identified in mosquitoes by NGS in recent years [8–13]. One of the first mosquito-associated RNA viruses to be discovered using this technology was Negev virus, the prototype virus of a newly proposed taxon, designated Negevirus, which also includes Dezidougou virus (DEZV) and Wallerfield virus (WALV) [8, 12]. Negeviruses have been isolated exclusively from mosquitoes and phlebotomine sandflies and are assumed to have insect-restricted host ranges [8, 12, 14–21]. Phylogenetic analyses have shown that negeviruses separate into two monophyletic clades (Nelorpivirus and Sandewavirus) and the amount of genetic diversity between viruses of different clades is sufficient to allow each clade to be elevated to the level of genus [20]. Negeviruses have a close phylogenetic relationship with plant viruses of the genera Cilevirus, Higrevirus and Blunervirus, some of which are arthropod-borne, and therefore it has been postulated that negeviruses possess the capacity to replicate in plants, although direct experimental evidence is lacking [12, 19, 20, 22].
Shi and colleagues recently discovered over a thousand novel putative RNA viruses in the largest NGS investigation of viruses ever performed on invertebrates [23]. This groundbreaking study provides valuable insights into virus biodiversity and evolution. More than 220 species of terrestrial and marine invertebrates from 9 metazoan phyla collected in China were examined. Among the numerous viruses discovered were multiple unclassified RNA viruses designated as Hubei noda-like virus 1 to 26. Phylogenetic studies revealed that Hubei noda-like viruses are closely related to viruses in the family Nodaviridae, a group of non-enveloped, bipartite RNA viruses that are classified according to their host associations. The family contains two established genera, Alphanodavirus (insect-associated) and Betanodavirus (fish-associated), and one proposed genus, Gammanodavirus (decapod-associated), besides a large number of other divergent but unclassified proposed members [23–25].
In the present study, mosquitoes of multiple species from the Yucatan Peninsula of Mexico were tested for the presence of novel and recognized RNA viruses. Mosquitoes were assayed by virus isolation in mosquito cell culture and then total RNA was extracted from all cultures, regardless of whether cytopathic effect (CPE) occurred, and examined for viral sequences using metagenomics. The replicative abilities of each novel and recognized virus were assessed using multiple vertebrate and invertebrate cell lines to determine their in vitro host ranges.
Results
Mosquito collections and virus discovery
This study was performed using 1359 mosquitoes of 7 species and 5 genera collected in the Yucatan Peninsula of Mexico in 2007 and 2008 (Table 1). The mosquitoes were sorted into 37 pools and homogenized, and then an aliquot of each homogenate underwent 2 blind passages in Aedes albopictus (C6/36) cells. Eight homogenates caused cell death and seven others caused mild to severe clumping or rounding of cells. Total RNA was extracted from all second passage cultures, regardless of whether CPE was observed, and analysed by RNA-seq for viral sequences. Two novel viruses, designated Uxmal virus (UXMV) and Mayapan virus (MYPV), were discovered (Table 2). The viruses were named after archaeological ruins in the Yucatan Peninsula of Mexico. Two recognized viruses were also isolated: Culex flavivirus (CxFV; genus Flavivirus, family Flaviviridae) and Houston virus (HOUV; a variant of the species Alphamesonivirus-1 of the genus Alphamesonivirus, family Mesoniviridae). An acronym has not been previously assigned to Houston virus, but HOUV is used in this paper. Virus sequences were not recovered from several samples that produced CPE, indicating that the metagenomics analysis did not identify all viruses.
Table 1. Mosquitoes tested by virus isolation in cell culture and viral metagenomics.
| Species | No. of mosquitoes | No. of pools |
|---|---|---|
| Aedes aegypti | 62 | 4 |
| Aedes taeniorhynchus | 764 | 16 |
| Aedes trivittatus | 30 | 2 |
| Anopheles vestitipennis | 16 | 1 |
| Culex quinquefasciatus | 305 | 10 |
| Mansonia titillans | 89 | 2 |
| Psorophora ferox | 93 | 2 |
| Total | 1359 | 37 |
Table 2. Novel and recognized RNA viruses isolated from mosquitoes collected in the Yucatan Peninsula of Mexico.
| Virus | No. of isolations | Isolate name(s) | Taxonomic classification | Study site | Mosquito species |
|---|---|---|---|---|---|
| Culex flavivirus | 1 | CxFV-T1123 | Flavivirus (Flaviviridae) | Tixkokob | Cx. quinquefasciatus |
| Houston virus | 2 | HOUV-M742, HOUV-M2668 | Alphamesonivirus (Mesoniviridae) | Merida |
Cx. quinquefasciatus, Ae. taeniorhynchus |
| Mayapan virus | 2 | MYPV-H44, MYPV-H56 | aNodaviridae? | Tzucacab | Ps. ferox |
| Uxmal virus | 3 | UXMV-M985, UXMV-M1000, UXMV-M2038 | Negevirus | Merida | Ae. taeniorhynchus |
a, The closest known relatives of Mayapan virus are unclassified viruses that phylogenetically group with viruses in the family Nodaviridae.
Uxmal virus
UXMV was isolated from three pools of female Aedes (Ochlerotatus) taeniorhynchus collected in Merida (Table 2). All second-passage C6/36 cell cultures displayed 40–60 % CPE at 3 to 4 days post-inoculation, as indicated by the presence of elongated cells, clumping of cells into a chain-like formation and cell death. The in vitro host range experiments confirmed that C6/36 cells are permissive to UXMV replication and revealed that Culex tarsalis (CT) cells also support replication (Table 3). Unlike for the C6/36 cells, CPE was not observed in the CT cells. Anopheles, lepidopteran and vertebrate cells did not support virus replication. Homogenates prepared from 268 male Ae. taeniorhynchus in 15 pools were also tested by reverse transcription/polymerase chain reaction (RT-PCR) for UXMV, and all were negative.
Table 3. In vitro host ranges of novel and recognized viruses isolated from mosquitoes in the Yucatan Peninsula of Mexico.
| Cell line | Virus | |||
|---|---|---|---|---|
| Culex flavivirus | Houston virus | Mayapan virus | Uxmal virus | |
| Mosquito cells | ||||
| C6/36 | a+ | a+ | + | a+ |
| CT | − | + | − | + |
| Sua 4.0 | − | − | − | − |
| Lepidopteran cells | ||||
| High Five | − | − | − | − |
| Sf9 | − | − | − | − |
| Vertebrate cells | ||||
| BSR-T7/5 | − | − | − | − |
| DEF | − | − | − | − |
| HeLa | − | − | − | − |
| LLC-MK2 | − | − | − | − |
| Murine microglia | − | − | − | − |
| Vero | − | − | − | − |
a, CPE was observed.
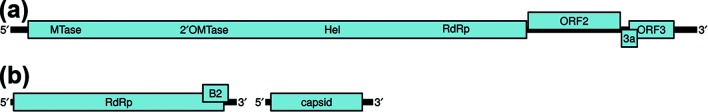
The genome of one isolate (designated UXMV-M985) was fully sequenced (GenBank accession no. MH719095). The sequence consists of 9227 nt, excluding the polyadenylate tail, and is most closely related to the genomes of WALV (69.5 % nt identity with 67.4 % coverage) and DEZV (66.9 % nt identity with 70.0 % coverage). The genomic organization of UXMV is characteristic of viruses in the taxon Negevirus, with the genome predicted to encode three major open reading frames (ORFs) that are separated by short (22 to 119 nt) intergenic regions and flanked by 5′ and 3′ untranslated regions (UTRs) of 235 nt and 286 nt, respectively (Fig. 1a).
Fig. 1.
Predicted genomic organization of (a) Uxmal virus and (b) Mayapan virus. The predicted viral methyltransferase, 2′-O-methyltransferase, helicase and RNA-dependent RNA polymerase domains located within ORF 1 of Uxmal virus are denoted as MTase, 2′OMTase, Hel and RdRp, respectively.
ORF1 encodes a putative replicase protein of 2239 residues that is most closely related to the corresponding translation products of WALV (60.2 % identity and 77.6 % similarity, with 100 % coverage) and DEZV (60.8 % identity and 77.2 % similarity, with 98.6 % coverage). The putative protein contains predicted viral methyltransferase (pfam01660), 2′-O-methyltransferase (pdb3R24), helicase (pfam01443) and RNA-dependent RNA polymerase (RdRp; pfam00978) domains at residues 82 to 413, 775 to 965, 1312 to 1579 and 1783 to 2220, respectively. These domains are highly conserved among the replicase proteins of negeviruses [12, 19].
ORF 2 is predicted to encode a membrane-associated protein of 413 residues. The putative protein is most closely related to the corresponding translation products of WALV (44.9 % identity and 64.7 % similarity, with 100 % coverage) and DEZV (35.3 % identity and 54.4 % similarity, with 98.3 % coverage). The putative protein is predicted to contain an amino-terminal signal peptide (residues 1 to 16), in addition to three carboxy-terminal transmembrane domains (residues 337 to 356, 368 to 389 and 395 to 410) and two carboxy-terminal cytoplasmic domains (residues 357 to 367 and 411 to 413), which is consistent with the ORF 2-encoded translation products of other negeviruses [16, 17, 26]. Three potential N-linked glycosylation sites are also present (residues 178, 244 and 269).
ORF 3 is predicted to encode a membrane-associated protein of 200 residues. The putative protein is most closely related to the corresponding translation products of WALV (63.0 % identity and 82.5 % similarity, with 100 % coverage) and DEZV (62.1 % identity and 75.6 % similarity, with 99.5 % coverage). The putative protein is predicted to contain four transmembrane domains (residues 49 to 71, 101 to 121, 133 to 158 and 165 to 183) and two cytoplasmic domains (residues 72 to 100 and 159 to 164). Two potential N-linked glycosylation sites are also present (residues 42 and 72). An additional ORF, which we have called ORF3a, spans nucleotide positions 8236 to 8448. The ORF overlaps the 5′ end of ORF3 and is conserved in sandewaviruses (except the divergent Hubei virga-like virus 7 and Lodeiro virus) and encodes a cysteine-rich putative protein of 70 residues (Fig. 2a). ORF3a mostly fills a previously presumed non-coding gap between ORFs 2 and 3 (Fig. 1a).
Fig. 2.
Alignment of (a) the translation products potentially encoded by ORF3a of Uxmal virus and selected related viruses and (b) deduced amino acid sequences of the putative B2 proteins of Mayapan virus and Hubei noda-like virus 5.
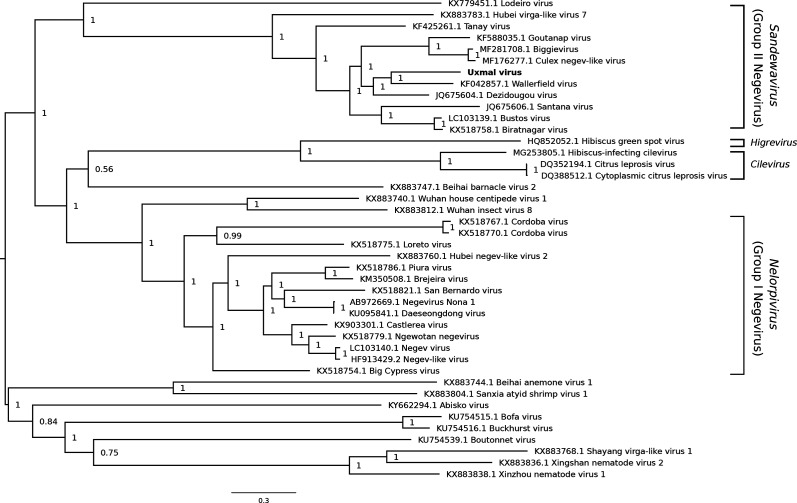
A phylogenetic tree was constructed with Bayesian methods using the deduced amino acid sequence of the RdRp of UXMV and the corresponding regions of related sequences (Fig. 3). UXMV is most closely related phylogenetically to WALV and DEZV, and the posterior support for this topological arrangement is 1.0. Most of the viruses used in the analysis belong to one of two previously established clades: Nelorpivirus (group I) and Sandewavirus (group II). UXMV is a group II negevirus.
Fig. 3.
Phylogenetic tree for Uxmal virus and selected related reference and non-reference sequences. The amino acid sequences of the ORF encoding the RdRp were aligned using muscle [60]. A maximum-likelihood phylogenetic tree was estimated using the Bayesian Markov chain Monte Carlo method implemented in MrBayes version 3.2.6 [61], sampling across the default set of fixed amino acid rate matrices with one million generations and discarding the first 25 % as burn-in. The figure was produced using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). The tree is midpoint-rooted and selected nodes are labelled with posterior probability values. GenBank accession numbers are indicated next to virus names.
Mayapan virus
MYPV was isolated from two pools of female Psorophora ferox collected in Tzucacab (Table 2). CPE was not observed in the first-or second-passage C6/36 cell cultures. The in vitro host range experiments revealed that MYPV has a narrow host range because only C6/36 cells were permissive to virus infection (Table 3). Male Ps. ferox were not tested for MYPV because none were available in our archived collections.
The genomic organization of MYPV has similarities with that of viruses in the family Nodaviridae. Viruses classified within this family have positive-sense, single-stranded, bipartite RNA genomes and the two segments are denoted as RNA1 and RNA2 [27]. RNA1 (3.1 kb) encodes the RdRp and RNA2 (1.4 kb) encodes the capsid protein. A subgenomic RNA (sgRNA3; 0.4 kb) is produced from RNA1 and encodes one or two small proteins, B1 and B2. The function of B1, which is identical to the C-terminus of the RdRp, is unknown, and B2 is a suppressor of RNA interference (RNAi) [28]. An additional subgenomic RNA (0.3 kb) is uniquely produced from RNA1 of mosinovirus, a nodavirus isolated from Culicidae spp. mosquitoes in Cote d’Ivoire [29].
Two non-overlapping MYPV sequences were recovered, indicating that the virus has a bipartite genome (GenBank accession nos MH719096 and MH719097; Fig. 1b). A less likely explanation is that we recovered genome segments from two distinct but related viruses. RNA1 consists of 3045 nt and is most closely related to the corresponding region of Hubei noda-like virus 5 (82.3 % identity with 98.6 % coverage), an unclassified Nodaviridae-like virus detected in spiders and insects in China [23]. The detection of Hubei noda-like virus 5 in spiders suggests that the virus naturally infects spiders or was acquired from recently eaten insects. RNA1 of MYPV is predicted to encode a large ORF flanked by 5′ and 3′ UTRs of 39 and 171 nt, respectively. The putative protein consists of 944 residues and is most closely related to the corresponding translation product of Hubei noda-like virus 5 (90.8 % identity and 95.2 % similarity, with 97.6 % coverage), followed by mosinovirus (43.2 % identity and 60.7 % similarity, with 96.3 % coverage). Domains characteristic of RdRps (cd01699) and dephospho-coenzyme A kinases (cd02022) are present at residues 469 to 685 and 283 to 349, respectively. A short ORF (nt 2591 to 2932) overlapping the 3′ end of the RdRp ORF is also conserved in Hubei noda-like virus 5 and might encode a B2 protein of 113 aa (Figs 1b and 2b).
RNA2 of MYPV consists of 1446 nt and is most closely related to the corresponding region of Hubei noda-like virus 11 (69.2 % identity and 12.1 % coverage), an unclassified Nodaviridae-like virus detected in spiders in China [23]. The sequence is predicted to encode a single ORF flanked by 5′ and 3′ UTRs of 71 and 142 nt, respectively. The putative protein consists of 410 residues and is most closely related to a corresponding translation product of Hubei noda-like virus 11 (44.5 % identity and 58.8 % similarity, with 89.6 % coverage). A domain homologous to the alphanoda-like capsid protein is present at residues 55 to 364.
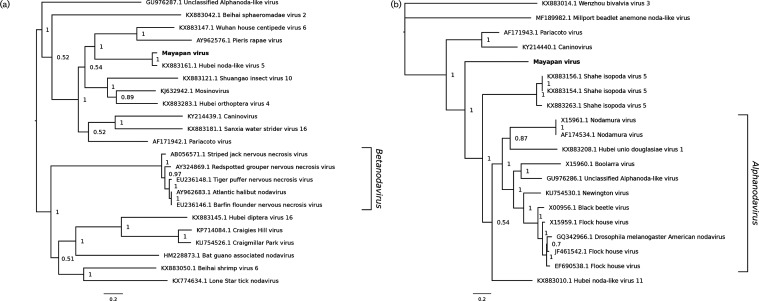
Phylogenetic trees were constructed with Bayesian methods using the deduced amino acid sequences of the RdRp and capsid protein of MYPV and the corresponding regions of related viruses (Fig. 4). In the analysis performed using RdRp sequences, it was demonstrated that MYPV is most closely related to Hubei noda-like virus 5. The posterior support for this topological arrangement is 1.0. MYPV and Hubei noda-like virus 5 belong to a clade that includes selected other unclassified Nodaviridae-like viruses. A second major clade consists of additional unclassified Nodaviridae-like viruses, along with viruses in the genus Betanodavirus. Notably, the RdRp of MYPV appeared to be more closely related to RdRps of betanodaviruses than to RdRps of alphanodaviruses such as Flock House, black beetle and Nodamura viruses. Alphanodaviruses were not included in the tree because their RdRps have limited (about 25 %) amino acid identity (with 70–80 % coverage) to that of MYPV. In contrast, in the analysis performed using capsid protein sequences, MYPV was positioned basally to viruses in the genus Alphanodavirus. The alphanodavirus capsid protein is structurally very different from the betanodavirus capsid protein [30], which is why betanodaviruses do not appear in the capsid protein tree.
Fig. 4.
Phylogenetic tree for Mayapan virus and selected related reference and non-reference sequences. Amino acid sequences of the ORF encoding the (a) RdRp and (b) capsid protein were aligned using muscle [60]. A maximum-likelihood phylogenetic tree was estimated using the Bayesian Markov chain Monte Carlo method implemented in MrBayes version 3.2.6 [61], sampling across the default set of fixed amino acid rate matrices with one million generations and discarding the first 25 % as burn-in. The figure was produced using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). The tree is midpoint-rooted and selected nodes are labelled with posterior probability values. GenBank accession numbers are indicated next to virus names.
Recognized viruses
CxFV was isolated from a pool of male Cx. quinquefasciatus collected in Tixkokob 2008 (Table 2). C6/36 cells were permissive to CxFV replication, but the virus was unable to replicate in any other cell line (Table 3). The genome of the CxFV isolate (designated CxFV-T1123) was fully sequenced (GenBank accession no. MH719098) and shown to consist of 10 837 nt. blast analysis revealed that CxFV-T1123 has the greatest (99.8 %) nucleotide identity with CxFV-Mex07, an isolate of CxFV recovered from Cx. quinquefasciatus in Tixkokob in 2007 [31]. HOUV was isolated from two pools of female mosquitoes collected in Merida (Table 2). One isolate was recovered from Cx. quinquefasciatus and the other from Ae. taeniorhynchus. HOUV replicated in C6/36 and CT cells, with CPE only being observed in C6/36 cells (Table 3). The entire genome of one isolate (designated HOUV-M742) was fully sequenced and shown to consist of 20 129 nt, excluding the polyadenylate tail (GenBank accession number MH719099). blast analysis revealed that HOUV-M742 has the greatest (99.2 %) nucleotide identity with V3872, an isolate of HOUV recovered from Cx. quinquefasciatus in Houston in 2004 [32].
Discussion
We report the isolation and identification of two novel viruses and two recognized viruses in mosquitoes from the Yucatan Peninsula of Mexico. Sequences corresponding to a putative novel rhabdovirus (Merida virus) were identified in mosquitoes in the same area in an earlier metagenomics study, but an isolate was not recovered by virus isolation in C6/36 cells, even though the cell line supports the replication of numerous mosquito-associated viruses [33]. Other research groups have also discovered novel virus genomes in mosquitoes by NGS but could not recover isolates from inoculated C6/36 cells [13, 34]. The inability to culture some mosquito-associated viruses in C6/36 cells could be because these viruses are species-specific or have only adapted to a few mosquito species and lack the capacity to replicate in Ae. albopictus cells. The experimental approach used in this study was designed to avoid the situation in which novel virus-like sequences were identified but isolates were not recovered. To this end, the metagenomics analysis was preceded by virus isolation experiments and therefore an isolate was already on hand for every virus that was identified. Our experimental approach also allowed for the detection of both cytopathic and non-cytopathic viruses because NGS was not restricted to cultures that exhibited CPE.
Our data demonstrate that UXMV should be classified in the taxon Negevirus. This newly proposed monophyletic taxon is composed of viruses detected exclusively in mosquitoes and sandflies, with most being discovered through the use of metagenomics [8, 12, 14–20]. Negeviruses are assumed to have vertebrate-incompetent replication phenotypes because they cannot replicate in suckling mice or any vertebrate cell lines that have been tested. The most comprehensive in vitro host range experiments were performed by Vasilakis and colleagues, who assessed the replicative abilities of six negeviruses in three vertebrate cell lines [baby hamster kidney (BHK-21), human embryonic kidney (HEK-293) and Vero cells] and five mosquito cell lines [Ae. albopictus (C6/36 and C7/10), Anopheles albimanus, An. gambiae and Cx. tarsalis cells], in addition to Drosophila melanogaster and Phlebotomus papatasi cells [12]. All viruses replicated in every mosquito cell line, but none replicated in any other cell line. We provide additional evidence that negeviruses lack the capacity to replicate in vertebrate cells because UXMV could not replicate in any of the six vertebrate cell lines tested. A notable difference between the two studies is that all previously tested negeviruses replicated in Anopheles cells, while UXMV could not, suggesting that the mosquito host range of UXMV is not as broad as that of other negeviruses.
Negeviruses have a ubiquitous geographical distribution, having been reported in every continent with the exception of Antarctica [8, 12, 14–21]. The first negevirus to be identified in Mexico was Piura virus after its isolation from Aedes, Culex, Mansonia, Psorophora and Wyeomyia spp. mosquitoes in the southern state of Chiapas [20]. Piura virus belongs to the Nelorpivirus clade. UXMV is the only Sandewavirus clade virus known to occur in Mexico.
MYPV is a Nodaviridae-like virus that was recovered from Ps. ferox. Two pools of female Ps. ferox were tested and both yielded isolates, indicating that MYPV commonly infects this species in the study area. However, the sample size was small and additional females need to be tested to accurately determine the prevalence of MYPV in Ps. ferox. Adult males and immatures should also be tested to provide some indication of whether MYPV is maintained in nature by vertical transmission, but none are available in our archived collections. All vertebrate cell lines were refractory to virus replication, indicating that MYPV has a vertebrate-incompetent replication phenotype. The ability of Aedes, but not Anopheles or Culex, cells to support MYPV replication indicates that the virus has a narrow mosquito host range. C6/36 cells have a dysfunctional RNAi response and it remains to be determined whether MYPV can establish an infection in Aedes cells that possess a functional RNAi pathway [35, 36]. Recent studies have also revealed that CT cells possess a dysfunctional RNAi response [37].
The isolation of CxFV from Cx. quinquefasciatus was not unexpected because we have previously reported a high prevalence of CxFV in mosquitoes of this species in the Yucatan Peninsula of Mexico [31, 38, 39]. CxFV is assumed to have an insect-restricted host range because it has been isolated exclusively from mosquitoes and cannot replicate in any vertebrate cell lines that have been tested [40–42]. Our study provides additional evidence that the virus is insect-specific, with several previously untested vertebrate cell lines being refractory to infection. The inability of CT cells to support CxFV infection was not unexpected because the cell line is persistently infected with Calbertado virus, a closely related insect-specific flavivirus, which is potentially suppressing the replication of CxFV by superinfection exclusion [43].
HOUV was originally isolated from Ae. albopictus and Cx. quinquefasciatus in the United States and was then isolated from Cx. quinquefasciatus in Xkaladzonot, a rural town in the Yucatan Peninsula of Mexico [32, 44]. HOUV is a variant of the species Alphamesonivirus-1, which also includes Cavally virus from Cote d’Ivoire, Nam Dinh virus from Vietnam and China, and Ngewotan virus from Indonesia and Australia [45–49]. Alphamesonivirus-1 belongs to the recently established family Mesoniviridae, which consists exclusively of viruses assumed to have insect-restricted host ranges [32, 50]. The findings from our in vitro host range experiments support this assumption and indicate that the mosquito host range of HOUV is restricted to Culicinae mosquitoes because it replicated in Aedes and Culex, but not Anopheles, cell cultures.
In summary, we report the isolation and sequence characterization of both novel and recognized RNA viruses from mosquitoes collected in the Yucatan Peninsula of Mexico. None of the viruses could replicate in any of the vertebrate cell lines tested, suggesting that they all possess insect-restricted host ranges. Despite their apparent vertebrate-incompetent replication phenotypes, further investigation into these viruses is warranted because coinfection experiments have revealed that insect-specific viruses can alter the replication and transmission of pathogenic viruses in mosquitoes [51–54]. Viral sequences were not recovered from some C6/36 cell cultures that exhibited CPE, indicating that the metagenomics analysis failed to detect some viruses. One explanation for this observation is that these viruses are highly divergent from all known viruses and their genomes have no significant identity with any sequences in the GenBank database.
Methods
Cell culture
The cell lines used in this study were as follows: Aedes albopictus (C6/36), African green monkey kidney (Vero), Anopheles gambiae (Sua 4.0), baby hamster kidney (BSR-T7/5), Culex tarsalis (CT), duck embryonic fibroblast (DEF), human epithelial (HeLa), murine microglia, rhesus macaque (LLC-MK2), Spodoptera frugiperda (Sf9) and Trichoplusia ni (High Five) cells. Cells were cultured in Dulbecco’s modified Eagle’s medium (all mammalian cells), Eagle’s minimum essential medium (DEF cells), Express Five medium (High Five cells), Liebovitz L15 medium (C6/36 cells), Schneider’s Drosophila medium (CT and Sua 4.0 cells) and SF-900 medium (Sf9 cells) (all cell culture media were obtained from Thermo Fisher Scientific, Massachusetts, USA). All media were supplemented with 10 % foetal bovine serum, 2 mM l-glutamine, 100 units ml−1 of penicillin and 100 µg ml−1 of streptomycin, with the exception of the Express Five and SF-900 media, which were serum-free. Vertebrate cells were cultured at 37 °C with 5 % CO2 and invertebrate cells were cultured at 28 °C.
Mosquitoes
Mosquitoes were collected at study sites in Merida, Tixkokob and Tzucacab in the Yucatan Peninsula of Mexico. Detailed descriptions of the study sites and the protocols used for the collection, identification and homogenization of mosquitoes have been provided elsewhere [31, 38, 55]. Briefly, collections were made in 2007 and 2008 using Mosquito Magnets Pro-Liberty (American Biophysics Corp., Rhode Island, USA) baited with propane and octenol. Mosquitoes were transported alive to the laboratory, frozen at −80 °C and sorted into pools of up to 50 according to species, sex, date of collection and study site. Mosquitoes were placed in polypropylene, round-bottom 5 ml tubes with 1.8 ml of CO2-independent cell culture medium (Thermo Fisher Scientific) and four 4.5mm diameter copper-clad steel beads (BB-calibre airgun shot) and then homogenized by vortexing for 30 s. The homogenates were centrifuged (12 000 g, 10 min, 4 °C) and supernatants were collected.
Virus isolation in C6/36 cell culture
An aliquot (100 µl) of each supernatant was added to 2 ml of Liebovitz L15 medium supplemented with 2 % foetal bovine serum, l-glutamine, penicillin and streptomycin. Samples were filtered and inoculated onto subconfluent monolayers of C6/36 cells in 25 cm2 flasks. Another 4 ml of the aforementioned medium was added to each flask and the cells were incubated at 28 °C and monitored regularly for CPE. Supernatants were collected when 40–60 % of the cell monolayer displayed CPE. If CPE was not observed, supernatants were collected at 7 days post-inoculation. A second blind passage was performed and then the cell monolayers and supernatants were harvested.
Library preparation
Total RNA was extracted from all second-passage C6/36 cell cultures using Trizol Reagent (Thermo Fisher Scientific), fragmented using RNase III (Thermo Fisher Scientific) and assessed for quality using an Agilent 2100 Bioanalyser (Agilent, CA, USA). Libraries were constructed using the Ion Total RNA-Seq Kit v2 (Thermo Fisher Scientific) and barcoded using the Ion Xpress RNA-Seq Barcode 1–16 kit (Thermo Fisher Scientific). Libraries were assessed for quality and analysed at the Genomic Technologies Facility at Iowa State University using an Ion Proton Sequencer (Thermo Fisher Scientific).
Bioinformatics
Ion-proton reads were analysed using the FastX Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) to remove bar-codes and low-quality ends (Phred quality score ≥33). Duplicate reads were identified and removed using Cdhit-454 (http://weizhongli-lab.org/cd-hit/). Host sequences were depleted by mapping the remaining reads to Ae. aegypti, Ae. albopictus, An. gambiae and/or Cx. quinquefasciatus genomes using Bowtie 2 [56]. Unmapped reads were analysed using the sortMeRNA program to remove ribosomal RNA-related reads [57]. The remaining reads were subjected to de novo SPAdes assembly (version 3.5.0) [58]. Contigs were aligned by blastn, blastx and tblastx to the NCBI nucleotide database (downloaded August 2017) using an e-value of <10−5. Unaligned contigs were translated into all six reading frames and aligned to the HMM-FRAME, pfam, CDD and TIGRFAM databases (downloaded August 2017). The data were transformed by Python programming (https://www.python.org/).
RT-PCR and Sanger sequencing
RT-PCR and Sanger sequencing were performed to verify the virus sequences identified by NGS and to close gaps between contigs. RT-PCR was also used to test cell cultures for the presence of viral RNA in the host range experiments. Complementary DNAs were generated using Superscript III reverse transcriptase (Thermo Fisher Scientific) and PCRs were performed using high-fidelity Taq polymerase (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions. Primers were designed from the sequences generated by NGS and are available upon request. RT-PCR products were purified using the PureLink gel extraction kit (Thermo Fisher Scientific) and sequenced using a 3730×1 DNA sequencer (Applied Biosystems, CA, USA).
5′ and 3′ rapid amplification of cDNA ends
The 5′ and 3′ ends of virus genomes were identified using 5′ and 3′ rapid amplification of cDNA ends, respectively. Briefly, a DNA adaptor (5′-rApp/TGGAATTCTCGGGTGCCAAGGT/ddC-3′) was ligated to the viral genomic and anti-genomic RNAs using T4 RNA ligase (New England BioLabs, MA, USA). Complementary cDNAs were created using SuperScript III (Thermo Fisher Scientific) and an adapter-specific primer. PCRs were performed using adapter- and gene-specific primers, and amplicons were purified and subjected to Sanger sequencing.
Sequence alignments and identification of conserved domains
The nucleotide and deduced amino acid sequences of each virus were compared to other sequences in the GenBank database by the application of blastn and blastp, respectively [59]. Percentage amino acid identities and similarities were calculated using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). Conserved domains were identified using blastp, InterProScan (http://www.ebi.ac.uk/interpro/search/sequence-search) and HHpred (PMID 15980461). Potential N-linked glycosylation sites were identified using the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/) with the consensus sequence defined as Asn-X-Ser/Thr (where X is not Pro) in the context of specific surrounding sequences.
In vitro host range experiments
The in vitro host ranges of all novel and recognized viruses were assessed using 11 cell lines. Briefly, cell monolayers approaching confluency in 25 cm2 flasks were inoculated with 50 µl of virus-containing supernatant and incubated for 7 days or until 40–60 % of the cell monolayer displayed CPE. A total of five passages were performed, with supernatants being collected after each passage and tested by RT-PCR for viral RNA. A cell line was considered to support virus replication if RT-PCR products were detected after every passage.
Funding information
Mosquito collections were made possible by grant 5R21AI067281 from the National Institutes of Health. The metagenomics analysis was supported by an intramural grant from Iowa State University. Host range experiments were supported by grant R01AI114720 from the National Institutes of Health. A. E. F. and C. T. are supported by Wellcome Trust grant 106207 and European Research Council grant 646891 to A. E. F.
Acknowledgements
The authors thank Kristina Larsen and Sean Johnston for technical assistance.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
No human subjects or vertebrate animals were used in this study
Footnotes
Abbreviations: 2′OMTase, 2′-O-methyltransferase; CPE, cytopathic effect; CT, Culex tarsalis; CxFV, Culex flavivirus; DEZV, Dezidougou virus; Hel, helicase; HOUV, Houston virus; MTase, methyltransferase; MYPV, Mayapan virus; NGS, next-generation sequencing; ORF, open reading frame; RdRp, RNA-dependent RNA polymerase; RNAi, RNA interference; RT-PCR, reverse transcription/polymerase chain reaction; UTR, untranslated region; UXMV, Uxmal virus; WALV, Wallerfield virus.
References
- 1.Mokili JL, Rohwer F, Dutilh BE. Metagenomics and future perspectives in virus discovery. Curr Opin Virol. 2012;2:63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lipkin WI, Firth C. Viral surveillance and discovery. Curr Opin Virol. 2013;3:199–204. doi: 10.1016/j.coviro.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi M, Zhang YZ, Holmes EC. Meta-transcriptomics and the evolutionary biology of RNA viruses. Virus Res. 2018;243:83–90. doi: 10.1016/j.virusres.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roossinck MJ, Martin DP, Roumagnac P. Plant virus metagenomics: advances in virus discovery. Phytopathology. 2015;105:716–727. doi: 10.1094/PHYTO-12-14-0356-RVW. [DOI] [PubMed] [Google Scholar]
- 5.Liu S, Vijayendran D, Bonning BC. Next generation sequencing technologies for insect virus discovery. Viruses. 2011;3:1849–1869. doi: 10.3390/v3101849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brault AC, Blitvich BJ. Continued need for comprehensive genetic and phenotypic characterization of viruses: benefits of complementing sequence analyses with functional determinations. Am J Trop Med Hyg. 2018;98:1213. doi: 10.4269/ajtmh.18-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simmonds P, Adams MJ, Benkő M, Breitbart M, Brister JR, et al. Consensus statement: virus taxonomy in the age of metagenomics. Nat Rev Microbiol. 2017;15:161–168. doi: 10.1038/nrmicro.2016.177. [DOI] [PubMed] [Google Scholar]
- 8.Auguste AJ, Carrington CV, Forrester NL, Popov VL, Guzman H, et al. Characterization of a novel Negevirus and a novel Bunyavirus isolated from Culex (Culex) declarator mosquitoes in Trinidad. J Gen Virol. 2014;95:481–485. doi: 10.1099/vir.0.058412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cholleti H, Hayer J, Abilio AP, Mulandane FC, Verner-Carlsson J, et al. Discovery of novel viruses in mosquitoes from the Zambezi Valley of Mozambique. PLoS One. 2016;11:e0162751. doi: 10.1371/journal.pone.0162751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffey LL, Page BL, Greninger AL, Herring BL, Russell RC, et al. Enhanced arbovirus surveillance with deep sequencing: identification of novel rhabdoviruses and bunyaviruses in Australian mosquitoes. Virology. 2014;448:146–158. doi: 10.1016/j.virol.2013.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fauver JR, Grubaugh ND, Krajacich BJ, Weger-Lucarelli J, Lakin SM, et al. West African Anopheles gambiae mosquitoes harbor a taxonomically diverse virome including new insect-specific flaviviruses, mononegaviruses, and totiviruses. Virology. 2016;498:288–299. doi: 10.1016/j.virol.2016.07.031. [DOI] [PubMed] [Google Scholar]
- 12.Vasilakis N, Forrester NL, Palacios G, Nasar F, Savji N, et al. Negevirus: a proposed new taxon of insect-specific viruses with wide geographic distribution. J Virol. 2013;87:2475–2488. doi: 10.1128/JVI.00776-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colmant AMG, Etebari K, Webb CE, Ritchie SA, Jansen CC, et al. Discovery of new orbiviruses and totivirus from Anopheles mosquitoes in Eastern Australia. Arch Virol. 2017;162:3529–3534. doi: 10.1007/s00705-017-3515-x. [DOI] [PubMed] [Google Scholar]
- 14.Carapeta S, do Bem B, McGuinness J, Esteves A, Abecasis A, et al. Negeviruses found in multiple species of mosquitoes from southern Portugal: isolation, genetic diversity, and replication in insect cell culture. Virology. 2015;483:318–328. doi: 10.1016/j.virol.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 15.Fujita R, Kuwata R, Kobayashi D, Bertuso AG, Isawa H, et al. Bustos virus, a new member of the negevirus group isolated from a Mansonia mosquito in the Philippines. Arch Virol. 2017;162:79–88. doi: 10.1007/s00705-016-3068-4. [DOI] [PubMed] [Google Scholar]
- 16.Kawakami K, Kurnia YW, Fujita R, Ito T, Isawa H, et al. Characterization of a novel negevirus isolated from Aedes larvae collected in a subarctic region of Japan. Arch Virol. 2016;161:801–809. doi: 10.1007/s00705-015-2711-9. [DOI] [PubMed] [Google Scholar]
- 17.O'Brien CA, McLean BJ, Colmant AMG, Harrison JJ, Hall-Mendelin S, et al. Discovery and characterisation of Castlerea virus, a new species of Negevirus isolated in Australia. Evol Bioinform Online. 2017;13:117693431769126. doi: 10.1177/1176934317691269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira DD, Cook S, Lopes Â, de Matos AP, Esteves A, et al. Characterization of an insect-specific flavivirus (OCFVPT) co-isolated from Ochlerotatus caspius collected in southern Portugal along with a putative new Negev-like virus. Virus Genes. 2013;47:532–545. doi: 10.1007/s11262-013-0960-9. [DOI] [PubMed] [Google Scholar]
- 19.Nunes MRT, Contreras-Gutierrez MA, Guzman H, Martins LC, Barbirato MF, et al. Genetic characterization, molecular epidemiology, and phylogenetic relationships of insect-specific viruses in the taxon Negevirus. Virology. 2017;504:152–167. doi: 10.1016/j.virol.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kallies R, Kopp A, Zirkel F, Estrada A, Gillespie TR, et al. Genetic characterization of goutanap virus, a novel virus related to negeviruses, cileviruses and higreviruses. Viruses. 2014;6:4346–4357. doi: 10.3390/v6114346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nabeshima T, Inoue S, Okamoto K, Posadas-Herrera G, Yu F, et al. Tanay virus, a new species of virus isolated from mosquitoes in the Philippines. J Gen Virol. 2014;95:1390–1395. doi: 10.1099/vir.0.061887-0. [DOI] [PubMed] [Google Scholar]
- 22.Roy A, Hartung JS, Schneider WL, Shao J, Leon G, et al. Role bending: complex relationships between viruses, hosts, and vectors related to Citrus leprosis, an emerging disease. Phytopathology. 2015;105:1013–1025. doi: 10.1094/PHYTO-12-14-0375-FI. [DOI] [PubMed] [Google Scholar]
- 23.Shi M, Lin XD, Tian JH, Chen LJ, Chen X, et al. Redefining the invertebrate RNA virosphere. Nature. 2016:539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 24.Yong CY, Yeap SK, Omar AR, Tan WS. Advances in the study of nodavirus. PeerJ. 2017;5:e3841. doi: 10.7717/peerj.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naveenkumar S, Shekar M, Karunasagar I, Karunasagar I. Genetic analysis of RNA1 and RNA2 of Macrobrachium rosenbergii nodavirus (MrNV) isolated from India. Virus Res. 2013;173:377–385. doi: 10.1016/j.virusres.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Kuchibhatla DB, Sherman WA, Chung BY, Cook S, Schneider G, et al. Powerful sequence similarity search methods and in-depth manual analyses can identify remote homologs in many apparently "orphan" viral proteins. J Virol. 2014;88:10–20. doi: 10.1128/JVI.02595-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.ICTV . Family - Nodaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. London, UK: Elsevier Academic Press; 2012. pp. 1061–1067. (editors) [Google Scholar]
- 28.Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 29.Schuster S, Zirkel F, Kurth A, van Cleef KW, Drosten C, et al. A unique nodavirus with novel features: mosinovirus expresses two subgenomic RNAs, a capsid gene of unknown origin, and a suppressor of the antiviral RNA interference pathway. J Virol. 2014;88:13447–13459. doi: 10.1128/JVI.02144-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen NC, Yoshimura M, Guan HH, Wang TY, Misumi Y, et al. Crystal structures of a piscine betanodavirus: mechanisms of capsid assembly and viral infection. PLoS Pathog. 2015;11:e1005203. doi: 10.1371/journal.ppat.1005203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farfan-Ale JA, Loroño-Pino MA, Garcia-Rejon JE, Hovav E, Powers AM, et al. Detection of RNA from a novel West Nile-like virus and high prevalence of an insect-specific flavivirus in mosquitoes in the Yucatan Peninsula of Mexico. Am J Trop Med Hyg. 2009;80:85–95. doi: 10.4269/ajtmh.2009.80.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasilakis N, Guzman H, Firth C, Forrester NL, Widen SG, et al. Mesoniviruses are mosquito-specific viruses with extensive geographic distribution and host range. Virol J. 2014;11:97. doi: 10.1186/1743-422X-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charles J, Firth AE, Loroño-Pino MA, Garcia-Rejon JE, Farfan-Ale JA, et al. Merida virus, a putative novel rhabdovirus discovered in Culex and Ochlerotatus spp. mosquitoes in the Yucatan Peninsula of Mexico. J Gen Virol. 2016;97:977–987. doi: 10.1099/jgv.0.000424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shahhosseini N, Lühken R, Jöst H, Jansen S, Börstler J, et al. Detection and characterization of a novel rhabdovirus in Aedes cantans mosquitoes and evidence for a mosquito-associated new genus in the family Rhabdoviridae. Infect Genet Evol. 2017;55:260–268. doi: 10.1016/j.meegid.2017.09.026. [DOI] [PubMed] [Google Scholar]
- 35.Brackney DE, Scott JC, Sagawa F, Woodward JE, Miller NA, et al. C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl Trop Dis. 2010;4:e856. doi: 10.1371/journal.pntd.0000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott JC, Brackney DE, Campbell CL, Bondu-Hawkins V, Hjelle B, et al. Comparison of dengue virus type 2-specific small RNAs from RNA interference-competent and -incompetent mosquito cells. PLoS Negl Trop Dis. 2010;4:e848. doi: 10.1371/journal.pntd.0000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Göertz GP, Fros JJ, Miesen P, Vogels CB, van der Bent ML, et al. Noncoding subgenomic flavivirus RNA is processed by the mosquito RNA interference machinery and determines West Nile virus transmission by Culex pipiens mosquitoes. J Virol. 2016;90:10145–10159. doi: 10.1128/JVI.00930-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farfan-Ale JA, Loroño-Pino MA, Garcia-Rejon JE, Soto V, Lin M, et al. Detection of flaviviruses and orthobunyaviruses in mosquitoes in the Yucatan Peninsula of Mexico in 2008. Vector Borne Zoonotic Dis. 2010;10:777–783. doi: 10.1089/vbz.2009.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saiyasombat R, Dorman KS, Garcia-Rejon JE, Loroño-Pino MA, Farfan-Ale JA, et al. Isolation and sequence analysis of Culex flavivirus from Culex interrogator and Culex quinquefasciatus in the Yucatan Peninsula of Mexico. Arch Virol. 2010;155:983–986. doi: 10.1007/s00705-010-0665-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blitvich BJ, Firth AE. Insect-specific flaviviruses: a systematic review of their discovery, host range, mode of transmission, superinfection exclusion potential and genomic organization. Viruses. 2015;7:1927–1959. doi: 10.3390/v7041927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoshino K, Isawa H, Tsuda Y, Yano K, Sasaki T, et al. Genetic characterization of a new insect flavivirus isolated from Culex pipiens mosquito in Japan. Virology. 2007;359:405–414. doi: 10.1016/j.virol.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 42.Bolling BG, Eisen L, Moore CG, Blair CD. Insect-specific flaviviruses from Culex mosquitoes in Colorado, with evidence of vertical transmission. Am J Trop Med Hyg. 2011;85:169–177. doi: 10.4269/ajtmh.2011.10-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyler S, Bolling BG, Blair CD, Brault AC, Pabbaraju K, et al. Distribution and phylogenetic comparisons of a novel mosquito flavivirus sequence present in Culex tarsalis mosquitoes from western Canada with viruses isolated in California and Colorado. Am J Trop Med Hyg. 2011;85:162–168. doi: 10.4269/ajtmh.2011.10-0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cigarroa-Toledo N, Baak-Baak CM, Cetina-Trejo RC, Cordova-Fletes C, Martinez-Nuñez MA, et al. Complete genome sequence of houston virus, a newly discovered mosquito-specific virus isolated from Culex quinquefasciatus in Mexico. Microbiol Resour Announc. 2018;7 doi: 10.1128/MRA.00808-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi M, Neville P, Nicholson J, Eden JS, Imrie A, et al. High-resolution metatranscriptomics reveals the ecological dynamics of mosquito-associated RNA viruses in Western Australia. J Virol. 2017;91:e00680-17. doi: 10.1128/JVI.00680-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nga PT, Parquet MC, Lauber C, Parida M, Nabeshima T, et al. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog. 2011;7:e1002215. doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thuy NT, Huy TQ, Nga PT, Morita K, Dunia I, et al. A new nidovirus (NamDinh virus NDiV): Its ultrastructural characterization in the C6/36 mosquito cell line. Virology. 2013;444:337–342. doi: 10.1016/j.virol.2013.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhuang L, Zhang Z, An X, Fan H, Ma M, et al. An efficient strategy of screening for pathogens in wild-caught ticks and mosquitoes by reusing small RNA deep sequencing data. PLoS One. 2014;9:e90831. doi: 10.1371/journal.pone.0090831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zirkel F, Kurth A, Quan PL, Briese T, Ellerbrok H, et al. An insect nidovirus emerging from a primary tropical rainforest. MBio. 2011;2:e00077. doi: 10.1128/mBio.00077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lauber C, Ziebuhr J, Junglen S, Drosten C, Zirkel F, et al. Mesoniviridae: a proposed new family in the order Nidovirales formed by a single species of mosquito-borne viruses. Arch Virol. 2012;157:1623–1628. doi: 10.1007/s00705-012-1295-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hall-Mendelin S, McLean BJ, Bielefeldt-Ohmann H, Hobson-Peters J, Hall RA, et al. The insect-specific Palm Creek virus modulates West Nile virus infection in and transmission by Australian mosquitoes. Parasit Vectors. 2016;9:414. doi: 10.1186/s13071-016-1683-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goenaga S, Kenney JL, Duggal NK, Delorey M, Ebel GD, et al. Potential for co-infection of a mosquito-specific flavivirus, nhumirim virus, to block West Nile virus transmission in mosquitoes. Viruses. 2015;7:5801–5812. doi: 10.3390/v7112911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nasar F, Erasmus JH, Haddow AD, Tesh RB, Weaver SC. Eilat virus induces both homologous and heterologous interference. Virology. 2015;484:51–58. doi: 10.1016/j.virol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kent RJ, Crabtree MB, Miller BR. Transmission of West Nile virus by Culex quinquefasciatus say infected with Culex Flavivirus Izabal. PLoS Negl Trop Dis. 2010;4:e671. doi: 10.1371/journal.pntd.0000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Darsie RF. A survey and bibliography of the mosquito fauna of Mexico (Diptera: Culicidae) J Am Mosq Control Assoc. 1996;12:298–306. [PubMed] [Google Scholar]
- 56.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 58.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 60.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]