ABSTRACT
Background:
Overexpression of human epidermal growth factor receptor 2 (HER2) plays an important role in the development and progression in a variety of cancers and it is a novel therapeutic target for breast cancer and ovarian cancer. Euclea crispa (E. crispa) is a South African medicinal plant in the family Ebenaceae used in the management of different human diseases and disorders.
Aims:
The aim of this study was to evaluate the potential inhibitors against HER2 from hexane extract of E. crispa leaves.
Materials and Methods:
Chemical fingerprinting method was used to identify the presence of natural compounds from the extract whereas their inhibitory activities were analyzed by molecular docking analysis against HER2. Absorption, distribution, metabolism, and excretion (ADME) properties also predicted to establish the pharmacokinetics and pharmacodynamics profiles of the selected compounds.
Results:
The molecular docking analysis expressed that phenyl glucuronide, hydrocortisone acetate, and 6-(4,6-dioxo-1,4,5,6-tetrahydropyrimidin-2-yl-amino)hexanoic acid trifluoroacetate possess good inhibitory activities with good glide score of −6.63, −5.41, and −5.40 and glide energy of −35.03, −42.51, and −31.38 kcal/mol, respectively when compared with standard Food and Drug Administration–approved drug and other compounds. All the screened compounds were within the acceptable and permissible limits of ADME properties.
Conclusion:
Thus, from this study it can be concluded that, these screened natural compounds from E. crispa leaves may serve as potential inhibitors for HER2 and they might lead to development of new therapeutic agents against cancer and its associated complications.
Keywords: ADME properties, docking analysis, E. crispa, GC–MS analysis, HER2
INTRODUCTION
The ErbB family of receptor tyrosine kinases has played a vital role in the tumorigenesis of many types of solid tumors. Human epidermal growth factor receptor 2 (HER2), also known as CD340 (cluster of differentiation 340), proto-oncogene Neu, ErbB2 (rodent), or ErbB2 (human) is a protein in human encoded by the ErbB2 gene and it is a member of the human epidermal growth factor receptor (HER/EGFR/ERBB) family.[1] Overexpression of this oncogene has been shown to play an important role in the development and progression of certain types of breast cancer and ovarian cancer. HER2 initiates intracellular signaling pathways involved in cell proliferation, differentiation, migration, and apoptosis.[2] HER2 overexpression/amplification has been reported in ovarian cancer, but the exact percentage of HER2-positive tumors varies widely in the literature (between 8% and 66%).[3] Thus, the HER2 receptor is a novel and important therapeutic target in a variety of cancers including breast and ovarian cancer.[4]
Medicinal plants possessing a number of bioactive compounds are widely used in human disease management system.[5] A large proportion of the world population depends on the traditional medicine partly because it is comparatively effective with less/no side effects when compared with orthodox medicines and/or the bioactive compounds from the medicinal plants play a central role in the development of novel drug for the treatment and prevention of human diseases.[6]
Euclea crispa (E. crispa) is an afro-tropical plant species, commonly known as blue guarri (Eng.); bloughwarrie (Afr.); motlhaletsogane (Setswana); and iDungamuzi and umGwali (isiZulu). It is a hardy evergreen plant that usually forms a dense stand of shrubs, or grows to tree size. E. crispa is widespread and commonly found in the interior regions of Southern Africa.[7,8] This plant is used traditionally against wide range of ailments such as gonorrhea, leprosy, scabies, diarrhea, and wound infections. The leaf extracts of E. crispa also possess growth-inhibiting potential against both bacteria and fungi.[9] Hot water extracts of the root of this plant is used as antitussive and the infusion from the roots is used in the treatment of leprosy. Antibacterial properties of the crude as well as a semi-purified ethyl acetate fractions prepared from dried and ground E. crispa leaves were previously reported.[10] Therefore, the main aim of this study was to analyze the presence of natural compounds from hexane extract of E. crispa and to validate their inhibitory activities against HER2 protein, because this aspect of medicinal potential of this valuable plant has not been reported in any scientific literature.
MATERIALS AND METHODS
Plant collection
The fresh E. crispa leaves were collected from the area of Phuthaditjhaba, Qwaqwa, and authenticated by Prof. AOT Ashafa from University of the Free State, Qwaqwa Campus, South Africa, during the month of April 2017. The plant sample was deposited at University of the Free State herbarium following herbarium collection of Taylor and Van Wyk (1994) with reference number 6404000-400. The collected plant leaves were washed under running tap water to remove contaminants and foliar debris, air-dried, powdered, and stored in airtight container at 4°C for further studies.
Preparation of extract
Using exhaustive extraction procedure, the powdered plant material (100g) was soaked with hexane (500mL) and kept on the shaker (Labcon Platform Shaker, Johannesburg, South Africa) at 110rpm speed for 72 hours at room temperature. The extract was collected, filtered using Whatman No 1 filter paper, and concentrated to dryness using rotary evaporator set at 40°C. The dried extracts were stored at 4°C until further use.
Gas chromatography–mass spectrometry analysis
Gas chromatography–mass spectrometry (GC–MS) analysis of hexane extract of E. crispa was performed using Agilent Technologies 7890 A (DB-35ms Capillary Standard nonpolar column with dimensions of 30mm × 0.25mm ID × 0.25 μm film). Helium was used as carrier gas at low down of 1.0mL/min. The injector was functioned at 250°C and oven heat was maintained as follows: 60°C for 15 minutes, then slowly amplified to 280°C at 3 minutes. MS were taken at 70eV; a scan distance of 0.5 seconds and fragments starts from 50 to 650Da. Total GC operation period was 25 minutes. The comparative percentage amount of every module was calculated by evaluating its average peak area to the total areas; software adopted to handle mass spectra and chromatograms was Turbo Mass. The percentage composition of compounds in the plant extract was calculated. Interpretation of GC–MS was performed by the National Institute Standard and Technology (NIST) database and Wiley libraries in addition to comparison of their retention indices.[11]
Computational molecular analysis
Ligand selection preparation
The identified natural compounds selected from GC–MS analyzed hexane extract of E. crispa and Food and Drug Administration (FDA)–approved drug of cyclophosphamide (standard drug for comparison) were prepared using the LigPrep (Schrödinger, LLC, New York, NY, 2017) for molecular docking analysis. The structure of each ligands was optimized by means of OPLS_2005 force field using a default setting.
Preparation of protein structure
The HER2 protein three-dimensional structure was retrieved from the Protein Data Bank (PDB ID: 3BE1) and it was prepared by protein preparation wizards (standard methods) that are available in grid-based ligand docking with energetics (Protein Preparation Wizard; Epik, Schrödinger, LLC, New York, NY, 2017). Protein was optimized using sample water orientation and minimized by using RMSD 0.30 Å and OPLS_2005 force field.
Active site prediction
The HER2 protein active site and functional residues were identified and characterized by SiteMap (Schrödinger, LLC, New York, NY, 2017). SiteMap calculation begins with an initial search step that identifies or characterizes—through the use of grid points—one or more regions on the protein surface that may be suitable for binding ligands to the receptor. Contour maps were then generated; hydrogen binding possibilities, hydrophilic maps, and produced hydrophobic may guide the protein–ligand docking analysis.
Molecular docking analysis
All docking analysis were performed between HER2 protein and identified natural compounds (from hexane extract of E. crispa and standard drug) using the standard precision, which is the standard mode of Glide (Glide, Schrödinger, LLC, New York, NY, 2017), a grid-based ligand docking with energetic. All selected natural compounds were docked into the binding site of HER2 using Glide module. The scaling Vander Waals radii were 1.0 in the receptor grid generation. Grid was prepared with the bounding box set on 20 Å. The coordinates of this enclosing box with the help of the active site residues to be set as default. The force field used for the docking protocol was OPLS_2005. The docked lowest-energy complexes were found in the majority of similar docking conformations.
Absorption, Distribution, Metabolism, and Excretion properties prediction
The selected HER2 ligands were checked for their absorption, distribution, metabolism, and excretion (ADME) properties using QikProp (Schrödinger, LLC, New York, NY, 2017), which helps to analyze the pharmacokinetic and pharmacodynamics of the ligands by accessing the drug-like properties. The significant ADME properties such as molecular weight, H-bond donor, H-bond acceptor, and log P (O/W) were predicted.
RESULTS
On the basis of NIST library, GC–MS analysis [Figure 1] predicted 29 phytochemicals from hexane extract of E. crispa, and their retention time, percentage composition, molecular formula, molecular mass, and peak area were given in Table 1. The predicted compounds are namely tetracosane (14.98%); dodecane (10.76%); 2-ethyl-1-decanol (8.00%); tridecane (7.53%); 4,5,6,8-PTetramethoxy-2,3-dihydroindeno[1,2,3-ij]isoquinolin-9-ol (6.99%); diphenylvinylphosphine (6.38%); squalene (5.85%); triacontane (5.27%); 2,6-dimethylheptadecane (5.02%); docosane (3.68%); tetradecane (3.59%); 1-hepten-3-ol (2.63%); orthotolidine (2.31%); phenyl glucuronide (2.25%); 5-tridecylbenzene-1,3-diol (1.90%); benzoic acid 3-methyl-4-(1,3,3,3-tetrafluoro-2-methoxycarb onyl-propenylsulfanyl)-phenyl ester (1.76%); pentadecane (1.68%); 6-(4,6-dioxo-1,4,5,6-tetrahydropyrimidin-2-YL-amino)hexanoic acid trifluoroacetate (1.22%); benzhydrazide,3-chloro-N2-[3-(4-methoxyphenyl)-1-methyl-3-oxopropenyl- (1.09%); hydrocortisone acetate (0.95%); triacontane (0.95%); dioctyl phthalate (0.78%); phytol (0.66%); shogaol (0.49%); 1,1,3,3,5,5,7,7,9,9,11,11,13,13,15, 15-hexadecamethyloctasiloxane (0.47%); docosane (0.44%); tetradecamethyl hexasiloxane (0.42%); 3-(4-chlorophenyl)-5-styryl[1,2,4]oxadiazole (0.32%); and ephedrine (0.32%).
Figure 1.
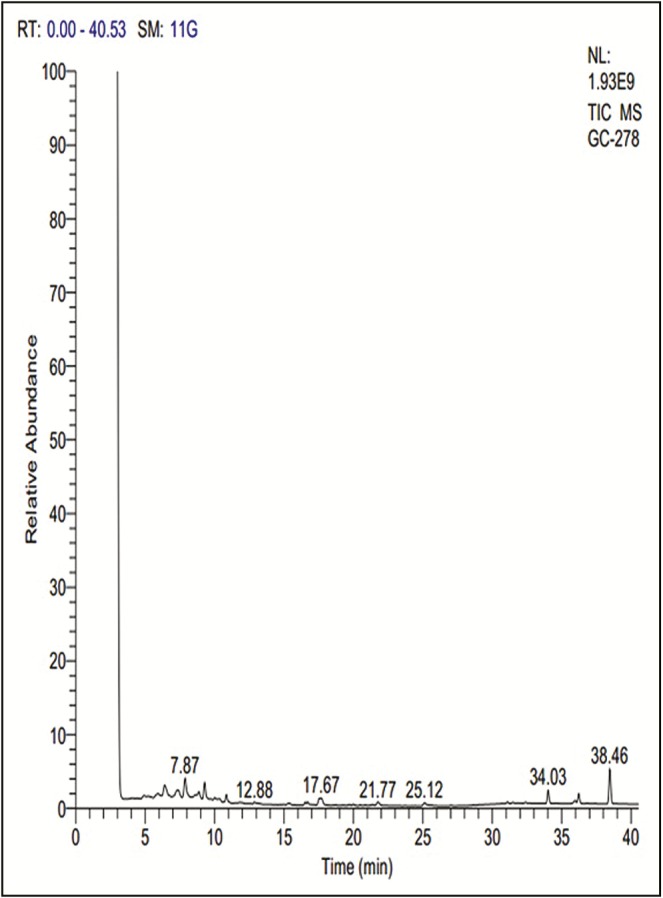
Gas chromatography–mass spectrometry analysis chromatogram of hexane extract of Euclea crispa leaves
Table 1.
Reported phytocompounds from GC–MS analysis result on hexane extract of E. crispa leaves
| S. No. | RT | Compounds | Molecular formula | Molecular weight | Peak area (%) |
|---|---|---|---|---|---|
| 1 | 4.11 | Hydrocortisone acetate | C23H32O6 | 404 | 0.95 |
| 2 | 4.92 | Docosane | C22H46 | 310 | 3.68 |
| 3 | 5.89 | 1-Hepten-3-ol | C7H14O | 114 | 2.63 |
| 4 | 6.40 | 2-Ethyl-1-decanol | C12H26O | 186 | 8.00 |
| 5 | 7.37 | 4,5,6,8-PTetramethoxy-2,3-dihydroindeno[1,2,3-ij] isoquinolin-9-ol | C19H19NO5 | 341 | 6.99 |
| 6 | 7.87 | Dodecane | C12H26 | 170 | 10.76 |
| 7 | 8.87 | 2,6-Dimethylheptadecane | C19H40 | 268 | 5.02 |
| 8 | 9.28 | Tridecane | C13H28 | 184 | 7.53 |
| 9 | 10.02 | Tetradecane | C14H30 | 198 | 3.59 |
| 10 | 10.38 | Benzoic acid 3-methyl-4-(1,3,3,3-tetrafluoro-2-methoxycarbonyl- propenylsulfanyl)-phenyl ester |
C19H14F4O4S | 414 | 1.76 |
| 11 | 11.83 | 6-(4,6-DIOXO-1,4,5,6-tetrahydropyrimidin-2-yl-amino)hexanoic acid trifluoroacetate | C12H16F3N3O6 | 355 | 1.22 |
| 12 | 12.88 | Pentadecane | C15H32 | 212 | 1.68 |
| 13 | 14.98 | 3-(4-Chlorophenyl)-5-styryl[1,2,4]oxadiazole | C16H11ClN2O | 182 | 0.32 |
| 14 | 15.36 | Benzhydrazide,3-chloro-N2-[3-(4-methoxyphenyl)-1-methyl-3- oxopropenyl- | C18H17ClN2O3 | 344 | 1.09 |
| 15 | 16.69 | Orthotolidine | C14H16N2 | 212 | 2.31 |
| 16 | 17.67 | Diphenylvinylphosphine | C14H13P | 212 | 6.38 |
| 17 | 19.69 | 1,1,3,3,5,5,7,7,9,9,11,11,13,13,15,15-hexadecamethyloctasiloxane | C16H50O7Si8 | 579 | 0.47 |
| 18 | 19.98 | Phytol | C20H40O | 296 | 0.66 |
| 19 | 21.39 | Shogaol | C17H24O3 | 276 | 0.49 |
| 20 | 21.77 | Phenyl glucuronide | C12H14O7 | 270 | 2.25 |
| 21 | 25.12 | 5-tridecylbenzene-1,3-diol | C19H32O2 | 292 | 1.90 |
| 22 | 29.43 | Ephedrine | C10H15NO | 165 | 0.32 |
| 23 | 30.70 | Tetradecamethyl hexasiloxane | C14H42O5Si6 | 458 | 0.42 |
| 24 | 31.10 | Triacontane | C30H62 | 422 | 0.95 |
| 25 | 31.48 | Dioctyl phthalate | C24H38O4 | 390 | 0.78 |
| 26 | 32.46 | Docosane | C22H46 | 310 | 0.44 |
| 27 | 34.03 | Triacontane | C30H62 | 422 | 5.27 |
| 28 | 36.22 | Squalene | C30H50 | 410 | 5.85 |
| 29 | 38.46 | Tetracosane | C24H50 | 338 | 14.98 |
RT = Retention time
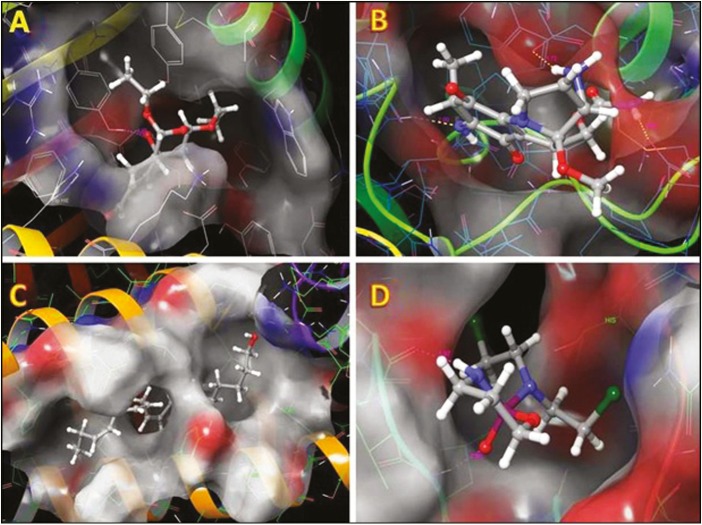
Significant binding affinities of the predicted phytochemicals and FDA-approved drug of cyclophosphamide against HER2 protein were seen using molecular docking analysis, and their binding properties were highlighted in Table 2. Phenyl glucuronide [Figure 2A], hydrocortisone acetate [Figure 2B], and 6-(4,6-dioxo-1,4,5,6-tetrahydropyrimidin-2-yl-amino)hexanoic acid trifluoroacetate [Figure 2C] possessed optimum glide score of −6.63, −5.41, and −5.40 and glide energy of −35.03, −42.51, and −31.38 kcal/mol, respectively when compared with standard FDA-approved drug [Figure 2D] and other compounds. These screened compounds were strongly bound with hydrophobic region of HER2 protein as predicted.
Table 2.
Glide scores and glide energies of HER2/ligand complexes
| S. No. | Compounds | Docking results |
|
|---|---|---|---|
| Glide score | Glide energy (Kcal/mol) | ||
| 1 | Hydrocortisone acetate | −5.417 | −42.515 |
| 2 | Docosane | Not docked | |
| 3 | 1-Hepten-3-ol | −2.174 | −17.726 |
| 4 | 2-Ethyl-1-decanol | −1.768 | −22.353 |
| 5 | 4,5,6,8-PTetramethoxy-2,3-dihydroindeno[1,2,3-ij]isoquinolin-9-ol | −3.739 | −35.888 |
| 6 | Dodecane | 1.051 | −19.325 |
| 7 | 2,6-Dimethylheptadecane | −0.451 | −26.551 |
| 8 | Tridecane | −0.118 | −21.009 |
| 9 | Tetradecane | 0.727 | −20.445 |
| 10 | SCHEMBL15979821 | −3.801 | −39.263 |
| 11 | 6-(4,6-dioxo-1,4,5,6-tetrahydropyrimidin-2-yl-amino)hexanoic acid trifluoroacetate | −5.401 | −31.389 |
| 12 | Pentadecane | 0.316 | −24.655 |
| 13 | 3-(4-Chlorophenyl)-5-styryl[1,2,4]oxadiazole | −3.035 | −36.153 |
| 14 | Benzhydrazide,3-chloro-N2-[3-(4-methoxyphenyl)-1-methyl-3- oxopropenyl- | Not docked | |
| 15 | Orthotolidine | −3.867 | −26.139 |
| 16 | Diphenylvinylphosphine | −1.51 | −22.8 |
| 17 | 1,1,3,3,5,5,7,7,9,9,11,11,13,13,15,15-hexadecamethyloctasiloxane | −1.475 | −38.258 |
| 18 | Phytol | −3.155 | −31.359 |
| 19 | Shogaol | −3.433 | −35.929 |
| 20 | Phenyl glucuronide | −6.635 | −35.039 |
| 21 | 5-tridecylbenzene-1,3-diol | −2.029 | −31.71 |
| 22 | Ephedrine | −3.933 | −24.321 |
| 23 | Tetradecamethyl hexasiloxane | Not docked | |
| 24 | Triacontane | Not docked | |
| 25 | Dioctyl phthalate | −3.361 | −38.176 |
| 26 | Docosane | Not docked | |
| 27 | Triacontane | Not docked | |
| 28 | Squalene | −2.14 | −31.436 |
| 29 | Tetracosane | Not docked | |
| 30 | Cyclophosphamide (FDA-approved drug) | −4.621 | −32.158 |
Highlighted bold terms indicated that, these compounds might inhibit the HER2 protein
Figure 2.
Docking analysis of selected complexes. (A) Human epidermal growth factor receptor 2 (HER2)/phenyl glucuronide, (B) HER2/hydrocortisone acetate, (C) HER2/6-(4,6-dioxo-1,4,5,6-tetrahydropyrimidin-2-yl-amino)hexanoic acid trifluoroacetate, and (D) HER2/cyclophosphamide
The ADME properties of the screened compounds were given in Table 3. These compounds come under acceptable and permissible range of Lipinski’s rule, which indicates that these properties make them as potential drug candidate for cellular transport through membrane and directly interacting with intracellular transcription factors in the drug delivery system.
Table 3.
ADME properties of screened phytocompounds and cyclophosphamide as predicted
| S. No. | Ligands | Molecular weight (g/mol) | H-bond donor | H-bond acceptor | Log P O/W |
|---|---|---|---|---|---|
| 1 | Hydrocortisone acetate | 404.503 | 2 | 6 | 2.2 |
| 2 | Phenyl glucuronide | 270.237 | 4 | 7 | 1.4 |
| 3 | 6-(4,6-dioxo-1,4,5,6-tetrahydropyrimidin-2-yl-amino) hexanoic acid trifluoroacetate | 355. 273 | 2 | 4 | 2.4 |
| 4 | Cyclophosphamide | 261.082 | 1 | 4 | 0.60 |
DISCUSSION
Medicinal plants are considered as rich resources of ingredients that can be used for drug development and synthesis.[12] Focus on the plant secondary metabolites for treating various ailments has been increasing over the past few years. These molecules are known to play a major role in the adaptation of plants to their environment and represent an important source of active pharmaceuticals.[13] GC–MS is a combined analytical method that use the features of gas–liquid chromatography and mass spectrometry to identify the active volatile compounds present in the plant solvent extracts.[14] In this study, a total of 29 volatile compounds were predicted from the hexane extract of E. crispa leaves by GC–MS analysis with tetracosane (14.98%); dodecane (10.76%); 2-ethyl-1-decanol (8.00%); tridecane (7.53%), 4,5,6,8-Ptetramethoxy-2,3-dihydroindeno[1,2,3-ij]isoquinolin-9-ol (6.99%); diphenylvinylphosphine (6.38%); squalene (5.85%); triacontane (5.27%); and 2,6-dimethylheptadecane (5.02%) were occupying the highest peak areas compared with rest of the compounds in gas chromatogram analysis. These phytochemicals possess many biological activities such as antioxidant, antimicrobial, antidiabetic, and anticancer.[15,16,17]
Molecular docking simulation is one of the significant methods to analyze the binding orientation of small molecule/target protein complexes. Furthermore, it can be used to know the mechanism of drug–receptor interactions.[18] It was performed to characterize the compounds on the basis of their ability to form favorable interactions within the active site of protein.[19] On the basis of site score and hydrophobic/hydrophilic areas of the substrate, the binding site of HER2 protein residues was predicted and the grid was generated for molecular docking approach. Among the 29 phytocompounds from the extract, the molecular docking simulation identified phenyl glucuronide, hydrocortisone acetate, and 6-(4,6-dioxo-1,4,5,6-tetrahydropyrimidin-2-yl-amino)hexanoic acid trifluoroacetate compounds as potential inhibitors for HER2 protein. This is because, these screened compounds strongly bind and interact with many hydrogen bonds on the hydrophobic and hydrophilic regions of HER2 proteins.
Optimization of the pharmacokinetics and pharmacological (ADME properties) properties of the drug molecule is one of the most complicated and challenging part of drug discovery and development process.[20] The ADME profile will also have a major impact on the likelihood of success of a drug, and it should be noted that the limitations of ADME properties are not more than 5 hydrogen bond donors, 10 hydrogen bond acceptors, and molecular mass of less than 500Da, as well as an octanol–water partition coefficient log P not exceeding 5.[21] These selected compounds of phenyl glucuronide, hydrocortisone acetate, and 6-(4,6-dioxo-1,4,5,6-tetrahydropyrimidin-2-yl-amino)hexanoic acid trifluoroacetate were within the acceptable and permissible limits of ADME properties.
CONCLUSION
This investigation identified 29 volatile phytochemicals that were present in the hexane extract of E. crispa, which hold many biological activities in the human disease and management system. Molecular docking analysis exposed that, when compared with identified compounds and FDA-approved drug, phenyl glucuronide, hydrocortisone acetate, and 6-(4,6-dioxo-1,4,5,6-tetrahydropyrimidin-2-yl-amino)hexanoic acid trifluoroacetate possess strong binding affinity with HER2 protein, which indicates they could be able to inhibit the HER2 protein and their related pathways in the numerous cancers. Pharmacokinetic properties of these compounds were within the acceptable and permissible limits. Therefore, on the basis of the results, this study can be concluded to establish the potential of the phytocompounds as prospective novel inhibitors for HER2 protein. On the other hand, further analysis is necessary to confirm the current findings.
Financial support and sponsorship
Financial support was given by Directorate Research Development, University of the Free State, South Africa (file no.: 2016435634).
Conflicts of interest
There are no conflicts of interest.
Acknowledgement
The authors are thankful to the Directorate Research Development of the University of the Free State, South Africa, for the postdoctoral fellowship award to Dr. Chella Perumal Palanisamy (file no.: 2016435634) and Alagappa University, Karaikudi, Tamil Nadu, India, for providing facilities and encouragement to complete this research work.
REFERENCES
- 1.Hsu JL, Hung MC. The role of HER2, EGFR, and other receptor tyrosine kinases in breast cancer. Cancer Metastasis Rev. 2016;35:575–88. doi: 10.1007/s10555-016-9649-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wee P, Wang Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers. 2017;9:52. doi: 10.3390/cancers9050052. (1–45) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Omar N, Yan B, Salto-Tellez M. HER2: An emerging biomarker in non-breast and non-gastric cancers. Pathogenesis. 2015;2:1–9. [Google Scholar]
- 4.Nida I, Naveed I. Human epidermal growth factor receptor 2 (HER2) in cancers: Overexpression and therapeutic implications. Mol Biol Int. 2014;2014:1–9. doi: 10.1155/2014/852748. Article ID: 852748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perumal PC, Sophia D, Raj CA, Ragavendran P, Starlin T, Gopalakrishnan VK. In vitro antioxidant activities and HPTLC analysis of ethanolic extract of Cayratia trifolia (L.) Asian Pac J Trop Dis. 2012;2:S952–6. [Google Scholar]
- 6.Poornima K, Chella Perumal P, Gopalakrishnan VK. Protective effect of ethanolic extract of Tabernaemontana divaricata (L.) R. Br. against DEN and fe NTA induced liver necrosis in Wistar Albino rats. Biomed Res Int. 2014;2014:1–9. doi: 10.1155/2014/240243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magama S, Pretorius JC, Zietsman PC. Antimicrobial properties of extracts from Euclea crispa subsp. crispa (Ebenaceae) towards human pathogens. S Afr J Bot. 2003;69:193–8. [Google Scholar]
- 8.Perumal PC, Ashafa AOT. Analysis of novel C-X-C chemokine receptor type 4 (CXCR4) inhibitors from hexane extract of Euclea crispa (Thunb.) leaves by chemical fingerprint identification and molecular docking analysis. J Young Pharm. 2018;10:173–7. [Google Scholar]
- 9.Alayande KA, Pohl CH, Ashafa AOT. Time-kill kinetics and biocidal effect of Euclea crispa leaf extracts against microbial membrane. Asian Pac J Trop Med. 2017;10:390–9. doi: 10.1016/j.apjtm.2017.03.022. [DOI] [PubMed] [Google Scholar]
- 10.Pretorius JC, Magama S, Zietsman PC. Purification and identification of antibacterial compounds from Euclea crispa subsp. crispa (Ebenaceae) leaves. S Afr J Bot. 2003;69:579–86. [Google Scholar]
- 11.Perumal PC, Sowmya S, Pratibha P, Vidya B, Anusooriya P, Starlin T, et al. Identification of novel PPAR? agonist from GC-MS analysis of ethanolic extract of Cayratia trifolia (L.): A computational molecular simulation studies. J App Pharm Sci. 2014;4:6–11. [Google Scholar]
- 12.Perumal PC, Sowmya S, Pratibha P, Vidya B, Anusooriya P, Starlin T, et al. Isolation, structural characterization and in silico drug-like properties prediction of a natural compound from the ethanolic extract of Cayratia trifolia (L.) Pharmacognosy Res. 2015;7:121–5. doi: 10.4103/0974-8490.147226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourgaud F, Gravot A, Milesi S, Gontier E. Production of plant secondary metabolites: A historical perspective. Plant Sci. 2001;161:839–51. [Google Scholar]
- 14.Liu CT, Zhang M, Yan P, Liu HC, Liu XY, Zhan RT. Qualitative and quantitative analysis of volatile components of zhengtian pills using gas chromatography mass spectrometry and ultra-high performance liquid chromatography. J Anal Methods Chem. 2016;2016:1206391. doi: 10.1155/2016/1206391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pratibha P, Sophia D, Perumal PC, Gopalakrishnan VK. In-silico docking analysis of Emilia sonchifolia (l.) dc. gas chromatography-mass spectroscopy derived terpenoid compounds against pancreatic cancer targets (AKT and BRCA2) World J Pharm Pharm Sci. 2014;3:1844–55. [Google Scholar]
- 16.Anusooriya P, Arul Raj CA, Perumal PC, Sowmya S, Vidya B, Pratibha P, et al. Screening of novel CXC chemokine receptor 4 inhibitors from ethyl acetate extract of Alpinia purpurata using GC-MS analysis and its molecular docking studies. Int J Pharm Phytochem Res. 2015;7:1–17. [Google Scholar]
- 17.Perumal PC, Pratibha P, Sowmya S, Oirere EK, Anusooriya P, Vidya B, et al. Discovery of novel inhibitors for HER2 from natural compounds present in Cayratia trifolia (l.): An in silico analysis. Int J Curr Pharm Rev Res. 2015;6:164–8. [Google Scholar]
- 18.Tripathi SK, Singh SK, Singh P, Chellaperumal P, Reddy KK, Selvaraj C. Exploring the selectivity of a ligand complex with CDK2/CDK1: A molecular dynamics simulation approach. J Mol Recognit. 2012;25:504–12. doi: 10.1002/jmr.2216. [DOI] [PubMed] [Google Scholar]
- 19.Srinivasan P, Perumal PC, Sudha A. Discovery of novel inhibitors for Nek6 protein through homology model assisted structure based virtual screening and molecular docking approaches. Scientific World J. 2014;2014:1–9. doi: 10.1155/2014/967873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perumal PC, Sowmya S, Velmurugan D, Sivaraman T, Gopalakrishnan VK. Assessment of dual inhibitory activity of isolated natural compound (Epifriedelanol) from Cayratia trifolia. (L.) against ovarian cancer. Bangladesh J Pharmacol. 2016;11:545–51. [Google Scholar]
- 21.Gilad Y, Nadassy K, Senderowitz H. A reliable computational workflow for the selection of optimal screening libraries. J Cheminform. 2015;7:61. doi: 10.1186/s13321-015-0108-0. [DOI] [PMC free article] [PubMed] [Google Scholar]