Key Points
Question
Are clinical sepsis phenotypes identifiable at hospital presentation correlated with the biomarkers of host response and clinical outcomes and relevant for understanding the heterogeneity of treatment effects?
Findings
In this retrospective analysis using data from 63 858 patients in 3 observational cohorts, 4 novel sepsis phenotypes (α, β, γ, and δ) with different demographics, laboratory values, and patterns of organ dysfunction were derived, validated, and shown to correlate with biomarkers and mortality. In the simulations using data from 3 randomized clinical trials involving 4737 patients, the outcomes related to the treatments were sensitive to changes in the distribution of these phenotypes.
Meaning
Four novel clinical phenotypes of sepsis were identified that correlated with host-response patterns and clinical outcomes and may help inform the design and interpretation of clinical trials.
Abstract
Importance
Sepsis is a heterogeneous syndrome. Identification of distinct clinical phenotypes may allow more precise therapy and improve care.
Objective
To derive sepsis phenotypes from clinical data, determine their reproducibility and correlation with host-response biomarkers and clinical outcomes, and assess the potential causal relationship with results from randomized clinical trials (RCTs).
Design, Settings, and Participants
Retrospective analysis of data sets using statistical, machine learning, and simulation tools. Phenotypes were derived among 20 189 total patients (16 552 unique patients) who met Sepsis-3 criteria within 6 hours of hospital presentation at 12 Pennsylvania hospitals (2010-2012) using consensus k means clustering applied to 29 variables. Reproducibility and correlation with biological parameters and clinical outcomes were assessed in a second database (2013-2014; n = 43 086 total patients and n = 31 160 unique patients), in a prospective cohort study of sepsis due to pneumonia (n = 583), and in 3 sepsis RCTs (n = 4737).
Exposures
All clinical and laboratory variables in the electronic health record.
Main Outcomes and Measures
Derived phenotype (α, β, γ, and δ) frequency, host-response biomarkers, 28-day and 365-day mortality, and RCT simulation outputs.
Results
The derivation cohort included 20 189 patients with sepsis (mean age, 64 [SD, 17] years; 10 022 [50%] male; mean maximum 24-hour Sequential Organ Failure Assessment [SOFA] score, 3.9 [SD, 2.4]). The validation cohort included 43 086 patients (mean age, 67 [SD, 17] years; 21 993 [51%] male; mean maximum 24-hour SOFA score, 3.6 [SD, 2.0]). Of the 4 derived phenotypes, the α phenotype was the most common (n = 6625; 33%) and included patients with the lowest administration of a vasopressor; in the β phenotype (n = 5512; 27%), patients were older and had more chronic illness and renal dysfunction; in the γ phenotype (n = 5385; 27%), patients had more inflammation and pulmonary dysfunction; and in the δ phenotype (n = 2667; 13%), patients had more liver dysfunction and septic shock. Phenotype distributions were similar in the validation cohort. There were consistent differences in biomarker patterns by phenotype. In the derivation cohort, cumulative 28-day mortality was 287 deaths of 5691 unique patients (5%) for the α phenotype; 561 of 4420 (13%) for the β phenotype; 1031 of 4318 (24%) for the γ phenotype; and 897 of 2223 (40%) for the δ phenotype. Across all cohorts and trials, 28-day and 365-day mortality were highest among the δ phenotype vs the other 3 phenotypes (P < .001). In simulation models, the proportion of RCTs reporting benefit, harm, or no effect changed considerably (eg, varying the phenotype frequencies within an RCT of early goal-directed therapy changed the results from >33% chance of benefit to >60% chance of harm).
Conclusions and Relevance
In this retrospective analysis of data sets from patients with sepsis, 4 clinical phenotypes were identified that correlated with host-response patterns and clinical outcomes, and simulations suggested these phenotypes may help in understanding heterogeneity of treatment effects. Further research is needed to determine the utility of these phenotypes in clinical care and for informing trial design and interpretation.
In this study, Sepsis-3 investigators use electronic health record and trial data from patients with sepsis within 6 hours of hospital presentation to define clinical phenotypes that correlate with host-response patterns, sepsis biomarkers, mortality, and treatment effects.
Introduction
Sepsis, defined as a dysregulated immune response to infection that leads to acute organ dysfunction, affects millions of individuals per year, and carries a high risk of death even when care is provided promptly.1,2 Although the understanding of the host immune response has advanced considerably, it has not translated into new therapies. A major barrier to progress is the overly broad definition of the syndrome, which encompasses a vast, multidimensional array of clinical and biological features. Different combinations of these features may naturally cluster into previously undescribed subsets or phenotypes that may have different risks for a poor outcome and may respond differently to treatments. However, efforts to determine such phenotypes have remained limited and have focused primarily on patients in the intensive care unit.3,4,5 In addition, these phenotypes must be identifiable at or soon after hospital presentation to guide treatment.
The objectives of this investigation, the National Institutes of Health–funded Sepsis Endotyping in Emergency Care (SENECA) project, were to develop and evaluate sepsis phenotypes. The first goal was to determine whether routine clinical information available at hospital presentation could be mathematically reduced to discrete, reproducible sepsis phenotypes. The second goal was to understand whether the different clinical phenotypes were associated both with patterns among biomarkers of the host immune response and with clinical outcomes. The third goal was to explore the heterogeneity of the treatment effects and the sensitivity of clinical trial results to the frequency distributions of these phenotypes. These mathematically derived phenotypes also were compared with traditional subgrouping strategies.
Methods
The project was approved by the University of Pittsburgh institutional review board and conducted under several data use agreements (PRO15110441, PRO19030218, PRO20061050, PRO010744, PRO12110516, PRO12020657, and PRO17120315). The data for the SENECA project were obtained under a waiver of informed consent and with authorization under the Health Insurance Portability and Accountability Act. Written informed consent was obtained for clinical trial data per published trial procedures.6,7,8
Overview
The study approach involved several data sets and statistical approaches. For the first goal (determining phenotypes), we derived the clinical phenotypes using unsupervised clustering methods that were applied to the data available at hospital presentation in a large database of hospital encounters. We then assessed phenotype reproducibility both by comparing phenotype derivation using alternative clustering methods in the initial data set and by exploring phenotype frequency distributions in several other cohort and clinical trial data sets (eFigure 1 in the Supplement). For the second goal (understanding the correlation of clinical phenotypes and biological markers of the host response with clinical outcome), we first examined correlations in several data sets between the clinical phenotypes and the concurrent patterns of biomarkers, reflecting different elements of the sepsis host response. We then assessed the association of phenotypes with mortality and other clinical outcomes. For the third goal (assessing the influence of phenotypes on clinical trial results), we explored traditional analyses of heterogeneity for treatment effects on observed clinical trial data and performed simulations on 3 trial data sets to understand the potential consequences of different phenotype frequency distributions on estimation of the treatment effects.
Data
We used data from 3 observational cohorts and 3 randomized clinical trials (RCTs)6,7,8,9 (Table 1). The first 2 cohorts (the SENECA derivation and validation cohorts) were drawn from electronic health record data on encounters at 12 community and academic hospitals within the UPMC health care system. We identified all adults (aged ≥18 years) who met sepsis criteria within the first 6 hours of presentation to the emergency department at the 12 hospitals during 2010 to 2012 for the derivation cohort and during 2013 to 2014 for the validation cohort.
Table 1. Characteristics of Cohorts and Clinical Trials.
| Sepsis Endotyping in Emergency Care (SENECA)a | Genetic and Inflammatory Markers of Sepsis (GenIMS)9 | A Controlled Comparison of Eritoran in Severe Sepsis (ACCESS)6 | Activated Protein C Worldwide Evaluation in Severe Sepsis (PROWESS)8 | Protocol-Based Care for Early Septic Shock (ProCESS)7 | ||
|---|---|---|---|---|---|---|
| Derivation Cohort | Validation Cohort | |||||
| No. of patients | 20 189 | 43 086 | 583 | 1706b | 1690 | 1341 |
| No. of sites | 12 | 12 | 28 | 197 | 164 | 31 |
| Enrollment period | 2010-2012 | 2013-2014 | 2001-2003 | 2006-2010 | 1998-2000 | 2008-2013 |
| Location of enrollment | ED | ED | ED | ED, medical ward, ICU | ED, medical ward, ICU | ED |
| Primary intervention | None | None | None | Eritoran (toll-like receptor 4 antagonist) vs placebo | Drotrecogin alfa (activated protein C) vs placebo | EGDT as standard care vs usual care |
| Primary outcome | 28-d mortalityc | 28-d mortalityc | 28-d mortality | 28-d mortality | 28-d mortality | 60-d mortality |
| Inclusion criteria | Sepsis-3 criteria (defined as both a body fluid culture and administration of antibiotics plus ≥2 SOFA points within first 6 h after ED arrival) | Sepsis-3 criteria (defined as both a body fluid culture and administration of antibiotics plus ≥2 SOFA points within first 6 h after ED arrival) | Community-acquired pneumonia with severe sepsis (defined by Sepsis-2 criteria and ≥3 SOFA points) | Severe sepsis and septic shock (defined as meeting ≥3 SIRS criteria, dysfunction in ≥1 organ, and APACHE II score of 21-37) | Severe sepsis and septic shock (defined as meeting ≥3 SIRS criteria and dysfunction in ≥1 organ) | Severe sepsis or septic shock (defined as meeting ≥2 SIRS criteria and either serum lactate level ≥4 mmol/L or refractory hypotension) |
Abbreviations: APACHE, Acute Physiology and Chronic Health Evaluation; ED, emergency department; EGDT, early goal-directed therapy; ICU, intensive care unit; SIRS, systemic inflammatory response syndrome; SOFA, Sequential Organ Failure Assessment.
There were 16 552 unique patients among 20 189 total patients in the derivation cohort and 31 160 unique patients among 43 086 total patients in the validation cohort.
Indicates total with available data.
Measured among unique patients in each cohort.
The third cohort was the Genetic and Inflammatory Markers of Sepsis (GenIMS) study. The GenIMS study was a multicenter, prospective cohort of patients with severe community-acquired pneumonia recruited from 4 regions in the United States (western Pennsylvania, Connecticut, Tennessee, and Michigan) within 1 hour of emergency department presentation, and for whom we had rich clinical information and a variety of biomarkers for the host immune response. The GenIMS study enrolled patients hospitalized at 28 sites from 2001 to 2003.9,10
All 3 RCTs were multicenter studies that involved patients with sepsis or septic shock and had rich clinical and biomarker data. The first trial called ACCESS (A Controlled Comparison of Eritoran in Severe Sepsis) compared eritoran (a highly specific myeloid differentiation protein 2 antagonist that inhibits toll-like receptor 4) vs placebo in patients with severe sepsis at 197 sites on 6 continents from 2006 to 2010 and reported no benefit for 28-day mortality.6 The second trial called PROWESS (Activated Protein C Worldwide Evaluation in Severe Sepsis) compared activated protein C (a commonly activated pleiotropic acute phase protein) vs placebo in patients with severe sepsis at 164 sites in 11 countries from 1998 to 2000 and reported improved survival, but increased bleeding adverse effects for 28-day mortality.8 The third trial called ProCESS (Protocol-Based Care for Early Septic Shock) compared early goal-directed therapy (a multicomponent resuscitation strategy) vs alternative resuscitation approaches in patients with septic shock at 31 sites in the United States from 2008 to 2013 and reported no benefit for 60-day inpatient mortality.7 The RCTs represent a range of RCT types from different clinical settings, testing different types of interventions, and reporting benefit, harm, or no effect (neutral).
Definitions of Sepsis
To identify patients with sepsis in the SENECA derivation cohort, the electronic health record was used to determine if a patient met the following Sepsis-3 criteria2 within the first 6 hours of hospital presentation: (1) evidence of a suspected infection and (2) presence of organ dysfunction. Evidence of a suspected infection was defined as the combination of administration of antibiotics (oral or parenteral) and a body fluid culture specimen obtained (blood, urine, or cerebrospinal fluid), the first of which was required within the first 6 hours of hospital presentation. The presence of organ dysfunction was defined as 2 or more Sequential Organ Failure Assessment (SOFA) points11 within the first 6 hours of hospital presentation. In the GenIMS cohort, the Sepsis-2 definition10 was used because it was available at the time. All patients in the 3 RCTs met variations of the Sepsis-2 criteria, and were therefore eligible for the current study (eMethods in the Supplement).
Candidate Clinical Variables for Phenotyping
We selected 29 candidate variables based on their association with sepsis onset or outcome, their incorporation in conceptual models of sepsis pathophysiology and host tolerance, and their availability in the electronic health record at hospital presentation.12,13,14 These included demographic variables (eg, age, sex, Elixhauser comorbidities), vital signs (eg, heart rate, respiratory rate, Glasgow Coma Scale score, systolic blood pressure, temperature, and oxygen saturation), markers of inflammation (eg, white blood cell count, premature neutrophil count [also called bands], erythrocyte sedimentation rate, and C-reactive protein), markers of organ dysfunction or injury (eg, alanine aminotransferase, aspartate aminotransferase, total bilirubin, blood urea nitrogen, creatinine, international normalized ratio, partial pressure of oxygen, platelets, and troponin), and serum levels of glucose, sodium, hemoglobin, chloride, bicarbonate, lactate, and albumin (eTable 1 in the Supplement). For each variable, we extracted the most abnormal value recorded within the first 6 hours of hospital presentation. In the SENECA derivation and validation cohorts, patient-reported race was derived from the UPMC registration system data using fixed categories consistent with the Centers for Medicare & Medicaid Services electronic health record meaningful use data set.
Biological Correlates and Clinical Outcomes
We studied 27 serum biomarkers measured at baseline in GenIMS, ACCESS, PROWESS, and ProCESS. All of the biomarkers are considered reflective of the host response for sepsis and are included broadly under the domains of inflammatory, endothelial, coagulation, and vital organ function (eTable 2 in the Supplement). The primary clinical outcome was 28-day mortality in the SENECA project derivation and validation cohorts and in the GenIMS, ACCESS, and PROWESS trials. The primary clinical outcome was hospital mortality truncated at 60 days in the ProCESS trial. One-year mortality was studied in the ACCESS, PROWESS, and ProCESS trials. Other outcomes included for exploratory analyses included intensive care unit admission during hospitalization, total days of administration of a vasopressor, and total days of mechanical ventilation during hospitalization.
Statistical Methods
To derive the phenotypes, we first assessed the candidate variable distributions, missingness, and correlation (eTable 3 in the Supplement). Multiple imputation with chained equations was used to account for missing data (eTable 4 and eMethods in the Supplement)15 and log transformation was used for nonnormal data. After evaluating correlation, we excluded highly correlated variables using rank-order statistics in the sensitivity analyses (eFigure 2 in the Supplement). Ordering points to identify the clustering structure (OPTICS) plots were used to determine the optimal clustering strategy.16 Based on these plots, we applied consensus k means clustering to 29 variables using a partitioning approach.17 To determine the optimal number of phenotypes with consensus k means clustering, we evaluated a combination of phenotype size, clear separation of the consensus matrix heatmaps, characteristics of the consensus cumulative distribution function plots, and adequate pairwise–consensus values between cluster members (>0.8). Once optimal phenotype number was determined, patterns of clinical variables were visualized in 3 ways: (1) t-distributed stochastic neighbor embedding plots (which show multidimensional data in 2 dimensions), (2) alluvial plots (which show the proportional distribution of phenotype members across specific variables), (3) chord diagrams (which show how phenotypes differ by major variable groups; eMethods in the Supplement), and (4) ranked plots of variables by the mean standardized difference between the phenotype pairs.18
To assess the reproducibility of the phenotypes, we first used a latent class analysis to derive the groups (eMethods in the Supplement).19 In the latent class analysis, the optimal phenotype number was confirmed using a combination of Bayesian information criteria, adequate size, high median probabilities of group membership within each phenotype, maximum entropy (a measure between 0 and 1 indicating better classification), and clinical features of potential groups. We also determined the proportion of patients with a probability of phenotype assignment on the margin, which was defined as between 45% and 55%. We assessed how robust the phenotypes were to sensitivity analyses of the derivation method, including (1) excluding variables with high missingness (eg, erythrocyte sedimentation rate, C-reactive protein, premature neutrophil count [bands]); (2) excluding both highly missing and highly correlated variables (sodium, hemoglobin, blood urea nitrogen, and alanine aminotransferase); and (3) using a 12-hour window of electronic health record data after hospital presentation (eMethods in the Supplement).
To determine the reproducibility in the external data, we used the SENECA validation cohort and rederived groups using consensus k means clustering. Then, in the GenIMS study and in the 3 RCTs, we predicted phenotype based on the clinical characteristics of typical cluster members in the SENECA derivation cohort. Predictions arose from the Euclidean distance from each patient to the centroid of each SENECA phenotype (eMethods in the Supplement). We studied the frequency and clinical characteristics of the predicted phenotype groups in the GenIMS study and in the 3 RCTs.
We determined the correlation of the phenotypes with 27 biomarkers of the host immune response and compared the mean (SD), the median (interquartile range [IQR]), and the ratio of biomarker distributions across phenotypes as appropriate. The χ2 test was used to compare in-hospital, 28-day, and 365-day mortality. The cumulative mortality was illustrated using probability plots and the differences were tested using the log-rank test.
To understand the implications of the phenotypes on the RCT estimates of the treatment effects, we conducted Monte Carlo simulations (10 000 iterations per simulation) in which the only variable modified was the proportion of phenotypes enrolled in the existing trial data set using random sampling with replacement. Six scenarios were created for each of the 3 trials (eMethods in the Supplement), in which the range of phenotypes was varied. The frequency for the range of phenotypes was informed in simulated trials using upper and lower bounds up to twice that observed across the hospitals in the SENECA derivation and validation cohorts. We also tested logistic regression models for 28-day and 365-day mortality using phenotype, treatment assignment, and their interaction as covariates (eMethods in the Supplement).
Several analyses were conducted to ensure the phenotypes were not simply recapitulations of more traditional clinical groups. First, we tested whether the phenotypes were explained by traditional measures of illness severity, such as the SOFA score or the Acute Physiology and Chronic Health Evaluation (APACHE) score. For the SENECA derivation cohort, alluvial plots were used to inspect whether the phenotypes overlapped with the SOFA score.11 We also determined the overlap of the phenotypes with the quartiles of APACHE and SOFA scores in the 3 RCTs.20 We further inspected the biomarker profiles and mortality by APACHE quartile in the ProCESS trial. Simulations in the ProCESS trial were also repeated, varying the proportions of the 4 severity-of-illness quartiles instead of the phenotypes, and comparing the potential causal relationship with the estimates of treatment benefit or harm (eMethods in the Supplement). Second, we explored whether the phenotypes were explained by the site of the infection. In the ACCESS trial, which includes independent adjudication of the source of infection, we generated alluvial plots and the proportions for infection sites across phenotypes. We measured the frequency of the phenotypes in a subset of patients with sepsis from a single source (bacteremia) among patients in the SENECA derivation cohort.
Data are presented as mean (SD) or median (IQR). For comparisons, we used analysis of variance and the Kruskal-Wallis test for continuous data and the χ2 test for categorical data. The threshold for statistical significance was less than .05 for 2-sided tests. There was no adjustment for the type I error rate due to multiple comparisons; therefore, the findings from these analyses should be considered exploratory. Analyses were performed with Stata version 14.2 (StataCorp) and R versions 3.4.1 and 3.5.0 (R Foundation for Statistical Computing).
Results
Patients
Among 1 309 025 patient encounters in the SENECA derivation cohort (eFigure 3 in the Supplement), 87 844 patients (6.7%) had suspected infection within 6 hours of hospital presentation and 20 189 met Sepsis-3 criteria (eTable 5 in the Supplement). The mean SOFA score was 3.9 (SD, 2.4) and the mean serum lactate level was 3.2 mmol/L (SD, 3.2 mmol/L). Among 1 119 388 encounters in the SENECA validation cohort, more patients had suspected infection (n = 103 259; 9.2%) and met Sepsis-3 criteria within 6 hours (n = 43 086); however, the demographic characteristics and SOFA scores were similar (eTables 5 and 6 in the Supplement). The SENECA validation cohort included 43 086 patients (mean age, 67 [SD, 17] years; 21 993 [51%] male; mean maximum 24-hour SOFA score, 3.6 [SD, 2.0]). Patients in the GenIMS cohort (total of 2320 enrolled, including 583 patients with sepsis) had more comorbidities and respiratory symptoms (eg, elevated respiratory rate, lower oxygen saturation). Across the SENECA derivation and validation cohorts and the GenIMS cohort, the in-hospital mortality ranged from 6% to 14% and from 16% to 23% among patients who required intensive care. In the 3 RCTs (eTable 7 in the Supplement), a total of 4737 patients (1706 in the trial on eritoran,6 1690 in the trial on activated protein C,8 and 1341 in the trial on early goal-directed therapy7) participated at 392 sites and short-term mortality (ie, at 28 days in 2 trials and at 60 days in 1 trial) ranged from 19% to 28%.
Derivation of Clinical Sepsis Phenotypes
In the SENECA derivation cohort, the consensus k means clustering models found that a 4-class model was the optimal fit with the 4 phenotypes of α, β, γ, and δ (eFigures 4 and 5 and eTable 8 in the Supplement). Consensus matrix plots and the relative change under the cumulative distribution function curve implied little statistical gain by increasing to a 5- or 6-class model. The size and characteristics of the phenotypes in the 4-class model appear in Table 2 and Figure 1. Phenotypes ranged in size (from 13% to 33% of the cohort) and differed broadly in clinical characteristics and organ dysfunction patterns. When ranking continuous variables by the standardized mean difference between phenotypes (Figure 2), patients with the α phenotype had fewer abnormal laboratory values and less organ dysfunction; those with the β phenotype were older, had greater chronic illness, and were more likely to present with renal dysfunction; those with the γ phenotype were more likely to have elevated measures of inflammation (eg, white blood cell count, premature neutrophil count [bands], erythrocyte sedimentation rate, or C-reactive protein), lower albumin level, and higher temperature; and those with the δ phenotype had elevated serum lactate levels, elevated levels of transaminases, and hypotension (eFigure 6-8 in the Supplement).
Table 2. Characteristics of the 4 Phenotypes.
| Characteristica | Total | Phenotype | |||
|---|---|---|---|---|---|
| α | β | γ | δ | ||
| No. of patients (%) | 20 189 (100) | 6625 (33) | 5512 (27) | 5385 (27) | 2667 (13) |
| Age, mean (SD), y | 64 (17) | 60 (18) | 71 (15) | 65 (16) | 63 (17) |
| Sex, No. (%) | |||||
| Male | 10 022 (50) | 3372 (51) | 2624 (48) | 2559 (48) | 1467 (55) |
| Female | 10 167 (50) | 3253 (49) | 2888 (52) | 2826 (52) | 1200 (45) |
| Race, No. (%) | |||||
| White | 15 640 (77) | 5165 (78) | 4221 (77) | 4269 (79) | 1985 (74) |
| Black | 2428 (12) | 805 (12) | 797 (14) | 539 (10) | 287 (11) |
| Otherb | 2121 (11) | 655 (10) | 494 (9) | 577 (11) | 395 (15) |
| Organ Dysfunction | |||||
| Elixhauser comorbidities, mean (SD)c | 1.8 (1.2) | 1.5 (1.1) | 2.4 (1.2) | 1.7 (1.1) | 1.7 (1.1) |
| Surgery, No. (%)d | 2727 (14) | 696 (11) | 786 (14) | 825 (15) | 420 (16) |
| Reached maximum within 24 h, mean (SD) | |||||
| SIRS criteriae | 1.8 (1.2) | 1.4 (1.1) | 1.2 (1.0) | 2.3 (1.0) | 2.5 (1.1) |
| SOFA scoref | 3.9 (2.4) | 3.0 (1.4) | 3.5 (1.7) | 4.0 (2.3) | 6.6 (3.7) |
| Inflammation | |||||
| Premature neutrophil count (bands), median (IQR), % | 7 (3-15) | 5 (2-11) | 4 (2-11) | 10 (4-18) | 14 (6-25) |
| C-reactive protein, median (IQR), mg/L | 6 (2-16) | 2 (0.4-6) | 5 (2-12) | 16 (9-32) | 13 (4-30) |
| Erythrocyte sedimentation rate, median (IQR), mm/h | 48 (25-88) | 28 (15-45) | 61 (38-99) | 92 (59-116) | 31 (14-55) |
| Temperature, mean (SD), °C | 37.0 (1.0) | 37.1 (0.9) | 36.7 (0.8) | 37.3 (1.0) | 36.7 (1.3) |
| White blood cell count, median (IQR), ×109/L | 10 (7-14) | 9 (6-12) | 9 (7-13) | 11 (7-16) | 12 (8-18) |
| Pulmonary | |||||
| Oxygen saturation, median (IQR), % | 94 (91-97) | 94 (91-97) | 95 (93-98) | 93 (90-96) | 95 (90-97) |
| Partial pressure of oxygen, mean (SD), mm Hg | 123 (89) | 100 (68) | 111 (75) | 98 (63) | 152 (106) |
| Respiratory rate, mean (SD), breaths/min | 22 (6) | 20 (4) | 20 (4) | 25 (7) | 25 (8) |
| Cardiovascular or Hemodynamic | |||||
| Bicarbonate, mean (SD), mEq/L | 25 (5) | 27 (4) | 25 (5) | 25 (5) | 20 (5) |
| Heart rate, mean (SD), beats/min | 97 (22) | 94 (19) | 84 (16) | 109 (21) | 108 (24) |
| Serum lactate, median (IQR), mmol/L | 1.5 (1.0-2.4) | 1.3 (1.0-1.9) | 1.2 (0.9-1.8) | 1.8 (1.2-2.7) | 3.3 (2.0-5.7) |
| Systolic blood pressure, median (IQR), mm Hg | 110 (93-128) | 118 (104-134) | 120 (103-138) | 99 (83-113) | 91 (77-109) |
| Troponin, median (IQR), ng/mL | 0.1 (0-0.1) | 0.1 (0-0.1) | 0.1 (0-0.1) | 0.1 (0-0.1) | 0.3 (0.1-1.4) |
| Renal | |||||
| Blood urea nitrogen, median (IQR), mg/dL | 24 (15-38) | 16 (11-22) | 38 (27-55) | 23 (15-34) | 32 (20-52) |
| Creatinine, median (IQR), mg/dL | 1.4 (1.0-2.2) | 1.1 (0.8-1.3) | 2.3 (1.6-3.9) | 1.2 (0.9-1.8) | 1.8 (1.2-2.8) |
| Hepatic | |||||
| Alanine transaminase, median (IQR), U/L | 30 (20-48) | 32 (22-49) | 25 (17-35) | 27 (18-40) | 69 (36-194) |
| Aspartate transaminase, median (IQR), U/L | 30 (20-53) | 28 (19-45) | 23 (17-35) | 30 (20-46) | 118 (59-276) |
| Bilirubin, median (IQR), mg/dL | 0.8 (0.5-1.3) | 0.8 (0.5-1.3) | 0.6 (0.4-0.9) | 0.8 (0.5-1.3) | 1.4 (0.8-3.3) |
| Hematologic | |||||
| Hemoglobin, mean (SD), g/dL | 12 (2) | 13 (2) | 11 (2) | 10 (2) | 11 (2) |
| International normalized ratio, median (IQR) | 1.3 (1.1-1.6) | 1.2 (1.1-1.3) | 1.2 (1.1-1.6) | 1.3 (1.2-1.7) | 1.7 (1.3-2.7) |
| Platelets, median (IQR), ×109/L | 188 (130-256) | 179 (128-246) | 200 (143-263) | 195 (131-269) | 164 (104-241) |
| Other | |||||
| Albumin, mean (SD), g/dL | 2.9 (0.7) | 3.5 (0.5) | 3.0 (0.6) | 2.4 (0.6) | 2.6 (0.7) |
| Chloride, mean (SD), mEq/L | 103 (7) | 103 (6) | 103 (7) | 101 (7) | 106 (8) |
| Glucose, median (IQR), mg/dL | 130 (105-179) | 121 (101-157) | 132 (105-184) | 134 (107-185) | 152 (115-227) |
| Sodium, mean (SD), mEq/L | 137 (5) | 137 (5) | 138 (5) | 136 (6) | 138 (7) |
| Glasgow Coma Scale score, mean (SD) | 11.4 (4.0) | 12.8 (3.0) | 13.6 (2.3) | 13.4 (2.6) | 10.5 (4.5) |
| Outcomes | |||||
| Mechanical ventilation, median (IQR), dd | 5 (2-10) | 4 (2-9) | 4 (2-9) | 6 (3-13) | 4 (2-9) |
| Administration of a vasopressor, median (IQR), dd | 3 (2-5) | 2 (2-4) | 3 (2-4) | 3 (2-5) | 3 (2-5) |
| Admitted to intensive care unit, No. (%)d | 9063 (45) | 1644 (25) | 1778 (32) | 3381 (63) | 2260 (85) |
| In-hospital mortality, No. (%) | 2082 (10) | 126 (2) | 286 (5) | 818 (15) | 852 (32) |
Abbreviations: IQR, interquartile range; SIRS, systemic inflammatory response syndrome; SOFA, Sequential Organ Failure Assessment.
SI conversion factors: To convert alanine transaminase and aspartate aminotransferase to μkat/L, multiply by 0.0167; bilirubin to μmol/L, multiply by 17.104; C-reactive protein to nmol/L, multiply by 9.524; creatinine to μmol/L, multiply by 88.4; glucose to mmol/L, multiply by 0.0555; lactate to mg/dL, divide by 0.111; urea nitrogen to mmol/L, multiply by 0.357.
Corresponds to minimum or maximum value (as appropriate) within 6 hours of hospital presentation. The variables in this Table were log transformed for modeling (eTable 3 in the Supplement). Comparisons across all 4 phenotypes were performed using the Kruskal-Wallis test, analysis of variance, or the χ2 test (P < .01 for all comparisons).
Includes Chinese, Filipino, Hawaiian, American Indian/Alaskan Native, Asian, Hawaiian/other Pacific Islander, Middle Eastern, Native American, not specified, or Pacific Islander.
A method of categorizing comorbidities of patients based on the International Classification of Diseases, Ninth Revision diagnosis codes found in administrative data. Scores range from 0 to 31.
At any time during hospitalization.
Indicates a scoring system that measures the inflammatory response. Scores range from 0 to 4 points.
Corresponds to the severity of organ dysfunction, reflecting 6 organ systems each. Scores range from 0 to 4 points for cardiovascular, hepatic, hematologic, respiratory, neurological, and renal. The total score range is from 0 to 24 points.
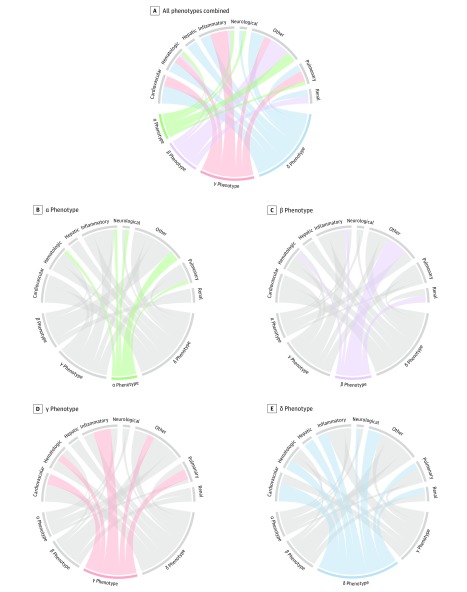
Figure 1. Chord Diagrams Showing Abnormal Clinical Variables by Phenotype.
In A, the ribbons connect from an individual phenotype to an organ system if the group mean is greater or lesser than the overall mean for the entire cohort. For example, the δ phenotype (light blue) is more likely to have members with abnormal cardiovascular and hepatic dysfunction (ribbons connect with these portions of the circle) vs β phenotype members (light purple) who are more likely to have kidney dysfunction and other abnormal variables (eg, increased age, comorbidity). In B-E, each phenotype is highlighted separately and the ribbons connect to the different patterns of clinical variables and organ system dysfunctions on the top of the circle.
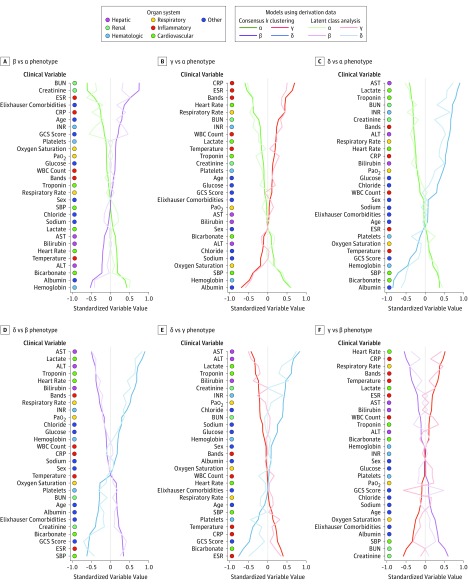
Figure 2. Comparison of Variables That Contribute to Clinical Phenotypes in the SENECA Derivation Cohort (n = 20 189).
In all panels, the variables are standardized such that all means are scaled to 0 and SDs to 1. A value of 1 for the standardized variable value (x-axis) signifies that the mean value for the phenotype was 1 SD higher than the mean value for both phenotypes shown in the graph as a whole. ALT indicates alanine transaminase; AST, aspartate transaminase; Bands, also known as premature neutrophil count; BUN, blood urea nitrogen; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; GCS, Glasgow Coma Scale; INR, international normalized ratio; Pao2, partial pressure of oxygen; SENECA, Sepsis Endotyping in Emergency Care; SBP, systolic blood pressure; WBC, white blood cell.
Variables such as sex, sodium level, glucose level, and white blood cell count contributed least to phenotype differences (eFigure 9 in the Supplement). Phenotypes also varied across the 12 SENECA hospitals as follows: α phenotype ranged from 24% to 42%; β phenotype ranged from 19% to 30%; γ phenotype ranged from 23% to 50%; and δ phenotype ranged from 5% to 23% (eFigure 10 in the Supplement). There was no difference across phenotypes in the rate of peripheral blood culture as the first body fluid culture after hospital presentation, whereas the rate of intravenous antibiotics (vs other routes of administration) ranged from 76% to 93% (eTable 9 and eFigure 11 in the Supplement).
Reproducibility
Latent class analysis confirmed the statistical fit of the 4-class model (Figure 2 and eFigure 12 and eTable 10 in the Supplement). Bayesian information criteria decreased as class number increased from 2 to 4 while entropy was preserved (>0.8). The clinical characteristics of the phenotypes were similar when derived using this method as well as by visualization with t-distributed stochastic neighbor embedding plots (eFigure 13 and eTable 11 in the Supplement). There was strong separation in the likelihood of membership for patients assigned to a given phenotype compared with those assigned to other phenotypes (eFigure 14 in the Supplement).
Phenotypes also were derived in the SENECA validation cohort and showed similar optimal phenotype numbers, frequency of phenotypes, and clinical characteristics as observed in the primary analysis (Figure 2; eFigures 15 and 16 and eTable 12 in the Supplement). No substantial changes were evident after excluding variables with high missingness (eTable 13 in the Supplement), after excluding variables with both high missingness and correlation (eTable 14 in the Supplement), and when the window for capturing data was expanded to 12 hours after hospital presentation (eTable 15 in the Supplement).
In the GenIMS cohort in which patients had sepsis due only to pneumonia (eMethods and eFigure 17 in the Supplement), all 4 phenotypes were present, albeit with slightly different frequencies compared with the SENECA derivation cohort. The clinical characteristics of the phenotypes were largely the same (eTable 16 in the Supplement). When the phenotypes were predicted in the 3 RCTs (eFigure 18-21 in the Supplement), the frequency distributions and clinical characteristics were also similar to the SENECA derivation cohort (eTable 17 for ACCESS, eTable 18 for PROWESS, and eTable 19 for ProCESS in the Supplement).
Correlation of Phenotypes With Biomarker Profiles
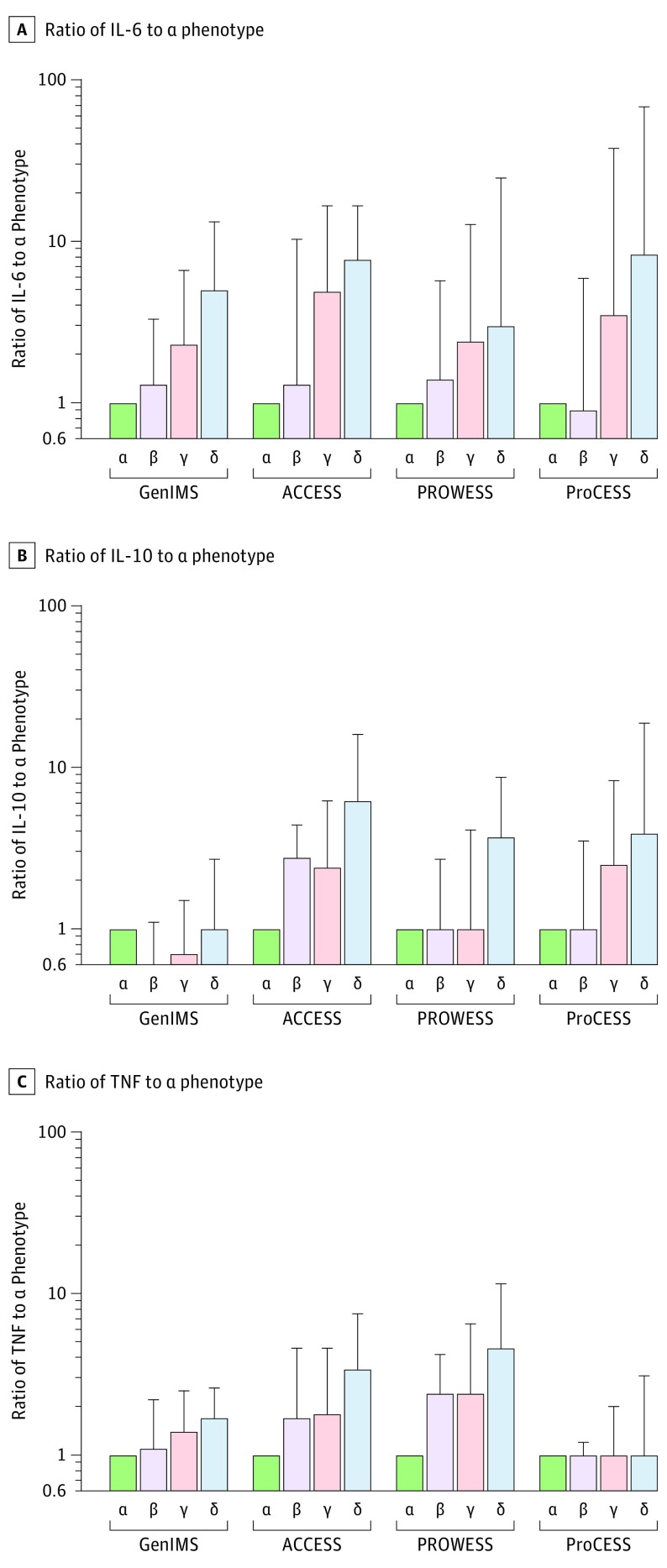
Broad differences were observed in the distributions of the host-response biomarkers across phenotypes (Figure 3). Of the 27 biomarkers measured in 4 studies, 23 were significantly different across phenotypes in at least 1 study (P < .05). In general, there was an increase in the markers of inflammation and in abnormal coagulation in both the γ and δ phenotypes compared with the α or β phenotypes (Figure 4). For example, in the GenIMS study, the ratio of the δ phenotype to the α phenotype for the median level of IL-6 was 5.0 (IQR, 1.6-13.2); in the ACCESS trial, it was 7.7 (IQR, 1.4-16.6); in the PROWESS trial, it was 3.0 (IQR, 0.7-24.6); and in the ProCESS trial, it was 8.3 (IQR, 1.4-67.7). Similar findings comparing the δ phenotype vs the α phenotype were present for IL-10 level (ranges of ratios across the studies for median level of IL-10, 1.3-6.2), but were less prominent for tumor necrosis factor (range of ratios across the studies for tumor necrosis factor, 1.0-4.6; Figure 3).
Figure 3. Inflammatory Cytokines Across Phenotypes.
Ratio of IL-6 was calculated as the cytokine value standardized by the median value for the α phenotype in each study (referent) illustrated on a log scale. All comparisons within data sets across phenotypes were significant (P < .001). Errors bars indicate the upper bound of the interquartile range of the biomarker standardized by the median value for the α phenotype. Across multiple cohorts and randomized trials, inflammatory cytokines IL-6, IL-10, and TNF measured at baseline were greater in the γ phenotype (pink) and δ phenotype (blue) compared with the α phenotype (green), suggesting a predominantly hyperinflammatory response. TNF indicates tumor necrosis factor.
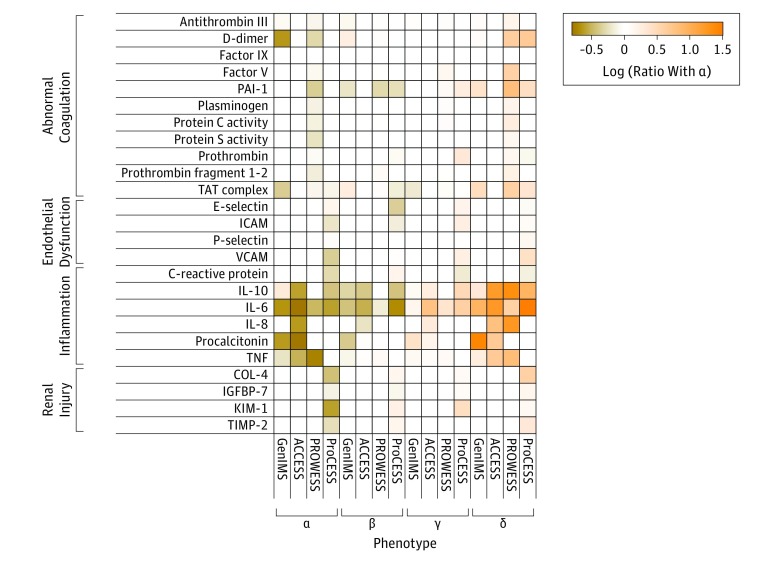
Figure 4. Ratio of Additional Biomarkers in Heatmap.
Heatmap shows the ratio of the median biomarker value for various markers of the sepsis host response grouped by those reflecting coagulation, endothelium, inflammation, and renal injury. Orange represents a greater median biomarker value for that phenotype compared with the median for the entire study, whereas colors in the tan to brown range represent lower median biomarker values compared with the median for the entire study. Empty cells are those for which the biomarker was not measured. The factor V, factor IX, plasminogen, protein C, and protein S biomarkers were reversed on the scale to coordinate the color map. The IL-1b and IL-12 biomarkers are not shown due to having less than 0.5-fold changes. ACCESS indicates A Controlled Comparison of Eritoran in Severe Sepsis; COL-4, collagen type 4; GenIMS, Genetic and Inflammatory Markers of Sepsis; ICAM, intercellular adhesion molecule 1; IGFBP-7, insulin-like growth factor–binding protein 7; KIM-1, kidney injury molecule 1; PAI-1, plasminogen activator inhibitor 1; ProCESS, Protocol-Based Care for Early Septic Shock; PROWESS, Activated Protein C Worldwide Evaluation in Severe Sepsis; TAT, thrombin-antithrombin; TIMP-2, tissue inhibitor of metalloproteinase 2; TNF, tumor necrosis factor; VCAM, vascular cell adhesion molecule.
Coagulation markers such as thrombin-antithrombin complex, plasminogen activator inhibitor 1, and D-dimer were significantly greater in the δ phenotype compared with the other phenotypes (P < .001; Figure 4 and eTables 20-23 in the Supplement). The levels of some markers of endothelial dysfunction (eg, intercellular adhesion molecule 1, E-selectin) were highest in the γ phenotype (P < .01), other markers were highest in the δ phenotype (eg, vascular cell adhesion molecule 1), and other markers were not different across groups (eg, P-selectin, P = .37). Markers of renal injury (eg, insulin-like growth factor–binding protein 7, collagen type 4, tissue inhibitor of metalloproteinase 2) were highest in both the β and δ phenotypes (P < .01).
Relationship With Mortality and Organ Support
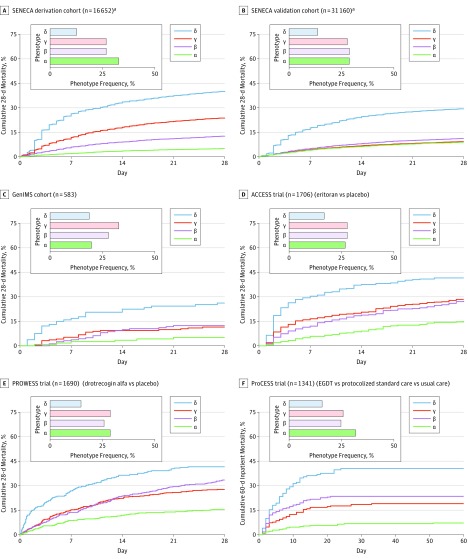
Phenotypes were associated with short- and long-term outcomes (eTables 24 and 25 in the Supplement). In the SENECA derivation cohort, the fewest in-hospital deaths occurred in the α phenotype (n = 126; 2%) compared with the β phenotype (n = 286; 5%), the γ phenotype (n = 818; 15%), and the δ phenotype (n = 852; 32%) (P < .001). Across all cohorts and trials, the 28-day mortality (Figure 5) and the 365-day mortality (eFigure 22 in the Supplement) were highest in the δ phenotype compared with the other phenotypes (P < .001). In the SENECA derivation cohort (n = 16 552 unique patients), cumulative 28-day mortality was 287 of 5691 (5%) for the α phenotype, 561 of 4420 (13%) for the β phenotype, 1031 of 4318 (24%) for the γ phenotype, and 897 of 2223 (40%) for the δ phenotype. In the SENECA validation cohort (n = 31 160 unique patients), cumulative 28-day mortality was 837 (9%) for the α phenotype, 923 (11%) for the β phenotype, 854 (9%) for the γ phenotype, and 1278 (29%) for the δ phenotype. Intensive care unit admission rates were higher in the δ phenotype compared with the other phenotypes (P < .01), whereas days of mechanical ventilation and administration of a vasopressor were variable across studies.
Figure 5. Short-term Mortality by Phenotype.
All panels show significant differences in mortality by phenotype (log-rank P < .001). In the SENECA derivation and validation cohorts, in the GenIMS cohort, and in the 3 randomized clinical trials, clinical phenotypes are associated with short-term mortality. This suggests that phenotypes are generalizable and prognostic across data sets with different severity, temporality, and definitions of sepsis and septic shock. ACCESS indicates A Controlled Comparison of Eritoran in Severe Sepsis; GenIMS, Genetic and Inflammatory Markers of Sepsis; ProCESS, Protocol-Based Care for Early Septic Shock; PROWESS, Activated Protein C Worldwide Evaluation in Severe Sepsis; SENECA, Sepsis Endotyping in Emergency Care.
aThe cumulative mortality data are only for unique patients in the SENECA derivation cohort (16 652 of 20 189 total patients) and in the SENECA validation cohort (31 160 of 43 086 total patients).
Differential Estimated Treatment Effects by Phenotype and Sensitivity of the Clinical Trial Results to Changes in Phenotype Distributions in the Trial Simulations
The estimated treatment effects by phenotype were variable in the observed data in the ACCESS, PROWESS, and ProCESS trials (eFigures 23-28 in the Supplement). Standard treatment × phenotype interactions were only significant in the ProCESS trial, but not for the other 2 trials based on the P < .05 criteria. The primary findings of the trial simulations appear in Figure 6 (more detailed examples appear in eFigures 29-31 in the Supplement). In general, the trials had similar baseline characteristics between simulation scenarios and original trial populations. For example, a doubling of the δ phenotype did not change the demographics and increased the mean baseline SOFA score from 7.2 (SD, 3.6) points to only 8.6 (SD, 3.6) points in the ProCESS trial (eTables 26-28 in the Supplement). The mortality rates for the control group were also stable across the simulations and were within the typically reported ranges (eTable 29 and eFigure 32 in the Supplement). For example, a doubling of the highly morbid δ phenotype was only associated with an increase in the mortality rate for usual care from 26% to 31% in the ACCESS trial, from 31% to 39% in the PROWESS trial, and from 19% to 26% in the ProCESS trial.
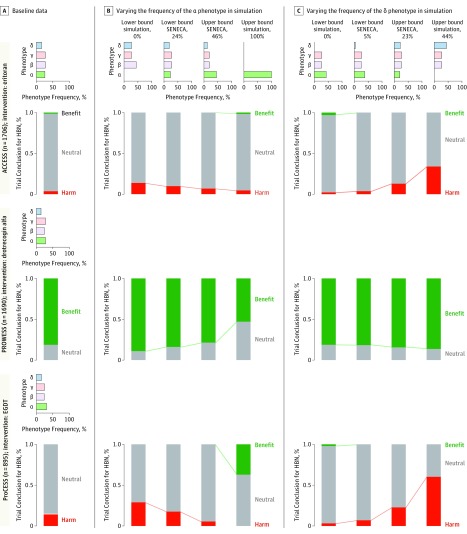
Figure 6. Sensitivity of Clinical Trial Results to the Relative Frequency of Phenotypes in Monte Carlo Simulation.
For each trial (ACCESS, PROWESS, and ProCESS), panel A shows the actual distribution of the 4 phenotypes in that trial (horizontal bar graph) and the observed proportion of trials concluding no difference (neutral), harm, or benefit in simulation (vertical stacked bar graph). Each simulation represents 10 000 iterations using sampling with replacement. Panel B shows how simulated trial results vary when the case mix is changed to the distributions shown in the top set of graphs by varying α (panel B) and δ (panel C). ACCESS indicates A Controlled Comparison of Eritoran in Severe Sepsis; EGDT, early goal-directed therapy; HBN, harm, benefit, or neutral; ProCESS, Protocol-Based Care for Early Septic Shock; PROWESS, Activated Protein C Worldwide Evaluation in Severe Sepsis; SENECA, Sepsis Endotyping in Emergency Care.
The trial conclusions about the treatment effects were relatively robust to large changes in the proportion of patients with the β and γ phenotypes. Despite modest changes to the baseline characteristics in the trial populations, the changes to the distributions for the α and δ phenotypes had substantial effects (Figure 6). For example, in the ProCESS trial, which under the baseline phenotype distribution had a 0% chance of finding benefit with early goal-directed therapy for 60-day inpatient mortality (and an 85% and 15% chance of finding no difference or harm, respectively), the chance of finding benefit increased to 35% when the α phenotype represented the majority of the population (eFigure 29 in the Supplement).
In contrast, when the δ phenotype was increased to 50% of the ProCESS trial population, there was a greater than 60% chance of finding that early goal-directed therapy was harmful. In the ACCESS trial (eFigure 30 in the Supplement), which under the baseline phenotype distribution had a 0% chance of finding benefit, a 91% chance of finding no difference, and a 9% chance of finding harm for 28-day mortality, an increase in the δ phenotype from 14% to 44% of the trial population resulted in 29% of simulated trials concluding eritoran caused harm. In the PROWESS trial, which had an 82% chance of finding a positive effect with the baseline phenotype distribution, 50% of the simulated trials showed no difference when the frequency of the α phenotype was increased to represent the majority of the trial population (eFigure 31 in the Supplement).
Comparison With Traditional Subgroups of Patients With Sepsis
The 4 phenotypes could not be described by severity of illness or site of infection alone. In the SENECA derivation cohort, all 4 phenotypes included both patients with and without organ dysfunction in all SOFA categories (Figure 1). The mean SOFA scores at hospital presentation were lower in patients with the α phenotype (3.0 [SD, 1.4]) and higher in patients with the δ phenotype (6.6 [SD, 3.7]), but overlapped in patients with the β phenotype (3.5 [SD, 1.7]) and in those with the γ phenotype (4.0 [SD, 2.3]) (Table 2 and eFigures 33 and 34 in the Supplement). In the ACCESS trial, although the δ phenotype had a greater proportion of patients with intraabdominal infections, there was a broad distribution for site of infection in each phenotype (eFigure 35 and eTable 30 in the Supplement). There was a similarly broad distribution for phenotypes among patients with sepsis due to bacteremia alone (n = 1714; eFigure 36 in the Supplement).
In the analyses to further explore whether the derived phenotypes were proxies for severity of illness, the pattern of baseline clinical variables and host-response biomarkers differed across the APACHE quartiles from the pattern for the 4 phenotypes (eTables 31 and 32 and eFigures 37 and 38 in the Supplement). The range of short-term mortality rates across the APACHE III quartiles was similar to the range across the 4 phenotypes (eFigure 38 in the Supplement). However, enrichment of the ProCESS trial using APACHE III quartiles was associated with smaller changes in the trial conclusions compared with phenotype enrichment (eFigure 39 in the Supplement).
Discussion
In this retrospective analysis of data sets from patients with sepsis, 4 clinical phenotypes of sepsis were derived using routinely available clinical data at the time of hospital presentation. The phenotypes were multidimensional, differed in their demographics, laboratory abnormalities, patterns of organ dysfunction, and were not homologous with traditional patient groupings such as by site of infection, organ dysfunction patterns, or severity of illness. The frequency and characteristics of the phenotypes were reproducible in additional cohorts and using different machine learning methods. The 4 sepsis phenotypes were strongly correlated with patterns of the host immune response, mortality, and other clinical outcomes. In simulations of 3 large, multicenter trials, conclusions about the estimated treatment benefit or harm were sensitive to phenotype distributions, especially the α and δ phenotypes.
These sepsis phenotypes can be identified at the time of patient presentation to the emergency department, and thus could be useful with regard to early treatment and enrollment in clinical trials. Only routinely available data were used in the clustering models, and the phenotypes were derived from a large observational cohort to ensure generalizability. Phenotype frequency distributions and characteristics were similar in studies with different definitions for sepsis. For example, the SENECA derivation and validation cohorts used electronic health record criteria for Sepsis-3,2 the GenIMS study used Sepsis-2,10 the ProCESS trial enrolled patients with early septic shock and used broad sepsis criteria, and both the ACCESS and PROWESS trials enrolled patients later in their clinical course and patients with more organ failure.
Of the 4 phenotypes identified, the δ phenotype was most strongly correlated with abnormal values of host-response biomarkers as well as clinical features of cardiovascular and liver dysfunction. These characteristics are similar to previously reported subclasses, including the hyperinflammatory subphenotype reported in acute respiratory distress syndrome, a condition most commonly caused by sepsis.18 The δ phenotype also resembles sepsis endotypes derived using transcriptomic analyses of circulating immune cells (such as the inflammopathic cluster, sepsis response signature 1, or the Molecular Diagnosis and Risk Stratification of Sepsis [MARS] 2 cluster) described in patients with sepsis in the intensive care unit.3,4,5 In contrast, the α phenotype had fewer laboratory abnormalities and less septic shock, which resembles the MARS 3 and sepsis response signature 2 endotypes reported in the same series, and which were found to have predominant expression of adaptive immune and B-cell development pathways.3,4,5 This concordance between clinical phenotypes and more computationally intensive transcriptomic endotypes could help identify subsets of patients most likely to benefit from particular immunomodulation strategies.
In RCT simulations, variations in the phenotypes had small changes in the distribution of average baseline characteristics, yet resulted in unstable trial conclusions. For example, the ACCESS trial found no benefit from eritoran on 28-day mortality. Yet, when the δ phenotype (the phenotype with the greatest proportion of intraabdominal infections) was increased to nearly half of the trial population, more than one-third of the simulated trials suggested harm from eritoran. This finding is consistent with animal models that suggest toll-like receptor 4 signaling aids bacterial clearance from the peritoneum in patients with intraabdominal sepsis.21 The high proportion with activated protein C and no benefit when the proportion of patients with the α phenotype was increased in the simulated PROWESS trial raises the possibility that such patients were also more common in the subsequent negative trials of activated protein C.22,23
The largest changes were seen in the ProCESS trial, which found no benefit from early goal-directed therapy compared with usual care. In simulations, when the δ phenotype was increased, early goal-directed therapy was harmful in more than half of the trials. This finding supports data from 2 RCTs conducted in low- to middle-income countries that found harm from early goal-directed therapy in select populations.24,25 Increases in the α phenotype suggested benefit from early goal-directed therapy, similar to the initial report by Rivers et al.26 These data highlight the importance of characterizing the heterogeneity of sepsis when comparing across trials with different conclusions.
These findings have additional implications. First, completed trials may have unrecognized heterogeneity in the treatment effects by clinical phenotype that were not apparent when analyzing (1) the entire cohort, (2) subgroups based on individual variables, or (3) stratification based on risk of death.27 However, a secondary analysis of treatment × phenotype interactions may be limited by small sample sizes. Second, these proof-of-concept clinical phenotypes could be incorporated prospectively in future study designs that test new biologically active therapeutics. Novel designs could enrich for a priori phenotypes as well as confirm the boundaries around predictive phenotypes during the trial.28
Limitations
This study has several limitations. First, only routinely available clinical data in the electronic health record were used to identify phenotypes, and the inclusion of other data such as clinicians’ impression, protein biomarkers, immune cell gene expression, or pathogen variables during derivation could change phenotype assignments. However, there appears to be some similarity between the clinical phenotypes derived in this study and those described in other series using such data.
Second, the statistical approach involved a variety of supervised decisions such as (1) the time window for capturing data at hospital presentation was 6 hours, (2) the selection of candidate variables, and (3) the handling of variable distributions. Changes to the initial assumptions, the time window for data capture, or the choice of optimal cluster number could alter the results. The findings were consistent when the electronic health record window of 12 hours was used.
Third, because missing data were common for some variables included in the clustering models, multiple imputation was used in the primary analysis. However, variables with high missingness were excluded from the sensitivity analyses and similar results were still found.
Fourth, differences in short- and long-term prognosis were present across phenotypes, perhaps due to different features of the validation cohorts, such as the definition of sepsis, demographics, or burden of organ dysfunction.
Fifth, characteristics of clinical phenotypes were derived initially from a single integrated health system in the United States at a single moment in clinical care. Although phenotypes were found to be generalizable in the other data sets examined, further exploration is necessary, especially using data from low- and middle-income countries, more recent clinical trials, and longitudinal cohorts.
Conclusions
In this retrospective analysis of data sets from patients with sepsis, 4 clinical phenotypes were identified that correlated with host-response patterns and clinical outcomes, and simulations suggested these phenotypes may help in understanding heterogeneity of treatment effects. Further research is needed to determine the utility of these phenotypes in clinical care and for informing trial design and interpretation.
Collaborators and acknowledgements
eMethods
eFigure 1. Study schematic
eFigure 2. Heatmap of correlation of phenotype variables
eFigure 3. Patient accrual in SENECA derivation cohort
eFigure 4. OPTICS plot for SENECA derivation data
eFigure 5. Consensus k clustering in SENECA derivation data
eFigure 6. Descriptive plots of phenotyping variables, age through chloride
eFigure 7. Descriptive plots of phenotyping variables, c-reactive protein through INR
eFigure 8. Descriptive plots of phenotyping variables, serum lactate through troponin
eFigure 9. Rank order of variables importance
eFigure 10. Frequency of phenotypes across 12 hospitals in SENECA derivation data and GenIMS
eFigure 11. Proportion of phenotypes without parenteral antibiotics or blood cultures as first intervention
eFigure 12. Histogram of latent class probabilities by phenotype
eFigure 13. t-SNE plots of phenotype assignments in SENECA derivation cohort
eFigure 14. Probability of assignment for phenotype members and non-members
eFigure 15. Consensus k means clustering results from SENECA validation cohort
eFigure 16. Mean standardized differences between variables across phenotype pairs in SENECA derivation and validation cohorts
eFigure 17 Euclidean distances by phenotype for GenIMS cohort study
eFigure 18. T-SNE plots of phenotype assignments in RCTs
eFigure 19. Euclidean distances by phenotype for ACCESS trial
eFigure 20. Euclidean distances by phenotype for PROWESS trial
eFigure 21. Euclidean distances by phenotype for ProCESS trial
eFigure 22. 365-day mortality by phenotype in ACCESS, PROWESS, and ProCESS trials
eFigure 23. Cumulative 28-day survival by treatment arm within phenotypes in the ACCESS trial
eFigure 24. Cumulative 365-day survival by treatment arm within phenotypes in the ACCESS trial
eFigure 25. Cumulative 28-day survival by treatment arm within phenotypes in the PROWESS trial
eFigure 26. Cumulative 365-day survival by treatment arm within phenotypes in the PROWESS trial
eFigure 27. Cumulative 28-day survival by treatment arm within phenotypes in the ProCESS trial
eFigure 28. Cumulative 365-day survival by treatment arm within phenotypes in the ProCESS trial
eFigure 29. Simulation of phenotype enrichment in the ProCESS trial
eFigure 30. Simulation of phenotype enrichment in the ACCESS trial
eFigure 31. Simulation of phenotype enrichment in the PROWESS trial
eFigure 32. Control group mortality rates in simulation compared to contemporary RCTs
eFigure 33. Alluvial plot of phenotypes by baseline SOFA score
eFigure 34. Distribution of phenotypes across APACHE quartiles in 3 RCTs
eFigure 35. Alluvial plot of phenotypes by infection site in the ACCESS trial
eFigure 36. Distribution of phenotypes among patients with bacteremia in SENECA derivation cohort
eFigure 37. Comparison of clinical variables between phenotypes and APACHE3 quartiles
eFigure 38. Comparison of biomarkers between phenotypes and APACHE3 quartiles
eFigure 39. Sensitivity analysis of enrichment by APACHE3 quartile in ProCESS trial
eTable 1. Clinical variables used in models to derive phenotypes
eTable 2. Biomarkers available in cohort and trial data
eTable 3. Range, direction, and transformation of variables for model in SENECA cohorts
eTable 4. Missing data
eTable 5. Characteristics of cohort studies
eTable 6. Characteristics of infection and organ dysfunction screening in SENECA derivation and validation cohorts
eTable 7. Characteristics of 3 randomized trials
eTable 8. Characteristics in derivation and validation data after multiple imputation
eTable 9. Blood culture rate and parenteral antibiotic administration by phenotype
eTable 10. Statistical measures of fit for latent class models
eTable 11. Clinical characteristics of phenotypes derived using latent class analysis
eTable 12. Clinical characteristics of phenotypes derived in SENECA validation cohort
eTable 13. Clinical characteristics of phenotypes after excluding variables with missing data
eTable 14. Clinical characteristics of phenotypes after excluding variables with missing data and high correlation
eTable 15. Clinical characteristics of phenotypes using 12-hour window of EHR data
eTable 16. Clinical characteristics of phenotypes predicted in the GenIMS cohort study
eTable 17. Clinical characteristics of phenotypes predicted in the ACCESS trial
eTable 18. Clinical characteristics of phenotypes predicted in the PROWESS trial
eTable 19. Clinical characteristics of phenotypes predicted in the ProCESS trial
eTable 20. Biomarkers by phenotypes in the GenIMS cohort study
eTable 21. Biomarkers by phenotypes in the ACCESS randomized trial
eTable 22. Biomarkers by phenotypes in the PROWESS randomized trial
eTable 23. Biomarkers by phenotypes in the ProCESS randomized trial
eTable 24. Primary and secondary outcomes by study
eTable 25. Primary and secondary outcomes by phenotype
eTable 26. Baseline characteristics of ACCESS trial in simulation scenarios
eTable 27. Baseline characteristics of PROWESS trial in simulation scenarios
eTable 28. Baseline characteristics of ProCESS trial in simulation scenarios
eTable 29. Control group mortality rate in phenotype simulations in 3 RCTs
eTable 30. Site of infection by phenotype in the ACCESS trial
eTable 31. Clinical characteristics by APACHE3 quartile in ProCESS
eTable 32. Biomarkers by APACHE3 quartile in ProCESS
eReferences
Section Editor: Derek C. Angus, MD, MPH, Associate Editor, JAMA (angusdc@upmc.edu).
References
- 1.Rhee C, Dantes R, Epstein L, et al. Incidence and trends of sepsis in US hospitals using clinical vs claims data, 2009-2014. JAMA. 2017;318(13):1241-1249. doi: 10.1001/jama.2017.13836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seymour CW, Liu VX, Iwashyna TJ, et al. Assessment of clinical criteria for sepsis: for the third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA. 2016;315(8):762-774. doi: 10.1001/jama.2016.0288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scicluna BP, van Vught LA, Zwinderman AH, et al. Classification of patients with sepsis according to blood genomic endotype. Lancet Respir Med. 2017;5(10):816-826. doi: 10.1016/S2213-2600(17)30294-1 [DOI] [PubMed] [Google Scholar]
- 4.Sweeney TE, Azad TD, Donato M, et al. Unsupervised analysis of transcriptomics in bacterial sepsis across multiple datasets reveals three robust clusters. Crit Care Med. 2018;46(6):915-925. doi: 10.1097/CCM.0000000000003084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davenport EE, Burnham KL, Radhakrishnan J, et al. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Respir Med. 2016;4(4):259-271. doi: 10.1016/S2213-2600(16)00046-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Opal SM, Laterre PF, Francois B, et al. ; ACCESS Study Group . Effect of eritoran, an antagonist of MD2-TLR4, on mortality in patients with severe sepsis. JAMA. 2013;309(11):1154-1162. doi: 10.1001/jama.2013.2194 [DOI] [PubMed] [Google Scholar]
- 7.Yealy DM, Kellum JA, Huang DT, et al. ; ProCESS Investigators . A randomized trial of protocol-based care for early septic shock. N Engl J Med. 2014;370(18):1683-1693. doi: 10.1056/NEJMoa1401602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernard GR, Vincent JL, Laterre PF, et al. ; Recombinant human protein C Worldwide Evaluation in Severe Sepsis (PROWESS) Study Group . Efficacy and safety of recombinant human activated protein C for severe sepsis. N Engl J Med. 2001;344(10):699-709. doi: 10.1056/NEJM200103083441001 [DOI] [PubMed] [Google Scholar]
- 9.Kellum JA, Kong L, Fink MP, et al. ; GenIMS Investigators . Understanding the inflammatory cytokine response in pneumonia and sepsis. Arch Intern Med. 2007;167(15):1655-1663. doi: 10.1001/archinte.167.15.1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy MM, Fink MP, Marshall JC, et al. ; SCCM/ESICM/ACCP/ATS/SIS . 2001 SCCM/ESICM/ACCP/ATS/SIS international sepsis definitions conference. Crit Care Med. 2003;31(4):1250-1256. doi: 10.1097/01.CCM.0000050454.01978.3B [DOI] [PubMed] [Google Scholar]
- 11.Vincent JL, Moreno R, Takala J, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. Intensive Care Med. 1996;22(7):707-710. doi: 10.1007/BF01709751 [DOI] [PubMed] [Google Scholar]
- 12.Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science. 2012;335(6071):936-941. doi: 10.1126/science.1214935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Angus DC, Linde-Zwirble WT, Lidicker J, et al. Epidemiology of severe sepsis in the United States. Crit Care Med. 2001;29(7):1303-1310. doi: 10.1097/00003246-200107000-00002 [DOI] [PubMed] [Google Scholar]
- 14.Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med. 2013;369(21):2063. [DOI] [PubMed] [Google Scholar]
- 15.Newgard CD, Haukoos JS. Advanced statistics: missing data in clinical research—part 2: multiple imputation. Acad Emerg Med. 2007;14(7):669-678. [DOI] [PubMed] [Google Scholar]
- 16.Ankerst M. OPTICS: ordering points to identify the clustering structure. SIGMOD Rec. 1999;28(2):49-60. doi: 10.1145/304181.304187 [DOI] [Google Scholar]
- 17.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572-1573. doi: 10.1093/bioinformatics/btq170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calfee CS, Delucchi K, Parsons PE, et al. ; NHLBI ARDS Network . Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med. 2014;2(8):611-620. doi: 10.1016/S2213-2600(14)70097-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rindskopf D, Rindskopf W. The value of latent class analysis in medical diagnosis. Stat Med. 1986;5(1):21-27. doi: 10.1002/sim.4780050105 [DOI] [PubMed] [Google Scholar]
- 20.Knaus WA, Wagner DP, Draper EA, et al. The APACHE III prognostic system: risk prediction of hospital mortality for critically ill hospitalized adults. Chest. 1991;100(6):1619-1636. doi: 10.1378/chest.100.6.1619 [DOI] [PubMed] [Google Scholar]
- 21.Deng M, Scott MJ, Loughran P, et al. Lipopolysaccharide clearance, bacterial clearance, and systemic inflammatory responses are regulated by cell type-specific functions of TLR4 during sepsis. J Immunol. 2013;190(10):5152-5160. doi: 10.4049/jimmunol.1300496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abraham E, Laterre PF, Garg R, et al. ; Administration of Drotrecogin Alfa (Activated) in Early Stage Severe Sepsis (ADDRESS) Study Group . Drotrecogin alfa (activated) for adults with severe sepsis and a low risk of death. N Engl J Med. 2005;353(13):1332-1341. doi: 10.1056/NEJMoa050935 [DOI] [PubMed] [Google Scholar]
- 23.Ranieri VM, Thompson BT, Barie PS, et al. ; PROWESS-SHOCK Study Group . Drotrecogin alfa (activated) in adults with septic shock. N Engl J Med. 2012;366(22):2055-2064. doi: 10.1056/NEJMoa1202290 [DOI] [PubMed] [Google Scholar]
- 24.Maitland K, Kiguli S, Opoka RO, et al. ; FEAST Trial Group . Mortality after fluid bolus in African children with severe infection. N Engl J Med. 2011;364(26):2483-2495. doi: 10.1056/NEJMoa1101549 [DOI] [PubMed] [Google Scholar]
- 25.Andrews B, Muchemwa L, Kelly P, Lakhi S, Heimburger DC, Bernard GR. Simplified severe sepsis protocol: a randomized controlled trial of modified early goal-directed therapy in Zambia. Crit Care Med. 2014;42(11):2315-2324. doi: 10.1097/CCM.0000000000000541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivers E, Nguyen B, Havstad S, et al. ; Early Goal-Directed Therapy Collaborative Group . Early goal-directed therapy in the treatment of severe sepsis and septic shock. N Engl J Med. 2001;345(19):1368-1377. doi: 10.1056/NEJMoa010307 [DOI] [PubMed] [Google Scholar]
- 27.Kent DM, Hayward RA. Limitations of applying summary results of clinical trials to individual patients: the need for risk stratification. JAMA. 2007;298(10):1209-1212. doi: 10.1001/jama.298.10.1209 [DOI] [PubMed] [Google Scholar]
- 28.Berry SM, Connor JT, Lewis RJ. The platform trial: an efficient strategy for evaluating multiple treatments. JAMA. 2015;313(16):1619-1620. doi: 10.1001/jama.2015.2316 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Collaborators and acknowledgements
eMethods
eFigure 1. Study schematic
eFigure 2. Heatmap of correlation of phenotype variables
eFigure 3. Patient accrual in SENECA derivation cohort
eFigure 4. OPTICS plot for SENECA derivation data
eFigure 5. Consensus k clustering in SENECA derivation data
eFigure 6. Descriptive plots of phenotyping variables, age through chloride
eFigure 7. Descriptive plots of phenotyping variables, c-reactive protein through INR
eFigure 8. Descriptive plots of phenotyping variables, serum lactate through troponin
eFigure 9. Rank order of variables importance
eFigure 10. Frequency of phenotypes across 12 hospitals in SENECA derivation data and GenIMS
eFigure 11. Proportion of phenotypes without parenteral antibiotics or blood cultures as first intervention
eFigure 12. Histogram of latent class probabilities by phenotype
eFigure 13. t-SNE plots of phenotype assignments in SENECA derivation cohort
eFigure 14. Probability of assignment for phenotype members and non-members
eFigure 15. Consensus k means clustering results from SENECA validation cohort
eFigure 16. Mean standardized differences between variables across phenotype pairs in SENECA derivation and validation cohorts
eFigure 17 Euclidean distances by phenotype for GenIMS cohort study
eFigure 18. T-SNE plots of phenotype assignments in RCTs
eFigure 19. Euclidean distances by phenotype for ACCESS trial
eFigure 20. Euclidean distances by phenotype for PROWESS trial
eFigure 21. Euclidean distances by phenotype for ProCESS trial
eFigure 22. 365-day mortality by phenotype in ACCESS, PROWESS, and ProCESS trials
eFigure 23. Cumulative 28-day survival by treatment arm within phenotypes in the ACCESS trial
eFigure 24. Cumulative 365-day survival by treatment arm within phenotypes in the ACCESS trial
eFigure 25. Cumulative 28-day survival by treatment arm within phenotypes in the PROWESS trial
eFigure 26. Cumulative 365-day survival by treatment arm within phenotypes in the PROWESS trial
eFigure 27. Cumulative 28-day survival by treatment arm within phenotypes in the ProCESS trial
eFigure 28. Cumulative 365-day survival by treatment arm within phenotypes in the ProCESS trial
eFigure 29. Simulation of phenotype enrichment in the ProCESS trial
eFigure 30. Simulation of phenotype enrichment in the ACCESS trial
eFigure 31. Simulation of phenotype enrichment in the PROWESS trial
eFigure 32. Control group mortality rates in simulation compared to contemporary RCTs
eFigure 33. Alluvial plot of phenotypes by baseline SOFA score
eFigure 34. Distribution of phenotypes across APACHE quartiles in 3 RCTs
eFigure 35. Alluvial plot of phenotypes by infection site in the ACCESS trial
eFigure 36. Distribution of phenotypes among patients with bacteremia in SENECA derivation cohort
eFigure 37. Comparison of clinical variables between phenotypes and APACHE3 quartiles
eFigure 38. Comparison of biomarkers between phenotypes and APACHE3 quartiles
eFigure 39. Sensitivity analysis of enrichment by APACHE3 quartile in ProCESS trial
eTable 1. Clinical variables used in models to derive phenotypes
eTable 2. Biomarkers available in cohort and trial data
eTable 3. Range, direction, and transformation of variables for model in SENECA cohorts
eTable 4. Missing data
eTable 5. Characteristics of cohort studies
eTable 6. Characteristics of infection and organ dysfunction screening in SENECA derivation and validation cohorts
eTable 7. Characteristics of 3 randomized trials
eTable 8. Characteristics in derivation and validation data after multiple imputation
eTable 9. Blood culture rate and parenteral antibiotic administration by phenotype
eTable 10. Statistical measures of fit for latent class models
eTable 11. Clinical characteristics of phenotypes derived using latent class analysis
eTable 12. Clinical characteristics of phenotypes derived in SENECA validation cohort
eTable 13. Clinical characteristics of phenotypes after excluding variables with missing data
eTable 14. Clinical characteristics of phenotypes after excluding variables with missing data and high correlation
eTable 15. Clinical characteristics of phenotypes using 12-hour window of EHR data
eTable 16. Clinical characteristics of phenotypes predicted in the GenIMS cohort study
eTable 17. Clinical characteristics of phenotypes predicted in the ACCESS trial
eTable 18. Clinical characteristics of phenotypes predicted in the PROWESS trial
eTable 19. Clinical characteristics of phenotypes predicted in the ProCESS trial
eTable 20. Biomarkers by phenotypes in the GenIMS cohort study
eTable 21. Biomarkers by phenotypes in the ACCESS randomized trial
eTable 22. Biomarkers by phenotypes in the PROWESS randomized trial
eTable 23. Biomarkers by phenotypes in the ProCESS randomized trial
eTable 24. Primary and secondary outcomes by study
eTable 25. Primary and secondary outcomes by phenotype
eTable 26. Baseline characteristics of ACCESS trial in simulation scenarios
eTable 27. Baseline characteristics of PROWESS trial in simulation scenarios
eTable 28. Baseline characteristics of ProCESS trial in simulation scenarios
eTable 29. Control group mortality rate in phenotype simulations in 3 RCTs
eTable 30. Site of infection by phenotype in the ACCESS trial
eTable 31. Clinical characteristics by APACHE3 quartile in ProCESS
eTable 32. Biomarkers by APACHE3 quartile in ProCESS
eReferences