Abstract
The precise cell type hosting latent human cytomegalovirus (HCMV) remains elusive. Here, we report that HCMV reprogrammes human haematopoietic progenitor cells (HPCs) into a unique monocyte subset to achieve latency. Unlike conventional monocytes, this monocyte subset possesses higher levels of B7-H4, IL-10 and inducible nitric oxide synthase (iNOS), a longer lifespan and strong immunosuppressive capacity. Cell sorting of peripheral blood from latently infected human donors confirms that only this monocyte subset, representing less than 0.1% of peripheral mononuclear cells, is HCMV genome-positive but immediate-early-negative. Mechanistic studies demonstrate that HCMV promotes the differentiation of HPCs into this monocyte subset by activating cellular signal transducer and activator of transcription 3 (STAT3). In turn, this monocyte subset generates a high level of nitric oxide (NO) to silence HCMV immediate-early transcription and promote viral latency. By contrast, the US28-knockout HCMV mutant, which is incapable of activating STAT3, fails to reprogramme the HPCs and achieve latency. Our findings reveal that via activating the STAT3–iNOS–NO axis, HCMV differentiates human HPCs into a longevous, immunosuppressive monocyte subset for viral latency.
HCMV is a widespread pathogen and can establish life-long latent infection in large populations1. Although HCMV reactivation from latency is normally asymptomatic in a healthy person, it can be fatal for immunocompromised individuals such as AIDS patients2 and organ transplant recipients3.
Previous studies have shown that HCMV latency is restricted to myeloid cells and the process of establishing dormancy probably involves the action of viral tegument proteins, as well as epi-genetic modifications of the viral genome4,5. The elegant work by Reeves and coworkers6 showed that circulating dendritic cells are sites of viral reactivation. However, not all myeloid linage cells can be the hosts of HCMV latency. Employing sensitive PCR, Bevan et al.7 were the first to detect the HCMV genome in whole peripheral blood from latently infected donors. Although conventional mature monocytes generally have a short life span, it has been demonstrated8 that HCMV infection could induce monocytes to differentiate into macrophages to obtain longevity. Since then, a number of researchers have established experimental latent systems in monocytes8–10. However, although monocytes are able to harbour experimental latency, it is widely accepted that HCMV achieves natural latency in CD34+ H PCs11–17. When primitive HPCs are infected with HCMV, the expression of viral lytic genes is rapidly suppressed, and the viral genome is maintained as a stable episome without producing new virions18,19. The host cells carrying latent viral genomes also have the capability to re-enter the active infection cycle in response to stimuli2. Despite increased research in the area of HCMV latent infection, much remains to be understood about the viral and cellular mechanisms that are involved in the establishment of latency.
HCMV infection can profoundly affect the infected cells, resulting in the modulation of cell metabolism, cell cycle, cell death and immune surveillance. These fundamental changes to the infected cells may contribute to the establishment of HCMV latency. Despite a much-restricted transcription profile, HCMV latent infection is associated with an active manipulation of host cell functions14,15,20,21,22, leading to optimization of the host cell for latent carriage and reactivation. Using next-generation sequencing to compare the transcriptome profile of HCMV latently infected CD14+ and CD34+ cells in experimental latency with that in natural latency, two long non-coding RNAs, RNA4.9 and RNA2.7 have been identified15, as well as the messenger RNAs encoding replication factors UL84 and UL44, which may contribute to mediating viral genome maintenance. It has also been reported14 that a smaller IE1 protein species expressed in latently infected HPCs is required for viral genome persistence and maintenance, suggesting that viral chromosome maintenance and replication in the host cells is dependent on certain HCMV latency-specific factors.
In the present study, through characterizing how establishment of latent infection with either a wild-type strain of HCMV or an HCMV US28-knockout mutant would impact CD34+ HPCs isolated from human bone marrow, we report that the clinical HCMV strain can reprogramme human HPCs into a unique monocyte subset necessary for establishing latency. Different from conventional monocytes, this monocyte subset possesses higher levels of B7-H4, IL-10 and inducible nitric oxide synthase (iNOS), a longer lifespan and a strong immunosuppressive capacity. Our mechanistic studies further show that a high nitric oxide (NO) level is critical for suppressing HCMV immediate-early (IE) gene expression and viral replication, and that HCMV-induced cellular STAT3 activity plays an essential role in reprogramming HPCs into this monocyte subset for viral latency.
Results
HCMV latent infection differentiates HPCs into an immunosuppressive monocyte subset.
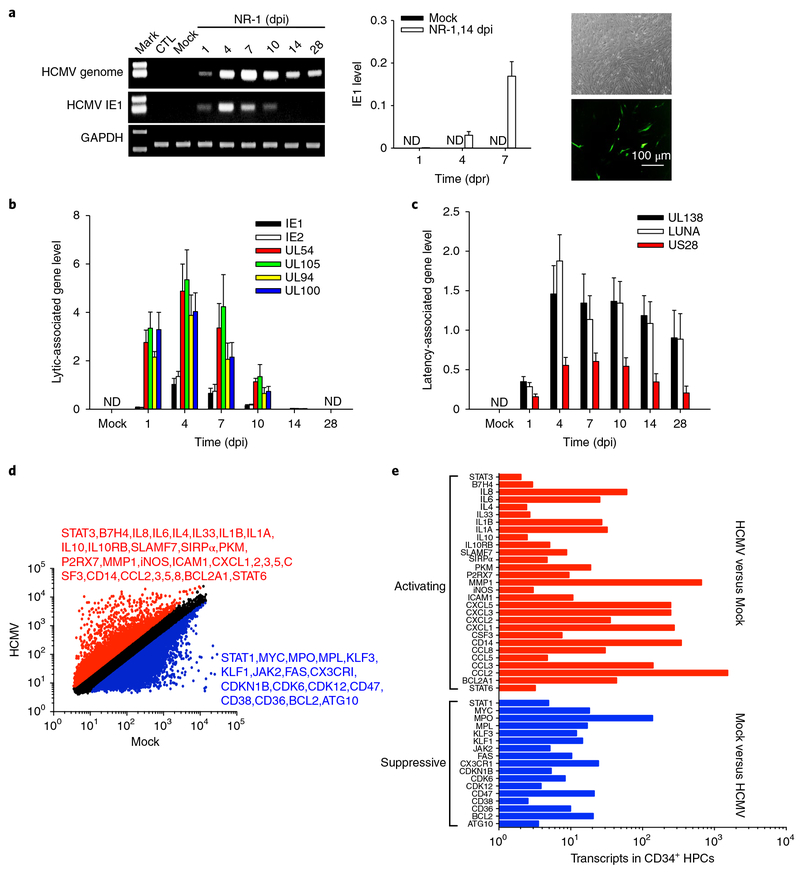
To identify the characteristics of HCMV latently infected cells, we infected CD34+ HPCs from human bone marrow with a clinical HCMV strain NR-1 (ref.23). The NR-1 strain contains a green fluorescent protein (GFP) reporter so the infected cells can be enriched through cell sorting23. In this experiment, bone marrow was collected from donors who were HCMV-negative as assessed using anti-HCMV antibody and HPCs were isolated as previously described17,23. Given that CD34+ HPCs are highly heterogeneous and contain haematopoietic stem cells (HSCs), HSC-derived multipotent progenitor (MPP), common myeloid progenitor (CMP) and granulocyte-macrophage progenitor, we used CD33 as a marker to sort CD33-positive common myeloid progenitor and granulocyte-macrophage progenitor from CD33-negative HSCs and MPP. In the present study, only more primitive progenitor cells, CD34+CD33− HSCs and MPP, were used for viral latent infection experiments. HPCs were infected with NR-1 at a multiplicity of infection (MOI) of two for the establishment of viral latency. Given a low efficiency of HCMV to HPCs at low MOI24, we sorted out the cells successfully infected with NR-123 at two days post infection (dpi). NR-1 latently infected HPCs were treated with 12-O-tetradecanoylphorbol-13-acetate (TPA) for 24 h and then incubated with human foreskin fibroblasts (HFF) to reactivate HCMV from latent infection. As shown in Fig. 1a–c, unlike laboratory Towne and Ad169 strains which failed to achieve latency in HPCs, NR-1 successfully achieved latency in HPCs after 14 dpi, as indicated by positive viral genome and latency-associated gene transcription but negative IE1 and lytic infection-associated gene transcription. Both PCR with reverse transcription and viral titre assays confirmed that viral latency could be reactivated by coculture with HFF (Supplementary Fig. 1). HCMV latent infection in CD34+CD33− HPCs after 14 dpi was confirmed by using another clinical isolate, VR-1814 (Supplementary Fig. 2).
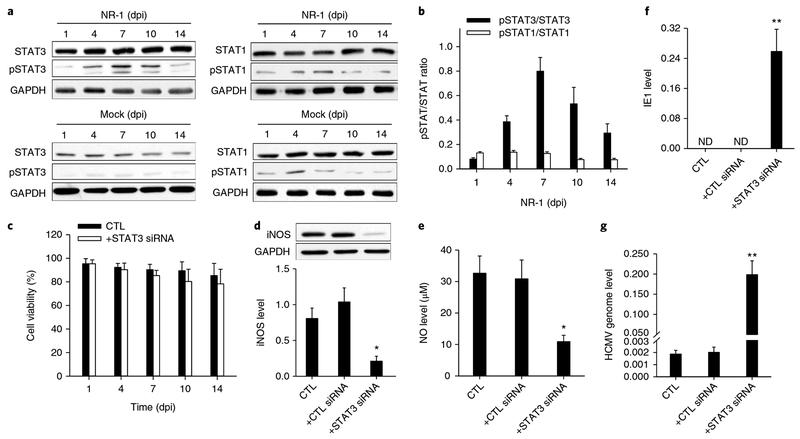
Fig. 1 |. HCMV NR-1 infection reprogrammes human CD34+ HPCs to achieve latent infection.
a, NR-1 successfully established latency in HPCs. HPCs isolated from bone marrow were infected with NR-1 or deactivated NR-1 (Mock) at a multiplicity of 2 p.f.u per cell. Virus was reactivated by TPA (20 ng ml−1) followed by coculture with HFF-1 cells. Left, levels of HCMV genome and IE1 in HPCs following NR-1 or Mock infection. Middle and right panels represent the quantitative PCR with reverse transcription result of IE1 expression and virus replication of GFP-expressing NR-1 after reactivating virus from latent infection in HPCs (NR-1, 14 dpi), respectively. b, Levels of HCMV IE1, IE2 and lytic infection-associated genes UL54, UL94, UL100 and UL105 in HPCs following the infection with NR-1 or Mock. c, Levels of HCMV latency-associated genes UL138, LUNA and US28 in HPCs following the infection with NR-1 or Mock. d, Alteration of transcription profiling of CD34+ HPCs by NR-1 latent infection (14 dpi). The red-coloured molecules are upregulated (fold change > 2) by NR-1 infection compared to Mock infection, whereas the blue-coloured molecules are downregulated (fold change < 0.5) by NR-1 infection compared to Mock infection. e, The activating (red-coloured) and suppressive (blue-coloured) signal pathways in NR-1 latently infected HPCs compared to those in Mock-infected HPCs. Data are presented as the mean ± s.e.m. of three independent experiments. CTL, control; ND, not detected; dpr, days post reactivation.
To define the cell type harbouring NR-1 latency, we compared the genome-wide transcription profiling in CD34+CD33− HPCs latently infected with NR-1 with that in Mock-infected CD34+CD33− HPCs. Affymetrix microarray data (GSE106879) showed that, compared to Mock infection, NR-1 latency upregulated 6,503 gene transcripts (fold change > 2, red) and downregulated 4,848 gene transcripts (fold change < 0.5, blue) in HPCs at 14 dpi (Fig. 1d). Specifically, compared to HPCs with Mock infection, the NR-1 latently infected HPCs expressed significantly lower levels of progenitor cell markers, such as CD34, Myc and KLF1/3, but higher levels of monocytic marker proteins, chemokines and adhesion molecules, including CD14, CD33, CCL2–8, ICAM1 and B7-H4 (Fig. 1e), suggesting that CD34+CD33− HPCs were reprogrammed into monocyte-like cells during HCMV latency.
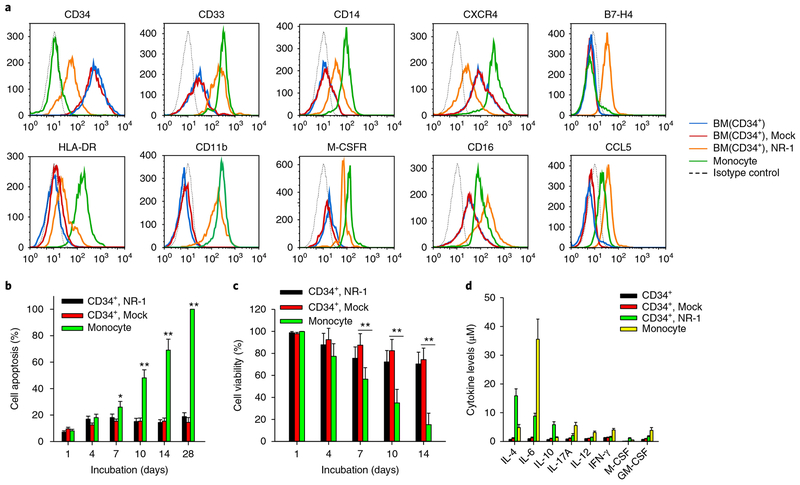
We next examined the expression profile of cellular surface marker proteins on the HPCs infected with NR-1 at 14 dpi by flow cytometry. As shown in Fig. 2a, NR-1 latent infection resulted in a significant loss of CD34 but gains of CD14, CD33, CD11b, CD16 and M-CSFR on HPCs, confirming that HPCs indeed differentiate into a monocyte-like cell subset. However, compared to mature monocytes, the cells harbouring NR-1 latency expressed lower levels of CD14 and HLA-DR but higher levels of M-CSFR and CD16. Moreover, NR-1-infected HPCs became B7-H4-positive, while mature monocytes remained B7-H4-negative. As the CD14loCD16+ monocyte subset, currently considered non-classical or ‘patrolling’ monocytes, exhibits a longer lifespan than classic CD14hiCD16− monocytes25, we tested whether the cells harbouring NR-1 latency also had a longer lifespan than mature monocytes. Cell apoptosis (Fig. 2b) and viability assays (Fig. 2c) both confirmed that NR-1 latently infected HPCs had significantly delayed apoptosis and increased cell viability compared to mature monocytes. Analysis of the cytokine profile of NR-1 latently infected HPCs confirmed that HPCs were reprogrammed by HCMV infection. As shown in Fig. 2d, the levels of IL-4, IL-6, IFN-γ, GM-CSF and IL-10 secreted by the latently infected cells were significantly higher than those secreted by HPCs with Mock infection. Compared to mature monocytes, NR-1 latently infected cells expressed higher levels of IL-4 and IL-10 but lower levels of IL-6, IFN-γ and GM-CSF.
Fig. 2 |. HCMV NR-1 infection reprogrammes human CD34+ HPCs into a long-life monocyte subset at the late stage of infection.
a, Expression of surface markers on conventional mature monocytes and HPCs infected with NR-1 or Mock at 14 dpi. BM, bone marrow. b,c, NR-1-infected HPCs at 14 dpi displayed delayed apoptosis (b) and increased cell viability (c) compared to mature monocytes and Mock-infected HPCs. d, Cytokine level in mature monocytes and HPCs infected with NR-1 or Mock at 14 dpi. Data are presented as the mean ± s.e.m. of three independent experiments. *P < 0.05, **P < 0.01 as determined by the two-tailed t-test (the P values are detailed in Supplementary Table 1).
We also tested gene expression of the infected CD34+CD33− HPCs at different time points following the infection. As shown in Supplementary Fig. 3, the CD34 mRNA level had rapidly decreased in HPCs infected with NR-1 compared to those with Mock infection. In contrast, the CD45 mRNA level started to dramatically increase in NR-1-infected CD34+CD33− HPCs after 7 dpi compared to that with Mock infection. The levels of CD33, B7-H4, CD16 and CD14 transcripts had increased in a linear fashion along the time course of infection in NR-1-infected HPCs compared to those with Mock infection. Comparison of CD34+CD33−CD14− HPCs to CD34+CD33+CD14− HPCs at the late stage of NR-1 infection showed that both HPCs exhibited similar levels of surface B7-H4, CD16 and CD14, suggesting that NR-1 infection results in a differentiation of both HPCs into the CD14loCD16+ monocyte subset (Supplementary Fig. 4).
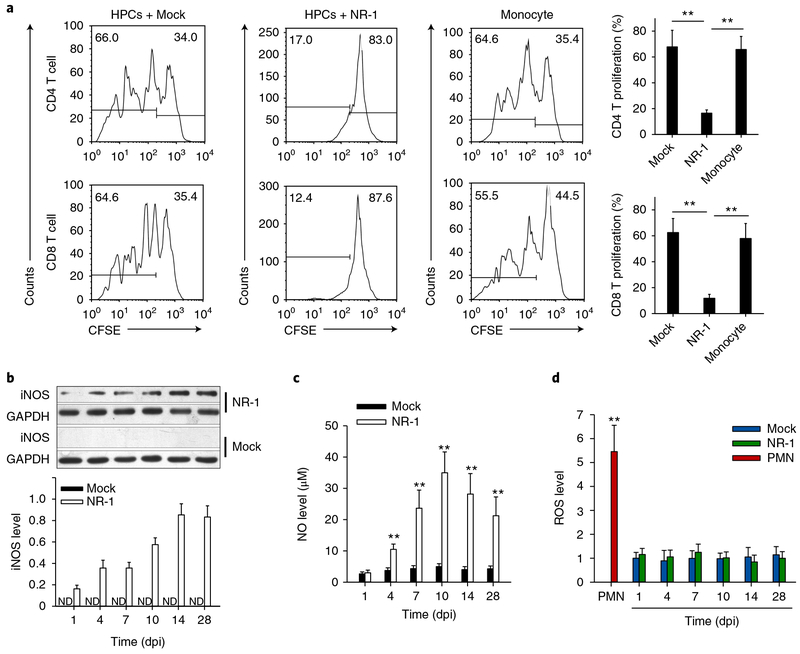
As evading the T-cell immune response is critical for HCMV latency, we tested whether the cells harbouring NR-1 latency can survive the attack by T cells. To our surprise, NR-1 latently infected HPCs strongly inhibited the proliferation of CD4+ and CD8+ T cells induced by concanavalin A (ConA), whereas either HPCs with Mock infection or mature monocytes showed a minimal inhibitory effect (Fig. 3a). Identification of the suppressive capacity of HCMV latently infected cells on T-cell proliferation is in agreement with the previous finding that HCMV UL111A can suppress CD4+ T-cell proliferation26. The findings that the cells harbouring NR-1 latency express CD33 and CD11b and possess immunosuppressive capability suggest that these cells may have acquired the phenotype of human myeloid-derived suppressor cells (MDSCs)27.
Fig. 3 |. NR-1-infected CD34+ HPCs at a late stage of infection (14 dpi or later) posess a strong immunosuppressive capacity to T-cell proliferation in a manner of Mo-MDSCs but not granulocytic MDSCs.
a, NR-1-infected HPCs (14 dpi) suppress CD4 and CD8 T-cell proliferation induced by ConA. The number in the top left corner of each plot represents the percentage of proliferated cells, and the number in the top right corner represents the percentage of non-proliferated cells. b–d, NR-1-infected HPCs expressed high levels of cellular iNOS (b) and NO (c) but not ROS (d) compared to Mock-infected HPCs. Data are presented as the mean ± s.e.m. of three independent experiments. **P < 0.01 as determined by the two-tailed t-test (the P values are detailed in Supplementary Table 1). ND, not detected.
Human MDSCs can be divided into two major populations: monocytic MDSCs (Mo-MDSCs) and granulocytic MDSCs27. Mo-MDSCs suppress T-cell proliferation through NO generated by iNOS, whereas granulocytic MDSCs suppress T-cell proliferation through releasing reactive oxygen species (ROS)27. To explore the mechanism by which the cells harbouring HCMV latency suppress T-cell proliferation, we assessed the levels of iNOS/NO and ROS at various dpi. As shown in Fig. 3b, NR-1 infection rapidly induced iNOS expression, whereas iNOS was barely detectable in HPCs with Mock infection. As expected, the NO level in NR-1 latently infected cells was also markedly elevated compared to HPCs with Mock infection (Fig. 3c). In agreement with this, increased levels of iNOS and NO were also detected in the HPCs latently infected with VR-1814 (Supplementary Fig. 5). By contrast, no increase of ROS level was observed in NR-1 latently infected cells (Fig. 3d). Compared to polymorphonuclear leukocytes that express high levels of ROS, HPCs with Mock or NR-1 infection displayed low ROS production. Comparison of NR-1-infected HPCs to conventional mature monocytes further showed that NR-1 latently infected cells are different from mature monocytes. As shown in Supplementary Fig. 6, NR-1 latently infected cells expressed significantly higher levels of CD16, CD86, CCR5, CD34 and iNOS but lower levels of CCR1 and CD33 than mature monocytes. These results demonstrate that the cells harbouring HCMV latency constitute a unique monocyte subset, exhibiting an immunosuppressive capacity to inhibit T-cell proliferation in a mechanism similar to Mo-MDSCs.
To validate whether this unique monocyte subset can serve as the host of latent HCMV, we directly isolated this monocyte subset from the peripheral blood of HCMV latently infected donors and determined the cellular levels of the HCMV genome and IE1. Peripheral blood mononuclear cells (PBMCs) were first isolated from donors (Supplementary Fig. 7a, left), followed by CD14-positive selection to gate out total monocytes (~13.7% of PBMCs) (Supplementary Fig. 7a, middle). Since this unique monocyte subset harbouring latent NR-1 is B7-H4-positive whereas mature monocytes are B7-H4-negative (Fig. 1c), we further sorted out the B7-H4-positive monocyte subset from whole CD14-positive monocytes by flow cytometry. As shown, this B7-H4-positive monocyte subset occupied only 0.78% of the total peripheral monocytes (Supplementary Fig. 7a, right). As expected, HCMV latency, indicated by viral genome-positive but IE-negative status, was confirmed in the whole PBMCs from HCMV antigen-positive donors but not HCMV antigen-negative donors (Supplementary Fig. 7b). Strikingly, higher levels of the HCMV genome were found in the small population of B7-H4-positive monocytes than in the large population of B7-H4-negative monocytes (Supplementary Fig. 7c). Given that the B7-H4-positive monocyte subset only occupies 0.78% of total monocytes and ~0.1% of whole PBMCs, the viral genome level per cell was significantly higher in B7-H4-positive monocytes than in B7-H4-negative conventional monocytes and the overall PBMCs (Supplementary Fig. 7d). Importantly, although the HCMV genome was positive in the B7-H4-positive monocyte subset, IE1 transcription was undetectable (Supplementary Fig. 7e). However, the level of viral IE1 had strongly increased following the reactivation of HCMV in the B7-H4-positive monocyte subset (Supplementary Fig. 7e,f).
High level of intracellular NO is key for establishing HCMV latency in progenitor cells.
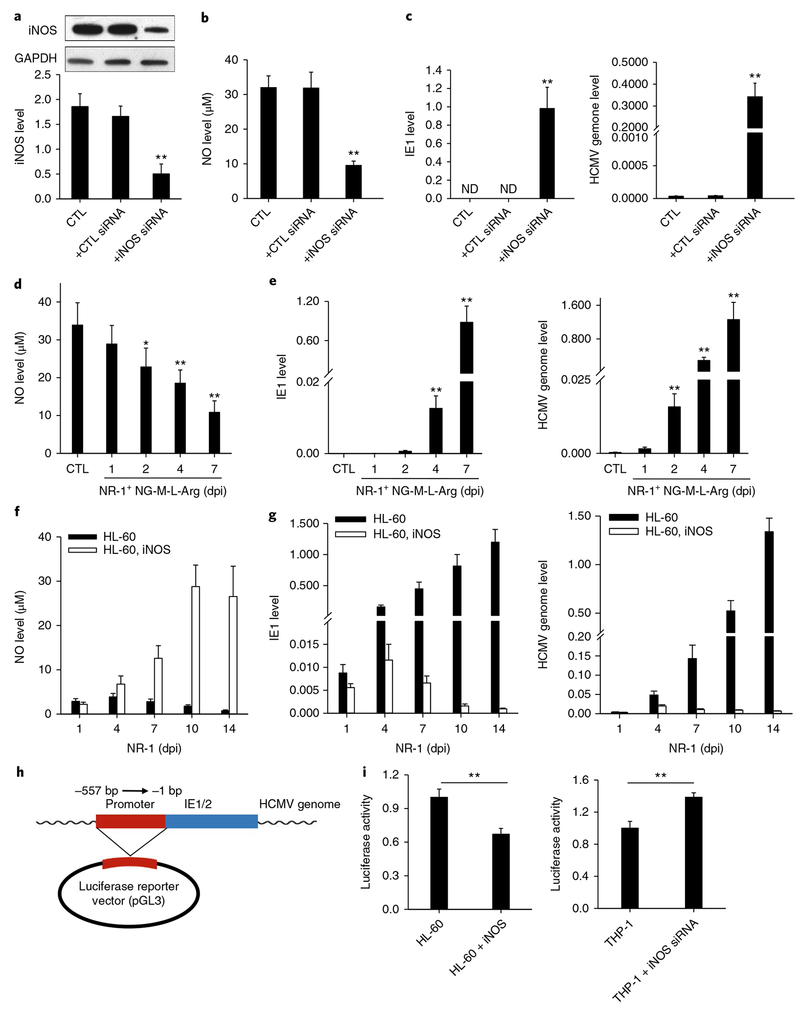
To understand how this B7-H4-positive monocyte subset contributes to HCMV latent infection, we explored the role of NO in modulating HCMV latency by both gain-of-function and loss-of-function assays. We first transfected NR-1-infected HPCs with iNOS short interfering RNA (siRNA) to decrease NO production or directly depleted cellular NO with NG-Methyl-L-arginine acetate salt (NG-M-L-Arg) in host cells. As shown in Fig. 4, decreasing cellular NO production via iNOS siRNA (Fig. 4a,b) in NR-1-infected HPCs dramatically increased HCMV IE1 expression and viral replication (Fig. 4c). Depletion of cellular NO by NG-M-L-Arg treatment in NR-1-infected HPCs (Fig. 4d) also strongly increased HCMV IE1 expression and viral replication (Fig. 4e). Moreover, we also observed that cellular NO level in the latently infected cells was decreased when the cells were treated with TPA28 for viral reactivation (Supplementary Fig. 8). Taken together, these results suggest that a high level of intracellular NO is critical for establishing and maintaining HCMV latency.
Fig. 4 |. Cellular iNOS/NO induced by STAT3 activity play a critical role in suppressing HCMV IE1 expression and viral replication in HPCs.
a,b, Levels of iNOS (a) and NO (b) in HPCs transfected with iNOS siRNA or scramble oligonucleotide. c, Increase of HCMV IE1 expression and genome replication in NR-1-infected HPCs after iNOS siRNA transfection. d, Depletion of cellular NO by NG-M-L-Arg. e, Increase of HCMV IE1 expression and genome replication in NR-1-infected HPCs after direct depletion of cellular NO by NG-M-L-Arg. f, Lentivirus-mediated iNOS overexpression in HL-60 cells increased intracellular NO level. g, Increase of NO level in HL-60 cells suppressed viral IE1 activity and genome replication. h,i, Inhibition of cellular NO on the activity of HCMV IE1 promoter. A luciferase reporter consisting of IE1/2 promoter region was constructed in pMIR-REPORT plasmid (h) and then transfected into HL-60 and THP-1 cells, respectively. HL-60 and THP-1 cells were treated with LV-iNOS or LV-iNOS siRNA to overexpress or knock down iNOS, respectively, prior to NR-1 infection at MOI of 2. Cellular luciferase activity (i) was assayed. Data are presented as the mean ± s.e.m. of three independent experiments. *P < 0.05, **P < 0.01 as determined by the two-tailed t-test (the P values are detailed in Supplementary Table 1). LV, lentivirus vector.
To further confirm the role of NO in HCMV latent infection, we overexpressed iNOS in promyelocytic cell line HL-60 with lentivirus. Since HL-60 cells produce a low level of NO and are unable to host HCMV latency, we tested whether increasing the NO level would make them suitable for viral latency. As shown in Fig. 4f, lentivirus-mediated iNOS expression in NR-1-infected HL-60 cells increased the production of NO, resulting in a strong suppression of both HCMV IE1 expression and viral replication (Fig. 4g). Increasing cellular NO level via iNOS overexpression (Supplementary Fig. 9a) also significantly alleviated the apoptosis of HL-60 cells infected with NR-1 (Supplementary Fig. 9b), suggesting that more viruses entered the quiescent rather than lytic stage.
To test whether intracellular NO can directly affect the transcription of the HCMV IE gene, we constructed a luciferase reporter plasmid consisting of the promoter region of the HCMV IE1/2 gene29 (Fig. 4h) and transfected it into HL-60 and THP-1 cells. As shown in Fig. 4i, the luciferase activity was strongly inhibited in the iNOS-overexpressing HL-60 cells, which suggests that cellular NO suppresses the IE1/2 promoter activity. Interestingly, in the THP-1 cells, the luciferase activity was significantly enhanced when iNOS was knocked down via iNOS siRNA. These results demonstrate that the high level of NO may contribute to the viral latent infection via directly controlling the activity of the HCMV IE1/2 promoter.
HCMV latency-induced progenitor cell reprogramming is dependent on STAT3 activity.
Next, we explored the mechanism underlying the reprogramming of HPCs to the Mo-MDSC-like monocyte subset following HCMV infection. Considering the role of STAT1 and STAT3 in regulating the differentiation of HPCs into Mo-MDSCs30 and the activation of STAT3 during HCMV infection31, we tested whether STAT1 and STAT3 were involved in the cellular reprogramming of the NR-1-infected HPCs. As shown in Fig. 5a,b, NR-1 infection induced a time-dependent increase of phosphorylated STAT3 (pSTAT3) level but not phosphorylated STAT1 (pSTAT1) level. A similar increased pSTAT3 level in HPCs was observed following latent infection with VR-1814 (Supplementary Fig. 10). These results suggest that STAT3 activity may play a critical role in HCMV-induced HPC differentiation to the Mo-MDSC-like monocyte subset, which produces a high level of NO. To confirm it, we knocked down STAT3 in HPCs using STAT3-specific siRNA and then infected the cells with NR-1 (Supplementary Fig. 11). The cell viability assay showed that treatment with lentivirus-mediated STAT3 siRNA or control oligonucleotide delivery did not affect the viability of HPCs (Fig. 5c). However, loss of STAT3 activity resulted in a significant reduction of iNOS expression (Fig. 5d) and NO level (Fig. 5e) in the NR-1-infected HPCs. Accordingly, the levels of HCMV IE1 transcript (Fig. 5f) and viral replication (Fig. 5g) were strongly increased in NR-1-infected HPCs when STAT3 was knocked down.
Fig. 5 |. Role of STAT3 signalling pathway in modulating HPC differentiation during HCMV latent infection.
a,b, Western blot images (a) and analysis (b) the levels of STAT3, pSTAT3, STAT1 and pSTAT1 in NR-1-infected HPCs. Note that NR-1 infection strongly induces STAT3 but not STAT1 activity in HPCs. c, Treatment with lentivirus-mediated STAT3 siRNA or control oligonucleotide delivery did not affect the viability of HPCs. d–g, Reduction of STAT3 and pSTAT3 via STAT3 siRNA decreased the cellular levels of iNOS (d) and NO (e), but enhanced IE1 expression (f) and viral replication (g) in NR-1-infected HPCs at 14 dpi. Data are presented as the mean ± s.e.m. of three independent experiments. *P < 0.05, **P < 0.01 as determined by the two-tailed t-test (the P values are detailed in Supplementary Table 1). CTL, control; ND, not detected.
To validate the critical role of STAT3 in HCMV-induced differentiation of HPCs into the immunosuppressive monocyte subset, we monitored HPCs following the infection by a mutated NR-1 strain that cannot activate STAT3. US28, a viral G protein-coupled receptor encoded by HCMV, has been found to be expressed in HCMV latently infected cells32. Although the exact role of US28 in HCMV latency remains unknown, previous studies showed that US28 constitutively activates signalling pathways linked to cell proliferation31,33. The discovery of colocalization of US28 with pSTAT3 in the vascular niche of the HCMV-associated tumour31 suggests that US28 may activate the JAK1–STAT3 signalling axis. As shown in Supplementary Fig. 12, US28 knockout did not impair the viral growth. However, western blot analysis showed that infection of US28-KO NR-1 strain failed to increase cellular pSTAT3 levels in CD34+ HPCs (Fig. 6a). In line with this, US28-KO infection failed to reprogramme CD34+ HPCs to the immunosuppressive monocyte subset, as shown by an absence of iNOS (Fig. 6b) and NO (Fig. 6c) induction. Different from NR-1 infection which induces B7-H4 and CD16 in CD34+ HPCs, US28-KO infection failed to increase the levels of these proteins but enhanced CD14 level (Fig. 6d). Furthermore, HPCs infected with US28-KO did not inhibit CD4 T-cell proliferation (Fig. 6e). In contrast to NR-1 infection establishing latency at 14 dpi, US28-KO infection in HPCs failed to achieve latency, as indicated by continuous viral replication and persistent IE1 expression (Fig. 6f,g).
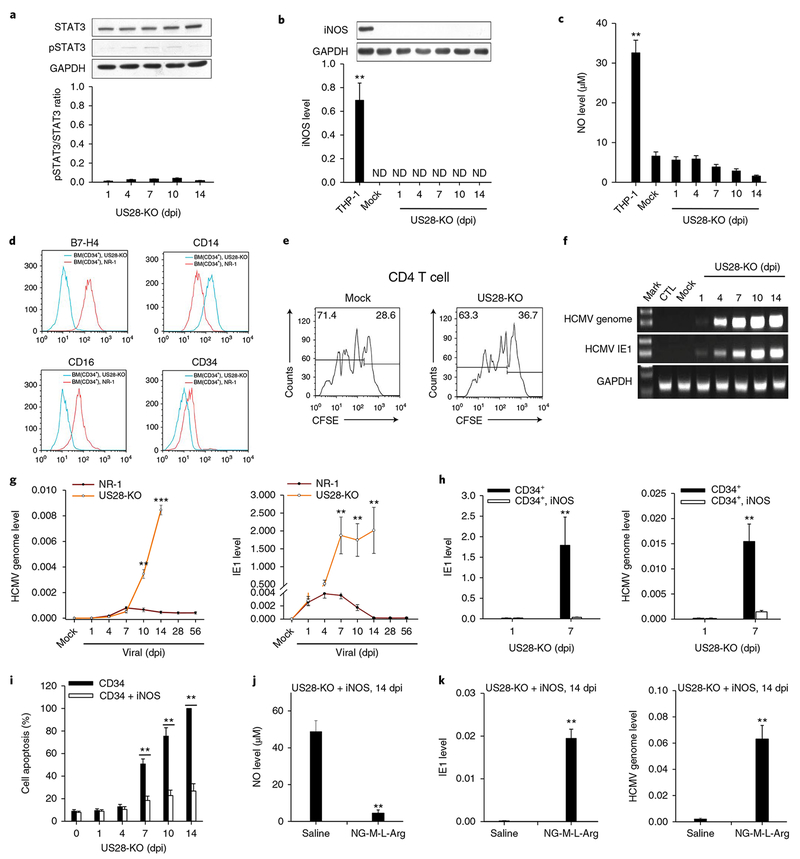
Fig. 6 |. US28-KO cannot establish latency in human CD34+ HPCs because it fails to activate the STAT3–iNOS–NO axis and reprogramme HPCs to an immunosuppressive monocyte subset.
a–c, US28-KO or Mock infection in human HPCs failed to activate STAT3 (a), or increase cellular levels of iNOS (b) and NO (c). d,e, US28-KO infection failed to reprogramme HPCs to the B7-H4+CD16+ immunosuppressive monocyte subset, as indicated by negative or low expression of B7-H4 and CD16 (d), as well as no suppression of CD4 T-cell proliferation (e). f, US28-KO infection failed to achieve latency in HPCs. g, Levels of viral genome and IE1 expression in HPCs infected with US28-KO or NR-1. h, NOS overexpression suppressed viral IE1 and genome expression in the HPCs infected with US28-KO. i, iNOS overexpression decreased the apoptotic rate of US28-KO-infected HPCs. j,k, Depleting cellular NO by NG-M-L-Arg abolished the effect of iNOS overexpression on facilitating US28-KO latency. NG-M-L-Arg depleted NO in US28-KO-infected HPCs that were overexpressed with iNOS (j), leading to significant increase of HCMV IE1 and genome levels (k). Data are presented as the mean ± s.e.m. of three independent experiments. **P < 0.01, ***P < 0.001 as determined by the two-tailed t-test (the P values are detailed in Supplementary Table 1).
Given that cellular NO produced by iNOS plays a key role in facilitating NR-1 latency in HPCs, failure to achieve US28-KO latent infection in HPCs may be due to a lack of iNOS and NO induction. To validate this hypothesis, we tested whether increasing the iNOS level can facilitate US28-KO to achieve latent infection. As shown in Supplementary Fig. 13, the levels of iNOS and NO in HPCs infected with US28-KO were successfully increased via lentivirus-mediated iNOS overexpression. Overexpression of iNOS via LV-iNOS transfection strongly suppressed the IE1 level as well as HCMV replication (Fig. 6h) in US28-KO-infected HPCs at 7 dpi. Overexpression of iNOS also significantly enhanced cell survival following US28-KO infection (Fig. 6i). In contrast, when cellular NO was depleted using NG-M-L-Arg in iNOS-overexpressed HPCs (Fig. 6j), the viral IE1 expression level and HCMV replication after US28-KO infection were strongly increased in US28-KO-infected HPCs at 14 dpi (Fig. 6k).
Discussion
As a critical component of innate immunity and host defence, NO stimulates antiviral activity against various viruses34,35. Here, we demonstrate that a high level of intracellular NO plays a key role in silencing HCMV replication, enabling the virus to establish latent infection. Although the molecular mechanism remains unclear, our results from a luciferase reporter assay show that NO strongly inhibits HCMV IE expression through suppressing IE1/2 promoter activity (Fig. 5h). In agreement with NO being crucial for suppressing Epstein–Barr virus lytic infection and B cell apoptosis, we have shown that rapid depletion of intracellular NO in HCMV latently infected cells results in cell apoptosis. NO may exert a direct effect in suppressing the process of apoptosis or an indirect effect through suppressing viral replication. Although it still remains speculative, several mechanisms have been proffered for how NO might directly suppress apoptosis. NO may impede apoptosis by switching the cell’s ATP production from oxidative phosphorylation to glycolysis which enables cell survival36.
We showed that STAT3 activity plays an important role in reprogramming HPCs into the immunosuppressive monocyte subset that harbours HCMV latency. Given the similarity between the cells harbouring NR-1 latency and Mo-MDSCs, we postulated that a similar molecular mechanism may be involved in STAT3-mediated iNOS upregulation and cell differentiation in HCMV-infected HPCs. Consistent with this hypothesis, a previous study37 identified STAT3 as a transcription factor associated with HCMV latency. The critical role of STAT3 activity in HCMV latency was also validated using the US28-KO HCMV mutant. Although the mechanism remains unclear, it has been shown that38 US28 is required for latent HCMV within HPCs. As shown in Fig. 6, we found that the US28-KO strain of HCMV failed to activate STAT3 in HPCs due to low iNOS expression and NO production. These results suggest that activation of the STAT3 signalling pathway, which requires the HCMV US28 gene, plays an essential role in establishing HCMV latency in HPCs. Considering that a previous report39 showed that latency-associated CMVIL-10 strongly activated STAT3 in immature dendritic cells, it would be interesting to know whether the US28 knockout would affect the expression of CMVIL-10. Our results also showed that NR-1 infection significantly increased CD33 expression in HPCs, supporting the notion that the cells harbouring NR-1 latency are CD33+-lineage-committed populations11. Comparing HCMV infection in various CD34+ cell subpopulations, Goodrum et al.16 previously found that only the CD34+/CD38− population and not the CD34+/c-kit+ population became the host cells for HCMV latent infection. In agreement with this finding, the transcript profiling (Fig. 1d,e) showed that CD38 transcript level in the cell populations harbouring NR-1 latency (14 dpi) was significantly decreased compared to that in HPCs with Mock infection.
Immunosuppressive function of the cells harbouring HCMV latent infection has been previously reported20. By analysing the secretome in cells carrying latent HCMV, it has been demonstrated20 that CC chemokine ligand CCL8 produced by HCMV latently infected cells could inhibit cytokine secretion by CD4+ T cells. Interestingly, our microarray analysis also showed that CCL8 transcript level was increased more than 30-fold in NR-1 latently infected HPCs compared to that in HPCs with Mock infection (Fig. 1d,e). Examining the literature on this unique monocyte subset also suggests it has a role in HCMV reactivation and dissemination. This immunosuppressive monocyte subset generally expresses cytokine receptors which enable it to migrate to and accumulate at inflammatory loci. Once at the site of inflammation, inflammatory cytokines or cell differentiating factors can induce it to differentiate into mature monocytes. As differentiation of this monocyte subset into mature monocytes reduces cellular iNOS and NO levels, HCMV in the latency phase would be reactivated. This is consistent with the observation that reactivation of latent HCMV generally occurs at inflammatory sites such as the lymph nodes as well as salivary and thymus glands40,41. Interestingly, it has been reported42 that a similar finding that MCMV could enhance the immune regulatory system to modulate host cell adaptive immunity via producing NO, a typical phenotype of the Mo-MDSC. This finding, in combination with our results, strongly argues that the B7-H4-positive, immunosuppressive monocyte subset is the true host of cytomegalovirus latency, and that the transport and differentiation of such a monocyte subset is tightly associated with cytomegalovirus reactivation and dissemination.
Based on these results, we propose a working model for HCMV achieving latency in human HPCs (Supplementary Fig. 14). According to this model, infection of CD34+ HPCs by HCMV immediately activates cellular STAT3. Activation of STAT3 then promotes the differentiation of HPCs into a unique monocyte subset that strongly expresses iNOS and produces NO. A high level of cellular NO, in turn, suppresses HCMV IE expression and viral replication, leading to HCMV latency. In line with this, the provocation that reactivates viruses from latency, such as TPA treatment, was associated with a reduction of cellular NO (Supplementary Fig. 8). Moreover, the NO-mediated immunosuppressive function of this monocyte subset also enables the cell to evade attack by T cells and thus facilitates HCMV latency. We further showed that latently infected HPCs are less susceptible to the induction of apoptosis and they exhibit greater longevity than classical monocytes. In conclusion, our findings provide evidence that HCMV reprogrammes human HPCs into a unique immunosuppressive monocyte subset to achieve latency. Mechanistic studies further show that the STAT3–iNOS–NO axis activated by viral infection plays an essential role in this reprogramming process. Identification of the membrane marker protein for cell type hosting HCMV latency, such as B7-H4, also opens up an avenue for developing strategies to eradicate the latently infected cells before organ transplantation.
Methods
Reagents, cells, virus and antibodies.
Bone marrow cells were obtained from materials collected from healthy donors in a standard health examination at Jiangsu Province People’s Hospital. The protocol was approved by the Institutional Review Board of Nanjing University. All participants received oral and written information about the study prior to written consent being obtained. Primary CD34+CD33− HPCs were isolated via MOFLO XDP SORTER (Beckman). Cultures of primary CD34+ HPCs were established as described previously23. Human HL-60, THP-1 and HFF-1 were purchased from the American Type Culture Collection (ATCC) and cultured according to the ATCC’s instruction. Antibodies were purchased from these sources: anti-iNOS, anti-STAT3, anti-pSTAT3, anti-STAT1 and anti-pSTAT1 antibodies (Cell Signalling); PE-conjugated antibodies against human CD4 and CD8 (Beckman); PE-conjugated anti-human CD34, CD11b,CD33, CD47, CD14, CXCR4, B7-H4, M-CSFR, HLA-DA, CCL5, IgG-isotype control antibodies and APC-conjugated anti-human HLA-DR (Biolegend); anti-GAPDH antibody (Santa Cruz). NR-1 and VR-1814 were HCMV clinical isolates while Towne and AD169 were laboratory-adapted strains23,43. HCMV NR-1BAC and TowneBAC were constructed by inserting a BAC sequence into the genome of HCMV clinical isolate NR-1 and laboratory-adapted strain Towne. Using the ‘scarless’ mutagenesis procedure, mutant US28-KO was derived from NR-1BAC by introducing a stop codon downstream from the US28 translation initiation site. The propagation of HCMV strains (for example, NR-1, NR-1BAC) in the infected cells has been described previously23,43.
Analysis of viral RNA and DNA.
The infected cells were collected and washed with PBS three times. For the viral and cell DNA extraction, a DNA extraction kit (QIAGEN) was used according to the manufacturer’s instructions. The quantity of viral genome was evaluated by quantitative PCR (qPCR) (norm alized to GAPDH) and semi-qPCR using the IE1 primer. In qP CR experiments, all samples were analysed in triplicate by an SYBR green probe using Applied Biosystems 7900 real-time PCR machine. All samples were analysed by PCR and agarose electrophoresis. The absolute levels of HCMV genome copy were determined according to a standard curve. Briefly, the IE1 open reading frame was synthesized and diluted to a series of concentrations. The samples were used to generate a concentration standard curve. The Ct (threshold cycle) value of cell-associated viral DNA was also determined with an equal loading amount of total infected cellular DNA (0.5 μg). Through a standard curve, the concentration of cell-associated viral DNA was determined. Intracellular viral RNA was isolated using Trizol Reagent (Invitrogen) according to the manufacturer’s instructions. Samples were treated with DNase using the DNA-free kit (Ambion). The concentration of RNA was determined and 0.5 μg was used for the reverse transcriptase reaction. IE1 complementary DNA was synthesized using the TaqMan reverse transcription kit with random hexamers according to the manufacturer’s protocol (Applied Biosystems). Equal amounts of cDNA were analysed by qPCR in triplicate using Applied Biosystems 7900 real-time PCR. The results were normalized to the level of GAPDH. The amounts of cDNA were also analysed by semi-qPCR, in which all samples were analysed by PCR of 28 cycles and agarose electrophoresis. The primers set for DNA and cDNA of IE1 is as follows: IE1, forward 5′-GCCT TCCCTAAGACCACCAAT-3′ and reverse 5′-ATTTTCTGGGCATAAGCCATAATC-3′. The primers sets for DNA and cDNA of GAPDH are: forward 5′-TCAGAAAAAGGGCCCTGACAACT-3′, reverse 5′-TCCCCTCTTCAAGGGGTCTACA-3′ and forward 5′-AGGCTAGGGACGGCCT-3′, reverse 5′-GCCATGGGTGGAATCATATTG-3′, respectively.
Viral infection and reactivation.
Bone marrow progenitor cells or HL-60 cells were infected with HCMV at a multiplicity of 2 plaque-forming units (p.f.u) per cell. The ultraviolet-inactivated virus was used as mock infection. One hour before infection, the cell culture media was changed to a serum-free fresh one. The infected cells were cultured for 2 h and then washed three times with PBS to remove cell-free virus. The infected cells were harvested at the indicated dpi. All cell media were changed every day. For the reactivation experiments, 20 ng ml−1 TPA (Sigma-Aldrich) was added to latently infected cell culture to induce viral lytic reactivation, then cells were cocultured with HFF-1. Cells were harvested after seven days. Virus particles were then harvested from the cells when they showed a complete cytopathic effect, and virus titres were measured with p.f.u assays of fibroblasts. HCMV was propagated in primary HFF cells in DMEM supplemented with 10% FBS and 100 U ml−1 each of penicillin and streptomycin. Virus stocks were stored in DMEM containing 10% FBS and 1.5% BSA in liquid nitrogen.
Microarray analysis.
Primary CD34+ HPCs were infected with HCMV at a multiplicity of 2 p.f.u per cell. The ultraviolet-inactivated virus was used as mock infection. At 14 dpi, the infected cells were harvested. Total RNA was extracted with TRIzol (Sigma-Aldrich) and subjected to the analysis with PrimeView Human Gene Expression Array (Affymetrix)44,45. A Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.) was used to measure the optical density values (A260/A280) of the samples, and electrophoresis of the 28S/18S bands was conducted to assess the quality of the total RNA samples. Following overnight hybridization, the chips were washed and stained according to the manufacturer’s protocols. The hybridized chips were scanned by Affymetrix GeneChip Scanner 3000 7G. The differentially expressed gene sets between experimental groups and controls were analysed by significance analysis of microarray. The gene sets were considered as significant if a false discovery rate < 0.05 and the fold changes (experimental groups versus controls) were larger than 2 or less than 0.5.
T-cell proliferation assays.
For the T-cell proliferation assay, peripheral blood was first separated with lymphocyte separation medium. Lymphocytes were then incubated with CFSE (5(6)-carboxyfluorescein diacetate succinimidyl ester of CFDA SE) for 15 min to label the cells according to the manufacturer’s instructions (Invitrogen). In the presence of ConA (Sigma-Aldrich)46, CFSE-labelled lymphocytes were cocultured at 2:1 ratios with the cells latently infected by HCMV or monocytes in 96-well flat-bottom plates. T-cell proliferation was analysed by flow cytometry on day 4.
Detection of ROS.
ROS was quantified using Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Molecular Probes) according to the manufacturer’s protocol. Briefly, cell supernatant was collected. After addition of 30 ng ml−1 TPA, the absorbance at 560 nm was measured using a microplate plate reader (Molecular Devices SpectraMax M2) at 37°C. Absorbance results were normalized to a standard curve generated by serial dilutions of 20 mM H2O2.
Measurement of cell supernatant cytokine levels.
Cell supernatant was collected. Levels of IL-4, IL-6, IL-10, IL-17A, M-CSF and GM-CSF were determined using ELISA kits (R&D Systems). The absorbance was measured using wavelength correction (A450) with a microplate reader (Bio-Rad).
Plasmid construction and transfection.
To generate luciferase reporter plasmids, full-length IE1 promoter was directly synthesized chemically and cloned into the pMIR-report plasmid (Ambion). Successful insertion was confirmed by sequencing. For luciferase reporter assays, 0.2 μg of firefly luciferase reporter plasmid and 0.1 μg of β-galactosidase expression vector (Ambion) were transfected into 239 T cells in 24-well plates using the Lipofectamine 3000 (Invitrogen). The β-galactosidase vector was used as a transfection control. At 24 h post-transfection, cells were analysed using a luciferase assay kit according to the manuals (Promega).
Infection with lentivirus.
Lentiviruses encoding STAT3, iNOS, STAT3 siRNA and iNOS siRNA genes were generated and confirmed by GenePharma. An empty-backbone lentivirus was used as a control. Cells were incubated with the lentiviruses at MOI of 5:1 along with 8 μg ml−1 Polybrene for 48 h before further treatment.
Apoptosis analysis.
Cell apoptosis was assayed by using the FITC-Annexin V Apoptosis Detection Kit I (BD Bioscience) as previously described47. In brief, cells were washed twice with cold PBS and then re-suspended in the binding buffer at a final density of 106 cells ml−1. Next, 100 μl of cell suspension containing 105 cells was taken for apoptosis measurement. FITC-annexin V and propidium iodide were added (5 μl of each) and the cell suspension was mixed and then incubated for 15 min in the dark. Finally, 400 μl of binding buffer was added to the cell suspension and the cells were analysed by flow cytometry using FACS Calibur (BD Bioscience) and Flowjo Software. The test was performed in triplicate under each condition.
Cell viability assay.
A 12 mM MTT (Sigma-Aldrich) stock solution was prepared by adding a 5 mg vial of MTT into 1 ml of sterile PBS. Cells were seeded at densities between 5,000 and 10,000 cells per well in phenol red-free medium (Gibco) and then cultured for 48 h. The microplates were centrifuged to pellet the cells and replaced with 100 μl of fresh medium. MTT stock solution (10 μl) and DMSO (50 μl) were added to each well and mixed thoroughly with a pipette. The microplates were then incubated at 37°C for 10 min. Each sample was mixed well and absorbance read at 540 nm. The cell-free wells containing 100 μl of medium and 10 μl MTT stock solution served as background controls.
Human PBMC and MDSC isolation.
Isolation of human PBMCs was performed as previously described48. Briefly, blood was drawn from healthy volunteers by venipuncture and centrifuged through a Percoll gradient (Sigma-Aldrich) at 200g for 30 min at room temperature. Mononuclear cells were collected and washed twice with PBS and 1 mM EDTA to remove platelets and residual Percoll. Cells were suspended in RPMI1640 supplemented with 10% FBS (Gibco) and then pelleted and re-suspended at 1 × 106 cells ml−1 in HBSS. PE-conjugated anti-human B7-H4 and FITC-conjugated anti-human CD14 (Biolegend) were added to cell suspensions at 4°C for 90 min, then immediately transferred to a 4°C centrifuge to pellet the cells. Cells were re-suspended in cold HBSS and maintained at 4°C. Stained cells (CD14+, B7-H4+) were placed on the MoFlo (Beckman Coulter Life Sciences) and sorted.
NO Measurement and NG-M-L-Arg treatment.
Cells (1 × 105) were lysed in 200 μl Cell and Tissue Lysis Buffer for Nitric Oxide Assay (Enzo), placed on ice (5 min) and then centrifuged at 12,000g for 5 min at 4°C. The supernatant fraction was collected and diluted 1:100 in Reaction Buffer. To assess NO production, the release of nitrite and nitrate, the stable breakdown product of NO, were analysed using a nitric oxide detection kit (Enzo). Measurements of known concentrations of nitrite were used to generate a standard curve between 0 and 100 μM of nitrite. To deplete cellular NO, cells were treated with 200 μM NG-Monomethyl-L-Arginine (Sigma-Aldrich).
Statistical analysis.
The variability within each group has been quantified with standard deviation and used for statistical comparison. All images of western blotting and qPCR with reverse transcription are representative of at least three independent experiments. Real-time PCR was performed in triplicate and each experiment was repeated several times. No statistical method was used to predetermine sample size. Data shown are presented as the mean ± s.e.m. of three independent experiments; differences are considered statistically significant at P < 0.05 using a Student’s t-test.
Life Sciences Reporting Summary.
Further information on experimental design is available in the Life Sciences Reporting Summary.
Data availability.
The microarray analysis data that support the findings of this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) and are accessible through the GEO Series accession number GSE106879. All uncropped western blot images that are presented in the main and in the supplementary figures are available in Supplementary Fig. 16a–c. All other data that support the findings of this study are available from the corresponding authors upon request.
Supplementary Material
Acknowledgements
The authors thank J.L. Littrell (Georgia State University, Atlanta, GA) for critical reading and constructive discussion of the manuscript. This work was supported by grants from the National Basic Research Program of China (973 Program) (2014CB542300), the National Natural Science Foundation of China (81101330, 31271378, 81250044 and 31600659), the Guangdong Innovative and Entrepreneurial Research Team Program (No. 2014ZT05S136), the Research Special Fund for Public Welfare Industry of Health (201302018) and the NIH (RO1-AI050468, RO1-DE023935 and RO1–025462).
Footnotes
Competing interests
The authors declare no competing interests.
Supplementary information is available for this paper at https://doi.org/10.1038/s41564-018-0131-9.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Loewendorf A & Benedict CA Modulation of host innate and adaptive immune defenses by cytomegalovirus: Timing is everything. J. Intern. Med 267, 483–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sinclair J & Sissons P Latency and reactivation of human cytomegalovirus. J. Gen. Virol 87, 1763–1779 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Sissons JG & Carmichael AJ Clinical aspects and management of cytomegalovirus infection. J. Infect 44, 78–83 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Reeves MB & Sinclair JH Analysis of latent viral gene expression in natural and experimental latency models of human cytomegalovirus and its correlation with histone modifications at a latent promoter. J. Gen. Virol 91, 599–604 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Saffert RT, Penkert RR & Kalejta RF Cellular and viral control over the initial events of humancytomegalovirus experimental latency in CD34+ cells. J. Virol 84, 5594–5604 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reeves MB & Sinclair JH Circulating dendritic cells isolated from healthy seropositive donors are sites of human cytomegalovirus reactivation in vivo. J. Virol 87, 10660–10667 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bevan IS, Daw RA, Day PJ, Ala FA & Walker MR Polymerase chain reaction for detection of human cytomegalovirus infection in a blood donor population. Br. J. Haematol 78, 94–99 (1991). [DOI] [PubMed] [Google Scholar]
- 8.Smith MS, Bentz GL, Alexander JS & Yurochko AD Human cytomegalovirus induces monocyte differentiation and migration as a strategy for dissemination and persistence. J. Virol 78, 4444–4453 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noriega VM et al. Human cytomegalovirus modulates monocyte-mediated innate immune responses during short-term experimental latency in vitro. J. Virol 88, 9391–9405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor-Wiedeman J, Sissons JG, Borysiewicz LK & Sinclair JH Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J. Gen. Virol 72, 2059–2064 (1991). [DOI] [PubMed] [Google Scholar]
- 11.Hahn G, Jores R & Mocarski ES Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc. Natl Acad. Sci. USA 95, 3937–3942 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendelson M, Monard S, Sissons P & Sinclair J Detection of endogenous human cytomegalovirus inCD34+ bone marrow progenitors. J. Gen. Virol 77, 3099–3102 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Reeves MB, Lehner PJ, Sissons JG & Sinclair JH An in vitro model for the regulation of human cytomegalovirus latency and reactivation in dendritic cells by chromatin remodelling. J. Gen. Virol 86, 2949–2954 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Tarrant-Elorza M, Rossetto CC & Pari GS Maintenance and replication of the human cytomegalovirus genome during latency. Cell Host Microbe 16, 43–54 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Rossetto CC, Tarrant-Elorza M & Pari GS Cis and trans acting factors involved in humancytomegalovirus experimental and natural latent infection of CD14+ monocytes and CD34+ cells. PLoS Pathog. 9, e1003366 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodrum F, Jordan CT, Terhune SS, High K & Shenk T Differential outcomes of human cytomegalovirus infection in primitive hematopoietic cell subpopulations. Blood 104, 687–695 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Goodrum FD, Jordan CT, High K & Shenk T Human cytomegalovirus gene expression during infection of primary hematopoietic progenitor cells: a model for latency. Proc. Natl Acad. Sci. USA 99, 16255–16260 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slobedman B & Mocarski ES Quantitative analysis of latent human cytomegalovirus. J. Virol 73, 4806–4812 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolovan-Fritts CA, Mocarski ES & Wiedeman JA Peripheral blood CD14+ cells from healthy subjects carry a circular conformation of latent cytomegalovirus genome. Blood 93, 394–398 (1999). [PubMed] [Google Scholar]
- 20.Mason GM, Poole E, Sissons JG, Wills MR & Sinclair JH Human cytomegalovirus latency alters the cellular secretome, inducing cluster of differentiation (CD)4+ T-cell migration and suppression of effector function. Proc. Natl Acad. Sci. USA 109, 14538–14543 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weekes MP et al. Latency-associated degradation of the MRP1 drug transporter during latent human cytomegalovirus infection. Science 340, 199–202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy JC, Fischle W, Verdin E & Sinclair JH Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 21, 1112–1120 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan C et al. Human cytomegalovirus miR-UL148D facilitates latent viral infection by targeting host cell immediate early response gene 5. PLoS Pathog. 12, e1006007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Connor CM & Murphy EA A myeloid progenitor cell line capable of supporting human cytomegalovirus latency and reactivation, resulting in infectious progeny. J. Virol 86, 9854–9865 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guilliams M et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat. Rev. Immunol 14, 571–578 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheung AK et al. The role of the human cytomegalovirus UL111A gene in down-regulating CD4+ T-cell recognition of latently infected cells: Implications for virus elimination during latency. Blood 114, 4128–4137 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Gabrilovich DI, Ostrand-Rosenberg S & Bronte V Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol 12, 253–268 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis TA et al. Phorbol esters induce differentiation of human CD34+ hemopoietic progenitors to dendritic cells: Evidence for protein kinase C-mediated signaling. J. Immunol 160, 3689–3697 (1998). [PubMed] [Google Scholar]
- 29.Addison CL, Hitt M, Kunsken D & Graham FL Comparison of the human versus murine cytomegalovirus immediate early gene promoters for transgene expression by adenoviral vectors. J. Gen. Virol 78, 1653–1661 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Yu H, Pardoll D & Jove R STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer 9, 798–809 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slinger E et al. HCMV-encoded chemokine receptor US28 mediates proliferative signaling through the IL-6-STAT3 axis. Sci. Signal 3, ra58 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Beisser PS, Laurent L, Virelizier JL & Michelson S Human cytomegalovirus chemokine receptor gene US28 is transcribed in latently infected THP-1 monocytes. J. Virol 75, 5949–5957 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langemeijer EV et al. Constitutive β-catenin signaling by the viral chemokine receptor US28. PLoS ONE 7, e48935 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Croen KD Evidence for antiviral effect of nitric oxide. Inhibition of herpes simplex virus type 1 replication. J. Clin. Invest 91, 2446–2452 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guidotti LG, McClary H, Loudis JM & Chisari FV Nitric oxide inhibits hepatitis B virus replication in the livers of transgenic mice. J. Exp. Med 191, 1247–1252 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Everts B et al. Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood 120, 1422–1431 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spencer JV, Cadaoas J, Castillo PR, Saini V & Slobedman B Stimulation of B lymphocytes by cmvIL-10 but not LAcmvIL-10. Virology 374, 164–169 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Humby MS & O’Connor CM HCMV US28 is important for latent infection of hematopoietic progenitor cells. J. Virol 90, 2959–2970 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raftery MJ et al. Shaping phenotype, function, and survival of dendritic cells by cytomegalovirus-encoded IL-10. J. Immunol 173, 3383–3391 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Crough T & Khanna R Immunobiology of human cytomegalovirus: From bench to bedside. Clin. Microbiol. Rev 22, 76–98 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halenius A & Hengel H Human cytomegalovirus and autoimmune disease. Biomed. Res. Int 2014, 472978 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daley-Bauer LP, Roback LJ, Wynn GM & Mocarski ES Cytomegalovirus hijacks CX3CR1(hi) patrolling monocytes as immune-privileged vehicles for dissemination in mice. Cell Host Microbe 15, 351–362 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunn W et al. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell Microbiol. 7, 1684–1695 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Bian Z et al. Cd47-Sirpalpha interaction and IL-10 constrain inflammation-induced macrophage phagocytosis of healthy self-cells. Proc. Natl Acad. Sci. USA 113, E5434–E5443 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi WL et al. Integrated miRNA and mRNA expression profiling in fetal hippocampus with Down syndrome. J. Biomed. Sci 23, 48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L et al. Role of myeloid-derived suppressor cells in glucocorticoid-mediated amelioration of FSGS. J. Am. Soc. Nephrol 26, 2183–2197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bundscherer A et al. Cell harvesting method influences results of apoptosis analysis by annexin V staining. Anticancer Res. 33, 3201–3204 (2013). [PubMed] [Google Scholar]
- 48.Liu Y et al. Signal regulatory protein (SIRPα), a cellular ligand for CD47, regulates neutrophil transmigration. J. Biol. Chem 277, 10028–10036 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray analysis data that support the findings of this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) and are accessible through the GEO Series accession number GSE106879. All uncropped western blot images that are presented in the main and in the supplementary figures are available in Supplementary Fig. 16a–c. All other data that support the findings of this study are available from the corresponding authors upon request.