Abstract
Targeted proteomics experiments based on selected reaction monitoring (SRM) have gained wide adoption in the use of clinical biomarkers, cellular modeling, and numerous other biological experiments due to their highly accurate and reproducible quantification. The quantitative accuracy in targeted proteomics experiments is reliant on the stable-isotope, heavy-labeled peptide standards that are spiked into a sample and used as a reference when calculating the abundance of endogenous peptides. Therefore, the quality of measurement for these standards is a critical factor in determining whether data acquisition was successful. With improved mass spectrometry (MS) instrumentation that enables the monitoring of hundreds of peptides in hundreds to thousands of samples, quality assessment is increasingly important and cannot be performed manually. We present Q4SRM, a software tool that rapidly checks the signal from all heavy-labeled peptides and flags those that fail quality-control metrics. Using four metrics, the tool detects problems with both individual SRM transitions and the collective group of transitions that monitor a single peptide. The program’s speed and simplicity enable its use at the point of data acquisition and can be ideally run immediately upon the completion of a liquid chromatography–SRM–MS analysis.
Keywords: quality control, quality assurance, targeted proteomics, SRM, MRM, software
Graphical Abstract
INTRODUCTION
Selected reaction monitoring (SRM), also known as multiple reaction monitoring (MRM), is a data acquisition technique used in targeted analysis of molecules, including targeted proteomic studies. It exploits the unique capability of triple-quadrupole (QQQ) mass spectrometers to monitor the predefined precursor and fragment ion pairs of specific molecules of interest throughout a liquid chromatography (LC) elution profile. Compared to shotgun proteomics, targeted proteomics using SRM has high selectivity, high sensitivity, and a wide linear dynamic range,1–3 which makes it especially useful in the accurate and reproducible quantification of low-abundance proteins in highly complex biological samples. SRM has been widely used in the fields of biomarker discovery,4–7 analysis of protein post-translational modifications8 and characterization of biological protein networks.4,9
In the recent years, multiple technical advances have greatly improved the throughput of SRM analyses, allowing for the quantification of hundreds of peptides in a single analysis.6,7,10 For example, a single 800-plex SRM assay (e.g., 400 unlabeled and heavy-labeled peptide pairs and 2400 transitions with retention time scheduling) using ultrahigh-performance liquid chromatography (UHPLC) has been developed to quantify proteins in plasma.11 Advanced labeling techniques utilizing in vitro proteome-assisted MRM for protein absolute quantification (iMPAQT) demonstrated the capability of SRM in genome-wide protein quantification of over 18 000 human proteins.12 Moreover, the scan speed of QQQ instrumentation has been greatly improved in recent releases of commercial instrumentation. The newly developed TSQ Altis (released in 2017) can scan more than 600 transitions per second, which is 6 times more than a traditional QQQ scan speed of 100 transitions per second. The breadth of measurement enabled by these technological improvements to QQQ mass spectrometry has increased the feasibility and popularity of large scale targeted proteomics studies,13 a major application of which will be in clinical studies in which up to hundreds of protein candidates need to be quantified in hundreds of clinical specimens.14
Quantitative accuracy is a primary motivating factor for utilizing a targeted proteomics protocol. The precise and reproducible absolute quantification produced by SRM assays is essential to many clinical and laboratory experiments.15,16 Because the abundance of an endogenous peptide is calculated from the measurement of the spiked-in reference standard, it is essential to assess the data quality of these references.17 In early applications of targeted proteomics, when instrument speed greatly limited the number of transitions that could be monitored, much of this quality assessment was done manually. However, recent improvements in instrument performance and experimental design have enabled a dramatic increase in the number of target peptides and associated SRM transitions, which makes manual quality assessment an untenable and laborious task.
A variety of computational tools assist in SRM experiment design and data analysis. The first task in creating an SRM experiment is the choice of proteins and representative peptides to monitor. Achieving a reliable protein abundance requires appropriately choosing peptides that have a strong signal and are free from interferences in the biological matrix. Numerous computational tools exist to facilitate assay design by identifying peptides and refining SRM transitions.18–21 To help share these assays and eliminate time spent designing the same transitions at multiple institutions, community portals have begun to host well-designed and vetted assays.22–24 Analyzing the experimental data requires significant computational effort to align files across replicates and experimental conditions, pick peaks and produce quantitative values, normalize data and perform statistical tests, etc.25–27
Among the many tools that are used in the SRM community, there remains a need for a tool to assist in quality assessment. In particular, the rapid quality assessment of reference transitions immediately following data acquisition lacks an easy-to-use tool. Although some tools exist, such as AuDIT,28 they are inadequate for the needs of large-scale clinical cohorts. Specifically, a primary concern is in the LC performance across thousands of runs. To identify systematic drift and column failure, we needed a new tool. Therefore, we have created Q4SRM, a software tool that rapidly checks the signal from all heavy-labeled target peptides and flags those that fail quality-control (QC) metrics.
METHODS AND IMPLEMENTATION
Q4SRM is a C# .NET application designed to perform quality assessment of transitions associated with the heavy-labeled reference peptides that are spiked into a sample. Because these peptides are spiked into every sample, their transitions are expected to be easily identified in each MS result file. The software is open source under the BSD license and available on GitHub at: https://github.com/PNNL-Comp-Mass-Spec/Q4SRM. The software expects two types of input. The first is a Thermo .RAW file representing the data acquisition from a triple quadrupole instrument, e.g., TSQ Vantage or TSQ Altis. To read this file format, the software utilizes the I/O codex that is part of the RawFileReader NuGet package distributed by Thermo (San Jose, CA); these dynamical-link library codes (DLLs) are included with the Q4SRM executable. The second input is a user-generated file that contains cutoffs and thresholds used in determining which data points are flagged with warnings. This is a simple tab-delimited text file where each row describes thresholds to be used for a specific peptide. An example file and walk-through tutorial are available in the project’s GitHub repository (https://github.com/PNNL-Comp-Mass-Spec/Q4SRM/wiki/Tutorial). If one desires uniform thresholds for all peptides, this file can be omitted and the thresholds specified directly in the interface.
The design of Q4SRM was influenced by our desire to have the program installed on instrument computers and run immediately and as quickly as possible following data acquisition. These goals led us to avoid requiring file conversion or using software that has difficult installation or requires third party packages. Q4SRM is designed to be as lightweight as possible. We also wanted to require minimal input from the user, to facilitate easy use. A file that contains both the instrument method information and the data acquisition output was preferred. The Thermo formatted RAW file contains information about the instrument “method”, which identifies the data acquisition settings to perform the SRM experiment. Because mzML conversions of the RAW file lack this method information (and would require Proteowizard to be installed), we have written Q4SRM to interact directly with the vendor format. To increase the compatibility of this software with various instruments, we created a version compatible with mzML input files. However, the interface is slightly different as the method information is not available in the same streamlined fashion as is available from Thermo instrument files.
To identify the transitions for reference and heavy peptides, Q4SRM looks for a keyword in the “Name” (TSQ Vantage) or “Compound Name” (TSQ Altis) field of the SRM Table (contained in the Instrument Method portion of the .RAW file); transitions lacking the keyword are ignored. It is customary in our lab to name the transitions associated with heavy peptides with the string “heavy” or “hvy”, e.g., “VSGVATDIQALK_heavy”. Note that multiple transitions for the same peptide have the same name. Because the program is open source, it is possible to adapt this parsing step for other keywords, if different conventions are used in other laboratories. The SRM Table portion of the .RAW file also provides a parent/precursor m/z, product m/z, and a start and stop time (in minutes) for each transition. The heavy transitions are then grouped according to name so that all transitions for each precursor can be associated appropriately in the output.
DATA EXTRACTION AND METRICS
For each heavy transition, we gather four pieces of information from the .RAW file: max intensity, time of max intensity, median intensity, and the sum total intensity during the scheduling window. With these pieces of information, we compute four metrics.
A pair of metrics are computed based on information relating to a single transition. The first metric, called peak position, calculates the time between the start or stop of transition acquisition and the time of the max intensity. The user defined cutoff (floating point number representing time in minutes) dictates what is considered an acceptable minimal value. The reason for this metric is to ensure that peaks fully elute within their expected scheduled time and are not clipped or truncated, which, in part, may be due to degradation of the LC column performance. Internally, the software scales the input values (time in minutes) to unit distance considering the entire time of a run in the range 0–1, analogous to the Normalized Elution Time strategy;29 however, this is not exposed to the user. The second metric, called the S/N heuristic, calculates the ratio of the maximal intensity to the median intensity. For the purpose of calculating the median value, all values below 5 are excluded. A user defined threshold (floating point number) dictates what is considered an acceptable minimal value. This approximates the signal-tonoise as the intensity of the transition relative to the background intensity of unrelated signal. We recognize that there are many ways to calculate a signal-to-noise ratio (S/N), and this heuristic is not intended to be a thorough calculation (which would involve a more statistical characterization of the noise). This heuristic is designed to quickly assess whether there is a strong and distinct peak relative to the other intensities within the acquisition window.
The last two metrics are computed based on information relating to all transitions belonging to the same peptide, i.e., a group of transitions. The first metric, called total signal, is the sum of the intensities for all transitions in the group. The user defined threshold (integer number) dictates what is considered an acceptable minimum value. The reason for this metric is to ensure that sufficient signal exists for all transitions in the group, which is required to have an accurate quantitative measurement. The second metric, called peak concurrence, calculates the difference between the time of the max intensity for each transition in the group, providing a warning for situations in which the transitions do not have a reasonably concurrent max intensity. Again, users specify a threshold (time in minutes) that defaults to 0.5 min or 30 s. It is expected that the time of max intensity of transitions for the same peptide should be identical; however, an interference in one transition may cause these values to be out of sync.
The program produces both text and graphical output. The text output file is a tab-delimited text file that contains the four computed metrics as well as other associated information for each peptide. An example of the output is in the GitHub repository. For graphical output, the program produces an image of each group of transitions, similar to what is seen in Figures 1 and 2. There is also a summary image that shows which data points give warnings and which pass QC.
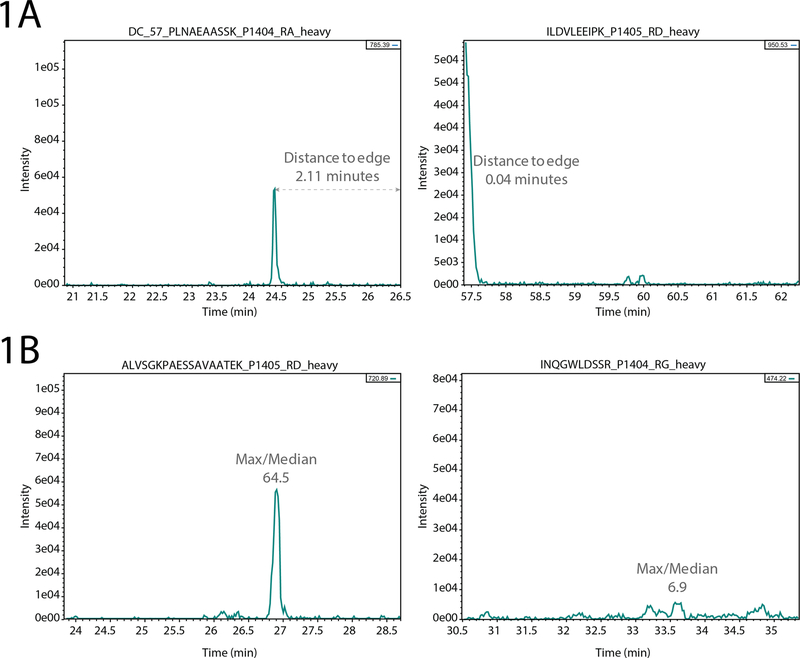
Figure 1.
Single-transition metrics. (A) The distance of a peak to the edge of the scheduled time window indicates whether the peak was potentially incompletely measured (right). (B) The ratio of the maximal intensity to the median indicates whether the transition has a strong intensity relative to the background signal. Low ratios (right) suggest a poorly defined signal and may not be reliable when used as a reference to measure endogenous peptide abundance.
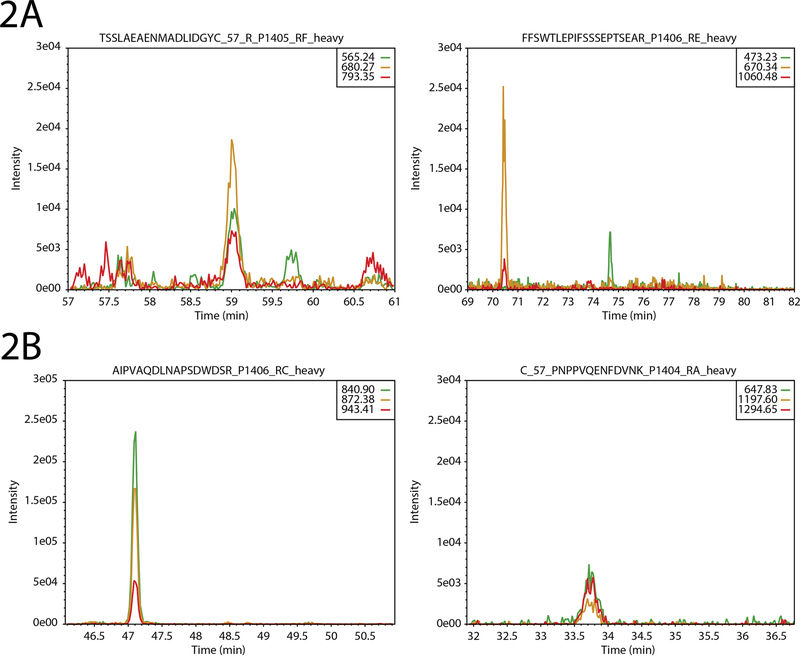
Figure 2.
Multitransition metrics. (A) Transitions that belong to the same peptide should have identical elution profiles. On the left is shown a set of transitions that all elute at the same time; on the right, we see that the peak of the yellow and red transition peaks (~70 min) are distant from the green transition peak (~74 min). (B). Transitions should have sufficient intensity as a group. On the left is a strong and intense set of transitions (note the y-axis scale); on the right is a set of low-intensity transitions. Depending on the user-defined threshold for the total intensity metric, this set of transitions may yield a warning.
Q4SRM is available with both a graphical user interface and a command-line interface. The graphical user interface version facilitates the selection of input files and the adjustment of settings and also provides a viewer mode in which the user can view the summary plot and get details on the different points. The command-line version provides access to the same settings as the graphical user interface version and facilitates use of Q4SRM with computational pipelines. A pictorial user guide that walks through download and use of both user interfaces is included on the GitHub repository wiki page (https://github.com/PNNL-Comp-Mass-Spec/Q4SRM/wiki).
RESULTS
Targeted proteomics experiments are rapidly growing in their capacity to measure a large number of peptide targets. Although a full analysis of the data will happen in the days and weeks that follow data acquisition and in the context of the entire experiment, it is essential to rapidly assess the quality of the data immediately as it is generated to determine whether the run was successful. To assist in point-of-acquisition quality assessment of liquid chromatography (LC)–SRM–mass spectrometry (MS) data sets, we have created the Q4SRM software package. This easy-to-use package rapidly checks transitions for each heavy-labeled peptide against a suite of essential QC metrics and provides simple and interpretable output to the operator, including a list of flagged transitions that need further manual inspection. Q4SRM is a lightweight software tool that can be installed on the computers that control MS instrumentation. Even for files with thousands of scheduled transitions, the software takes less than 1 min to analyze a single Thermo .RAW file.
A pair of metrics assessed by Q4SRM report information on individual transitions (Figure 1). First, the program measures the distance from the maximal peak intensity to the edge of the scheduled acquisition window. This metric flags a transition with a warning when the peak maximum is too close to the edge of the window, signaling that this peak is potentially clipped (Figure 1A). This would mean that the quantitation will not be accurate because some of the peptide’s elution profile was not measured. The user specified threshold should be set in relation to the schedule window size, the expected LC peak widths and the operator’s personal tolerance. The second metric derived from data for a single transition is an approximate measure of signal to noise (Figure 1B). Although the S/N may be calculated a variety of ways, our goal here is to quickly determine whether there is a problem with the data. Therefore, the S/N heuristic calculated by Q4SRM is the ratio between the peak maximum intensity and the median intensity. For example, a value of 50 means that the peak maximum is 50 times greater than the median intensity during the schedule window, thus indicating that the peak is strongly intense above background signal. With these two metrics, users can be confident that the measured transition of the heavy labeled standard is both clearly within the scheduled LC time window and sufficiently intense to serve as a reference in the calculation of an accurate quantitative value for the endogenous peptide.
A pair of metrics assessed by Q4SRM report information about the group of transitions related to the same peptide (Figure 2). Despite the good performance of individual transitions, it is necessary that the group performs as expected. The first metric measures how close in elution the transitions are to each other, also known as peak concurrence. Because each transition is intended to measure the same peptide, they are expected to have an identical elution profile. However, due to potential interferences or missing signal, the transitions may appear out of sync with each other. Figure 2A shows two sets of transitions. One transition has acceptable peak concurrence despite being low-abundance (Figure 2A, left); in the other set of transitions, one of the peaks is clearly several minutes after the elution of the other two (Figure 2A, right). The second metric, total ion intensity, simply measures the total intensity of all transitions associated with a peptide (Figure 2B). This metric can be set to a different threshold for each peptide because each peptide and transition is expected to have a different characteristic response during the LC–SRM–MS analysis. Failures of this metric can signal a few different challenges. First, there might be a problem with the spike-in level during sample preparation. Second, there might be an instrument performance problem causing low signal. Finally, it is possible that the peptide was completely out of range of the schedule window (due to LC column problems).
Q4SRM compares each of the four metrics against user-specified input. For some of the metrics, acceptable values may be broadly similar across laboratories. For example, many chromatography systems are set up to produce peak widths of ~30 s. Therefore, the peak concurrence metric default is reasonable for many laboratories to use without change. Similarly, the peak position metric might be used without changing the default, depending on the length of the acquisition window. However, the two metrics that relate to signal intensity are expected to be highly specific to each peptide and each lab’s sample preparation. For this reason, it is advised that users identify a meaningful value from their own data. A convenient method for setting these values is to take the intensity values from a data acquisition when the peptides were spiked into either a blank background or a sample matrix. By averaging values over several initial testing runs, a threshold can be set that is appropriate for the observed range of response.
CONCLUSIONS
Before data analysis begins in earnest, assessing the quality of the acquired data is essential. For experiments that contain many samples, and where the time between data acquisition and data analysis is long, this quality assurance (QA) step should not be delayed; rather, QC/QA should happen immediately upon data acquisition to give feedback as soon as possible to the instrument operator.30 To fill this need in targeted proteomics studies, we created Q4SRM, which can analyze the heavy labeled reference peptides in an LC–SRM–MS data file within 1 min. It quickly computes a set of four essential QC metrics that helps to identify low-quality SRM transitions. The number of flagged transitions for any data set depends on user-specified thresholds and instrument performance. We have found it to be an essential tool for maintaining high data quality and instrument health. To assist in long-term monitoring of QC metrics, Q4SRM’s text output contains the metrics on all transitions. With this information, users can collate and compare output across many different acquisition files using data analytic platforms such as R or Jupyter.
ACKNOWLEDGMENTS
We thank Geremy Clair for assistance with figures. This work was supported by the NIH National Institute of General Medical Sciences grant no. P41GM103493 (Richard Smith, PNNL), the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC) grant nos. 1U24CA210972 (S.H.P.) and 5U24CA210955 (T.L.), the NCI Early Detection Research Network (EDRN) Interagency Agreement no. ACN15006-001 (T.L.), and the MoTrPAC consortium no. U24DK112349 (Joshua Adkins, PNNL). Additional support was provided by the TEDDY Study Group, which is funded by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), the National Institute of Allergy and Infectious Diseases (NIAID), the National Institute of Child Health and Human Development (NICHD), the National Institute of Environmental Health Sciences (NIEHS), the Centers for Disease Control and Prevention (CDC), and the Juvenile Diabetes Research Foundation (JDRF) and supported in part by the National Center for Advancing Translational Sciences Clinical and Translational Science (NCATS) Awards to the University of Florida and the University of Colorado. Work was performed in the Environmental Molecular Science Laboratory, a U.S. Department of Energy (DOE) national scientific user facility at Pacific Northwest National Laboratory (PNNL) in Richland, WA. Battelle operates PNNL for the DOE under contract no. DE-AC05-76RLO01830.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Lange V; Picotti P; Domon B; Aebersold R Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol 2008, 4, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Picotti P; Bodenmiller B; Aebersold R Proteomics meets the scientific method. Nat. Methods 2013, 10, 24–27. [DOI] [PubMed] [Google Scholar]
- (3).Shi T; et al. Advances in targeted proteomics and applications to biomedical research. Proteomics 2016, 16, 2160–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Picotti P; Bodenmiller B; Mueller LN; Domon B; Aebersold R Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 2009, 138, 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Hüttenhain R; et al. Reproducible quantification of cancer-associated proteins in body fluids using targeted proteomics. Sci. Transl. Med 2012, 4, 142ra94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kennedy JJ; et al. Demonstrating the feasibility of large-scale development of standardized assays to quantify human proteins. Nat. Methods 2014, 11, 149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Duriez E; et al. Large-Scale SRM Screen of Urothelial Bladder Cancer Candidate Biomarkers in Urine. J. Proteome Res 2017, 16, 1617–1631. [DOI] [PubMed] [Google Scholar]
- (8).Shi T; et al. Sensitive targeted quantification of ERK phosphorylation dynamics and stoichiometry in human cells without affinity enrichment. Anal. Chem 2015, 87, 1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Shi T; et al. Conservation of protein abundance patterns reveals the regulatory architecture of the EGFR-MAPK pathway. Sci. Signaling 2016, 9, rs6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kim Y; Jeon J; Mejia S; Yao CQ; Ignatchenko V; Nyalwidhe JO; Gramolini AO; Lance RS; Troyer DA; Drake RR; et al. Targeted proteomics identifies liquid-biopsy signatures for extracapsular prostate cancer. Nat. Commun 2016, 7, 11906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Burgess MW; Keshishian H; Mani DR; Gillette MA; Carr SA Simplified and efficient quantification of low-abundance proteins at very high multiplex via targeted mass spectrometry. Mol. Cell. Proteomics 2014, 13, 1137–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Matsumoto M; et al. A large-scale targeted proteomics assay resource based on an in vitro human proteome. Nat. Methods 2017, 14, 251–258. [DOI] [PubMed] [Google Scholar]
- (13).Zhang M; et al. Sensitive, High-Throughput, and Robust Trapping-Micro-LC-MS Strategy for the Quantification of Biomarkers and Antibody Biotherapeutics. Anal. Chem 2018, 90, 1870–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Surinova S; et al. On the development of plasma protein biomarkers. J. Proteome Res 2011, 10, 5–16. [DOI] [PubMed] [Google Scholar]
- (15).Schiess R; Wollscheid B; Aebersold R Targeted proteomic strategy for clinical biomarker discovery. Mol. Oncol 2009, 3, 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Simicevic J; et al. Absolute quantification of transcription factors during cellular differentiation using multiplexed targeted proteomics. Nat. Methods 2013, 10, 570–576. [DOI] [PubMed] [Google Scholar]
- (17).Carr SA; et al. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol. Cell. Proteomics 2014, 13, 907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Brusniak M-YK; et al. ATAQS: A computational software tool for high throughput transition optimization and validation for selected reaction monitoring mass spectrometry. BMC Bioinf. 2011, 12, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Aiyetan P; Thomas SN; Zhang Z; Zhang H MRMPlus: an open source quality control and assessment tool for SRM/MRM assay development. BMC Bioinf. 2015, 16, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zauber H; Kirchner M; Selbach M Picky: a simple online PRM and SRM method designer for targeted proteomics. Nat. Methods 2018, 15, 156–157. [DOI] [PubMed] [Google Scholar]
- (21).Mohammed Y; et al. PeptidePicker: a scientific workflow with web interface for selecting appropriate peptides for targeted proteomics experiments. J. Proteomics 2014, 106, 151–161. [DOI] [PubMed] [Google Scholar]
- (22).Whiteaker JR; et al. CPTAC Assay Portal: a repository of targeted proteomic assays. Nat. Methods 2014, 11, 703–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Sharma V; et al. Panorama: a targeted proteomics knowledge base. J. Proteome Res 2014, 13, 4205–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Bhowmick P; Mohammed Y; Borchers CH MRMAssayDB: an integrated resource for validated targeted proteomics assays. Bioinformatics 2018, 34, 3566–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).MacLean B; et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Choi M; et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 2014, 30, 2524–2526. [DOI] [PubMed] [Google Scholar]
- (27).Rost HL; et al. TRIC: an automated alignment strategy for reproducible protein quantification in targeted proteomics. Nat. Methods 2016, 13, 777–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Abbatiello SE; Mani DR; Keshishian H; Carr SA Automated detection of inaccurate and imprecise transitions in peptide quantification by multiple reaction monitoring mass spectrometry. Clin. Chem 2010, 56, 291–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Norbeck AD; et al. The utility of accurate mass and LC elution time information in the analysis of complex proteomes. J. Am. Soc. Mass Spectrom 2005, 16, 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Stanfill BA; et al. Quality Control Analysis in Real-time (QC-ART): A Tool for Real-time Quality Control Assessment of Mass Spectrometry-based Proteomics Data. Mol. Cell. Proteomics 2018, 17, 1824–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]