ABSTRACT
Background
Individual differences in human perception of sweetness are partly due to genetics; however, which genes are associated with the perception and the consumption of sweet substances remains unclear.
Objective
The aim of this study was to verify previous reported associations within genes involved in the peripheral receptor systems (i.e., TAS1R2, TAS1R3, and GNAT3) and reveal novel loci.
Methods
We performed genome-wide association scans (GWASs) of the perceived intensity of 2 sugars (glucose and fructose) and 2 high-potency sweeteners (neohesperidin dihydrochalcone and aspartame) in an Australian adolescent twin sample (n = 1757), and the perceived intensity and sweetness and the liking of sucrose in a US adult twin sample (n = 686). We further performed GWASs of the intake of total sugars (i.e., total grams of all dietary mono- and disaccharides per day) and sweets (i.e., handfuls of candies per day) in the UK Biobank sample (n = ≤174,424 white-British individuals). All participants from the 3 independent samples were of European ancestry.
Results
We found a strong association between the intake of total sugars and the single nucleotide polymorphism rs11642841 within the FTO gene on chromosome 16 (P = 3.8 × 10−8) and many suggestive associations (P < 1.0 × 10−5) for each of the sweet perception and intake phenotypes. We showed genetic evidence for the involvement of the brain in both sweet taste perception and sugar intake. There was limited support for the associations with TAS1R2, TAS1R3, and GNAT3 in all 3 European samples.
Conclusions
Our findings indicate that genes additional to those involved in the peripheral receptor system are also associated with the sweet taste perception and intake of sweet-tasting foods. The functional potency of the genetic variants within TAS1R2, TAS1R3, and GNAT3 may be different between ethnic groups and this warrants further investigations.
Keywords: sweet taste, perception, preference, sugar intake, genome-wide association scan, BMI, FTO, taste receptor
Introduction
The perception of and liking for a given degree of sweetness vary between individuals. Twin studies have shown that heritabilities (h2) of preferences for sucrose solution and sweet foods range from low (1, 2) to moderate (3, 4) (h2 = 0.23–0.40). Our twin study (5) showed moderate heritability on the perceived intensities of the monosaccharides glucose (h2 = 0.31) and fructose (h2 = 0.34) and of the high-potency sweeteners neohesperidin dihydrochalcone (NHDC; h2 = 0.31) and aspartame (h2 = 0.30); a common genetic factor accounted for >75% of the genetic variance in each sweet taste. However, the particular genes and genetic variants that are responsible for variation in human sweet taste perception and the intake of sweet foods remain largely underexplored.
Previous studies reported that genetic variants within or near sweet taste receptor genes TAS1R2 (6) and TAS1R3 (7), and a downstream gene, GNAT3 (8), are associated with the variance of sucrose sensitivity (i.e., the ability to discriminate between sucrose solutions of different concentrations). Variants within TAS1R2 were also shown to be associated with the intake of sugars and sweet foods in both children (9–11) and adults (6, 12–14). Although variants within TAS1R3 were not associated with the intake of sweet foods (9, 12, 15), they were found to be associated with the preference for sucrose solutions (16). Other genetic variants reported to be associated with sugar consumption are within the glucose transporter gene GLUT2 (13, 17) (also known as SLC2A2) and the fibroblast growth factor gene FGF21 (18–21). Notably, except for the association with FGF21 which was identified in large genome-wide association studies, these associations with both sweet taste perception and intake of sweet substances were reported in studies of small sample sizes (n ≤ 160 and 1037, respectively) and some studies (7, 8, 13, 17) were even comprised of mixed ethnicities (i.e., Caucasians, Asians, and African Americans) so they remain to be validated in larger independent samples. A summary of these associations is presented in Table 1.
TABLE 1.
Genetic variants previously reported to be associated with the perception and intake of sweet substances1
| Gene | SNP:allele | Study population | Association with sweet phenotypes |
|---|---|---|---|
| TAS1R2 | rs12033832:A | P1-1: 696 young adults2 from the Toronto Nutrigenomics and Health study completed dietary assessment and a subset (n = 95) completed sensory tests (6) | Higher sucrose sensitivity (lower detection threshold) and lower sugar intake (grams per day) among those with BMI ≥ 25; opposite associations among those with BMI < 25. |
| P2: 30 young adults2 from an Australian cohort (12) | Higher percentage energy intake from carbohydrate in an ad libitum meal session ≤ 40 min. | ||
| P3: 144 unrelated individuals (92 Europeans, 37 Asians, 15 Africans) (7) | No association with sucrose sensitivity. | ||
| rs3935570:T | P1-1 (6) | Higher sucrose sensitivity (lower detection threshold) among those with BMI ≥ 25; no association with sugar intake (grams per day) regardless of BMI. | |
| rs35874116:A | P1-2: 1037 young adults (482 whites, 362 East Asians, 114 South Asians, 79 others) from the Toronto Nutrigenomics and Health study (13) | Higher intake of carbohydrate (grams per day) and sugar (grams per day) among those with BMI ≥ 25. | |
| P4: 100 individuals from the Canadian Trial of Carbohydrate in Diabetes multicenter intervention study2 (13) | Higher intake of sugar (grams per day). | ||
| P5: 312 children (43.2% white) from a Brazilian cohort (9) | Higher sugar intake (kilocalories per day) at age 3.9 y; no association at ages 1.1 and 7.7 y. | ||
| P2 (12) | Higher intake of sweets (grams) in an ad libitum meal session ≤ 40 min. | ||
| P6: 441 adults2 from a Mexican cohort (14) | Lower intake of carbohydrate (grams per day) and percentage energy intake from carbohydrate. | ||
| P7: 47 children (87.5% Caucasian) from the Guelph Family Health Study (10) | Higher percentage energy intake from snacks. | ||
| P1-1 (6) | No association with sucrose sensitivity and sugar intake (grams per day). | ||
| P3 (7) | No association with sucrose sensitivity. | ||
| rs121377303 | P8: 65 adults (85% Caucasian) and 60 children (81% Caucasian) from the Guelph Family Health Study (11) | Association with sucrose suprathreshold among adults. | |
| rs75346183 | P8 (11) | Association with preference for sucrose solution among children. | |
| rs97017963 | P8 (11) | Association with preference for sucrose solution and percentage energy intake from added sugar among children. | |
| TAS1R3 | rs307355:C | P3 (7) | Higher sucrose sensitivity. |
| P2 (12) | No association with percentage energy intake from carbohydrate or intake of sweets (grams) in an ad libitum meal session ≤ 40 min. | ||
| rs35744813:C | P3 (7) | Higher sucrose sensitivity. | |
| P9: 76 mothers (32.9% white, 52.6% black, 5.3% Hispanic/Latino/Latina, 1.3% Asian, 7.9% others) and 101 children (31.7% white, 42.6% black, 8.9% Hispanic/Latino/Latina, 2% Asian, 14.9% others) (16) | Preference for a sucrose solution of a lower concentration among mothers; no association in children. | ||
| P10: 235 children (46 whites, 136 blacks, 2 Asians, 51 others) (15) | No association with sucrose detection threshold and percentage energy intake from added sugar. | ||
| P5 (9) | No association with sugar intake (kilocalories per day). | ||
| P2 (12) | No association with percentage energy intake from carbohydrate or intake of sweets (grams) in an ad libitum meal session ≤ 40 min. | ||
| GNAT3 | rs7792845:T | P11: 160 unrelated individuals (103 Caucasians, 41 Asians, 16 Africans) (8) | Higher sucrose sensitivity. |
| rs940541:T | P11 (8) | Higher sucrose sensitivity. | |
| rs1107660:T | P11 (8) | Higher sucrose sensitivity. | |
| rs1107657:T | P11 (8) | Higher sucrose sensitivity. | |
| rs1524600:C | P11 (8) | Higher sucrose sensitivity. | |
| rs6467217:T | P11 (8) | Higher sucrose sensitivity. | |
| rs6970109:C | P11 (8) | Higher sucrose sensitivity. | |
| rs6975345:T | P11 (8) | Higher sucrose sensitivity. | |
| rs10242727:A | P11 (8) | Higher sucrose sensitivity. | |
| rs6467192:G | P11 (8) | Higher sucrose sensitivity. | |
| rs6961082:C | P11 (8) | Higher sucrose sensitivity. | |
| GLUT2 | rs5400:C | P1-2 (17) | Higher intake of carbohydrate (grams per day) and sucrose (grams per day). |
| P4 (13) | Higher sugar intake (grams per day). | ||
| FGF21 | rs838133:A | P12: 33,533 Europeans from the DietGen Consortium (18) | Lower percentage energy intake from carbohydrate. |
| P13: 6515 adults from a Danish cohort2 (20) | Higher weekly intake of sweet snacks and candies. | ||
| P14: 176,989 Europeans from the UK Biobank (21) | Higher percentage energy intake from carbohydrate. | ||
| rs838145:G | P15: 38,360 Europeans from the CHARGE Consortium (19) | Higher percentage energy intake from carbohydrate. |
1SNP:allele, SNP and the effect allele. Each study population is given a number (e.g., P1) along with reporting its sample size and ethnicity (as provided in the original study) when it is referred to the first time. P1-1 and P1-2 are from the same study cohort but the sample size of P1-2 is bigger. SNP, single nucleotide polymorphism.
2Unknown ethnicity.
3Unknown effect allele.
To verify previously reported associations and reveal novel loci, we performed genome-wide association scans (GWASs) of sweet taste perception (primary outcome variables) in an Australian adolescent twin sample (n = 1757) and a US adult twin sample (n = 686), and the intake of sugars and sweets (i.e., candies) (secondary outcome variables) in the UK Biobank sample (n = ≤174,424) (22). We also looked for evidence for genetic variation in biological pathways and compared the expression levels of associated genes in different human tissues including taste buds and brain tissues.
Methods
This article used the STrengthening the REporting of Genetic Association Studies (STREGA) reporting guidelines (23).
Samples
The Australian sample was a subset of participants from the Brisbane Adolescent Twin Study (24), also referred to as the Brisbane Longitudinal Twin Study, who performed the taste test between 2003 and 2014. Most twins performed the taste test at age 14 y but with older siblings the mean ± SD age of the sample was older (16.1 ± 2.7 y). Zygosity was determined from genome-wide genotyping. The US sample was a subset of adult twins who performed the taste test at the annual Twins Days Festival at Twinsburg, Ohio, between 2009 and 2015. Only Caucasians were included in the present study. Zygosity was assessed with 3 methods: self-reported identity, experimenter ratings of photographs for physical similarity, and genome-wide genotyping. The UK Biobank recruited 502,650 participants (aged 37–73 y; 54.4% female; 5% of those invited) from 21 centers across England, Wales, and Scotland between 2006 and 2010 (22). Intake of total sugars was available for 211,051 participants and intake of sweets was available for a subsample of 25,533 participants.
Ethical statement
The Australian study was approved by the QIMR Berghofer Medical Research Institute Human Research Ethics Committee. The US study was approved by the Institutional Review Board (#4) of the University of Pennsylvania. The UK Biobank study was approved by the UK National Health Service National Research Ethics Service. Written consent was obtained from both the participants and their parents (for subjects younger than 18 y old).
Taste test in the Australian and US samples
For the Australian sample, detailed information on the taste test and data processing has been described previously (5). Briefly, the taste battery included duplicated presentations of 4 sweet (0.60 M glucose, 0.30 M fructose, 8.0 × 10−5 M NHDC, and 1.4 × 10−3 M aspartame) and 5 bitter (propylthiouracil, sucrose octaacetate, quinine, caffeine, and denatonium benzoate) solutions, plus 2 water solutions as controls. The 20 samples were presented in random order. Participants were instructed to rate their perceived intensity for each solution using a general labeled magnitude scale (gLMS) (25), with labels of no sensation (0 mm), barely detectable (2 mm), weak (7 mm), moderate (20 mm), strong (40 mm), very strong (61 mm), and strongest imaginable (114 mm). Mean intensity ratings from the duplicate presentations for each stimulus were used in the present study. Because the perceived intensity of the 4 sweeteners was highly correlated at the genetic level (genetic correlation [rg] = 0.78–0.89) and most of the phenotypic variance (71% for glucose, 77% for fructose, 64% for NHDC, and 59% for aspartame) was accounted for by a common factor, as shown in our previous multivariate variance component analysis (5), we further calculated a general sweet factor score (gSweet, a mean intensity of the 4 sweet tastes weighted by loadings from the common factor). A total of 5 phenotypes—perceived intensity of glucose, perceived intensity of fructose, perceived intensity of NHDC, perceived intensity of aspartame, and gSweet—from the Australian sample were analyzed. For the US sample, as described previously (2), participants tasted sweet (sucrose), bitter (phenylthiocarbamide and quinine), sour (citric acid), and salty (sodium chloride and potassium chloride) solutions, and other tastes (e.g., vegetable juice) and rated perceived sweetness, bitterness, sourness, saltiness, burn, intensity, and liking using a 7.7-cm visual analog scale (VAS). A total of 3 phenotypes—perceived intensity, perceived sweetness, and liking of the 0.35 M (i.e., 12% wt:vol) sucrose solution—were included in the present study. The 3 phenotypes were weakly to moderately correlated (phenotypic correlation [rp] = 0.21–0.43) (2).
Intake of total sugars and sweets in the UK Biobank sample
UK Biobank is a large long-term biobank study from the United Kingdom that aims to identify the contribution of genetic and environmental factors to disease. Detailed information on phenotyping and genotyping is presented elsewhere (22). Self-reported dietary data were collected via online 24-h dietary recall questionnaires. Intake of total sugars (grams per day) was calculated based on food and beverage consumption. Total sugars represent all dietary mono- and disaccharides, including those from milk, fruit, and vegetables but excluding any supplements. Intake of sweets (handfuls of candies per day) was only collected from participants who reported consuming any biscuits (i.e., cookies), chocolate, or sweets (i.e., candies). They were asked, “How many handfuls of sweets (hard and soft, e.g., peppermints, toffees, fudge, fruit flavoured sweets) did you have?” Answers included quarter, half, 1, 2, 3, 4, and ≥5, which were converted to 0.25, 0.5, 1, 2, 3, 4, and 5 before analysis.
Genotyping, imputation, and quality control
For the Australian sample, genotyping was performed with the Illumina 610-Quad BeadChip for 1255 participants and HumanCoreExome-12 version 1.0 BeadChip for 502 participants. Single nucleotide polymorphisms (SNPs) were phased using ShapeIT (26) and then imputed using Minimac3 (27) and the Haplotype Reference Consortium of Caucasian European ancestry (Release 1) (28), with 7,035,128 SNPs passing quality control (QC) as outlined previously (29). To ensure SNPs were imputed with high data quality, we excluded those with a call rate < 90%, minor allele frequency < 0.05, imputation score < 0.3, and Hardy–Weinberg equilibrium score of P < 1.0 × 10−6, with a total of 4,381,914 SNPs remaining. Individuals who were >6 SDs from the centroid of the first 2 genetic principal components (PCs) PC1 and PC2 were excluded, so the sample was of exclusively European ancestry. For the US sample, genotyping was performed using the HumanOmniExpressExome-8 version 1-2 Chip and imputed using 1000 Genome Phase 1 integrated Haplotype, with 6,175,124 SNPs passing QC. With the same post-imputation QC criteria, 5,833,901 SNPs were used in the analyses. The Australian and US samples had a total of 4,001,140 SNPs in common. All UK Biobank participants have been genotyped using the Affymetrix UK BiLEVE Axiom array or Affymetrix UK Biobank Axiom array comprising 805,426 markers in the official release. Imputations were performed using IMPUTE2 and UK10K haplotype and Haplotype Reference Consortium reference panels, as described elsewhere (22). SNPs with a call rate < 90%, minor allele frequency < 0.01, imputation score < 0.3, and Hardy–Weinberg equilibrium score of P < 1.0 × 10−6 were excluded, with a total of 10,353,649 SNPs remaining in the analyses. Only participants who classified as white British, determined by similarity of genetic PC values (PC1, PC2), were included in the present study.
Statistical analysis
The genome-wide association analysis for each of the 5 phenotypes from the Australian study and 3 phenotypes from the US study was conducted using genome-wide efficient mixed-model association (30). As described previously (31), this method fits a linear mixed model for each SNP and uses the genetic relatedness matrix, calculated from genome-wide genotyping data, to account for the relatedness of individuals (i.e., twins and siblings from the same family). For the Australian sample, the covariates included age, sex, and the history of ear infection, previously shown to be associated with taste intensity ratings of this sample (5), and the first 5 genetic PCs. Before modeling, a square root transformation was applied to each of the 5 intensity scores to obtain a more normal distribution (32). For the US sample, the covariates included age, sex, and the first 2 genetic PCs. A meta-analysis of the perceived intensity was performed using the Australian gSweet scores and the US sucrose intensity scores and the P value–based approach weighted by sample size implemented in the software METAL (33). The GWASs of the intake of total sugars and sweets in the UK Biobank were performed using BOLT-LMM (34). This fits a linear mixed model by including a genetic relation matrix derived from genotyped SNPs as a random effect to correct for population structure and cryptic relatedness. Covariates included age, sex, and the first 10 genetic PCs. Because the associations between TAS1R2 and the intake of sugars were suggested to depend on BMI (in kg/m2) (6, 13), we also performed analyses including BMI as an extra covariate or stratifying the sample into 2 groups based on a cutoff of BMI = 25. To ensure that any potential technical or population stratification artifacts had a negligible impact on the results, we calculated the genomic inflation factor (λ) for the Australian and the US phenotypes (λ ranged between 0.99 and 1.02). As λ can be inflated by a large sample size, we calculated the linkage disequilibrium (LD) score regression intercepts (35) for the UK Biobank phenotypes; intercepts of 1.05 and 1.01 for the intake of total sugars and sweets, respectively, indicated little or no population stratification. See Supplemental Figure 1 for Q–Q plots. A priori suggestive and significance thresholds were set at P = 1.0 × 10−5 and 5.0 × 10−8, respectively. Given that the sweet phenotypes were correlated within each sample (2, 5), we estimated the number of independent tests to be 2.62, 2.66, and 1.49 for the Australian, the US, and the UK Biobank sample, respectively, using a matrix spectral decomposition algorithm (36); the Bonferroni-corrected significance thresholds became P = 1.9 × 10−8, 1.9 × 10−8, and 3.4 × 10−8, respectively. Manhattan and Q–Q plots were created using FUMA-GWAS (37). Regional associational plots were created using LocusZoom (38).
Gene-based association analyses were performed by summarizing the SNP association results at the gene level. The contribution of each SNP to the gene-based association was adjusted by the LD between SNPs within a gene. This was done using the function “fastBAT” implemented in the software GCTA (39). With 24,765 genes provided in the Genome Reference Consortium Human Build 37 (GRCh37/hg19) and the number of independent phenotypes for each of the Australian, the US, and the UK Biobank samples, conservative Bonferroni-corrected thresholds were set at P = 7.7 × 10−7, 7.6 × 10−7, and 1.4 × 10−6, respectively.
Further, we examined the associations between phenotypes (predicted by GWAS results) and gene expression levels (predicted by expression quantitative trait loci, i.e., the associations between SNPs and gene expression levels, results from the GTEx project [40]) using MetaXcan (41). With a library of 44 tissue models (including the “cross-tissue” model) from the GTEx project (40), the maximum number of 11,440 genes expressed per tissue, and the number of independent phenotypes for each of the Australian, the US, and the UK Biobank samples, conservative Bonferroni-corrected thresholds were set at P = 3.8 × 10−8, 3.7 × 10−8, and 6.6 × 10−8, respectively.
Tissue enrichment and pathway analyses were performed with methods implemented in DEPICT version 1.1 (42). The preference was to use only genome-wide significant SNPs if there were ≥10 independent associations with P < 5.0 × 10−8. Owing to the lack of genome-wide significant signals, we used SNPs with P < 1.0 × 10−5. For traits with <10 independent loci with P < 1.0 × 10−5 (i.e., perceived intensity of glucose, fructose, aspartame, and the meta-analysis results), we included SNPs with P < 1.0 × 10−4. We set the Bonferroni-corrected thresholds for tissue-enrichment analysis at P = 9.1 × 10−5, 9.0 × 10−5, and 1.6 × 10−4 for the Australian, the US, and the UK Biobank sample, respectively, and for pathways analysis at P = 1.3 × 10−6, 1.3 × 10−6, and 2.3 × 10−6 for the Australian, the US, and the UK Biobank sample, respectively (considering the number of independent phenotypes in each sample and assuming that gene expressions in all 209 tissue/cell samples are independent, and all 14,463 pathways are independent), and the false discovery rate at <0.05.
We calculated the genetic correlations between the intake of total sugars and sweets, and BMI in the UK Biobank sample using the online platform LD hub (43), which takes the GWAS summary results to perform LD score regression and estimate the genetic correlation between 2 traits (44).
Gene expression
For genes near the identified SNPs or genes identified in gene-based or pathway analyses, we examined their expression levels in 31 different tissues using our in-house data of human taste tissues and the publicly available data from GTEx (version 7, 30 general tissue types) (40). For the in-house assay, we collected 6 taste samples from human fungiform papillae of adults (5 men and 1 woman) using published procedures (45) and isolated the RNA following the manufacturer's directions for processing the taste tissue with Quick-RNA MiniPrep R1054 (Zymo Research). We evaluated RNA quality expressed as an RNA integrity number using the Agilent 2200 TapeStation system (Agilent Technologies). The 6 samples with sufficient RNA quality, as determined by the Next-Generation Sequencing Core of the University of Pennsylvania (RNA integrity number > 7), underwent library preparation and sequencing (100 base pairs single-end) on the HiSeq 4000 sequencer (Illumina) using the manufacturer's sequencing protocols. We mapped reads to the reference genome (GRCh38.p10) after the raw sequence data in fastq format passed quality filters of Trimmomatic (46) and we normalized the counts using the R (The R foundation for Statistical Computing) package ballgown (47), with the median expression level for each gene used in the analyses. The expression levels (e.g., reads per kilo base per million mapped reads and transcript per million) for all genes in all tissue types were winsorized at 1 and 50 and then log2 transformed and presented in a heatmap.
Results
Sample characteristics
The main characteristics of the 3 samples of European ancestry are summarized in Table 2, together with the sweet phenotypes measured in each sample. The Australian adolescent sample was younger than the US and the UK Biobank samples. There were more female than male participants in all 3 samples. For the Australian sample, the mean intensity ratings for all sweet solutions were between “moderate” and “strong” on the gLMS. The US adults rated perceived intensity of the sucrose solution slightly below the middle point on the VAS and the perceived sweetness and liking of the same solution toward the upper end of the scale. Among all UK Biobank participants who reported their intake data, ∼83% were white British and these were included in the GWAS. See Supplemental Figure 2 for participant flowcharts showing the selection process for the participants from each sample.
TABLE 2.
Characteristics of the Australian, the US, and the UK Biobank samples of European ancestry1
| Mean ± SD (range) | ||
|---|---|---|
| Australian | ||
| n = 1757 from 942 families, comprised of 222 complete MZ pairs, 427 complete DZ twin pairs, and 459 unpaired twins and siblings; 54.1% female | Age, y | 16.1 ± 2.7 (12.0–25.8) |
| Glucose intensity | 31.6 ± 16.1 (2.5–111.0) | |
| Fructose intensity | 32.0 ± 17.7 (2.0–114.0) | |
| NHDC intensity | 34.7 ± 18.9 (2.0–114.0) | |
| Aspartame intensity | 26.6 ± 16.4 (0–112.5) | |
| gSweet | 31.4 ± 15.2 (4.4–102.6) | |
| US | ||
| n = 686 from 347 families, comprised of 310 complete MZ pairs, 29 complete DZ twin pairs, and 8 unpaired twins and siblings; 74.5% female | Age, y | 36.8 ± 15.5 (18.0–80.0) |
| Sucrose intensity | 3.4 ± 1.4 (0–7.6) | |
| Sucrose sweetness | 5.0 ± 1.7 (0–7.6) | |
| Sucrose liking | 4.8 ± 1.5 (0–7.6) | |
| UK Biobank | ||
| n = 174,424 for intake of total sugar; n = 21,447 for intake of sweets;2 54.5% female3 | Age,3 y | 56.41 ± 7.9 (39.0–72.0) |
| Intake of total sugars, g/d | 121.1 ± 50.7 (0–1156.9) | |
| Intake of sweets,2 handfuls/d | 1.0 ± 1.1 (0.25–5.0) | |
1DZ, dizygotic; gSweet, the general sweet factor (a weighted mean of ratings of glucose, fructose, NHDC, and aspartame); MZ, monozygotic; NHDC, neohesperidin dihydrochalcone. The Australian sample rated perceived intensity of sweet solutions using a general labeled magnitude scale; the US sample rated perceived intensity, sweetness, and liking for sucrose solution using a visual analog scale.
2Intake of sweets was only collected from participants who reported consuming any biscuits (cookies), chocolate, or sweets (candies).
3Based on the sample of 174,424 participants with intake of total sugars.
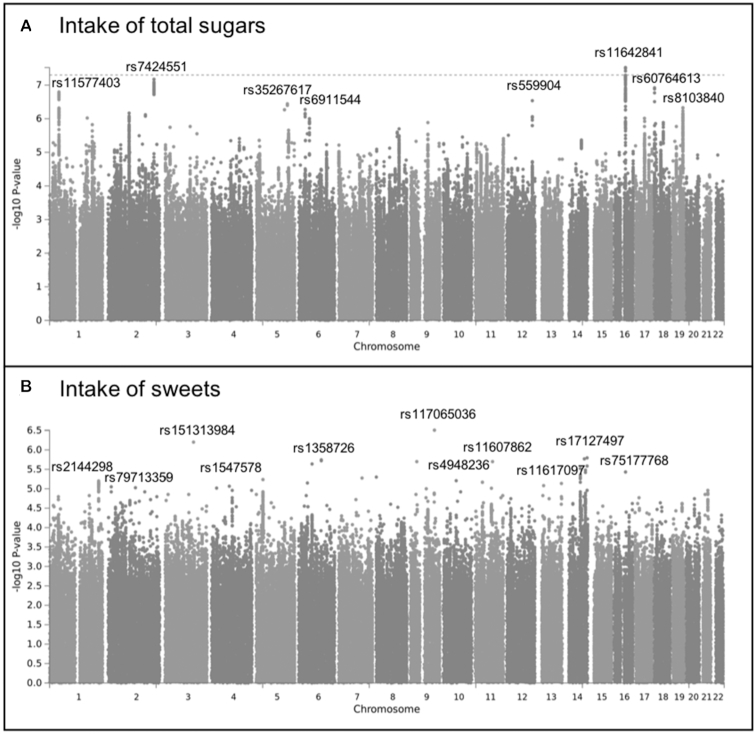
GWAS
The strongest association was between the intake of total sugars in the UK Biobank sample and rs11642841 within the FTO gene on chromosome 16 (Figure 1, Supplemental Table 1). This variant within FTO was one of the first variants to be associated with BMI in genome-wide analyses and has been repeatedly reported to be robustly associated with BMI and related traits (48, 49). The rs11642841 C allele associated positively with intake of total sugar (β = 0.928 g/d, SE = 0.169, P = 3.8 × 10−8), inversely with BMI (β = −0.301 kg/m2, SE = 0.010, P = 6.1 × 10−211), but not with the intake of sweets (β = 0.002 handfuls/d, SE = 0.010, P = 0.84), in the UK Biobank sample. The association between rs11642841 and the intake of total sugar remained strong after including BMI as a covariate (β = 0.772 g/d, SE = 0.169, P = 4.9 × 10−6). We further examined the associations between the 3 traits and found that the intake of total sugars was negatively associated with BMI (rp = −0.04; 95% CI: −0.04, −0.03; rg = −0.23; 95% CI: −0.29, −0.17), whereas the intake of sweets was positively associated with BMI (rp = 0.09; 95% CI: 0.07, 0.10; rg = 0.67; 95% CI: −0.27, 1.00) at both phenotypic and genetic levels (Table 3).
FIGURE 1.
Manhattan plots displaying the association -log10P value for each SNP in the genome and the intake of total sugars (grams per day; n = 174,424) (A) and the intake of sweets (handfuls of candies per day; n = 21,447) (B) in the UK Biobank white-British participants. Only the top SNP with P < 1.0 × 10−5 for each chromosome is labeled. SNP, single nucleotide polymorphism.
TABLE 3.
Genetic and phenotypic correlations with 95% CIs between the intake of total sugars and sweets and BMI in the UK Biobank sample1
| Intake of sweets | BMI | ||
|---|---|---|---|
| Genetic correlations | Intake of total sugars, g/d | −0.27 (−0.85, 0.32) | −0.23 (−0.29, −0.17) |
| Intake of sweets, handfuls/d | — | 0.67 (−0.27, 1.00) | |
| Phenotypic correlations | Intake of total sugars, g/d | 0.34 (0.33, 0.35) | −0.04 (−0.04, −0.03) |
| Intake of sweets, handfuls/d | — | 0.09 (0.07, 0.10) |
1Genetic correlations were calculated using genome-wide association summary results and LD score regression. BMI in kg/m2.
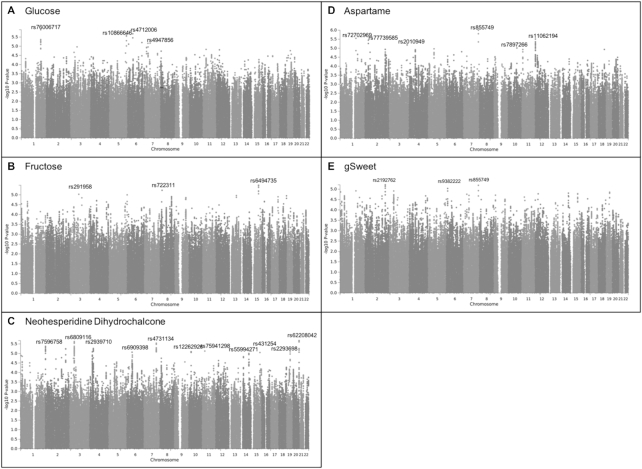
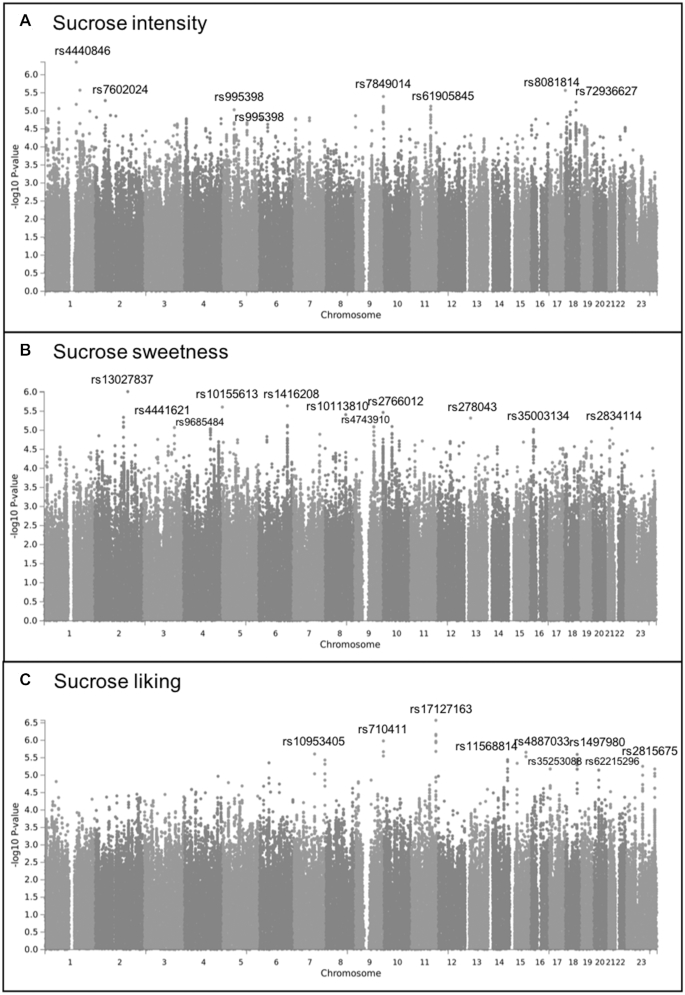
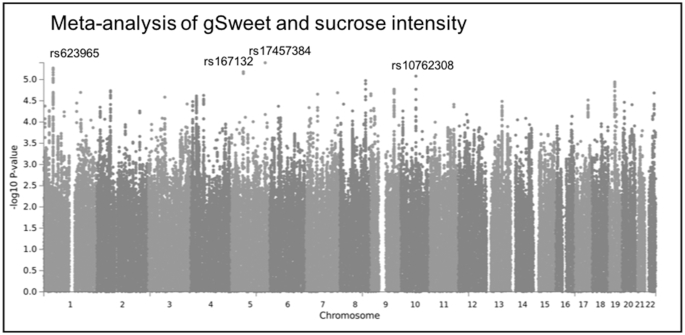
There were several suggestive associations for each of the sweet taste phenotypes (P < 1.0 × 10−5). In the Australian sample these included 7 independent associations for glucose intensity, 3 for fructose intensity, 13 for NHDC intensity, 6 for aspartame intensity, and 3 for gSweet (Figure 2, Supplemental Table 2). In the US sample there were 10 associations for sucrose intensity, 14 for sucrose sweetness, and 13 for sucrose liking (Figure 3, Supplemental Table 3). The meta-analysis of gSweet for the Australian sample and sucrose intensity for the US sample showed 4 additional suggestive independent associations (Figure 4, Supplemental Table 4). We also found 53 suggestive associations for the intake of total sugars and 14 for the intake of sweets in the UK Biobank sample (Figure 1, Supplemental Table 1). Information on each SNP including its location (chromosome and base pair), effect allele and non-effect allele, effect size (β, SE, percentage of variance accounted for), P value, and the nearest gene is reported in Supplemental Tables 1–4.
FIGURE 2.
Manhattan plots displaying the association -log10P value for each SNP in the genome and the perceived intensity of glucose (A), fructose (B), NHDC (C), aspartame (D), and gSweet (E) in 1757 Australian adolescent twins and their siblings. Only the top SNP with P < 1.0 × 10−5 for each chromosome is labeled. gSweet, general sweet factor (a weighted mean of ratings of glucose, fructose, NHDC, and aspartame); NHDC, neohesperidin dihydrochalcone; SNP, single nucleotide polymorphism.
FIGURE 3.
Manhattan plots displaying the association -log10P value for each SNP in the genome and the perception of sucrose reported via ratings of intensity (A), sweetness (B), and liking (C) in 686 US adult twins and unpaired individuals. Only the top SNP with P < 1.0 × 10−5 for each chromosome is labeled. SNP, single nucleotide polymorphism.
FIGURE 4.
Manhattan plot for the meta-analysis of the perceived intensity of gSweet (Figure 2E) and sucrose (Figure 3A). The top single nucleotide polymorphism with P < 1.0 × 10−5 for each chromosome is labeled. gSweet, general sweet factor (a weighted mean of ratings of glucose, fructose, neohesperidin dihydrochalcone, and aspartame).
We examined the cross-associations between all suggestively associated SNPs and sweet phenotypes. Whereas the SNPs identified in the Australian twins were associated with all 5 phenotypes in the same sample, and in the same direction, their associations with phenotypes in the US and the UK Biobank samples were mostly null (Figure 5, Supplemental Table 5). Similarly, all SNPs identified in the US sample were associated with all 3 US phenotypes in the same direction but their associations with the phenotypes in the Australian and the UK Biobank samples were close to the null. Among the 4 SNPs identified in the meta-analysis of the gSweet in the Australian sample and sucrose intensity in the US sample, rs17457384 was associated with the intake of sweets in the UK Biobank sample (P = 0.004), with the allele associated with higher sweet intensity ratings being associated with a higher intake of sweets. Lastly, the SNPs identified in the UK Biobank sample were weakly or null associated with all other phenotypes in any sample (Supplemental Table 6).
FIGURE 5.
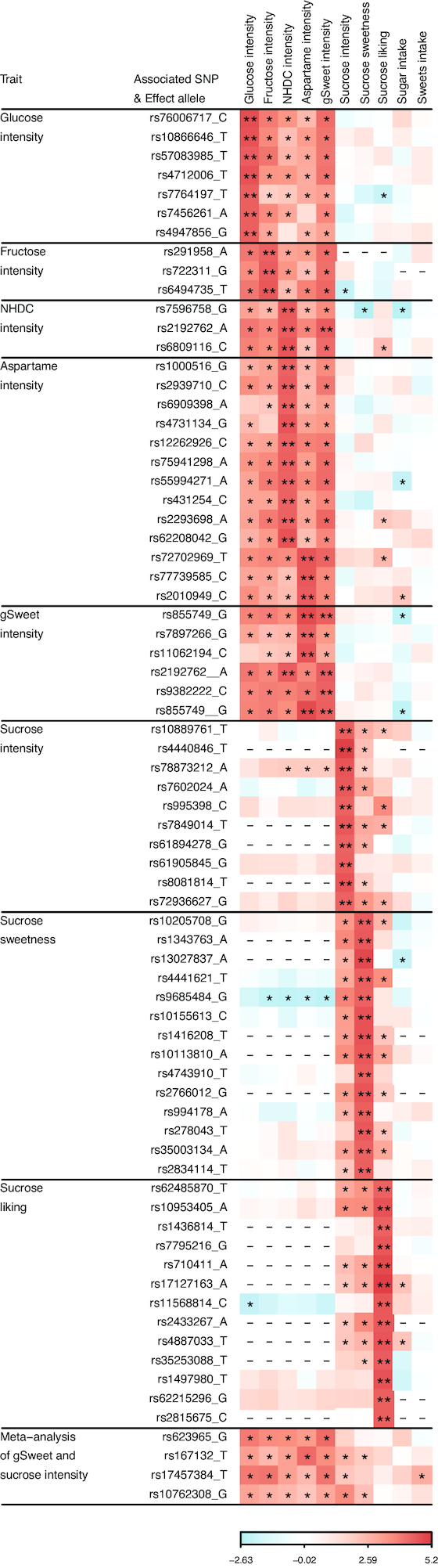
Heatmap showing the effect of SNP on the perceived intensity of glucose, fructose, NHDC, aspartame, and gSweet in the Australian sample, the perceived intensity, sweetness, and liking for sucrose in the US sample, and the intake of sugars (grams per day) and sweets (handfuls of candies per day) in the UK Biobank. Effect sizes are presented as β/SE (see Supplemental Table 5 for details) and range from −2.63 to 5.2, with negative and positive values coded in blue and red colors, respectively. SNPs identified in the Australian sample affect all 5 Australian phenotypes in the same direction and those identified in the US sample affect all 3 US phenotypes in the same direction. Although these effects appear to be sample-specific, meta-analysis identified 4 additional SNPs that have similar effects in both samples, of which rs17457384 is associated with sweets intake in the UK Biobank, with the allele for higher intensity ratings leading to a lower intake. gSweet, general sweet factor (a weighted mean of ratings of glucose, fructose, NHDC, and aspartame); NHDC, neohesperidin dihydrochalcone; SNP, single nucleotide polymorphism. * , P < 0.05; **, P < 1.0 × 10 −5; −, SNP not available.
In further functional analyses we found that 12 of the 139 suggestively associated SNPs (8 for the intake of total sugars, 1 for the intake of sweets, 1 for NHDC intensity, 1 for sucrose sweetness, and 1 for the meta-analysis of gSweet and sucrose intensity) were in high LD (r2 ≥ 0.6) with nonsynonymous variants within protein coding (exonic) regions (Supplemental Table 7). All 12 SNPs were correlated with missense mutations; and 2 SNPs, rs62060920 and rs603985, which were suggestively associated with the intake of total sugars, were further correlated with nonsense mutations that create stop codons within the same gene (i.e., MAPT and FUT2, respectively).
Gene-based tests using fastBAT did not identify any significant association (see Supplemental Table 8 for the associations with P < 1.0 × 10−5), whereas the results from MetaXcan showed a significant association between the intake of total sugars and the expression of FTO in the muscle tissue (P = 2.62 × 10−8; see Supplemental Table 9 for the top associations). Pathway analyses showed that the perceived intensity of sucrose was associated with the stomach inflammation pathway (P = 2.2 × 10−13 and false discovery rate < 0.05). Genes involved in this pathway were SLC50A1, AL645568.1 (RP11-296O14.3), CCDC68, GPBP1, EFNA1, NALT1 (RP11-611D20.2), B3GNTL1, CNTN5, FSHR, and WLS. No association in the tissue enrichment analyses reached the statistical significance threshold. The strongest associations (P < 0.01) were found within the genitalia, muscles, connective tissues, pharynx, immune system, and central nervous system (Supplemental Table 10).
Associations with TAS1R2, TAS1R3, GNAT3, GLUT2, and FGF21
SNPs within TAS1R2, TAS1R3, and GNAT3 previously reported to be associated with the taste sensitivity of sucrose solution were not associated with any of the sweet perception or preference phenotypes in the Australian or US samples (P > 0.05 for all SNP associations, except for the association between TAS1R3 rs307055 and sucrose intensity ratings with P = 0.03; Supplemental Table 11; Supplemental Figure 3). However, the TAS1R3 alleles associated with higher sucrose sensitivity tended to decrease ratings for all Australian and US sweet phenotypes.
SNPs within TAS1R2 and TAS1R3 were not associated with the intake of total sugars and sweets in the UK Biobank sample (P > 0.1). Because the associations between TAS1R2 rs35874116 and the intake of sugars were suggested to depend on BMI (6, 13), we followed previous studies by dividing the UK Biobank sample into 2 groups based on a BMI score of 25 (n = 109,507 and 64,554 for BMI ≥ 25 and < 25, respectively) or including BMI as a covariate, but no association was detected in any of the tests (P > 0.12). Six SNPs in high LD within the GNAT3 gene (rs1524600, rs6467217, rs6970107, rs6975345, rs10242727, and rs6467192; r2 > 0.6 between the 6 SNPs) were marginally associated with the intake of total sugars (P < 0.05, Supplemental Table 11), with alleles associated with higher sucrose sensitivity leading to a lower intake. The GLUT2 rs5400 was not associated with the intake of total sugars and sweets (Supplemental Figure 4) and the null association remained after we included BMI as a covariate or stratified the UK Biobank sample by a BMI score of 25 (P > 0.05). We successfully replicated the association between FGF21 and intake, with the rs838133 A allele (β = 0.860 g/d, SE = 0.170, P = 4.8 × 10−7) and rs838145 G allele (β = 0.782 g/d, SE = 0.167, P = 2.7 × 10−6) leading to higher intake of total sugars.
Gene-based tests using fastBAT did not identify any significant associations between TAS1R2, TAS1R3, GNAT3, and GLUT2 and all phenotypes (P > 0.05), but there was a strong association between FGF21 and the intake of total sugar in the UK Biobank sample (P = 2.71 × 10−6). MetaXcan results showed some marginal associations with P < 0.05 (Supplemental Table 9) and the strongest association was between the intake of total sugar and expression of TAS1R3 in brain tissue (P = 3.15 × 10−4).
Because the associations with TAS1R2, TAS1R3, GNAT3, and GLUT2 were reported previously in samples of unknown or mixed ethnicities, we wondered whether they were driven by population stratification, i.e., there are mean differences in the same sweet phenotype between populations and an artificial association is created due to the difference in the allele frequency between populations. We looked up their allele frequencies using the 1000 Genomes reference panel (50) and found that some of the allele frequencies, particularly for SNPs within TAS1R3, GNAT3, and GLUT2, are very different between African Americans and others (Table 4). Using the full sample of the UK Biobank, which is comprised of mixed ethnicities, we reproduced these associations with the intake of total sugars and sweets and even BMI, but the associations became null after adjusting for genetic PCs (Table 4) or after the sample was stratified by ethnic group (Supplemental Table 12). We showed an unexpected association between rs5400 and BMI (P = 1.04 × 10−8 after adjusting for genetic PCs) and the association remained strong within the white-British-only subset of the UK Biobank (P = 2.7 × 10−7).
TABLE 4.
Associations between TAS1R1, TAS1R3, GNAT3, and GLUT2 SNPs and the intake of total sugars and sweets and BMI before and after adjusting for genetic PCs using all participants from the UK Biobank sample of all ancestries1
| Allele frequency | P Intake of total sugars | P Intake of sweets | P BMI | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP:allele | AFR | AMR | ASN | EUR | UKB | No PC | PC adjusted | No PC | PC adjusted | No PC | PC adjusted |
| TAS1R2 | rs12033832:A | 0.23 | 0.23 | 0.48 | 0.31 | 0.35 | 0.86 | 0.47 | 0.76 | 0.60 | 8.94 × 10−3 | 0.87 |
| rs3935570:T | 0.30 | 0.19 | 0.12 | 0.25 | 0.27 | 0.49 | 0.31 | 0.72 | 0.63 | 0.04 | 0.52 | |
| rs35874116:A | 0.66 | 0.69 | 0.90 | 0.68 | 0.67 | 0.15 | 0.15 | 0.81 | 0.74 | 0.15 | 0.72 | |
| rs12137730:A | 0.83 | 0.60 | 0.81 | 0.65 | 0.64 | 0.76 | 0.41 | 0.03 | 0.05 | 0.03 | 0.60 | |
| rs7534618:T | 0.77 | 0.76 | 0.52 | 0.67 | 0.63 | 0.23 | 0.83 | 0.86 | 0.67 | 7.10 × 10−4 | 0.41 | |
| rs9701796:G | 0.21 | 0.17 | 0.27 | 0.21 | 0.21 | 0.59 | 0.85 | 0.20 | 0.15 | 0.29 | 0.06 | |
| TAS1R3 | rs307355:C | 0.48 | 0.84 | 0.82 | 0.92 | 0.92 | 2.39 × 10−7 | 0.57 | 9.74 × 10−3 | 0.14 | 8.85 × 10−35 | 0.13 |
| rs35744813:C | 0.32 | 0.83 | 0.83 | 0.92 | 0.92 | 6.38 × 10−12 | 0.57 | 4.11 × 10−3 | 0.17 | 1.20 × 10−62 | 0.11 | |
| GNAT3 | rs7792845:T | 0.15 | 0.37 | 0.31 | 0.42 | 0.40 | 0.17 | 0.85 | 0.98 | 0.69 | 1.73 × 10−7 | 0.39 |
| rs940541:T | 0.11 | 0.33 | 0.24 | 0.37 | 0.35 | 0.23 | 0.79 | 0.80 | 0.92 | 1.55 × 10−8 | 0.13 | |
| rs1107660:T | 0.11 | 0.34 | 0.26 | 0.37 | 0.36 | 0.18 | 0.88 | 0.74 | 0.96 | 4.35 × 10−8 | 0.28 | |
| rs1107657:T | 0.11 | 0.34 | 0.26 | 0.37 | 0.36 | 0.19 | 0.85 | 0.75 | 0.96 | 2.85 × 10−8 | 0.24 | |
| rs1524600:C | 0.45 | 0.90 | 0.87 | 0.89 | 0.88 | 7.32 × 10−6 | 0.55 | 1.00 × 10−3 | 0.02 | 1.67 × 10−30 | 0.53 | |
| rs6467217:T | 0.52 | 0.92 | 0.87 | 0.89 | 0.88 | 3.36 × 10−4 | 0.69 | 2.00 × 10−3 | 0.02 | 7.39 × 10−21 | 0.73 | |
| rs6970109:C | 0.53 | 0.92 | 0.87 | 0.89 | 0.88 | 6.28 × 10−4 | 0.77 | 2.45 × 10−3 | 0.02 | 1.06 × 10−19 | 0.75 | |
| rs6975345:T | 0.24 | 0.87 | 0.86 | 0.87 | 0.87 | 1.59 × 10−7 | 0.84 | 3.55 × 10−4 | 0.02 | 1.98 × 10−47 | 0.93 | |
| rs10242727:A | 0.24 | 0.87 | 0.86 | 0.87 | 0.87 | 2.34 × 10−7 | 0.87 | 4.07 × 10−4 | 0.02 | 1.11 × 10−46 | 0.95 | |
| rs6467192:G | 0.29 | 0.88 | 0.86 | 0.87 | 0.87 | 4.31 × 10−7 | 0.70 | 8.46 × 10−4 | 0.03 | 7.40 × 10−41 | 0.83 | |
| rs6961082:C | 0.86 | 0.97 | 0.87 | 0.95 | 0.94 | 0.18 | 0.42 | 0.31 | 0.39 | 0.66 | 0.39 | |
| GLUT2 | rs5400:C | 0.54 | 0.81 | 0.98 | 0.86 | 0.87 | 9.20 × 10−3 | 0.49 | 0.64 | 0.71 | 2.05 × 10−51 | 1.04 × 10−82 |
1Allele frequencies for AFR, AMR, ASN, and EUR are based on the 1000 Genome reference panel. n = 206,551 for the intake of total sugar; n = 20,532 for the intake of sweets; n = 485,476 for BMI. See Supplemental Table 12 for the ancestral demography of the UK Biobank. AFR, African; AMR, Admixed American (i.e., Colombian, Puerto Rican, and Mexican individuals); ASN, Asian; EUR, European; P, association P value for an SNP and a trait; PC, principal component; SNP:allele, single nucleotide polymorphism and the allele used to calculate allele frequency; UKB, UK Biobank.
2 P = 2.7 × 10−7 in all the Europeans from the UK Biobank (n = 407,708).
Gene expression
We examined the expression levels of genes nearby the suggestively associated SNPs (Supplemental Tables 1–4), genes containing nonsynonymous variants (Supplemental Table 7), those involved in the stomach inflammation pathway, and the 5 previously reported sweet taste genes (i.e., TAS1R2, TAS1R3, GNAT3, GLUT2 [SLC2A2], and FGF21). The expression levels of 133 genes and 121 genes were available in the in-house assays for the taste tissue and in the GTEx database for other tissues, respectively (presented as a heatmap in Supplemental Figure 5). In general, expression levels of the same gene were similar across all tissues. The 2 genes FUT2 and MAPT, which contain both missense and nonsense mutations, were expressed particularly higher in the salivary gland and the brain, respectively. The former was also expressed in the taste tissue at a moderate level, whereas the latter was barely expressed in the taste tissue. The sweet taste receptor gene TAS1R3 was weakly expressed across all tissues; the information on TAS1R2 and GNAT3 was only available in the in-house data on taste tissue and they were barely expressed.
Discussion
To our knowledge, this is the first large-scale GWAS of the perception, liking, and consumption of sweet substances using 3 independent population samples of Australian adolescents and US and UK adults. We found a strong association between the FTO gene and sugar intake, and suggestive associations for both the perception and intake of sweet substances. Whereas we replicated the association between the FGF21 gene and sugar intake, we found limited support for the previously reported associations within the TAS1R2, TAS1R3, GNAT3, and GLUT2 genes.
The SNP associated with sugar intake (rs11642841) is highly correlated with the repeatedly reported BMI-associated SNP rs9939609 within the FTO gene (48) (r2 = 0.73 in the UK Biobank white-British sample). The BMI-increasing allele has been shown to increase the intake of energy (51, 52), dietary fat (53), and protein (54); the risk of cardiometabolic disorders (e.g., type 2 diabetes, coronary artery disease, and hypertension); and fasting glucose and insulin concentrations (55). Counterintuitively, in the present study the BMI-increasing allele was associated with a lower intake of reported total sugars. We further found negative genetic and phenotypic correlations between sugar intake and BMI in the UK Biobank sample, in agreement with an earlier observational study (56). These negative associations could be because obese people derive a greater proportion of energy from protein and fat and less from carbohydrates (57). There may also be reporting bias, with those who have greater BMI tending to underreport sugar intake, or the possibility that our result is a chance finding because we did not find information to replicate these associations in other independent studies. In contrast, the intake of sweets, which contributed to a part of the total sugars, and BMI were positively correlated. This suggests that people who are overweight or obese reduce calories by reducing the intake of total carbohydrates but they still eat more sweets than people with normal weight.
We showed evidence of nonsynonymous variants associated with both perception and intake of sweet substances. Particularly, 2 SNPs (rs62060920 and rs603985) associated with sugar intake were correlated with not only amino acid–changing variants but also stop codon–creating variants within the same genes, i.e., MAPT and FUT2, respectively. Expression levels of these 2 genes MAPT and FUT2 were higher in the brain and the salivary gland, respectively, suggestive of their roles in central and peripheral mechanisms regulating sugar intake. Another SNP, rs623965, identified in the meta-analysis of sweet intensity and which is correlated with a nonsynonymous variant within the KDMA4 gene, was recently shown to be associated with educational attainment (P = 3.8 × 10−7) (58). This suggests a shared molecular mechanism for the sweet taste perception and cognitive function, which can be indirectly supported by our recent finding of the phenotypic correlation between intelligence and gSweet (rp = −0.07, P < 0.05) (32). These results highlight the potential significance of the brain in the perception and intake of sweet substances.
We failed to replicate the associations between sweet taste perception and variants within the TAS1R2, TAS1R3, and GNAT3 genes in all 3 samples of European ancestry. Previous studies reported these associations in smaller samples (n = 95 [6], 144 [7], and 160 [8]); moreover, 2 of these samples (7, 8) were comprised of mixed ethnicities (i.e., Europeans, Asians, and African Americans), in which the allele frequencies vary (Table 4), and 1 was of unknown ethnicity (6). Their analyses did not include genetic PCs to account for population stratification because no genome-wide genotyping data were available and it was unclear whether ethnicity was included as a covariate. Using the full UK Biobank sample including mixed ethnicities, we reproduced the associations with TAS1R3 and GNAT3; however, the associations became null after adjusting for genetic PCs, suggesting that spurious associations may arise for these particular SNPs if population stratification is not considered appropriately. This is why we could not replicate these associations in the European-only populations. Nevertheless, the effect of TAS1R3 on sucrose preference was observed in an independent adult female sample primarily composed of African Americans (16), indicating that the TAS1R3 SNPs could have functional potency for sweet taste perception within other ethnic groups. It is noteworthy that the same genetic variants might affect sweet perception and intake differently in different populations of the same genetic ancestry owing to cultural differences, especially those populations having traditional compared with obesogenic foodways. Therefore, an ideal study design needs to consider the effect of genetic ethnicity as well as the gene × environment interactions.
In agreement with previous studies we observed no association between the intake of sugars and sweet food and TAS1R3, but we failed to replicate the associations with TAS1R2 (6, 9–14) and GLUT2 (13, 17) in the large UK Biobank sample. As some of these study samples were of mixed ethnicities, we demonstrated that an artificial association with sugar intake and even BMI could be created by population stratification (Table 4). We found unexpectedly an association between GLUT2 rs5400 and BMI (P = 2.7 × 10−7 among white-British individuals in the UK Biobank sample). However, this association is relatively weak in an equally large European sample (P = 0.019) (59) and is not in the top 751 independent associations in today's largest GWAS meta-analysis of BMI (n = ∼700,000 Europeans including participants from the UK Biobank) (60), so its effect on BMI requires further investigation.
Among the SNPs suggestively associated with sweet taste perception, rs17457384 near the GABRB2 gene was associated with the intake of sweets in the UK Biobank sample. This SNP was identified in the meta-analysis of sweet intensity, with the T allele being associated with a higher perceived intensity rating of gSweet (Australian sample) and sucrose (US sample) and a higher intake of sweets (UK Biobank sample). The GABRB2-encoded protein is a receptor of neurotransmitters that mediate synaptic transmission in the central nervous system and it is highly expressed in the brain. This finding again signifies the neurological influence on sweet consumption.
Differences in the association patterns between the Australian adolescent and the US adult samples could be due to differences in their study characteristics, including the sample age (i.e., mean ages were 16.1 compared with 36.8 y) and the measuring scale (i.e., gLMS compared with VAS). We previously showed that the perceived intensity of sweet solutions decreased from adolescents to adults at a rate of 2–5%/y between ages 12 and 25 y (5). Measuring sweet taste phenotypes using gLMS and VAS could lead to different data distributions (61). These together make replications more difficult when the sample sizes are not big enough. Notably, the genome-wide significant associations for the bitter taste perception (i.e., rs713598, rs1726866, and rs10246939 for propylthiouracil/phenylthiocarbamide and rs10772420 for quinine [29, 62, 63]) were consistent between the 2 samples (Supplemental Table 13). This indicates that the genetics of sweet taste is more complicated and our findings require further validation in larger samples.
Pathway analyses showed that the cellular process related to stomach inflammation is associated with the perceived intensity of sucrose in the US adult sample. Alteration in taste is one of the symptoms of stomach gastritis (64, 65) and the association might be linked via reflux or vomiting, which expose taste receptors in the oral cavity to gastric acid (66). Mouse studies have shown that taste receptors expressed in the stomach can modify gastric motility and food intake (67, 68). As human sweet taste receptors (i.e., TAS1R3; Supplemental Figure 5) are also expressed along the gastrointestinal tract, perhaps molecules involved in stomach inflammation could regulate the function of sweet taste receptors, which in turn influences taste perception.
Despite our samples being considerably larger than any previous studies of the associations we have examined, the present study may still lack statistical power. In particular, we were unable to detect genome-wide significant associations for sweet taste perception. This suggests that sweet taste is a polygenic trait and its variation is influenced by many SNPs, each with a small effect. Genetically informative data on taste perception are relatively limited compared with disease traits or other sensory traits, such as vision (69) or hearing (70). This is presumably due to variation in taste perception posing no immediate threat to life, or to a lesser extent. There are large genetic studies collecting food intake data (e.g., UK Biobank), which may be used as proxies of taste perception. However, our results indicate that the genetic profiles of intake of sweets can be different from those of sweet perception. Therefore, using the dietary data may not accurately capture individual differences in taste perception and is less likely to detect the true underlying genes, which highlights the value of our data sets.
In conclusion, this GWAS study showed an association between FTO and the intake of total sugars, suggestive of shared biology between the sugar consumption and BMI-related traits. Findings from the present study were solely based on Europeans, so whether they can be generalized to other ethnic groups requires further investigation. The failure to replicate the associations with TAS1R2, TAS1R3, GNAT3, and GLUT2 warrants future studies using people with different ancestral backgrounds, such as Africans and Asians. Lastly, our results suggest that genes apart from those within the peripheral sweet receptor system are also associated with the perception and intake of sweet substances and those involved in brain functions and stomach inflammation could be a direction for future research.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kirsten J Mascioli, Christopher Tharp, Fujiko Duke, Deborah Lee, and Corrine Mansfield from the Monell Chemical Senses Center for manufacturing the taste tests; and from the QIMR Berghofer, Marlene Grace, Ann Eldridge, Natalie Garden, and Kerrie McAloney for project coordination, data collection, and data entry, and David Smyth for computer support. In particular, we thank twins and their families for participating in the taste tests with the Monell Chemical Senses Center and the QIMR Berghofer Medical Research Institute.
The authors’ responsibilities were as follows—L-DH, PASB, MJW, NGM, and DRR: designed the research; SDG: cleaned and imputed the Australian genotyping data; SM: obtained the UK Biobank data; L-DH, CL, PG, GC-P, J-SO, JA, and GZ: analyzed the data; L-DH: drafted the initial manuscript; PG, GC-P, DAL, PASB, MJW, NGM, and DRR: critically reviewed the manuscript; L-DH: had primary responsibility for final content; and all authors: read and approved the final manuscript. DAL has received support from Roche Diagnostics and Medtronic for research unrelated to that presented here. None of the other authors reported a conflict of interest related to the study.
Notes
Supported by NIH grants DC02995 (to PASB) and DC004698 (to DRR), Australian National Health and Medical Research Council (NHMRC) grants 241944 and 1031119 (to NGM), and Australian Research Council grants DP1093900 and DP0664638 (to NGM and MJW). LDH has been financially supported by NHMRC project grant GNT1125200. We collected genotype data from equipment purchased in part with NIH funds from grant OD018125. DAL works in a unit that is supported by the University of Bristol and the UK Medical Research Council (grant MC_UU_00011/6). This research has been conducted using the UK Biobank Resource (application number 25331).
Supplemental Tables 1–13 and Supplemental Figures 1–5 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/ajcn/.
Abbreviations used: gLMS, general labeled magnitude scale; gSweet, mean intensity of the 4 sweet tastes weighted by loadings from the common factor; GWAS, genome-wide association scan; h2, heritability; LD, linkage disequilibrium; NHDC, neohesperidin dihydrochalcone; PC, principal component; QC, quality control; rg, genetic correlation; rp, phenotypic correlation; SNP, single nucleotide polymorphism; VAS, visual analog scale.
References
- 1. Keskitalo K, Silventoinen K, Tuorila H, Perola M, Pietilainen KH, Rissanen A, Kaprio J. Genetic and environmental contributions to food use patterns of young adult twins. Physiol Behav. 2008;93(1–2):235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Knaapila A, Hwang LD, Lysenko A, Duke FF, Fesi B, Khoshnevisan A, James RS, Wysocki CJ, Rhyu M, Tordoff MG et al.. Genetic analysis of chemosensory traits in human twins. Chem Senses. 2012;37(9):869–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Keskitalo K, Knaapila A, Kallela M, Palotie A, Wessman M, Sammalisto S, Peltonen L, Tuorila H, Perola M. Sweet taste preferences are partly genetically determined: identification of a trait locus on chromosome 16. Am J Clin Nutr. 2007;86(1):55–63. [DOI] [PubMed] [Google Scholar]
- 4. Smith AD, Fildes A, Forwood S, Cooke L, Llewellyn C. The individual environment, not the family is the most important influence on preferences for common non-alcoholic beverages in adolescence. Sci Rep. 2017;7(1):16822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hwang LD, Zhu G, Breslin PA, Reed DR, Martin NG, Wright MJ. A common genetic influence on human intensity ratings of sugars and high-potency sweeteners. Twin Res Hum Genet. 2015;18(4):361–7. [DOI] [PubMed] [Google Scholar]
- 6. Dias AG, Eny KM, Cockburn M, Chiu W, Nielsen DE, Duizer L, El-Sohemy A. Variation in the TAS1R2 gene, sweet taste perception and intake of sugars. J Nutrigenet Nutrigenomics. 2015;8(2):81–90. [DOI] [PubMed] [Google Scholar]
- 7. Fushan AA, Simons CT, Slack JP, Manichaikul A, Drayna D. Allelic polymorphism within the TAS1R3 promoter is associated with human taste sensitivity to sucrose. Curr Biol. 2009;19(15):1288–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fushan AA, Simons CT, Slack JP, Drayna D. Association between common variation in genes encoding sweet taste signaling components and human sucrose perception. Chem Senses. 2010;35(7):579–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Melo SV, Agnes G, Vitolo MR, Mattevi VS, Campagnolo PDB, Almeida S. Evaluation of the association between the TAS1R2 and TAS1R3 variants and food intake and nutritional status in children. Genet Mol Biol. 2017;40(2):415–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chamoun E, Hutchinson JM, Krystia O, Mirotta JA, Mutch DM, Buchholz AC, Duncan AM, Darlington G, Haines J, Ma DWL et al.. Single nucleotide polymorphisms in taste receptor genes are associated with snacking patterns of preschool-aged children in the Guelph Family Health Study: a pilot study. Nutrients. 2018;10(2):153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chamoun E, Carroll NA, Duizer LM, Qi W, Feng Z, Darlington G, Duncan AM, Haines J, Ma DWL; Guelph Family Health Study. The relationship between single nucleotide polymorphisms in taste receptor genes, taste function and dietary intake in preschool-aged children and adults in the Guelph Family Health Study. Nutrients. 2018;10(8):990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Han P, Keast RSJ, Roura E. Salivary leptin and TAS1R2/TAS1R3 polymorphisms are related to sweet taste sensitivity and carbohydrate intake from a buffet meal in healthy young adults. Br J Nutr. 2017;118(10):763–70. [DOI] [PubMed] [Google Scholar]
- 13. Eny KM, Wolever TM, Corey PN, El-Sohemy A. Genetic variation in TAS1R2 (Ile191Val) is associated with consumption of sugars in overweight and obese individuals in 2 distinct populations. Am J Clin Nutr. 2010;92(6):1501–10. [DOI] [PubMed] [Google Scholar]
- 14. Ramos-Lopez O, Panduro A, Martinez-Lopez E, Roman S. Sweet taste receptor TAS1R2 polymorphism (Val191Val) is associated with a higher carbohydrate intake and hypertriglyceridemia among the population of west Mexico. Nutrients. 2016;8(2):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Joseph PV, Reed DR, Mennella JA. Individual differences among children in sucrose detection thresholds: relationship with age, gender, and bitter taste genotype. Nurs Res. 2016;65(1):3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mennella JA, Finkbeiner S, Lipchock SV, Hwang LD, Reed DR. Preferences for salty and sweet tastes are elevated and related to each other during childhood. PLoS One. 2014;9(3):e92201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eny KM, Wolever TM, Fontaine-Bisson B, El-Sohemy A. Genetic variant in the glucose transporter type 2 is associated with higher intakes of sugars in two distinct populations. Physiol Genomics. 2008;33(3):355–60. [DOI] [PubMed] [Google Scholar]
- 18. Chu AY, Workalemahu T, Paynter NP, Rose LM, Giulianini F, Tanaka T, Ngwa JS, Qi Q, Curhan GCCHARGE Nutrition Working Group; et al.; CHARGE Nutrition Working Group Novel locus including FGF21 is associated with dietary macronutrient intake. Hum Mol Genet. 2013;22(9):1895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tanaka T, Ngwa JS, van Rooij FJ, Zillikens MC, Wojczynski MK, Frazier-Wood AC, Houston DK, Kanoni S, Lemaitre RN, Luan J et al.. Genome-wide meta-analysis of observational studies shows common genetic variants associated with macronutrient intake. Am J Clin Nutr. 2013;97(6):1395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Soberg S, Sandholt CH, Jespersen NZ, Toft U, Madsen AL, von Holstein-Rathlou S, Grevengoed TJ, Christensen KB, Bredie WLP, Potthoff MJ et al.. FGF21 is a sugar-induced hormone associated with sweet intake and preference in humans. Cell Metab. 2017;25(5):1045–53..e6. [DOI] [PubMed] [Google Scholar]
- 21. Frayling TM, Beaumont RN, Jones SE, Yaghootkar H, Tuke MA, Ruth KS, Casanova F, West B, Locke J, Sharp S et al.. A common allele in FGF21 associated with sugar intake is associated with body shape, lower total body-fat percentage, and higher blood pressure. Cell Rep. 2018;23(2):327–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O'Connell J et al., The UK Biobank resource with deep phenotyping and genomic data, Nature, 2018;562(7726):203–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, von Elm E, Khoury MJ, Cohen B, Davey-Smith G, Grimshaw J et al.. STrengthening the REporting of Genetic Association Studies (STREGA)—an extension of the STROBE statement. Genet Epidemiol. 2009;33(7):581–98. [DOI] [PubMed] [Google Scholar]
- 24. Wright MJ, Martin NG. Brisbane adolescent twin study: outline of study methods and research projects. Aust J Psychol. 2004;56(2):65–78. [Google Scholar]
- 25. Green BG, Shaffer GS, Gilmore MM. Derivation and evaluation of a semantic scale of oral sensation magnitude with apparent ratio properties. Chem Senses. 1993;18(6):683–702. [Google Scholar]
- 26. O'Connell J, Gurdasani D, Delaneau O, Pirastu N, Ulivi S, Cocca M, Traglia M, Huang J, Huffman JE, Rudan I et al.. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 2014;10(4):e1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2015;31(5):782–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dachet C, Poirier O, Cambien F, Chapman J, Rouis M. New functional promoter polymorphism, CETP/−629, in cholesteryl ester transfer protein (CETP) gene related to CETP mass and high density lipoprotein cholesterol levels: role of Sp1/Sp3 in transcriptional regulation. Arterioscler Thromb Vasc Biol. 2000;20(2):507–15. [DOI] [PubMed] [Google Scholar]
- 29. Reed DR, Zhu G, Breslin PA, Duke FF, Henders AK, Campbell MJ, Montgomery GW, Medland SE, Martin NG, Wright MJ. The perception of quinine taste intensity is associated with common genetic variants in a bitter receptor cluster on chromosome 12. Hum Mol Genet. 2010;19(21):4278–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44(7):821–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hwang LD, Gharahkhani P, Breslin PAS, Gordon SD, Zhu G, Martin NG, Reed DR, Wright MJ. Bivariate genome-wide association analysis strengthens the role of bitter receptor clusters on chromosomes 7 and 12 in human bitter taste. BMC Genomics. 2018;19(1):678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hwang LD, Breslin PA, Reed DR, Zhu G, Martin NG, Wright MJ. Is the association between sweet and bitter perception due to genetic variation?. Chem Senses. 2016;41(9):737–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Loh PR, Tucker G, Bulik-Sullivan BK, Vilhjalmsson BJ, Finucane HK, Salem RM, Chasman DI, Ridker PM, Neale BM, Berger B et al.. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47(3):284–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, Daly MJ, Price AL, Neale BM; Schizophrenia Working Group of the Psychiatric Genomics Consortium. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74(4):765–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bakshi A, Zhu Z, Vinkhuyzen AA, Hill WD, McRae AF, Visscher PM, Yang J. Fast set-based association analysis using summary data from GWAS identifies novel gene loci for human complex traits. Sci Rep. 2016;6:32894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA, Compton CC, DeLuca DS, Peter-Demchok J, Gelfand ET et al.. A novel approach to high-quality postmortem tissue procurement: the GTEx project. Biopreserv Biobank. 2015;13(5):311–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barbeira A, Dickinson SP, Bonazzola R, Zheng J, Wheeler HE, Torres JM, Torstenson ES, Shah KP, Garcia T, Edwards T et al.. Exporing the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun. 2018;9(1):1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pers TH, Karjalainen JM, Chan Y, Westra HJ, Wood AR, Yang J, Lui JC, Vedantam S, Gustafsson S, Esko T et al.. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6:5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, Hemani G, Tansey K, Laurin C, Early G et al.. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33(2):272–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, Duncan Let al. ReproGen Consortium, Psychiatric Genomics Consortium, Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3, An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Spielman AI, Pepino MY, Feldman R, Brand JG. Technique to collect fungiform (taste) papillae from human tongue. J Vis Exp. 2010;(42):e2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Frazee AC, Pertea G, Jaffe AE, Langmead B, Salzberg SL, Leek JT. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat Biotechnol. 2015;33(3):243–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW et al.. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Loos RJ, Yeo GS. The bigger picture of FTO: the first GWAS-identified obesity gene. Nat Rev Endocrinol. 2014;10(1):51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cecil JE, Tavendale R, Watt P, Hetherington MM, Palmer CN. An obesity-associated FTO gene variant and increased energy intake in children. N Engl J Med. 2008;359(24):2558–66. [DOI] [PubMed] [Google Scholar]
- 52. Speakman JR, Rance KA, Johnstone AM. Polymorphisms of the FTO gene are associated with variation in energy intake, but not energy expenditure. Obesity (Silver Spring). 2008;16(8):1961–5. [DOI] [PubMed] [Google Scholar]
- 53. Park SL, Cheng I, Pendergrass SA, Kucharska-Newton AM, Lim U, Ambite JL, Caberto CP, Monroe KR, Schumacher F, Hindorff LA et al.. Association of the FTO obesity risk variant rs8050136 with percentage of energy intake from fat in multiple racial/ethnic populations: the PAGE study. Am J Epidemiol. 2013;178(5):780–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ahmad T, Lee IM, Pare G, Chasman DI, Rose L, Ridker PM, Mora S. Lifestyle interaction with fat mass and obesity-associated (FTO) genotype and risk of obesity in apparently healthy U.S. women. Diabetes Care. 2011;34(3):675–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fall T, Hagg S, Magi R, Ploner A, Fischer K, Horikoshi M, Sarin AP, Thorleifsson G, Ladenvall C, Kals M et al.. The role of adiposity in cardiometabolic traits: a Mendelian randomization analysis. PLoS Med. 2013;10(6):e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Macdiarmid JI, Vail A, Cade JE, Blundell JE. The sugar–fat relationship revisited: differences in consumption between men and women of varying BMI. Int J Obes Relat Metab Disord. 1998;22(11):1053–61. [DOI] [PubMed] [Google Scholar]
- 57. Ortega RM, Requejo AM, Andrés P, López-Sobaler AM, Redondo R, González-Fernández M. Relationship between diet composition and body mass index in a group of Spanish adolescents. Br J Nutr. 1995;74(6):765–73. [DOI] [PubMed] [Google Scholar]
- 58. Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA, Turley P, Chen GB, Emilsson V, Meddens SF et al.. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533(7604):539–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Turcot V, Lu Y, Highland HM, Schurmann C, Justice AE, Fine RS, Bradfield JP, Esko T, Giri A, Graff M et al.. Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat Genet. 2018;50(1):26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, Frayling TM, Hirschhorn J, Yang J, Visscher PM et al.. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum Mol Genet. 2018;27(20):3641–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hayes JE, Allen AL, Bennett SM. Direct comparison of the generalized Visual Analog Scale (gVAS) and general Labeled Magnitude Scale (gLMS). Food Qual Prefer. 2013;28(1):36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299(5610):1221–5. [DOI] [PubMed] [Google Scholar]
- 63. Ledda M, Kutalik Z, Souza Destito MC, Souza MM, Cirillo CA, Zamboni A, Martin N, Morya E, Sameshima K, Beckmann JS et al.. GWAS of human bitter taste perception identifies new loci and reveals additional complexity of bitter taste genetics. Hum Mol Genet. 2014;23(1):259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Veldhuyzen van Zanten SJ, Tytgat KM, Pollak PT, Goldie J, Goodacre RL, Riddell RH, Hunt RH. Can severity of symptoms be used as an outcome measure in trials of non-ulcer dyspepsia and Helicobacter pylori associated gastritis?. J Clin Epidemiol. 1993;46(3):273–9. [DOI] [PubMed] [Google Scholar]
- 65. Cecchini MP, Pellegrini C, Bassetto MA, Osculati F, Sbarbati A, Marcolini L, Pegoraro M, Fontana R, De Franceschi L. Might Helicobacter pylori infection be associated with distortion on taste perception?. Med Hypotheses. 2013;81(3):496–9. [DOI] [PubMed] [Google Scholar]
- 66. Sipiora ML, Murtaugh MA, Gregoire MB, Duffy VB. Bitter taste perception and severe vomiting in pregnancy. Physiol Behav. 2000;69(3):259–67. [DOI] [PubMed] [Google Scholar]
- 67. Farre R, Tack J. Food and symptom generation in functional gastrointestinal disorders: physiological aspects. Am J Gastroenterol. 2013;108(5):698–706. [DOI] [PubMed] [Google Scholar]
- 68. Janssen S, Laermans J, Verhulst PJ, Thijs T, Tack J, Depoortere I. Bitter taste receptors and α-gustducin regulate the secretion of ghrelin with functional effects on food intake and gastric emptying. Proc Natl Acad Sci U S A. 2011;108(5):2094–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gharahkhani P, Burdon KP, Fogarty R, Sharma S, Hewitt AW, Martin S, Law MH, Cremin K, Bailey JNC, Loomis SJ et al.. Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat Genet. 2014;46(10):1120–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hoffmann TJ, Keats BJ, Yoshikawa N, Schaefer C, Risch N, Lustig LR. A large genome-wide association study of age-related hearing impairment using electronic health records. PLoS Genet. 2016;12(10):e1006371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.