Abstract
Optimal mitochondrial function is critical for healthy cellular activity, particularly in cells that have high energy demands like those in the nervous system and muscle. Consistent with this, mitochondrial dysfunction has been associated with a myriad of neurodegenerative diseases and aging in general. Caenorhabditis elegans have been a powerful model system for elucidating the many intricacies of mitochondrial function. Mitochondrial respiration is a strong indicator of mitochondrial function and recently developed respirometers offer a state-of-the-art platform to measure respiration in cells. In this protocol, we provide a technique to analyze live, intact C. elegans. This protocol spans a period of ~7 days and includes steps for (1) growing and synchronization of C. elegans, (2) preparation of compounds to be injected and hydration of probes, (3) drug loading and cartridge equilibration, (4) preparation of worm assay plate and assay run, and (5) post-experiment data analysis.
Keywords: Biology, Issue 144, C. elegans, respiration, oxygen consumption rate (OCR), respirometer, mitochondria, metabolism
Introduction
Adenosine triphosphate (ATP), the main source of cellular energy, is produced in the mitochondria by enzymes in the electron transport chain (ETC) located in the inner mitochondrial membrane. Pyruvate, a key metabolite utilized for mitochondrial ATP production, is imported into the mitochondrial matrix where it is decarboxylated to produce acetyl coenzyme A (CoA). Subsequently, acetyl CoA enters the citric acid cycle resulting in the generation of nicotinamide adenine dinucleotide (NADH), a key electron carrier molecule. As electrons from NADH are passed to oxygen via the ETC, protons build up in the mitochondrial intermembrane space, which results in the generation of an electrochemical gradient across the membrane. These protons will then flow from the intermembrane space across this electrochemical gradient back into the mitochondrial matrix through the proton pore of the ATP synthase, driving its rotation and the synthesis of ATP1 (Figure 1).
Figure 1. Schematic of the major players involved in cellular respiration and the effect of FCCP and sodium azide.
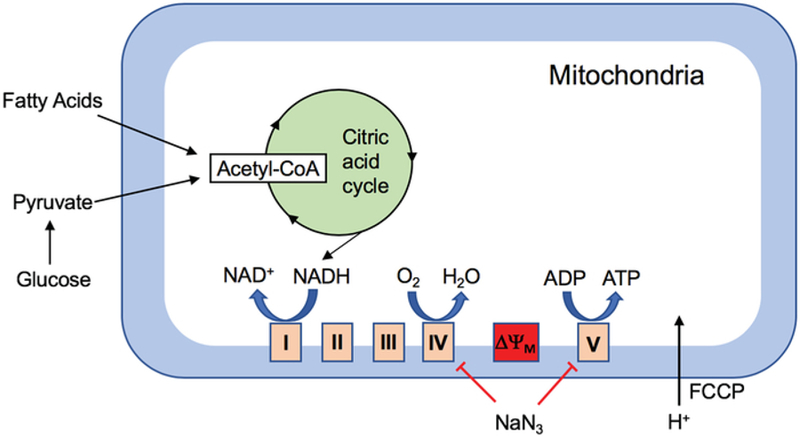
Transfer of electrons from NADH to the ETC complex I results in the generation of an electrochemical gradient across the inner mitochondrial membrane as protons get pumped across it. Protons flowing back into the mitochondrial matrix from the intermembrane space via complex V results in ATP synthesis. Addition of FCCP results in the uncoupling of this process by disrupting the mitochondrial membrane potential and thereby ATP synthesis, while oxygen consumption continues, allowing for the measurement of maximal OCR. Sodium azide (NaN3) is an inhibitor of complexes IV and V, thereby allowing for the measurement of non-mitochondrial respiration.
Mitochondrial function is not limited to energy production but is also crucial for calcium homeostasis, reactive oxygen species (ROS) scavenging, and apoptosis, critically positioning their function in organismal health2. Mitochondrial function can be assessed using a variety of assays, including but not limited to analyses that measure mitochondrial membrane potential, ATP and ROS levels, and mitochondrial calcium concentrations. However, these assays provide a single snapshot of mitochondrial function and therefore might not provide a comprehensive view of mitochondrial health. Since oxygen consumption during ATP generation is reliant on a myriad of sequential reactions, it serves as a superior indicator of mitochondrial function. Interestingly, variations in oxygen consumption rates have been observed as a result of mitochondrial dysfunction3,4,5.
Oxygen consumption rates (OCR) of living samples can be measured using techniques that can be broadly divided into two groups: amperometric oxygen sensors and porphyrin-based phosphors that can be quenched by oxygen6. Amperometric oxygen sensors have been used extensively to measure OCR in cultured cells, tissues, and in model systems, such as C. elegans. However, porphyrin-based phosphors containing respirometers possess the following advantages: (1) they allow for a side by side comparison of two samples in triplicate, (2) they require smaller sample size (e.g., 20 worms per well versus ~2,000−5,000 worms in the chamber)7, and (3) the respirometer can be programmed to do four different compound injections at desired times throughout the experimental run, eliminating the need for manual application.
In this protocol, steps involved in using a porphyrin-based oxygen-sensing respirometer to measure OCR in live, intact C. elegans are described. While there is a written protocol for the use of the large format, high throughput respirometer8, this protocol has been adapted for use with a more budget friendly, accessible, and smaller scale instrument. This protocol is particularly useful for assessing the difference in OCR between two strains, where high-throughput screening is not required and its use would be excessive.
Protocol
NOTE: Figure 2 provides a schematic overview of the full protocol.
Figure 2. Schematic of steps involved in the measurement of OCR in C. elegans.
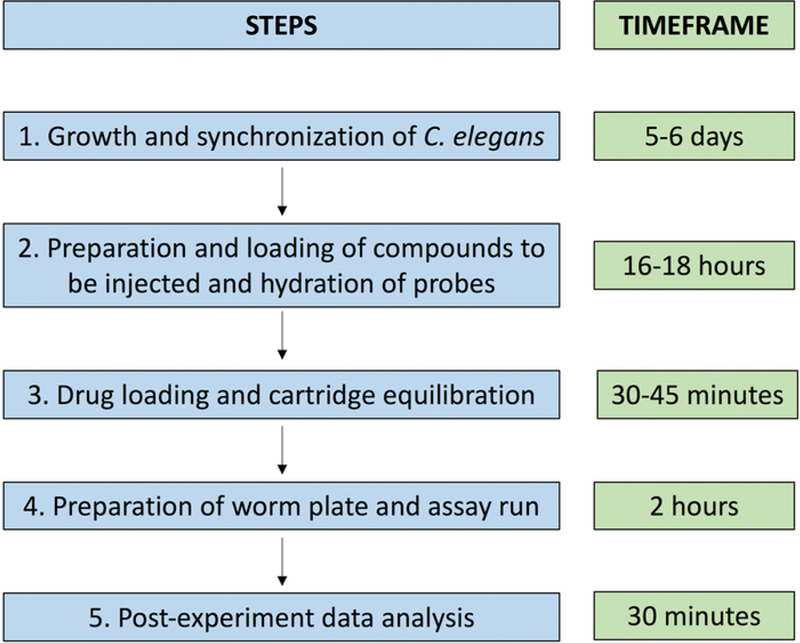
The five steps involved in the assay setup and run (left) and a timeframe for each step (right).
1. Growth and synchronization of nematode population9,10
Transfer L4 larvae of desired genetic backgrounds (e.g., N2 [wild type] and sel-12 animals) onto nematode growth media (NGM) plates (see Table 1 for recipe) freshly seeded with a lawn of Escherichia coli (OP50)11. Use at least two 100 mm or three 60 mm plates for each strain. Incubate the worms at the appropriate growth temperature (between 15–25 °C) for 3–4 days or until plates are concentrated with large number of eggs and gravid worms.
Wash the eggs and worms off the plates using approximately 6 mL of M9 buffer (see Table 1 for recipe) per 100 mm plate of nematodes and transfer them with glass Pasteur pipettes into individual 15 mL centrifuge tubes for each strain. Spin these tubes down for 3 min at 6,180 x g and aspirate out the M9 buffer, retaining just the animals and eggs pellet.
Add 3–4 mL of bleach solution (see Table 1 for recipe) to each tube and intermittently vortex for 6 min. Add M9 buffer to fill each tube and spin at 6,180 x g for 1 min and aspirate supernatant. Repeat this wash with M9 three times and move the egg pellet to a fresh 15 mL tube containing approximately 9 mL of M9 buffer.
Synchronize the freshly hatched worms by nutating at 20 °C for 16–48 h. Spin these tubes down at 6,180 x g for 1.5 min and put the synchronized L1 animals down on individual NGM plates freshly seeded with a lawn of OP50 (approximately 6,000–10,000 animals per 100 mm plate) and keep at 20 °C.
-
These animals will reach the L4 larval stage after ~42 h. At this time, move the L4 larvae using a platinum pick to OP50 seeded NGM plates containing 0.5 mg/mL 5-fluoro-2’-deoxyuridine (FUdR) to prevent them from producing progeny.
NOTE: Make sure that within an experiment all strains are synchronized for a similar amount of time. FUdR treatment sterilizes the animals and prevents egg laying and progeny production, which could influence OCR. These sterilized animals will be analyzed the next day (day 1 adult animals at ~66 h) for the assay. FUdR has been reported to impact physiology and lifespan of certain mutant worms. Therefore, this should be taken into consideration when the drug is used for sterilization12,13,14 Worms can also be sterilized by the means of feminizing mutations such as fem-1(hc17) or fem-3(e2006). However, these mutations might impact mitochondrial function.
Table 1:
Recipes for NGM plates, M9 buffer, and bleach solution.
| M9 buffer (1 L) | |||
|---|---|---|---|
| dH2O | 1,000 mL | ||
| NaCl | 5 g | ||
| KH2PO4 | 3 g | ||
| Na2HPO4 | 6 g | ||
| 1 M MgSO4 | 1 mL *add after autoclaving | ||
| Split between 2 bottles: 500 mL each. Autoclave on liquid cycle (15 min exposure) | |||
| Bleach solution (50 mL) | |||
| dH2O | 36 mL | ||
| Bleach | 14 mL | ||
| 10 N NaOH | 800 μL | ||
| Standard Nematode Growth Media (NGM) plates | |||
| 1 L | 0.5 L | 0.25 L | |
| NaCl | 3 g | 1.5 g | 0.75 g |
| Bacto-agar | 17 g | 8.5 g | 4.25 g |
| Bacto-peptone | 2.5 g | 1.25 g | 0.625 g |
| a. Autoclave on liquid cycle (45 min exposure), allow to cool to ~60 °C and then use sterile technique and add the following: | |||
| 1 L | 0.5 L | 0.25 L | |
| 1 M CaCl2 | 1 mL | 0.5 mL | 0.25 mL |
| 1 M MgSO4 | 1 mL | 0.5 mL | 0.25 mL |
| 1 M KPO4 | 25 mL | 12.5 mL | 6.25 mL |
| 5 mg/mL cholesterol | 1 mL | 0.5 mL | 0.25 mL |
| b. Swirl to mix thoroughly after each addition. After all additions are made, pour plates | |||
2. Preparation of compounds to be injected and hydration of probes
NOTE: During the assay run, both basal and maximal respiration rates of the nematodes are measured. Maximal respiration is triggered in the animals upon the addition of carbonyl cyanide-4 (trifluormethoxy)phenylhydrazone (FCCP), an uncoupling ionophore that disturbs the mitochondrial membrane potential and thus ATP synthesis by transporting protons through the mitochondrial membrane, while allowing proton pumping, electron transport, and oxygen consumption to proceed4,15 uncoupled from ATP synthesis (Figure 1). The final step in the assay involves the addition of sodium azide (NaN3), a drug that inhibits complexes IV and V in the ETC, allowing one to determine non-mitochondrial respiration16 (Figure 1). The following steps can be performed the day before the actual assay run.
-
Prepare 1 mL stock solutions of the FCCP (10 mM in dimethyl sulfoxide [DMSO]: 1000x the final assay concentration) and NaN3 (400 mM in dH2O: 10x final assay concentration) and store at −20 °C.
NOTE: Run a concentration curve to optimize the concentration of FCCP required to elicit maximal OCR response for each instrument and laboratory setting.
-
Hydrate the sensor cartridge by adding 200 µL of dH2O to each well (and the surrounding reservoir) in the plate, ensuring that the sensor probes are submerged in the dH2O and store overnight at room temperature.
NOTE: The cartridge probes can be left submerged for up to 72 h but in the case of a prolonged hydration, the plates should be wrapped in paraffin film and stored at 4 °C. If overnight hydration is not possible, the sensor cartridge should be hydrated for at least 4 h before assay.
-
Turn off the heating element within the respirometer interface and store the instrument inside a 15 °C incubator overnight to lower core temperature within the instrument to prevent the animals from overheating during the assay run.
NOTE: The respirometer is equipped with a heating element but cannot be cooled. Within a 15 °C incubator, the respirometer will have a stable temperature between 18–22 °C, which is a healthy temperature for C. elegans maintenance.
3. Drug loading and cartridge equilibration
Pipette out and discard the dH2O used to hydrate the sensor probes and replace it with 200 µL of the calibrant solution (pH 7.4) in each well. Dilute the FCCP stock solution to 100 µM FCCP in dH2O and add 20 µL of the diluted solution to the injection port A in the sensor cartridge. Add 22 µL of 400 mM sodium azide to port B of the senor cartridge.
Turn the respirometer on and on the home screen select Start; the templates page will appear. On the templates page, select Blank or a previously designed template; the Groups page will appear. On the Groups page, select the wells A and H as the background wells and assign the remaining 6 wells into appropriate groups according to the experimental plan.
-
Push the arrow on the lower right corner to go to the Protocol page. On the Protocol page, ensure that the Equilibrate, Basal and Injections 1 and 2 buttons are selected. On this page, adjust the number of readings of basal OCR, as well as maximal OCR (after injection 1 with FCCP) and non-mitochondrial OCR (after injection 2 with sodium azide).
NOTE: Each measurement is preceded by a mix and wait step and the time frames for these parameters are shown in Table 2.
-
Select the arrow on the lower right corner and a prompt to load the sensor cartridge plate will appear. Ensure that the sensor cartridge plate is loaded in the right orientation, following the instructions on the reminder prompt screen. The respirometer will now equilibrate the cartridge.
NOTE: This equilibration provides ample time to place the animals to be assayed into appropriate wells of the cell plate.
Table 2:
Characteristic assay parameters in C. elegans respirometry.
| Calibration | Basal | FCCP | Sodium Azide |
|---|---|---|---|
| Port(s): A | Port(s): B | ||
| Equilibration: Yes | Mix: 00:02:00 | Mix: 00:02:00 | Mix: 00:02:00 |
| Wait: 00:00:30 | Wait: 00:00:30 | Wait: 00:00:30 | |
| Measure: 00:02:00 | Measure: 00:02:00 | Measure: 00:02:00 | |
| Cycles: 5 | Cycles: at least 5 | Cycles: 2−5 | |
| Duration: 00:22:30 | Duration: 00:22:30 | Duration: 00:09:00 |
4. Preparation of worm plate and assay run
Add 200 µL of M9 buffer into each of the 8 wells of the cell plate and into the reservoirs surrounding the wells.
-
Pick ~100 worms from each strain onto unseeded NGM plates and allow to rest for 2–3 min. Wet the end of the platinum pick with M9 buffer and pick 20 age-synchronized animals into wells B-G, leaving the background wells A and H empty.
NOTE: After loading each well, wait approximately 2 min to allow the animals to settle into the bottom of the wells. It is also advisable that a worm number curve be run to optimize the worm number per well for each instrument and laboratory setting.
By now, the respirometer should be calibrated and by clicking OK on the screen, the plate containing calibration buffer is ejected and can be removed from the respirometer, while the sensor cartridge stays inside the instrument. Replace the calibrant plate with the plate containing animals. Load plate and close the door by hitting Continue and allow the assay to run.
5. Post-experiment data analysis
Once the assay run is complete, follow the prompts on the screen and remove the cell plate and sensor cartridge and insert a flash drive into the USB port to save the run data in the .war format. Remove the sensor cartridge and allow the animals to settle to the bottom of the wells for approximately 2 min. Place the cell plates under a stereo dissecting microscope and count the number of animals per well.
-
Open the analysis software on the computer and open the normalization tab to normalize OCR to animal number. Apply appropriate labels for the various groups under the modify tab and export the file as a Prism file for further analysis.
NOTE: The average of the first five measurements, before the addition of FCCP is the basal OCR, while the average of the five measurements after FCCP addition is the maximal OCR and the average of the last five measurements (minimum of two measurements should be done) after sodium azide addition is the non-mitochondrial respiration rate. The assay should be repeated three times to ensure reproducibility.
Representative Results
Using the protocol described herein, OCR of wild type animals and three different sel-12 mutant strains were determined. sel-12 encodes the C. elegans ortholog of presenilin17. Mutations in human presenilin are the most common genetic aberration associated with the development of familial Alzheimer’s disease18. Our studies have shown elevated mitochondrial calcium levels in sel-12 mutant animals compared to wild type animals3. Since calcium dysregulation can result in altered mitochondrial function3,19,20, OCR in sel-12 mutant and wild type animals were measured to examine the effect of sel-12 mutations on mitochondrial function and health. Wild type animals consistently showed basal OCR rates below 5 pmol/min/worm, while all three strains of sel-12 mutants showed significantly elevated OCR of ~7 pmol/min/worm (Figure 3 and Figure 4). Upon the addition of FCCP, as expected, there was an increase in the OCR of wild type as well as sel-12 mutants (Figure 3 and Figure 5). Wild type animals showed maximum OCR of ~7 pmol/min/worm, while sel-12 mutants had OCR of ~10 pmol/min/worm (Figure 5).
Figure 3. Characteristic OCR profile in C. elegans respirometry.
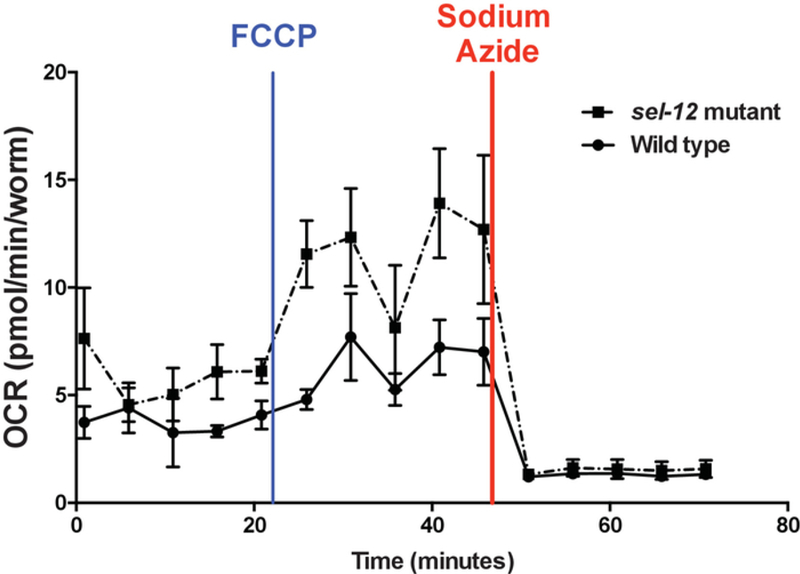
The five initial readings show the basal respiration, followed by five readings of maximal and five readings of non-mitochondrial respiration after FCCP and sodium azide injections, respectively. Error bars represent the standard error of measurement (SEM).
Figure 4. Basal respiration in wild type and various sel-12 mutants.
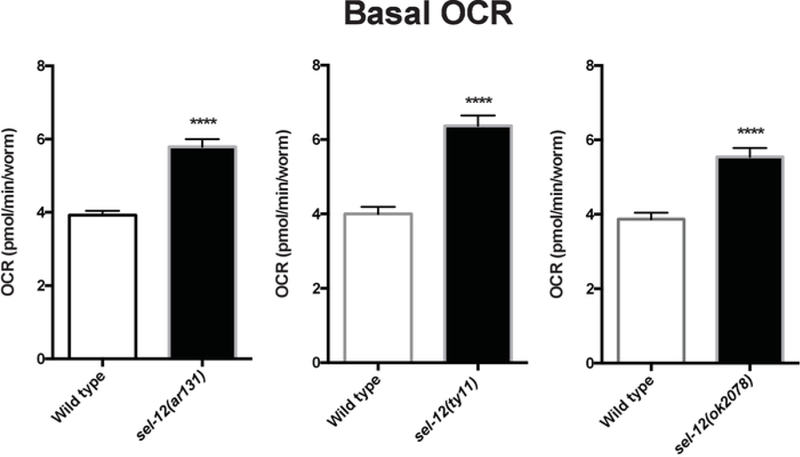
Average basal respiration in Day 1 adult age-matched wild type and sel-12 mutant animals. Data compiled from three assay repeats. Error bars represent SEM and **** indicates p <0.0001. p values were calculated using a two-tailed t-test.
Figure 5. Maximal respiration in wild type and various sel-12 mutants.
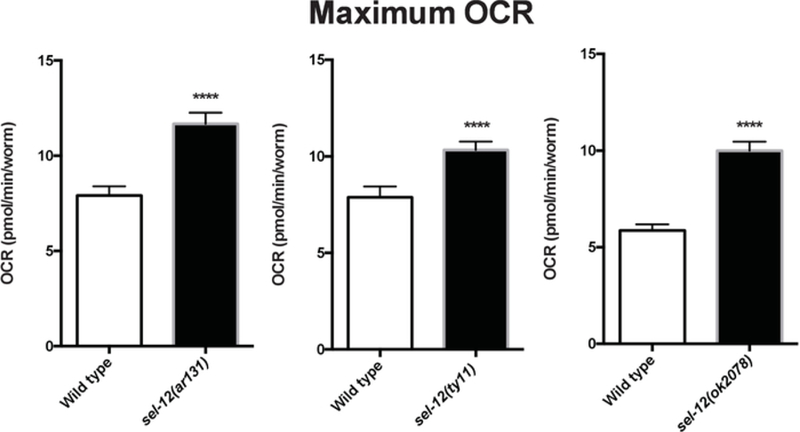
Average maximal respiration in Day 1 adult age-matched wild type and sel-12 mutant animals after FCCP injection. Data compiled from three assay repeats. Error bars represent SEM and **** indicates p <0.0001. p values were calculated using a two-tailed t-test.
Discussion
Mitochondrial respiration is an insightful indicator of mitochondrial function; therefore, being able to measure the oxygen consumption rates in a biological system, whether in vitro or in vivo is highly valuable. Respirometers sense oxygen levels using porphyrin-based phosphors that get quenched by oxygen or via amperometric oxygen sensors that rely on the generation of an electric current proportional to oxygen pressure. Clark electrode falls into the latter category and has been used extensively in literature, especially while analyzing respiration in C. elegans. However, the need for a large sample size and the inability to assess more than one sample at a time makes amperometric oxygen sensors inefficient.
This protocol provides a simple guide to measure OCR in live, intact C. elegans without isolating mitochondria, a process that could potentially impact the mitochondrial membrane potential and, therefore, the OCR. Given that animals exhibit different OCR through various life stages, animals used in this protocol should be age synchronized. Younger animals have lower OCR compared to adult animals and the OCR levels can drop again as animals age further3. C. elegans are also sterilized by the use of FUdR to ensure that OCR are not confounded by the presence of progeny. Nevertheless, if animals of varying sizes are to be examined, strategies for normalization need to be addressed.
This protocol utilizes a platinum pick to transfer animals to the assay plate. In contrast to liquid transfer of animals, this manipulation enables the researcher to carefully examine and determine the health of the animals before and during the transfer. Also, it allows for better control of worm number and prevents the introduction of eggs and carcasses. Since the animals are alive and active during the assay run, variability is likely to arise from probe positioning. Assays should therefore be done in triplicate and should be repeated a minimum of three times. Also, double checking animal count post analysis is important for normalizing to animal number. A major drawback of this protocol is the inability to compare more than two samples at once, without compromising the number of replicates within a single assay. Despite this limitation, this protocol can be a very powerful research tool in analyzing OCR when comparing two genotypes or conditions. Moreover, this protocol could be easily adapted to examine the OCR of animals grown on different food sources, supplements, or drug treatments.
Acknowledgments
The authors would like to acknowledge Dr. Kevin Bittman for his guidance in establishing the Seahorse XFp in the lab. National Institutes of Health grant GM088213 supported this work.
Footnotes
Video Link
The video component of this article can be found at https://www.jove.com/video/59277/
Disclosures
The authors have nothing to disclose.
References
- 1.Nelson DL, Cox MM in Lehninger Principles of Biochemistry (ed Ahr K) Ch. 19, 707–772 W. H. Freeman and Company; (2008). [Google Scholar]
- 2.Marchi S et al. Mitochondrial and endoplasmic reticulum calcium homeostasis and cell death. Cell Calcium 69, 62–72 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Sarasija S et al. Presenilin mutations deregulate mitochondrial Ca(2+) homeostasis and metabolic activity causing neurodegeneration in Caenorhabditis elegans. eLife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luz AL et al. Mitochondrial Morphology and Fundamental Parameters of the Mitochondrial Respiratory Chain Are Altered in Caenorhabditis elegans Strains Deficient in Mitochondrial Dynamics and Homeostasis Processes. PLoS One 10, e0130940 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryu D et al. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nature Medicine 22, 879–888 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Perry CG, Kane DA, Lanza IR, Neufer PD Methods for assessing mitochondrial function in diabetes. Diabetes 62, 1041–1053 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulz TJ et al. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metabolism 6, 280–293 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Koopman M et al. A screening-based platform for the assessment of cellular respiration in Caenorhabditis elegans. Nature Protocols 11, 1798–1816 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarasija S, Norman KR Analysis of Mitochondrial Structure in the Body Wall Muscle of Caenorhabditis elegans. Bio-protocol 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarasija S, Norman KR Measurement of ROS in Caenorhabditis elegans Using a Reduced Form of Fluorescein. Bio-protocol 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaudhuri J, Parihar M, Pires-daSilva A An introduction to worm lab: from culturing worms to mutagenesis. Journal of Visualized Experiments (47) (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aitlhadj L, Sturzenbaum SR The use of FUdR can cause prolonged longevity in mutant nematodes. Mechanisms of Ageing and Development 131, 364–365 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Rooney JP et al. Effects of 5’-fluoro-2-deoxyuridine on mitochondrial biology in Caenorhabditis elegans. Experimental Gerontology 56, 69–76 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Raamsdonk JM, Hekimi S FUdR causes a twofold increase in the lifespan of the mitochondrial mutant gas-1. Mechanisms of Ageing and Development 132, 519–521 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heytler PG, Prichard WW A new class of uncoupling agents--carbonyl cyanide phenylhydrazones. Biochemical and Biophysical Research Communications 7, 272–275 (1962). [DOI] [PubMed] [Google Scholar]
- 16.Massie MR, Lapoczka EM, Boggs KD, Stine KE, White GE Exposure to the metabolic inhibitor sodium azide induces stress protein expression and thermotolerance in the nematode Caenorhabditis elegans. Cell Stress Chaperones 8, 1–7 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levitan D, Greenwald I Facilitation of lin-12-mediated signalling by sel-12, a Caenorhabditis elegans S182 Alzheimer’s disease gene. Nature 377, 351–354 (1995). [DOI] [PubMed] [Google Scholar]
- 18.Sherrington R et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature 375, 754–760 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Glancy B, Balaban RS Role of mitochondrial Ca2+ in the regulation of cellular energetics. Biochemistry 51, 2959–2973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarasija S, Norman KR A gamma-Secretase Independent Role for Presenilin in Calcium Homeostasis Impacts Mitochondrial Function and Morphology in Caenorhabditis elegans. Genetics 201, 1453–1466 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]


