Abstract
Organisms have evolved different strategies to seclude certain molecules to specific locations of the cell. This is most pronounced in eukaryotes with their extensive intracellular membrane systems. Intracellular compartmentalization is particularly critical in genome containing organelles, which due to their bacterial evolutionary ancestry still maintain protein-synthesis machinery that resembles more their evolutionary origin than the extant eukaryotic cell they once joined as an endosymbiont. Despite this, it is clear that genome-containing organelles such as the mitochondria are not in isolation and many molecules make it across the mitochondrial membranes from the cytoplasm. In this realm the import of tRNAs and the enzymes that modify them prove most consequential. In this review we discuss two recent examples of how modifications typically found in cytoplasmic tRNAs affect mitochondrial translation in organisms that forcibly import all their tRNAs from the cytoplasm. In our view, the combination of tRNA import and the compartmentalization of modification enzymes must have played a critical role in the evolution of the organelle.
Keywords: Wybutosine, 1-methylguanosine, mitochondria, import, trypanosomes
INTRODUCTION
Organisms from all kingdoms of life require tRNAs for translation. Centrally, tRNAs play a critical role in connecting the genetic information found in DNA to that ultimately deposited in proteins, with mRNAs playing the relatively passive role of delivering the information to the ribosome. However, since tRNAs are not transcribed as fully mature molecules, they forcibly undergo a series of processing steps that renders them fully functional (Lopes et al., 2018). These include, trimming of extra sequences found at their 5’and 3’ ends, removal of introns when present, and addition of a CCA motif at their 3’ end. The latter universally becomes the end to which specific amino acids become covalently attached in the process of amino acylation. At any time during the maturation pathway, tRNAs may also undergo addition of sometimes numerous chemical groups to the four canonical nucleotides used for transcription; post-transcriptional chemical modifications may ensure proper folding of the tRNA molecule, but may also more directly affect decoding (Lorenz et al., 2017). Every available atom in the purine and pyrimidine rings have been seen naturally modified; the ribose sugar may also be the target of modifications. Thus far, over 100 different post-transcriptional modifications have been described and their types and numbers vary from organism to organism (Boccaletto et al., 2017); this variation hints at the possibility of a dynamic quality to modifications.
A major contributing factor to modification dynamics might be the degree of intracellular compartmentalization, which exists in all organisms. For example, although not always obvious, in some bacteria there is a well-defined developmental plan in the partitioning of the cell into mother cell and spore, each with its own requirements for tRNAs and protein synthesis. In other bacteria, such as members of the Planctomycetes, the genome is surrounded by a contiguous membrane, which forms a nucleoid body (Lonhienne et. al 2010). This may necessitate tRNA trafficking and depending on where a particular modification enzyme is localized may impact a tRNA’s modification set. By far, Eukarya offer the best documented and most numerous examples of intracellular compartmentalization, where genome-containing organelles are clearly separated from the rest of the cell by distinct membranes. In such cases, the existence of elaborate transport systems has been well established; some systems even specialize in the transport of large macromolecules such as proteins and RNA. Generally, in Eukarya, with the exception of some amitochondriate protists, there are 2–3 genome-containing compartments depending on the organism: nucleus and mitochondria in many eukaryotes; nucleus, mitochondria and plastids/chloroplast in plants and some protists. Although the nucleus encodes the bulk of the tRNA genes, the numbers in the other organelles may vary. For example, mammals encode ~21 different tRNAs in their mitochondrial genome. Based on wobble rules, it has been argued that the ~21 tRNAs represent a minimal set sufficient for decoding all the codons used in translation in a given organelle (Grosjean and Westhof 2016). However, such arguments rarely consider the impact of tRNA modifications and how these in turn affect decoding and importantly what roles modification may play in the evolution of mitochondrial genomes. In addition, although the majority of mitochondrial genomes may encode the minimal set of tRNAs mentioned above, a growing number of organisms encode less than a complete set. An extreme case occurs in kinetoplastids, including trypanosomatids, which have completely lost all of their tRNA genes from the mitochondrial genome (Alfonzo and Soll 2009). These organisms are therefore forced to import a complete set of tRNAs from the cytoplasm. Notably, mitochondrial tRNA import has been described in many plants (Michaud et al., 2011), protists (Simpson et al., 1989, Esseiva et al., 2004) and even mammals (Rubio et al., 2008). Clearly, the interplay between import and tRNA modification in such systems may play an important function in organellar translation.
The present review will highlight a few examples of how organellar tRNA transport across membranes and into the mitochondrion of T. brucei may have co- evolved with the dynamic nature of organellar genomes. Co-evolution may have at times provided a pre-existing mechanism permittingestablishment and maintenance of disparate events such as RNA editing and organellar tRNA import itself. In particular, we will focus on tRNA methylation at position 37 catalyzed by the TRM5 methyltransferase and the formation of the hypermodified nucleotide wyosine and its derivatives in Trypanosoma brucei. These modifications provide two recent examples of how tRNA partitioning and maturation may impact mitochondrial function in trypanosomatids but highlight broader themes and principles that may be relevant to other systems.
tRNA INTRACELLULAR TRANSPORT IN T. brucei
Since most tRNA genes in a eukaryotic cell are encoded in the nuclear genome, and the nucleus is not their main site of function, tRNAs have to be exported through the nuclear pore complex into to the cytosol. However, it is now widely accepted, that tRNA export from the nucleus is not unidirectional and tRNAs may shuffle between nucleus and cytosol. This is especially evident in S. cerevisiae and T. brucei, two organisms, where the tRNA splicing machinery localizes to the cytoplasm (Yoshihisa et al., 2003; Lopes et al., 2016) (Shaheen and Hopper 2005), yet some maturation steps, that only occur after splicing, are catalyzed by nucleus-localized enzymes. After cytoplasmic splicing, tRNAs are thus transported back to the nucleus by a mechanism termed tRNA retrograde import (Takano et al., 2005, Shaheen and Hopper 2005). It turns out that some modification enzymes that are critical for the maturation of particular tRNAs localize to the nucleus and are not able to recognize an intron-containing substrate. Following post-splicing nuclear modification, tRNAs must undergo a second round of export to the cytosol, but this time as mature or nearly mature molecules, where finally these tRNAs can engage in protein synthesis. It has been suggested, that this bidirectional movement can serve as a quality control step ensuring that immature tRNAs, which “escaped“ the nucleus through the primary export pathway still have a chance to be properly processed and fully modified (Kramer and Hopper, 2013).
Beyond the need for modifications, at least in S. cerevisiae cytosolic and nuclear tRNA pools are responsive to changes in nutritional conditions resulting in nuclear tRNA accumulation, which can be the result of either nuclear retention due to a slow down in primary exports after transcription or increased retrograde transport (Chatterjee et al., 2018). Taken together, tRNA dynamics between these two compartments may provide additional step to regulate cellular proteomes not only by the relative tRNA abundances in individual compartments during certain environmental conditions, but also by increasing the diversity of differentially modified tRNAs in cells.
Nevertheless, the cytoplasm does not have to be the “final destination“ for nucleus-encoded tRNAs. In many eukaryotes, the organellar genomes encode a less than complete set of tRNAs, which may not be sufficient for organellar protein synthesis. In such cases, a set of tRNAs has to be imported from the cytoplasm. Mitochondrial tRNA import has been well documented in numerous organisms. Recently, the increasing availability of complete mitochondrial genomes has revealed that mitochondrial tRNA import may be more widespread than previously thought and may in fact exist in all mitochondria-containing eukaryotes (Salinas-Giege et al., 2015). Although in cases where there is a complete lack of mitochondrial tRNA genes the need for import is obvious, there are examples where organisms that encode a seemingly complete set can also import cytosolic tRNAs as in the case of S. cerevisiae and human mitochondria (Kamenski et al., 2007; Rubio et al., 2008). This apparent redundancy may become particularly important under certain stress conditions (Kamenski et al., 2007). The most extreme case for the need of tRNA import from the cytoplasm occurs in kinetoplastid parasites (L. tarentolae and T. brucei) and other protists, where the mitochondrial genomes are completely devoid of tRNA genes and thus mitochondrial translation solely depends on the imported cytosolic tRNAs in order to maintain mitochondrial functions. Because alterations in cytosolic and nuclear tRNA pools as well as tRNA modification profiles may fluctuate in response to changes in environmental conditions, such changes would not only affect cytosolic translation, but also the translation of several essential proteins in the mitochondrion. Clearly, tracking tRNA modification and intracellular dynamics is not a trivial task in any organism, for example most eukaryotes have hundreds, if not thousands, of mitochondria. Thus for studies on how nucleo-cytoplasmic tRNA transport affects the modification and distribution of mitochondria-imported tRNAs, trypanosomes may offer an ideal playground for exploration. As it turns out, trypanosomes contain a single large mitochondrion per cell making it easier to track tRNAs. In the following sections, we provide several examples of how intracellular partitioning of tRNA and tRNA modification enzymes affects mitochondrial function in T. brucei.
Modification levels can be influenced by intrinsic mitochondrial functions. It has been argued that perhaps the most essential and evolutionarily conserved function of mitochondria is not simply to generate ATP via oxidative phosphorylation, but rather it is the generation of iron-sulfur clusters; required co- factors for many essential proteins. Indeed, the mitochondrion is the main site for iron sulfur cluster (ISC) assembly (Lill et al., 2014). A key component of the ISC assembly machinery is the cysteine desulfurase (Nfs), which together with three other proteins, IscU, Isd11, and frataxin, uses sulfur from cysteine to form FeS clusters (Braymer and Lill 2017). Interestingly, the Nfs complex is also involved in several tRNA modification pathways including thiolation (s2U)(Alfonzo and Lukeš, 2011). Formation of 2-thiouridine thiolation in tRNA requires both mitochondrial and cytosolic ISC machineries in yeast, as well as in trypanosomes (Nakai et al., 2004; Bruske et al., 2009; Wohlgamuth-Benedum et al., 2009). Generally, s2U is found in the first position of the anticodon (position 34) in tRNAGlnUUG, tRNAGluUUC and tRNALysUUU in most organisms, but in trypanosomes, including T. brucei, in addition is found at the adjacent position (U33) in the anitcodon loop of tRNATrp (Crain et al., 2002; Charrière et al., 2006). Remarkably, tRNATrp transits though the cytoplasm unthiolated and it is only modified once it enters the mitochondrion. This tRNA has a C at position 34, and therefore is not recognize as a substrate by the cytoplasmic thiolation machinery, which is specific for U34. The fact that it is then thiolated in the mitochondrion at an unusual position implies that the mitochondrial thiolation enzymes have evolved to recognize this alternative substrate. Remarkably, despite the fact that organisms use two different pathways for tRNA thiolation, one cytoplasmic and the other mitochondrial, because key components of the cytoplasmic thiolation machinery itself contains essential FeS clusters; even the cytoplasmic thiolation pathway still depends on the mitochondrial Nfs complex to make a functional cytoplasmic thiolation enzyme (Alfonzo and Lukeš, 2011). To complicate matters further, it has been reported that cytoplasmic thiolation of tRNAGlu serves as a negative determinant for import. In this system, only the unmodified tRNA (not containing s2U) may be the substrate for import, while the modified version is retained in the cytoplasm (Kaneko et al., 2003). However, the same may not be true of T. brucei (Paris et al., 2009). Thus in T. brucei, we suggest that mitochondrial tRNA import is quite robust with an unexpected lack of import selectivity for certain tRNAs. Namely, the import machinery cannot easily differentiate between some fully modified and partially or unmodified tRNA species; some of these as discussed could be potentially harmful for the proper function of the mitochondrion.
DEALING WITH HYPOMODIFIED tRNAS IN MITOCHONDRIA
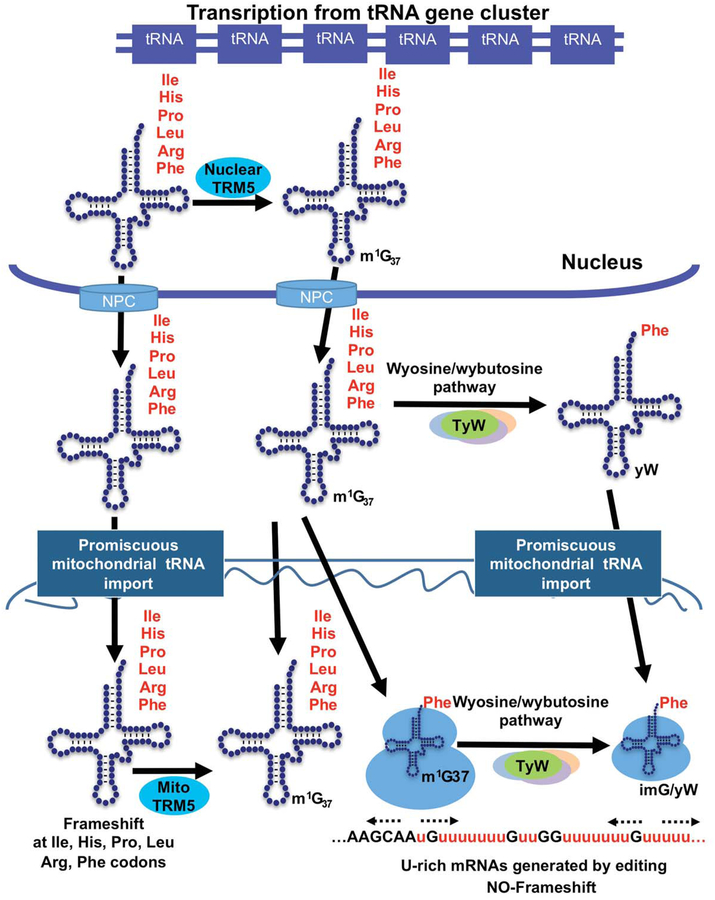
Because of the reported connection between modifications in tRNA and their intracellular distribution to various compartments, we investigated the nature of another tRNA modification, 1- methylguanosine (m1G) at position 37 in the anticodon loop in several tRNAs. Methylation at position 37 of tRNA is universally conserved and catalyzed by two evolutionarily unrelated enzymes: TRM5 in eukaryotes and TRMD in bacteria, classical examples of convergent evolution. m1G may positively regulate tRNA aminoacylation and is critically important for translational reading frame maintenance (Goto-Ito et al., 2017). During translation it prevents +1 frameshifting by the ribosome thus ensuring translational fidelity. In eukaryotes, TRM5 is dually localized to the nucleus and mitochondrion, and conversely responsible for m1G37 formation in nucleus-encoded and mitochondria-encoded tRNAs respectively (Goto-Ito et al., 2017). However, it was not clear what role, if any, mitochondrial TRM5 serves in organisms that do not encode tRNAs in their mitochondrial genomes. It was reasonable to expect that nuclear m1G methylation should be sufficient to supply methylated tRNAs to cytoplasmic and mitochondrial translation, the latter through the robust tRNA import pathway. Our interest was prompted by the initial observation of significant amounts of tRNAs lacking m1G37 in the mitochondrion of T. brucei, as documented by northern blots of cytosolic and mitochondrial fractions (Paris et al., 2013). A priori, it was clear that the import system could not distinguish between tRNAs that are fully methylated or unmethylated at position G37, raising again the question of how the mitochondrial translation system deals with the potentially harmful undermodified tRNAs, which may result in increased frameshifting. Despite, the aforementioned high levels of unmethylated tRNAs, cells in culture grow normally. However, down-regulation of TRM5 (TbTRM5) in T. brucei, led to a significant growth defect, which in retrospect is not unexpected, since m1G is present in 7 different tRNA species. Surprisingly, lack of m1G only had minor effects on cytoplasmic protein synthesis, suggesting that the presence of m1G37 in tRNA is far more important in mitochondria. Indeed lack of m1G37 leads to a decrease in mitochondrial protein synthesis, cytochrome C oxidase activity, associated reduction in respiration, and accumulation of reactive oxygen species (ROS) (Paris et al., 2013). These experiments suggest that in T. brucei, the potential translational problem caused by the presence of m1G37-lacking tRNAs in the mitochondrion is partly solved by the import of the TRM5 enzyme. TRM5 may then methylate any unmethylated tRNA that may find its way into the organelle, due to the relaxed selectivity of the import system, while keeping the ratio of methylated to unmethylated tRNA at an acceptable level (Fig. 1). These results also show a different tolerance for the unmethylated species in the cytosol and mitochondrion, respectively. In contrast to cytosolic translation, mitochondrial protein synthesis can cope with a relatively high amount of unmodified tRNA (~50%), but beyond that lack of methylation becomes a problem. This is reminiscent of mammalian mitochondria and the penetrance of certain mitochondrial disease-causing mutations. Several studies showed that the effects of certain mutations on energy metabolism depend on the ratio of wild type to mutated mitochondria. In other words depending on the mutation a certain percentage of mutated mitochondria was still acceptable but beyond that mitochondrial function was impacted (Rossignol et al., 2003). The results show, that in most cases, phenotypic manifestation of the genetic defect occurs only when a threshold level is exceeded, and this phenomenon has been named the ‘phenotypic threshold effect’. It tells us, that it is possible to considerably inhibit the activity of the respiratory chain complex up to a critical value without affecting the rate of mitochondrial respiration or ATP production to a degree that impairs the cells (Rossignol et al., 2003).
FIG 1. Intracellular compartmentalization of tRNAs and modification enzymes and their role in affecting mitochondria decoding and evolution.
Methylation at position 37 of certain tRNAs is important for translational fidelity and is also the precursor for another frameshift-preventing modification, wyosine and derivatives. Two versions of the latter exist in T. brucei one cytoplasmic and the other mitochondrial, but because a robust and promiscuous tRNA import pathway tRNAs modified to various extents have to be further modified to be fully functional. The unique wyosine formation pathway of T. brucei is proposed as part of the evolutionary adaption that permitted the organelle to cope with an abundance of U-rich sequences created by RNA editing, while preventing ribosomal frameshifting. NPC refers to the Nuclear Pore Complex, m1G37 refers to 1-methylguanosine found at position 37 of the anticodon of tRNAs Ile, His, Pro, Leu, Arg and Phe. In tRNAPhe this methylation is further modified to wybutosine (yW) or wyosine (imG) dependent on location.
A COMMON MODIFICATION IN AN UNUSUAL PLACE
In most eukaryotes, cytoplasmic tRNAPhe contains different derivatives of the hypermodified nucleotide wyosine. For example, wybutosine (yW) in yeast, hydroxywybutosine (OHyW) in mammals, chemically simpler forms of the modification are also found in Archaea, for example wyosine (imG) in Crenarcheota. The biosynthetic pathway for wyosine and derivatives has been recently reviewed (Perche-Letuvee et al., 2014) and involves 5 sequential chemical reactions in which S-adenosylmethionine (SAM or AdoMet) is utilized as a source of 3 different functional groups (a methyl group, an α-amino-a-carboxypropyl group, and a 5’-deoxyadenosine radical). In yeast and T. brucei, the initial step is localized to the nucleus and involves formation of m1G37 by the TRM5 methylase, followed by sequential reactions catalyzed by the enzymes TYW1, TYW2, TYW3 and TYW4 resulting in formation of wybutosine (Noma et al., 2006), which is the end product in most eukaryotes. The function of wybutosine has been most extensively studied in yeast and Archaea, where it serves to prevent frameshifting during decoding of UUU and UUC codons, which are particularly troubling for the ribosome in the context of U-rich sequences. In eukaryotes, wybutosine is solely a cytoplasmic modification, but recently, we reported that some of the genes in the pathway for wybutosine biosynthesis where duplicated in T. brucei (Sample et al., 2015). We found that the duplicated genes of wyosine biosynthesis (tyw1 and tyw3) could encode two differentially localized versions of their respective enzymes, a proposal that was confirmed by immunofluorescence with antibodies for each protein and further corroborated by cell fractionation followed by Western blots. In the case of tyw1, the two paralogs have marked size differences, where the larger of the two (tyw1L) is cytoplasmic and has all the functional domains characteristic of these types of enzymes in eukaryotes. The second smaller paralog (tyw1S) is mitochondrial and is missing the conserved FMN-binding domain and it is reminiscent of its ortholog from Archaea (de Crécy-Lagard et al., 2010). We obtained similar results with tyw3, with one paralog localizing to the cytoplasm (tyw3A) and the second to the mitochondria (tyw3B) (Sample et al., 2015). These observations led to the suggestion that T. brucei and perhaps related kinetoplastids all have two biosynthetic pathways for wyosine derivatives, one cytoplasmic and the other mitochondrial (Fig. 1). Some Archaea only encode tyw1 and 3 and in turn they only form wyosine in tRNA (de Crécy-Lagard et al., 2010). By using a combination of genetic analysis followed by mass spectrometry, we could show the presence of a wyosine-containing tRNAPhe in mitochondria and demonstrate that tyw1S, but not tyw1L, was involved in the reaction. Mass spectrometry analysis of total tRNA purified from T. brucei mitochondria surprisingly also revealed the presence of wybutosine-containing tRNAPhe. Likely, this species was the result of tRNA import from the cytoplasm, given that the remaining enzymes in the pathway (TYW2 and TYW4) do not localize to the mitochondria. This also implies that the substrate for wyosine biosynthesis for the mitochondria-localized tyw1S and tyw3 paralogs is either an m1G37-containing tRNAPhe, which escapes the cytosolic wybutosine pathway and is imported to mitochondria or an unmethylated tRNAPhe or both. Once again, these observations highlight the fact that as specific the import machinery may be for tRNA, it may not always be specific enough to distinguish between fully modified, partially modified or unmodified tRNAs.
WHY HAVE A STRICTLY CYTOPLASMIC MODIFICATION IN MITOCHONDRIA?
The combination of a robust mitochondrial tRNA import pathway and an import machinery unable to differentiate fully mature tRNAs from those that are not fully modified then raises the question of why. Answering this question, of course, is not trivial, given that no in vitro mitochondrial translation system exists. In addition, there is currently no proven method to introduce selectable markers into the trypanosome mitochondria; therefore, genetic approaches are out of the question. We have suggested that the need for wybutosine or wyosine in the T. brucei mitochondrion may be telling us something about a possible connection between the way mitochondria-encoded mRNAs are processed and ultimately used in translation. In T. brucei and all kinetoplastids, most of the protein-coding transcripts are synthesized as pre-mRNAs lacking fully translatable reading frames. It is now well established that these undergo extensive insertion and deletion of uridines catalyzed by the editosome in a manner that creates perfectly readable open reading frames (Benne et al., 1986). What has not been fully appreciated is that insertion of uridines also creates regions in a message that inevitably contain runs of uridines, which in other systems have a propensity to create problems with proper frame maintenance during translation. We have suggested that the trypanosome system has solved this problem by importing into the mitochondria the pathway for wybutosine; a pathway previously relegated to the cytoplasm of eukaryotes (Fig. 1). In terms of evolution, it is likely that the gene duplication that occurred in the kinetoplastid lineage may initially not have any fitness advantage to the cell, and that at least some of the proteins (i.e. tyw1S and tyw3B) had gained the ability to be imported to the mitochondria, thus forming wyosine (imG) in the organelle. Wyosine formation, although initially a purely neutral step in evolution, became important with the advent of U-insertion editing (Benne et al., 1986). This provided a scenario whereby as mRNA editing evolved, potentially lethal translational vagaries created by the U-insertion process (Farabaugh and Björk 1999), such as increased propensity for ribosomal frameshifting and occurrence of other translational errors could be easily remediated by the pre-existence of a common modification at an unusual location.
In a broader sense, it is interesting to note that expectedly, in organisms that import from the cytoplasm all of the tRNAs used in mitochondrial translation, there must be a level of co-evolution between the nuclear and mitochondrial genomes. For instance, one would predict that the two would become nearly identical in terms of their genetic code; such mitochondria will tend to use a more universal code. Obviously, in trypanosomes the use of UGA for tryptophan remains an exception. However, if one compares codon usage between the trypanosome mitochondrial genome and the nuclear genome, it is evident that whereas the T. brucei nuclear genome has a G/C bias at the 3rd position of codons, the mitochondrial genome has an A/T bias. Indicating that despite the fact that all the tRNAs, needed for mitochondrial translation, are imported, still the genetic code, at least in terms of codon usage, has not co-evolved. The mitochondrial genome is still under A/T mutational pressure at the 3rd codon position, which is characteristic of most mitochondria. One thing then is clear, that additional selective pressures exist to maintain such bias and these go beyond whatever fitness effect importing tRNAs may provide. Regardless, the import of the wyosine pathway into mitochondria, as a potential way to maintain translational fidelity, is only made possible byThis scenario, again, is only made possible by the availability of a relatively promiscuous mitochondrial tRNA import machinery, whose evolution would have then also predated the appearance of the editing pathway.
ACKNOWLEDGEMENTS
This work was supported in part by NIH grants (GM084065 and AI131348) to JDA and from ERDF/ESF project Centre for research of pathogenicity and virulence of parasites (No. CZ.02.1.01/0.0/0.0/16_019/0000759) to ZP.
References
- Alfonzo JD, and Lukeš J (2011) Assembling Fe/S-clusters and modifying tRNAs: Ancient co-factors meet ancient adaptors. Trends Parasitol 27: 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonzo JD, and Söll D (2009) Mitochondrial tRNA import--the challenge to understand has just begun. Biol Chem. 390:717–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R, Van den Burg J, Brakenhoff J, Sloof P, Van Boom J, Tromp M (1986) Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA Cell 46: 819–826. [DOI] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, Piątkowski P, Bagiński B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A, Helm M (2017) MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Research 46: D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braymer JJ, Lill R (2017) Iron-sulfur cluster biogenesis and trafficking in mitochondria. J Biol Chem. 292:12754–12763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruske EI, Sendfeld F, and Schneider A (2009) Thiolated tRNAs of Trypanosoma brucei are imported into mitochondria and dethiolated after import. J Biol Chem 284: 36491–36499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrière F, Helgadóttir S, Horn EK, Söll D, and Schneider A (2006) Dual targeting of a single tRNATrp requires two different tryptophanyl-tRNA synthesis in Typanosoma brucei. Proc Natl Acad Sci U S A 103: 6847–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee K, Nostramo RT, Wan Y, and Hopper AK (2018) tRNA dynamics between the nucleus, cytoplasm and mitochondrial surface: Location, location, location. Biochim Biophys Acta - Gene Regul Mech 1861: 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crain PF, Alfonzo JD, Rozenski J, Kapushoc ST, McCloskey JA, and Simpson L (2002) Modification of the universally unmodified uridine-33 in a mitochondria-imported edited tRNA and the role of the anticodon arm structure on editing efficiency. RNA 8: 752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crécy-Lagard V, Brochier-Armanet C, Urbonavicius J, Fernandez B, Phillips G, Lyons B, Noma A, Alvarez S, Droogmans L, Armengaud J, Grosjean H (2010) Biosynthesis of wyosine derivatives in tRNA: an ancient and highly diverse pathway in Archaea. Mol Biol Evol. 27:2062–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esseiva AC,Naguleswaran A, Hemphill A, Schneider A (2004) Mitochondrial tRNA import in Toxoplasma gondii. J Biol Chem. 279:42363–8. [DOI] [PubMed] [Google Scholar]
- Farabaugh PJ, Björk GR (1999) How translational accuracy influences reading frame maintenance. EMBO J. 18:1427–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto-Ito S, Ito T, and Yokoyama S (2017) Trm5 and trmd: Two enzymes from distinct origins catalyze the identical trna modification, m1g37. Biomolecules 7: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H, and Westhof E (2016) An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 44: 8020–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenski P, Kolesnikova O, Jubenot V, Entelis N, Krasheninnikov IA, Martin R, and Tarassov I (2007) Evidence for an Adaptation Mechanism of Mitochondrial Translation via tRNA Import from the Cytosol. Mol Cell 26: 625–637. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Suzuki TT, Kapushoc ST, Rubio MA, Ghazvini J, Watanabe K, et al. (2003) Wobble modification differences and subcellular localization of tRNAs in Leishmania tarentolae: implication for tRNA sorting mechanism. EMBO J 22: 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EB, and Hopper AK (2013) Retrograde transfer RNA nuclear import provides a new level of tRNA quality control in Saccharomyces cerevisiae. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R, Srinivasan V, Mühlenhoff U, and Muhlenhoff U (2014) The role of mitochondria in cytosolic-nuclear iron–sulfur protein biogenesis and in cellular iron regulation. Curr Opin Microbiol 22: 111–119. [DOI] [PubMed] [Google Scholar]
- Lonhienne TG, Sagulenko E, Webb RI, Lee KC, Franke J, Devos DP, Nouwens A, Carroll BJ, Fuerst JA (2010) Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci USA 107:12883–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes RR, Kessler AC, Polycarpo C, Alfonzo JD (2015) Cutting, dicing, healing and sealing: the molecular surgery of tRNA. Wiley Interdiscip Rev RNA 6:337–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes RRS, Silveira GDEO, Eitler R, Vidal RS, Alfonzo JD, Polycarpo C, et al. (2016) The essential function of the Trypanosoma brucei Trl1 homolog in procyclic cells is maturation of the intron-containing tRNA Tyr. RNA 1190–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz C, Lünse CE, Mörl M, (2017) tRNA Modifications: Impact on Structure and Thermal Adaptation. Biomolecules 7: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud M, Cognat V, Duchên AM, Maréchal‐Drouard L (2011) A global picture of tRNA genes in plant genomes. Plant J 66: 80–93. [DOI] [PubMed] [Google Scholar]
- Nakai Y, Umeda N, Suzuki T, Nakai M, Hayashi H, Watanabe K, and Kagamiyama H (2004) Yeast Nfs1p Is Involved in Thio-modification of Both Mitochondrial and Cytoplasmic tRNAs. J Biol Chem 279: 12363–12368. [DOI] [PubMed] [Google Scholar]
- Noma A, Kirino Y, Ikeuchi Y, and Suzuki T (2006) Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J 25: 2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris Z, Horáková E, Rubio MAT, Sample P, Fleming IMC, Armocida S, et al. (2013) The T. brucei TRM5 methyltransferase plays an essential role in mitochondrial protein synthesis and function. RNA 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris Z, Rubio MAT, Lukes J, Alfonzo JD (2009) Mitochondrial tRNA import in Trypanosoma brucei is independent of thiolation and the Rieske protein. RNA 15: 1398–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perche-Letuvee P, Molle T, Forouhar F, Mulliez E, and Atta M (2014) Wybutosine biosynthesis: structural and mechanistic overview. RNA Biol 11: 1508–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol R, Faustin B, Rocher C, Malgat M, Mazat J, and Letellier T (2003) Mitochondrial threshold effects. 762: 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio MAT, Rinehart JJ, Krett B, Duvezin-Caubet SS, Reichert AS, Soll D, et al. (2008) Mammalian mitochondria have the innate ability to import tRNAs by a mechanism distinct from protein import. Proc Natl Acad Sci U S A 105: 9186–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas-Giege T, Giege R, and Giege P (2015) tRNA biology in mitochondria. Int J Mol Sci 16: 4518–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sample PJ, Koreny L, Paris Z, Gaston KW, Rubio MAT, Fleming IMC, et al. (2015) A common tRNA modification at an unusual location: the discovery of wyosine biosynthesis in mitochondria. Nucleic Acids Res 43: 4262–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen HH, Hopper AK, (2005) Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 102:11290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AM, Suyama Y, Dewes H, Campbell DA, Simpson L (1989) Kinetoplastid mitochondria contain functional tRNAs which are encoded in nuclear DNA and also contain small minicircle and maxicircle transcripts of unknown function. Nucleic Acids Res 17:5427–5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano A, Endo T, Yoshihisa T (2005) tRNA actively shuttles between the nucleus and cytosol in yeast. Science 309:140–142. [DOI] [PubMed] [Google Scholar]
- Wohlgamuth-Benedum JM, Rubio MAT, Paris Z, Long S, Poliak P, Lukes J, and Alfonzo JD (2009) Thiolation controls cytoplasmic tRNA stability and acts as a negative determinant for tRNA editing in mitochondria. J Biol Chem 284: 23947–23953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T, Yunoki-Esaki K, Ohshima C, Tanaka N, and Endo T (2003) Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol Biol Cell 14: 3266–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]