Abstract
During organogenesis in all multi-cellular organisms, axial patterning is required to transform a single layer organ primordium into a three-dimensional organ. The Drosophila eye model serves as an excellent model to study axial patterning. Dorso-ventral (DV) axis determination is the first lineage restriction event during axial patterning of the Drosophila eye. The early Drosophila eye primordium has a default ventral fate, and the dorsal eye fate is established by onset of dorsal selector gene pannier (pnr) expression in a group of cells on the dorsal eye margin. The boundary between dorsal and ventral compartments called the equator is the site for Notch (N) activation that triggers cell proliferation and differentiation. This review will focus on (a) chronology of events during DV axis determination; (b) how early division of eye into dorsal and ventral compartments contributes towards the growth and patterning of the fly retina, and (c) functions of DV patterning genes.
Keywords: Drosophila eye, Eye development, Axial patterning, Dorso-ventral patterning, Dorsal selector, ventral eye genes, Lobe, Serrate, Pannier, Homothorax, Teashirt, Wingless
INTRODUCTION
During development, the patterning signals progressively restrict cell fates by subdividing a large developing field into smaller fields with limited developmental potential. These smaller fields that correspond to the domains of expression of selector genes are referred to as compartments. The selective spatio-temporal expression pattern of the cell fate selector genes is responsible for the formation of compartments (Blair, 2001; Curtiss et al., 2002; Held, 2002; Dahmann et al., 2011). The boundary between the compartments, where two different cell types are juxtaposed, is responsible for generating new signaling centers to regulate patterning, growth and differentiation of a developing field (Meinhardt, 1983; Blair, 2001). Thus, formation of the developmental boundaries is crucial for maintaining the downstream patterning events (Blair, 2001; Curtiss et al., 2002; Dahmann et al., 2011). Therefore, an important question in developmental biology is how these boundaries are generated and maintained during development. The aim of this review is to provide an overview of recent advances in our understanding of generation of the boundary between the dorsal and ventral compartment of the eye and its implications on development of the eye as an organ. This process is referred to as Dorso-Ventral (DV) patterning of the Drosophila eye. The Drosophila eye, an ideal model system for studying organogenesis, has been extensively used to investigate tissue patterning and cell-cell communication during axial patterning. Furthermore, Drosophila eye serves as an excellent model to understand the genetic mechanism responsible for division of a developing field into several smaller fields with positional fate restrictions (Singh et al., 2005b). The genetic machinery that controls Drosophila eye development closely resembles that of higher vertebrates, suggesting conservation of certain genetic pathways throughout evolution (Wawersik and Maas, 2000; Gehring, 2005; Erclik et al., 2009; Kumar, 2009). The axial pattern of the eye disc has not been well studied until recently due to the complexity of eye development.
Axial patterning, which is essential for organogenesis in all the multi-cellular organisms, involves formation of Antero-Posterior (AP), Dorso-Ventral (DV) and Proximo-Distal (PD) compartments (Cohen et al., 1993; Cohen, 1993). During axial patterning of the wing- and the leg-imaginal discs, the sequence of events involves the division of a field into anterior and posterior compartments of independent cell lineages, followed by subdivision of these imaginal discs into dorsal and ventral compartments. Interestingly, this sequence of division is not followed in the developing eye imaginal disc because it does not have analogous anterior-posterior axis. Instead, DV patterning is the first lineage restriction in the developing eye imaginal disc (Singh and Choi, 2003; Singh et al., 2005b). Despite the differences in the sequence of events, evidence suggests that some aspects of the DV patterning mechanism are highly conserved in the developing eye and the wing. An important common conclusion is that the border between DV compartments is a center for organizing the growth and patterning of the disc. In the subsequent sections of the review, we will focus on the mechanism involved in generation of DV domains in the developing eye, and the genetic basis for the establishment of the DV pattern.
2. Eye imaginal disc
In Drosophila, a holometabolous insect, the primordia of all the adult structures are sequestered in the larva as epidermal invaginations that are called imaginal discs (Bodentstein, 1950; Ferris, 1950; Atkins and Mardon, 2009). The adult eye, antenna, head cuticle, and head structures develop from a common developing field called an eye-antennal imaginal disc (Cohen, 1993; Held, 2002). The regions of the eye-antennal imaginal disc, which give rise to head structures including ptilinum, frons and maxillary palpus, originate from five embryonic segments and the acron (Jurgens and Hartenstein, 1993; Younossi-Hartenstein and Hartenstein, 1993). The monolayer epithelium does not accurately reflect the sac-like anatomy of the imaginal discs (Gibson and Schubiger, 2001). Drosophila imaginal discs are a contiguous cell sheet of flattened epithelial cells with two opposing surfaces, a columnar epithelium called disc proper (DP) and a squamous peripodial epithelium called as peripodial membrane (PM) (McClure and Schubiger, 2005; Atkins and Mardon, 2009). The Drosophila retina develops from the DP while the PM of the eye-antennal disc gives rise to the adult head structures (Figure 1; Milner et al., 1983; Haynie and Bryant, 1986; Atkins and Mardon, 2009). The eye-antennal imaginal disc emerges from an embryonic imaginal primordium, which is an anterior-dorsal sac comprising of approximately 20 cells that are set aside during mid- embryogenesis (Poulson, 1950; Garcia-Bellido and Merriam, 1969; Yamamoto, 1996). The embryonic precursors for imaginal discs grow asynchronously from the rest of the developing embryo (Anderson, 1972b; Anderson, 1972a; Crick and Lawrence, 1975; Cohen, 1993; Held, 2002; Kumar, 2011). In the first two larval stages, the eye imaginal disc cells divide and grow. The distinction between the developing antenna and the eye field begins to appear during the second instar larvae (Kumar and Moses, 2001; Kenyon et al., 2003; Dominguez and Casares, 2005; Atkins and Mardon, 2009). The developing eye field is further divided into precursors for the eye proper, head cuticle and the ocelli while the antennal field divides into precursors for the antenna and head cuticle. Retinal differentiation begins from the posterior margin of the eye imaginal disc during late second instar or early third instar stage of larval development (Ready et al., 1976). Since the retinal differentiation is synchronous in nature, it appears like a wave of differentiation initiating at the posterior margin of eye imaginal disc, which then proceeds anteriorly. The wave of differentiation is referred to as the morphogenetic furrow (MF, Figure 1A, A′ arrowhead); it results in transition of an undifferentiated epithelium to differentiated cell types comprising of regularly spaced photoreceptor clusters (Ready et al., 1976; Wolff and Ready, 1993). Posterior to the furrow, photoreceptor clusters are generated by a sequence of events including the selection of the R8 founder neuron and recruitments of additional photoreceptor precursors in the order of R2/5, R3/4, and R1/6/7 (Wolff and Ready, 1993; Kumar, 2011). Thus, if the eye imaginal disc is largely undifferentiated until second instar of development, an interesting issue is how compartments are identified in the Drosophila eye imaginal disc.
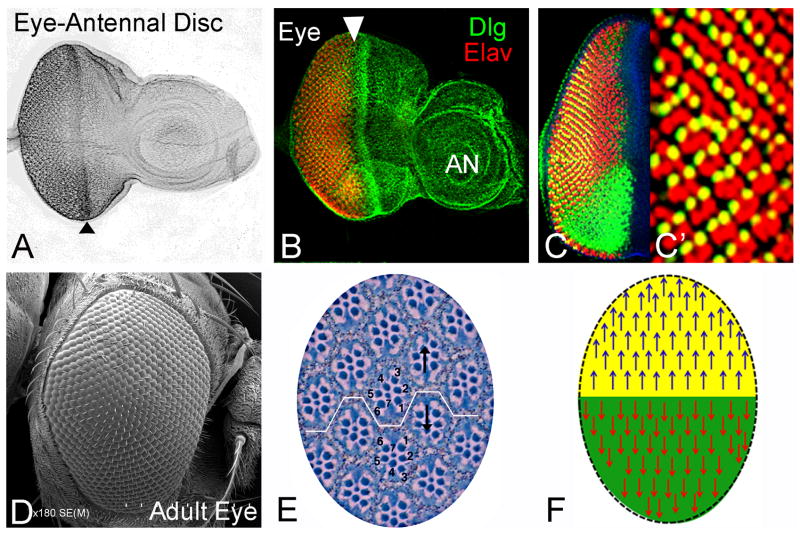
Figure 1. Drosophila eye is a highly organized structure.
(A–C) Eye antennal imaginal disc of a third instar larva. (B) Eye imaginal disc stained for membrane marker Disc large (Dlg: green) and Elav (red), a pan-neuronal marker that marks the photoreceptor neurons are shown. Arrowhead marks the position of the morphogenetic furrow (MF). (C, C′) Eye imaginal disc stained with Bar (B: green) to mark DV polarity. (D) Scanning electron micrograph (SEM) of the wild-type Drosophila eye. Adult eye is a compound eye which is made up of 750–800 unit eyes, each termed as an ommatidium (Wolff and Ready, 1993). All ommatidia are organized as a regularly spaced hexagon arranged in mirror image symmetry along the dorso-ventral (DV) axis. (E) Cross section of the adult compound eye showing arrangement of ommatidial clusters along DV axis (Wolff and Ready, 1993). Note that in each ommatidium, eight rhabdomeres are organized in an asymmetric trapezoidal fashion. All these ommatidia within a compound eye are organized in two basic clusters based on the orientation of the trapezoid. If the R3 rhabdomere points up, it is dorsal, whereas the reverse is ventral. (F) Cartoon showing the mirror image symmetry of ommatidial cluster orientation in the dorsal and ventral half of the adult eye. All the blue arrows mark the ommatidia in the dorsal half of the eye, while the red ones mark the ventral half of the eye. All images are oriented as Dorsal (up), Ventral (down), Anterior (right), and Posterior (left). [AN: antenna]
3. Dorsal and ventral compartments and the equator
The adult compound eye is comprised of approximately 800 unit eyes or ommatidia (Figure 1D). Each ommatidium consists of eight photoreceptor neurons assembled in an asymmetric trapezoidal pattern, and when viewed from the top it resembles a honeycomb-like, hexagonal facet. The surrounding cell types are non-neuronal and include pigment, cone cells, and mechanosensory bristles (Figure 1E, F; Wolff and Ready, 1993). This pattern of organization is repeated in all ommatidia of the eye. However, despite the similarity in the cellular composition of each ommatidium, the spatial arrangement of ommatidia in the eye is organized in two orientations (Figure 1E, F). These two orientations also serve as a marker to distinguish the dorsal and the ventral halves of adult eye. The photoreceptors are arranged in a trapezoidal fashion within an ommatidium. The ommatidial clusters within an eye are organized in a mirror asymmetry as they are polarized in the opposite directions (Figure 1E, F). The boundary between the ommatidia of the dorsal half and their mirror image ventral ommatidia is referred to as an equator (Figure 1E, F). This mirror symmetry, which corresponds to DV axis or compartments of the adult eye has been described in many insect eyes (Dietrich, 1909). Since the developmental mechanisms underlying the DV pattern have not been studied in detail, it raises an interesting question of how the dorsal and ventral pattern is established.
The pioneering studies to discern the relation between the equator and the DV compartmental boundary in the Drosophila eye suggested that the equator is not determined as the boundary between the dorsal and ventral cell lineages (Ready et al., 1976). Even though, the result from this study does not exclude the possibility that the dorsal and the ventral domains of the eye derive from two independent cell lineages, the lineage boundary may not precisely correspond to the equator. A series of elegant genetic analysis experiments involving a large number of mosaic clones in the adult eye and the head supported this idea (Baker, 1978). These experiments demonstrated that clones strictly follow the DV boundary, and they do not intermingle near the DV border (Held, 2002). These results validated the hypothesis that Drosophila eye derives from DV compartments. The wing imaginal disc is divided into anterior and posterior groups of cells in its early stage of development, which is followed by further partitioning into dorsal and ventral compartments (Lawrence and Morata, 1976). To analyze whether the eye and the head are also subdivided into different domains by sequential compartmentalization as in the wing, another mosaic analysis was carried out. Nearly all clones (96%) were restricted to either dorsal or ventral domains of the eye, thereby conforming the presence of a boundary between the dorsal and ventral cells. Thus, clones generated in the dorsal or ventral compartment did not cross the DV boundary. A few clones (4%) did cross the DV border, which was probably due to the fact that such clones might have been induced prior to compartmentalization (formation of dorsal and ventral compartment boundary) or two independent dorsal and ventral clones might have juxtaposed at the equator region thereby giving a false notion of a single clone not respecting the DV boundary (Baker, 1978). The DV lineage restriction observed in the adult eye was also confirmed in the developing eye imaginal disc where large clones do not cross the DV midline and showed sharp outline along the DV midline and the clones located within the dorsal or ventral domain had wiggly borders (Dominguez and de Celis, 1998). Unlike the DV lineage restriction that is established in the first instar larval eye imaginal discs; there are evidences that anterior-posterior restriction occurs a little later in the second instar eye imaginal disc. Thus, the significance of this anterior-posterior restriction remains to be seen as the morphological distinction between the anterior and posterior regions. In the subsequent sections we discuss the role of DV patterning genes, and the generation of pattern along the DV axis of the eye.
4. Genesis of the eye
The DV boundary has been suggested as the site for the activation of Notch (N) signaling in the eye imaginal disc to promote growth (de Celis et al., 1996; Go et al., 1998; Baonza and Garcia-Bellido, 2000). However, if DV patterning occurs as late as seen in the adult eye based on the orientation of the photoreceptors, then it may not be crucial for the growth of the eye. Thus, efforts were channeled towards investigating whether DV patterning takes place earlier during eye development. The seminal papers from three different groups established that DV lineage restriction takes place earlier during larval eye development due to domain specific expression of the DV patterning genes (Cho and Choi, 1998; Dominguez and de Celis, 1998; Papayannopoulos et al., 1998). The DV patterning genes are a class of genes involved in generating and maintaining the DV lineage in the eye. These reports identified a new time line for the initiation of DV patterning to early larval development. They also identified the genes whose expression and/or function was restricted either to the dorsal or ventral compartment of the eye. Thus, it was proposed that DV patterning in the eye is generated by the domain-restricted expression of the dorsal and the ventral eye genes (Cho and Choi, 1998; Dominguez and de Celis, 1998; Papayannopoulos et al., 1998; Cho et al., 2000).
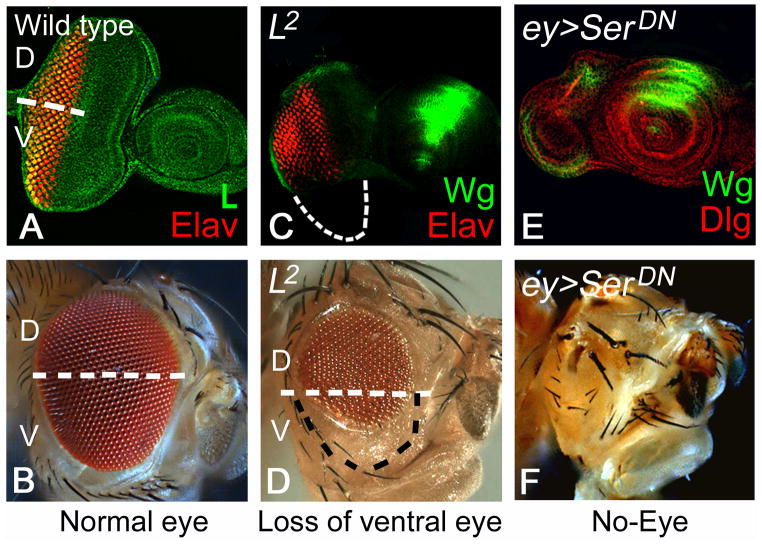
The basic question was what is the default state of the early eye primordium? To answer this question, several groups directed their efforts to identify the genes that are expressed in the early eye primordium. During embryonic development, the eye primordium begins as a homogenous group of cells that continue to grow during first larval instar to form the eye imaginal disc. All the cells of the first instar eye imaginal disc uniformly express Lobe (L), a gene known to be involved in ventral eye development (Singh and Choi, 2003). These studies revealed that until the late first instar of larval eye development, the entire eye primordium is ventral in fate and depends on the function of ventral genes like L and its downstream target Serrate (Ser) (Singh and Choi, 2003; Singh et al., 2005b; Kumar, 2011). Loss-of-function of the L gene, which is expressed ubiquitously in the eye imaginal disc (Figure 3A), results in selective growth defects in the ventral half of the eye (Figure 2, 3C, D). The loss-of-function studies suggested that the requirement of L function evolves along the temporal axis (Singh and Choi, 2003; Singh et al., 2005b). During early eye development, loss-of-function of L results in the complete loss of the eye field (Figure 3B). However, loss of L gene function later during eye development causes selective loss of ventral half of eye (Singh et al., 2005b). Loss-of-function of Ser also results in the similar loss of ventral eye phenotype (Table 1; Singh and Choi, 2003; Singh et al., 2005b). Interestingly, the timing of restriction of L/Ser functional domain from the entire developing eye field (Fig. 3D, E) to only the ventral half of eye (Figure 3B, C) corresponds to the onset of pannier (pnr) gene expression along the dorsal margin of the eye (Table 1; Figure 2). During late first instar of eye development, the entire homogenous population of the ventral cells of the eye primordium transition into two distinct dorsal and ventral lineages with the onset of pnr expression on the dorsal eye margin (Singh and Choi, 2003). This suggests that the ventral fate is the ground state of the larval eye imaginal disc, and L and Ser are essential for survival and/or maintenance of this ventral state (Singh and Choi, 2003; Singh et al., 2005b; Singh et al., 2006).
Figure 3. Lobe (L) and Serrate (Ser) are required for cell survival in developing eye imaginal disc.
(A) In wild-type eye imaginal disc, L (green) expression is ubiquitous. Elav (red) marks the photoreceptor neurons. (B) wild-type adult eye. (C, D) Loss of L results in the preferential loss of ventral half of the (C) developing eye imaginal disc, and (D) the adult eye. (C) Eye imaginal discs stained for Wg (green) to identify dorsal versus ventral eye imaginal disc compartment. The boundary of the eye field is as outlined in C (white) and D (black) showing preferential loss of ventral eye. (E, F) Early loss-of-function of Ser by misexpressing dominant negative form of Ser in the entire eye imaginal (Kumar and Moses, 2001; Singh and Choi, 2003) using an ey-Gal4 driver (Hazelett et al., 1998; Singh and Choi, 2003) results in complete loss of eye field both in (E) the eye imaginal disc, and (E) the adult.
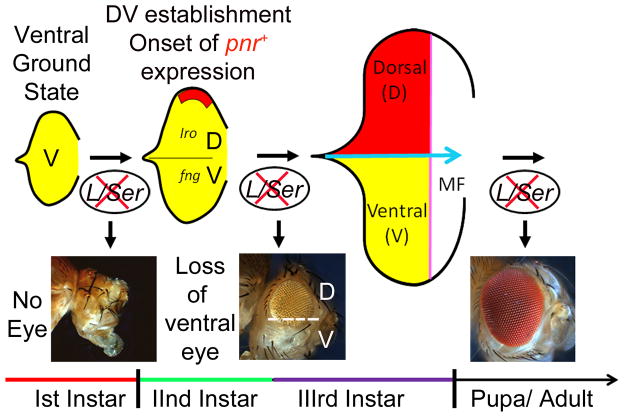
Figure 2. Ventral is the default state of the early eye imaginal disc.
Larval eye primordium comprising of a few cells require the function of ventral genes L/Ser for growth and proliferation (Singh and Choi, 2003). Loss-of-function phenotype of L/Ser in the developing eye imaginal disc evolves along the temporal scale. During early first instar of larval development, loss-of-function of L/Ser results in complete loss of the eye field. However, after the onset of pnr expression during early second larval instar, which results in generation of DV lineage in the developing eye imaginal disc, loss of L/Ser results in loss of only the ventral half of the eye. However, in late third instar stage of development when the retinal differentiation is almost complete loss of L/Ser does not have significant affect on the overall adult eye morphology. Based on these results it was proposed that the entire early eye primordium, prior to onset of pnr expression, is ventral in fate (Singh and Choi, 2003).
Table 1.
Genes involved in dorso-ventral patterning and domain specific expression and growth.
| Drosophila | Vertebrate Homolog | Nature | Function in eye | References |
|---|---|---|---|---|
| Ventral Genes | ||||
| fringe (fng) | Lunatic fringe | Glycosyl transferase | Secreted signaling protein, DV boundary formation | (Irvine and Wieschaus, 1994; Cho and Choi, 1998; Dominguez and de Celis, 1998; Papayannopoulos et al., 1998) |
| Serrate (Ser) | Jagged-1 | Ventral N ligand | Ventral eye growth and development | (Speicher et al., 1994; Cho and Choi, 1998; Dominguez and de Celis, 1998; Papayannopoulos et al., 1998; Cho et al., 2000) |
| Chip | Nli/Ldb1/Clim-2 | Transcription co-factor | Define ventral eye boundary | (Roignant et al., 2010) |
| Sloppy paired (Slp) | BF-1 (not complete homology) | Forkhead transcription factor | Ventral eye growth | (Sato and Tomlinson, 2007) |
| decapentaplegic (dpp) | BMP | TGF-β | Ventral growth | (Chanut and Heberlein, 1997; Singh et al., 2005b) |
| Dorsal Genes | ||||
| pannier (pnr) | GATA-4 | Zinc finger, GATA family | Dorsal eye fate selector | (Ramain et al., 1993; Maurel-Zaffran and Treisman, 2000; Gomez-Skarmeta and Modolell, 2002; Singh et al., 2005b; Oros et al., 2010) |
| araucan (ara) | Irx 1, 3 | homeodomain | Dorsal eye fate selector | (Gomez-Skarmeta and Modolell, 1996; Cavodeassi et al., 1999; Pichaud and Casares, 2000; Gomez-Skarmeta and Modolell, 2002) |
| Caupolican (caup) | Irx2, 5 | homeodomain | Dorsal eye fate selector | (Gomez-Skarmeta and Modolell, 1996; Cavodeassi et al., 1999; Pichaud and Casares, 2000; Gomez-Skarmeta and Modolell, 2002) |
| mirror (mirr) | Irx 4, 6 | homeodomain | Dorsal eye fate selector | (McNeill et al., 1997; Heberlein et al., 1998; Kehl et al., 1998; Yang et al., 1999; Singh et al., 2005b) |
| Delta (Dl) | Delta like 3 (DLL3) | Transmembrane Notch Ligand | Dorsal Notch (N) Ligand | (Cho and Choi, 1998; Dominguez and de Celis, 1998; Papayannopoulos et al., 1998; Cho et al., 2000; Dominguez and Casares, 2005; Singh et al., 2005b) |
| Genes with DV asymmetric response | ||||
| homothorax (hth) | Meis | homeodomain | Ventral eye suppression | (Rieckhof et al., 1997; Pai et al., 1998; Ryoo et al., 1999; Pichaud and Casares, 2000; Dominguez and Casares, 2005; Singh et al., 2005b; Bessa et al., 2008) |
| teashirt (tsh) | TSH1, TSH2, TSH3 | C2H2 zinc finger transcription factor | Dorsal eye growth, ventral eye suppression | (Fasano et al., 1991; Pan and Rubin, 1998; Bessa et al., 2002; Singh et al., 2002; Singh et al., 2004; Datta et al., 2009) |
| Lobe (L) | PRAS40 (Ortholog) | Proline rich Akt substrate | Ventral eye growth, no affect on dorsal eye | (Chern and Choi, 2002; Singh and Choi, 2003; Singh et al., 2005a; Singh et al., 2005b; Singh et al., 2006; Wang and Huang, 2009) |
| Expression on both margins | ||||
| optomotor blind (omb) | Tbx5 | Transcription factor | Expressed on dorsal and ventral eye margin. Not known. | (Calleja et al., 1996; Singh et al., 2004) |
5. DV patterning in the eye
5.1 Genes regulating ventral eye growth
L, a gene involved in ventral eye development (Table 1) was first reported in 1925, as a gene required for eye growth (Morgan et al., 1925) and was cloned in 2002 (Chern and Choi, 2002). L encodes an ortholog of PRAS40 (Oshiro et al., 2007; Vander Haar et al., 2007; Wang and Huang, 2009), and it is required during all stages of larval eye development (Chern and Choi, 2002; Singh et al., 2005b). L protein is expressed in both dorsal and ventral domains throughout the eye imaginal disc development (Figure 3A). Even though L is expressed uniformly in the entire eye field, L function is not required for growth and differentiation in the dorsal region of the eye (Chern and Choi, 2002; Singh et al., 2005b). Evidence for this ‘domain-specific’ function of L (ventral specificity) came from genetic mosaic analysis using a null mutation (Chern and Choi, 2002; Singh and Choi, 2003). Loss-of-function clones of L show an eye suppression phenotype specifically in the ventral eye; however, the dorsal clones do not suppress eye fate and exhibit well-organized photoreceptor clusters (Chern and Choi, 2002; Singh and Choi, 2003). Further genetic analysis revealed that this domain specific L function in growth was downstream to N-signaling, mediating N function either in the same or parallel pathway (Chern and Choi, 2002). Thus, the growth of early eye disc is controlled asymmetrically in the dorsal and ventral domains. Expression of L in both dorsal and ventral domains is puzzling since its function is only required for growth of the ventral domain. It is possible that an unidentified partner that is expressed in dorsal domain of the eye imaginal disc antagonizes the L function, or L may be selectively activated by some yet to be identified partner in the ventral domain.
Another candidate gene that may be contributing to ventral eye development is decapentaplegic (dpp), a member of TGF-β family of proteins, which acts as a long range secreted morphogen (Table 1; Nellen et al., 1996; Chanut and Heberlein, 1997). Dpp forms a gradient in the early eye anlage (anterior brain and eye field) that transverses from dorsal to ventral (Chang et al., 2001). In the early eye imaginal disc, Dpp is preferentially expressed in the ventral eye domain (Cho et al., 2000). In dpp mutants, the ventral part of early eye disc exhibits similar pattern defects as seen in L mutants. This dpp phenotype may be an outcome of ectopic induction of pnr or wg expression in the ventral domain as observed in L mutants (Singh et al., 2005a). In the DP of early eye imaginal disc Dpp, Hedgehog (Hh) and Wg signaling from the peripodial membrane is required to trigger N activation. Similar to limb patterning and development (Brook and Cohen, 1996; Penton and Hoffmann, 1996; Theisen et al., 1996), during eye imaginal disc development, Dpp antagonizes Wg. This antagonistic interaction occurs in the peripodial membrane across the DV border (Cho et al., 2000). Thus, Dpp signaling plays a role in inducing DV polarity from peripodial membrane.
Ser is the N ligand in the ventral domain of the eye imaginal disc (Table 1; Speicher et al., 1994; Cho and Choi, 1998; Dominguez and de Celis, 1998; Papayannopoulos et al., 1998; Dominguez and Casares, 2005). Ser acts downstream of L in the ventral eye as Ser transcription is repressed in early eye discs from Lsi homozygous larvae. Evidence for this conclusion comes from the Ser-lacZ reporter expression. Interestingly, Ser-lacZ expression in the posterior medial region remains at significant level. Loss-of-function clones of L cause strong reduction of Ser in the ventral eye whereas increased levels of L using the “flp out” approach induce Ser expression even in the dorsal domain of eye imaginal disc. Thus, L that acts downstream to N may be involved in inducing the Ser expression in the developing eye imaginal disc (Chern and Choi, 2002). Hypomorphic alleles of Ser exhibit reduced eye size suggesting a role for Ser in eye development. However, Ser loss-of-function clones do not exhibit any phenotypes in the eye (Sun and Artavanis-Tsakonas, 1996; Papayannopoulos et al., 1998; Chern and Choi, 2002), but misexpression of dominant negative form of Ser (SerDN) causes severe growth defects in eye imaginal disc (Kumar and Moses, 2001; Singh and Choi, 2003). It is possible that Ser function may be compensated by another factor, or Ser may somehow be secreted or transendocytosed into neighboring cells as shown in experiments performed using cell culture system (Klueg and Muskavitch, 1999; Kumar and Moses, 2001; Singh et al., 2005b). This may explain the apparent lack of phenotype in Ser mutant clones. Misexpression of SerDN in early eye imaginal disc using ey-Gal4 (Hazelett et al., 1998) results in either preferential loss of ventral eye or loss of the entire eye (Singh and Choi, 2003). Misexpression of SerDN in random gain-of-function clones generated by “flp-out” method (Pignoni and Zipursky, 1997) results in suppression of eye fate in the ventral half of eye. The similar phenotypes of SerDN misexpression and L mutants in the eye disc further validate that L and Ser work in the same pathway to regulate the growth of ventral eye domain.
It is known that Ser can activate N only at the DV border since Ser-N interaction is prevented by Fringe (Fng) in the ventral domain cells away from the DV border of the eye imaginal disc. Fng is a glucosaminyltransferase that elongates O-linked fucose residues to the EGF domains of N (Okajima and Irvine, 2002). Fng is known to bind N to promote N-Delta (Dl) interaction and is required to restrict N activation at the DV border (Irvine and Wieschaus, 1994; Kim et al., 1995; Fleming et al., 1997). Contrary to the positive function of Fng in N-Dl interaction, Fng inhibits Ser-N interaction when it is bound to N protein (Ju et al., 2000; Singh et al., 2005b). As a result, the N activation by Dl is enhanced only at the DV border. The expression pattern of these DV patterning genes changes dynamically in the developing eye imaginal disc, thereby showing striking differences before and after the initiation of retinal differentiation. Initially, fng is expressed in the ventral domain of eye imaginal disc, but as the eye imaginal disc undergoes retinal differentiation and the morphogenetic furrow proceeds anteriorly, fng expression further evolves. At this stage, fng exhibits preferential localization anterior to the furrow in both dorsal and ventral eye domain. These results validate the conclusion of genetic mosaic studies, which suggested that DV pattern is established during early eye development prior to retinal differentiation. The essential role of Fng in DV patterning was demonstrated by analysis of fng mutant clones. In loss-of-function clones of fng in the ventral eye, DV polarity is reorganized near the ectopic fng+/fng− border resulting in non-autonomous polarity reversals. This leads to the generation of de novo equators and ectopic localized activation of N at the fng+/fng− boundary (de Celis et al., 1996; Cho and Choi, 1998; Go et al., 1998; Baonza and Garcia-Bellido, 2000).
The DV axis in the wing imaginal disc is inverted in comparison to the eye imaginal disc. In the eye imaginal disc, Dl and Ser are preferentially expressed in the dorsal and ventral domains respectively. However, the localization of Dl and Ser preferential expression is reversed in the developing wing imaginal disc. The inversion of the DV axis in the eye and the wing disc may be due to the fact that the eye disc rotates 180° during embryogenesis (Struhl, 1981). Therefore, Ser functions as an N ligand in the dorsal cells, whereas Dl is the N ligand in the ventral cells. Not surprisingly, fng is ventral-specific in the eye but dorsal-specific in the wing imaginal disc.
Other candidate genes involved in ventral eye development are Chip and sloppy paired (slp) (Table 1). Chip, a transcriptional co-factor, is required for the ventral eye development (Roignant et al., 2010). Chip, a ubiquitous transcriptional co-factor, interacts with classes of transcription factors during neural development. Chip has been reported to establish the ventral boundary of the eye and the head tissue (Roignant et al., 2010). Slp belongs to forkhead family of transcription factors, which are required for embryonic patterning (Grossniklaus et al., 1992). Slp locus has two transcription units. Both of them are expressed in the ventral eye and are functionally redundant (Sato and Tomlinson, 2007). Slp and Iro-C proteins have been shown to repress each other at the DV midline. N signaling at the DV midline suppress Slp at the midline (Sato and Tomlinson, 2007).
5.2 Dorsal selector genes
It is known that compartment boundaries are defined by the spatio-temporal expression of genes (Blair, 2001; Curtiss et al., 2002; Dahmann et al., 2011). For example, engrailed (en) and apterous (ap) are expressed in the posterior and the dorsal compartments of wing imaginal disc, respectively (Brower, 1986; Cohen et al., 1992; Hidalgo, 1998; Held, 2002). Thus, “Selector” genes were identified that assign a unique property to the cells within their expression domains, which results in the formation of unique territories (Blair, 2001; Curtiss et al., 2002). Moreover, loss-of-function of these selector genes results in the loss of that particular fate. In the Drosophila eye, the presence of these selector genes (which exhibit dorsal or ventral domain specific expression) became apparent in the earlier enhancer trap screens (Bier et al., 1989; Bhojwani et al., 1995; Sun et al., 1995). Enhancer trap lines containing mini-white (w) and lacZ reporter gene (P-lacW) (Bellen et al., 1989; Bier et al., 1989; Wilson et al., 1989; Bhojwani et al., 1995; Sun et al., 1995) that show domain specific expression in the eye were isolated in the screens. Interestingly, some of these lines have w+ expression restricted only to the dorsal half of the adult eye. These enhancer trap lines have made significant contributions towards understanding the DV patterning in the eye (Choi et al., 1996; Sun and Artavanis-Tsakonas, 1996; McNeill et al., 1997; Kehl et al., 1998; Morrison and Halder, 2010). Most of these dorsal specific P insertion lines were mapped to the chromosomal region 69CD, identifying this region as a hot spot for P-lacW insertions that show dorsal eye specific expression. The molecular characterization of this 69CD chromosomal region revealed the existence of a cluster of homeobox genes, araucan (ara), caupolican (caup), and mirror (mirr) (Gomez-Skarmeta and Modolell, 1996; McNeill et al., 1997; Grillenzoni et al., 1998; Heberlein et al., 1998; Kehl et al., 1998; Singh et al., 2005b). This cluster of homeobox genes that are located within an approximately 140Kb region (Netter et al., 1998) are expressed in the dorsal half of the eye. They are referred to as Iroquois –complex (Iro-C) because the mutation in these genes lack lateral thoracic bristles and resemble the hair style of the Indian tribe, the Iroquois, also referred to as Mohawks (a native tribe which shaved all but a medial stripe of hairs on the head) (Gomez-Skarmeta and Modolell, 1996; Leyns et al., 1996). They named the genes Araucan and Caupolican in honor of Amerindian tribes: Aracaunians and one of their heroes Caupolican.
The members of Iro-C are highly conserved, essential genes and exhibit significant differences in their expression pattern (Gomez-Skarmeta and Modolell, 2002). Mirr is strongly and dynamically expressed in the CNS, (Netter et al., 1998; Urbach and Technau, 2003) and it is essential for follicle cell patterning (Jordan et al., 2000) while Ara and Caup are preferentially expressed in mesodermal tissues in the embryos (Netter et al., 1998). However, in the eye imaginal disc, all three Iro-C members are expressed in the dorsal half (Figure 4E), raising a possibility that they might be functionally redundant. Loss-of-function of the mirre48 allele shows weak but significant defects of non-autonomous DV polarity reversals in comparison to mirr+ ommatidia in the dorsal half of the eye (McNeill et al., 1997). Compartments of different cell lineages do not intermingle due to differences in cell identities and affinities (Garcia-Bellido et al., 1973; Irvine, 1999; Dahmann et al., 2011). Somatic clones of cells lacking mirr function in dorsal half of the eye exhibit smooth clone borders, indicating that cells lacking mirr avoid mixing with the neighboring mirr expressing cells. However, the clones in the ventral half where mirr is not expressed show wiggly clone borders (Yang et al., 1999). This analysis suggests that mirr functions as a dorsal fate selector. Since the phenotype of mirr clones was not strong enough, it raised the possibility that ara and caup, the other two members of Iro-C, can partly compensate for the loss of mirr function in the eye. The issue of functional redundancy got resolved when a deficiency iroDMF3, which uncovers all three Iro-C genes by the deletion of ara and caup as well as a 5′-region of mirr (Gomez-Skarmata et al., 1996; Diez del Corral et al., 1999), was employed for clonal analysis. Loss-of-function clones of iroDMF3 in the eye showed repolarization of the ommatidial polarity in the dorsal clones along with dorsal eye enlargement or formation of an ectopic eye field on the dorsal margin. There was no phenotype in the ventral half of the eye (Figure 4F, G). These results further highlighted the importance of boundary between the dorsal and ventral cell types. These results strongly support that the three Iro-C genes are partially redundant, and the Iro-C as a whole is required for organizing the DV polarity pattern and growth of the eye.
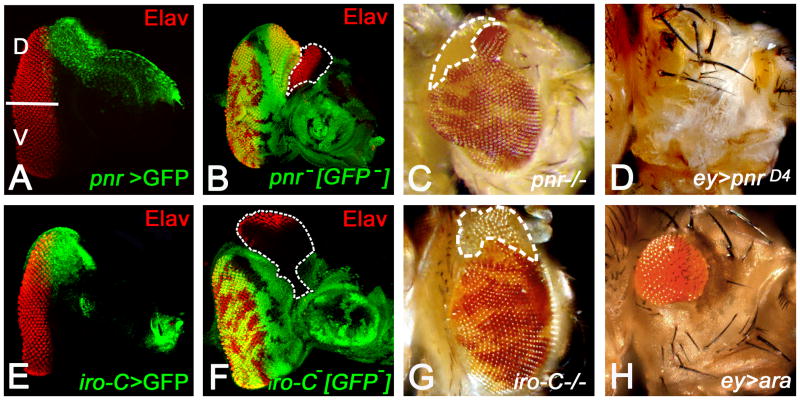
Figure 4. Pnr and Iro-C members function as dorsal eye fate selectors.
(A) Pnr expression (green) is restricted to the dorsal eye margin of developing eye imaginal disc (Maurel-Zaffran and Treisman, 2000; Pichaud and Casares, 2000; Singh and Choi, 2003). Elav (red) marks the photoreceptor neurons. (B, C) Loss-of-function clones of pnr results in the enlargement of existing dorsal eye field (e.g., in the clone outlined in B) in eye imaginal disc (B) and adult eye(C). (B) Note that there is a non-autonomous eye enlargement in eye imaginal disc, which is attributed to generation of de novo equator in the dorsal compartment of eye imaginal disc (Maurel-Zaffran and Treisman, 2000; Oros et al., 2010). (D) Misexpression of pnr (ey>pnrD4) in the eye imaginal disc suppresses the eye fate validating a late function of pnr in defining the eye field boundary (Oros et al., 2010). (E) The expression domain of the members of Iroquois complex (Iro-C>GFP, green) spans the dorsal region of the eye imaginal (G, H) Loss-of-function of Iro-C causes dorsal eye enlargements in the (G) eye imaginal disc and in (H) adult eye. These phenotypes are similar to the (B, C) pnr loss-of-function phenotypes. (H) Misexpression of ara, a member of Iro-C, in entire eye imaginal disc (ey>ara) results in small eye. [D: Dorsal, V: Ventral]
Loss-of-function of iroDMF3 also suggested that Iro-C genes function as dorsal selectors for head structures as well since mutant clones in the dorsal region induce the formation of ventral head structures (Cavodeassi et al., 2000). Ectopic ventral head tissues that resulted from loss of Iro-C genes are cell-autonomous and therefore, accompanied by loss of corresponding dorsal structures. In contrast, ectopic ventral eyes are generated non-cell autonomously since reversals of DV ommatidial polarity are detected in Iro-C+ wild-type region adjacent to the mutant clones. This also supports the idea that the DV boundary is an organizing center for DV pattern and growth in the eye imaginal disc. Furthermore, DV patterning of the eye occurs in earlier larval stages than the head patterning. In the Drosophila eye, pnr, another dorsal gene, which is expressed in the dorsal eye margin (Figure 4A), exhibits similar loss-of-function (Table 1; Figure 4B, C) and gain-of-function (Figure 4D) phenotypes as observed with Iro-C in the eye and the head (Maurel-Zaffran and Treisman, 2000; Pichaud and Casares, 2000; Singh et al., 2005b; Oros et al., 2010). Pnr, a GATA-1 transcription factor, plays an important role in the dorsal eye development, and acts as a selector for the dorsal eye fate (Ramain et al., 1993; Maurel-Zaffran and Treisman, 2000; Pichaud and Casares, 2000; Dominguez and Casares, 2005; Singh et al., 2005b; Oros et al., 2010). In the hierarchy of dorsal genes, pnr is the top most gene, and it induces Wingless (Wg) which, in turn induces the expression of downstream target genes mirr in the dorsal half of the eye (Maurel-Zaffran and Treisman, 2000; Dominguez and Casares, 2005; Singh et al., 2005b). During later stages of development which corresponds to the retinal differentiation stage in late second instar and third instar of larval eye development, pnr is involved in defining the dorsal eye margin by regulating the retinal determination (RD) genes (Oros et al., 2010).
Wg, a secretory protein and a morphogen, is expressed along the antero-lateral margins of the third instar eye imaginal disc (Table 1; Baker, 1988). Wg plays multiple roles during eye development. One of these roles of Wg is to promote growth of early eye imaginal disc. During early eye development, Wg expression is restricted to the dorsal eye domain (Cho et al., 2000; Chang et al., 2001). During retinal differentiation stage, Wg is known to prevent ectopic induction of retinal differentiation from the lateral eye imaginal disc margin (Ma and Moses, 1995; Treisman and Rubin, 1995). Thus, Wg that acts as a negative regulator of eye during retinal differentiation functions as a dorsal eye fate gene. Dl, an N ligand in the dorsal eye imaginal disc, has been assigned to the dorsal gene category in the early eye imaginal disc (Table 1). Dl is preferentially expressed in dorsal domain of eye imaginal discs during first and second instar stages.
6. Genes with domain specific growth response: Role of teashirt (tsh) and homothorax (hth)
In addition to the genes that exhibit DV domain specific expression during patterning, there is a group of genes, which show differential functions in the dorsal-ventral compartments, but are not expressed in a DV specific pattern. These genes can be broadly classified into two groups: (I) Genes expressed uniformly in the eye imaginal disc but their functional domain is restricted only to the ventral half of the eye, for example L (described in Section 4, 5.1) and hth (Table 1). (II) Genes that are expressed uniformly in the early eye imaginal disc function differently in the dorsal and ventral half of the eye, for example, tsh (Table 1).
Hth: Hth a vertebrate homolog of murine proto-oncogene MEIS1 (Moskow et al., 1995), encodes a homeodomain transcription factor of the three-amino-acid extension loop (TALE) sub-family (Rieckhof et al., 1997). Like L, hth is expressed in the entire early eye primordium. However, with the onset of differentiation in the eye, Hth expression gets restricted to the cells anterior to the furrow (Figure 5A, A′ Pai et al., 1998; Pichaud and Casares, 2000; Bessa et al., 2002; Singh et al., 2002a). Even though hth is expressed anterior to the furrow both in the dorsal and ventral half of the eye imaginal disc (Figure 5A, A′), the loss-of-function of hth causes eye enlargement only in the ventral eye margin (Figure 5B, B′). However, loss-of-function clones of hth in the dorsal compartment do not show any phenotype in the eye imaginal disc. Furthermore, hth mutant cells do not survive in the anterior eye (Pichaud and Casares, 2000; Bessa et al., 2002; Bessa et al., 2008). As expected, misexpression of hth suppresses the eye fate (Pai et al., 1998). Moreover, eye suppression function of Hth is independent of any domain constraint. Hth is involved in multiple functions during development, and it is required for nuclear localization of a homeoprotein Extradenticle (Exd). hth encodes a protein with nuclear localization signal (NLS) and two conserved domains: the N terminal evolutionarily conserved MH domain (for Meis and Hth), and a C-terminal region including the homeodomain (HD) (Rieckhof et al., 1997; Kurant et al., 1998; Pai et al., 1998; Noro et al., 2006). Alternative splicing is known to provide additional complexity to the genes encoding the Hth transcription factors (Glazov et al., 2005; Noro et al., 2006). Hth forms a heterodimer with Exd through its MH domain and translocates into the nucleus to regulate transcription (Ryoo et al., 1999; Jaw et al., 2000; Stevens and Mann, 2007). Since Exd is present in the entire eye, the ventral specific function of Hth has been proposed through its interaction with Wg and Tsh. Together they are involved in suppression of eye fate on the ventral margin.
Tsh: The homeotic gene tsh encodes a C2H2 zinc-finger transcription factor with three widely spaced Zinc finger domains (Fasano et al., 1991). Tsh plays an important role during Drosophila eye development (Pan and Rubin, 1998; Bessa et al., 2002; Singh et al., 2002a; Singh et al., 2005b; Datta et al., 2009; Kumar, 2009; Kumar, 2010; Kumar, 2011). tsh is expressed both in dorsal and ventral eye anterior to the furrow, and it exhibits a DV constraint in its function. In the ventral eye, tsh acts as repressor of eye fate, whereas in the dorsal eye, it promotes eye development (Singh et al., 2002b; Singh et al., 2004). Interestingly, the DV constraint in tsh function in the eye stems from the partners with which it collaborates in the dorsal or the ventral eye disc (Singh et al., 2004). It was shown that Tsh cooperates with Iro-Complex members and Dl in the dorsal eye for it growth promotion function (Singh et al., 2004). The function of tsh in the ventral eye is dependent on Hth and Ser. The expression of tsh overlaps with hth in the eye imaginal disc, and like hth, tsh expression also evolves during larval eye development. Initially, in first instar eye imaginal disc, tsh is expressed in the entire eye imaginal disc but its expression retracts anteriorly to nearly three- quarters of the eye imaginal disc when the retinal differentiation begins (Bessa et al., 2002; Singh et al., 2002a). Furthermore, Tsh and Hth physically interact with each other [along with Pax-6 homolog, Eyeless (Ey)] to repress the expression of downstream target genes (Bessa et al., 2002; Dominguez and Casares, 2005). Further insights into the potential mechanism of tsh and hth in regulating growth and differentiation in the eye came initially from analysis of expression patterns of the retinal determination (RD) gene network members (Bessa et al., 2002). It has been proposed that Tsh, Hth, and Ey coexpress in the proliferating cells anterior to furrow to block precocious retinal differentiation and promote cell proliferation (Bessa et al., 2002; Singh et al., 2002a; Dominguez and Casares, 2005). All these studies suggest that DV patterning genes contribute towards the growth of the eye field.
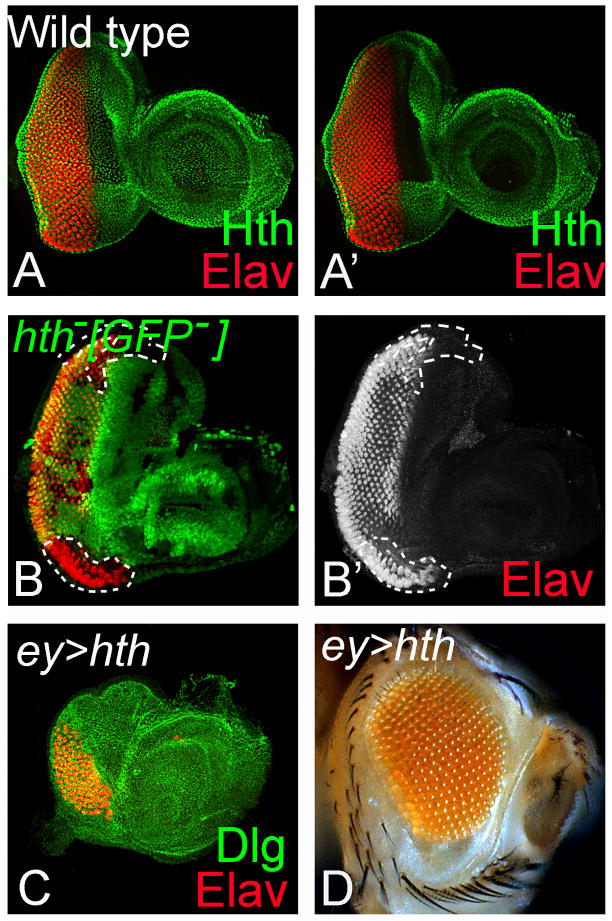
Figure 5. Domain specific function of hth is restricted to the ventral eye margin.
(A, A′) Hth (green) is expressed in the entire peripodial membrane (PM) whereas disc proper specific expression of Hth is restricted anterior to the furrow both in the dorsal as well as ventral domain of the eye imaginal disc (Rieckhof et al., 1997; Pai et al., 1998; Pichaud and Casares, 2000; Bessa et al., 2002; Singh et al., 2002a).. ELAV (red), a pan neural marker, marks the photoreceptors neurons in the eye imaginal disc. (B, B′) Loss-of-function clones of hth marked by the absence of the GFP reporter (clonal boundary marked by white dotted line) in the ventral eye results in eye enlargement, whereas in the dorsal eye these clones do not have any effect. (C, D) Misexpression of hth in the entire eye using ey-Gal4 driver (ey>hth) results in reduced eye field as seen in (C) eye imaginal disc and (D) adult eye (Pai et al., 1998).
7. Boundary formation during organogenesis
One of the important questions is how organ size and growth are regulated by DV patterning genes in the eye. The dorsal selector genes assign a dorsal fate, and thereby, generate a group of cells with unique properties that makes them different from the default ventral state cells of the developing eye disc. Interestingly, the boundary between the dorsal and ventral cells is maintained by the antagonistic interactions between the genes required for the growth and development of the dorsal and ventral domains of the eye (Singh et al., 2005a). It has been shown that L is essential for growth of the ventral eye tissue, but it is dispensable in the dorsal region specified by pnr function (Singh and Choi 2003). In addition to a boundary between the dorsal and ventral compartment within the eye, a boundary is defined between the developing eye field and the surrounding head cuticle on the dorsal and ventral margins. Since the adult eye, head cuticle, and other mouthparts are generated from the eye-antennal imaginal disc, there is a sequential fate restriction between the developing eye and head cuticle. Interestingly, these DV patterning genes play an important role of defining the boundary of the eye field on the dorsal and the ventral margins (Oros et al., 2010 ).
The boundary between the eye field and the head cuticle on the dorsal margin is regulated by pnr. It has been shown that pnr function evolves during eye development. During early second instar of development, pnr is required for defining the dorsal lineage, before the onset of retinal differentiation, by inducing Wg and members of the Iro-C complex (Maurel-Zaffran and Treisman, 2000; Singh and Choi, 2003; Singh et al., 2005b; Oros et al., 2010). However, later during the late second- instar stage of eye development, when the morphogenetic furrow (MF) is initiated, pnr suppresses the photoreceptor differentiation at the dorsal eye margin (Oros et al., 2010). The endogenous expression of pnr is only in the peripodial membrane of the dorsal eye margin which gives rise to the adult head cuticle. Loss-of-function clones of pnr exhibit ectopic dorsal eyes, which are restricted within the clones, suggests that absence of pnr function promotes ectopic eye formation in the dorsal eye margin. Thus, Pnr defines the boundary between the head cuticle and the dorsal margin of the developing eye field (Oros et al., 2010). Since pnr is not expressed in the ventral eye, there is a different mechanism to define the boundary of eye field on the ventral margin. The boundary of eye field on the ventral eye margin is defined by the antagonistic interaction of L with hth (Singh et al., 2011, in press). In the ventral eye, transcriptional co-factor Chip interacts with the LIM-homeodomain proteins to define the boundary of the eye field (Roignant and Treisman, 2009). Interestingly, Chip mediated regulation of the ventral eye boundary is independent of hth (Roignant and Treisman, 2009). Thus, the genetic cascade that regulates the boundary of eye field on the dorsal and the ventral margin of the eye is different.
8. Concluding Remarks
Our understanding of the axial patterning of the Drosophila eye is far from complete. In this review, we have described the overview of key developmental events and genes involved in early DV patterning. The DV compartment formation is a key to initiate patterning and growth in the early eye imaginal disc. The present information clearly illustrates that DV patterning is required to initiate the generation of heterogeneous population (dorsal and ventral cell fate) of cells within a homogenous (default ventral fate) early eye primordial. Although our knowledge on the DV patterning in the eye has dramatically increased in recent years, we still do not know the molecular interactions important for the regulation of DV patterning. Moreover, many more genes (both known and novel) are expected to be involved in DV patterning, and future studies using novel genetic and bioinformatics approaches should help in defining the full complement of genes involved in this intricate process. Identification and functional analysis of more molecular players involved in this process will help provide a better picture of how a small number of cells in the disc primordium grow to form a precise pattern of mirror symmetry in the compound eye. Furthermore, the possibility of crosstalk of the DV patterning pathway with other signaling pathways to regulate growth during early phase of eye development cannot be ruled out. All this information will lay foundation about understanding the process of organogenesis, as loss-of-function of the genes involved in DV patterning results in the loss of the eye field or a part of the eye field. The complexity and precision of the neural connectivity in the adult visual system has fascinated researchers for a long time. The DV polarity of the retina is responsible for controlling the targeting of the retinal axon projections to the brain in humans and other higher vertebrates. Thus, DV patterning genes also contribute towards the wiring of the brain to the retina. How all these different facets work together to define the final form of this complex structure eye is an open question and is of fundamental importance.
8.1 Similarities with vertebrate eye
The basic sensory epithelium design of the vertebrate and most invertebrate eyes including Drosophila eye is similar (Charlton-Perkins and Cook, 2010; Sanes and Zipursky, 2010). The morphogenetic furrow (MF) in the fly eye is analogous to the wave of neurogeneis in the vertebrate retina (Neumann and Nuesslein-Volhard, 2000; Hartenstein, 2002). Recent studies in the vertebrate visual systems have identified several genes that are expressed in a DV domain specific manner in the retina. BMP4, a TGF-β closely related to Dpp, has been implicated in development of progenitor cells in the dorsal half of the eye and in establishment of the DV axis of the retina in Xenopus (Papalopulu and Kintner, 1996). The dorsal selectors of the vertebrate eye, BMP-4 and TbX5, act to restrict the expression of Vax2 and Pax2 to the ventral domain of the eye (Koshiba-Takeuchi et al., 2000; Mui et al., 2002; Peters, 2002; Peters and Cepko, 2002). These DV expression domains correspond to the developmental compartments (Peters, 2002; Peters and Cepko, 2002). The DV patterning plays an important role in retinotectal projection pattern (Koshiba-Takeuchi et al., 2000; McLaughlin et al., 2003). The R-cell projections form a precise topographic connection with the optic lobe, and are referred to as retinotopy, which is common to both the vertebrate and the insect visual system (Gaul, 2002). Furthermore, Jagged-1(Jag1), a vertebrate homolog of the Drosophila ventral eye gene Ser, shows a DV asymmetric expression pattern in the retina. Moreover, the loss-of-function of Jag1 results in Alagille’s syndrome, which also affects the eye (Oda et al., 1997; Xue et al., 1999; Kim and Fulton, 2007). Interestingly, it has been shown that mouse retina also begins with a default ventral like state (Murali et al., 2005). Therefore, the DV boundary may play conserved roles in organizing growth and pattern of visual system in higher animals, and studies in Drosophila will further our knowledge in the area of animal development mechanisms and help to unravel the genetic underpinnings of developmental defects caused by mutations in human homologs of Drosophila DV patterning genes.
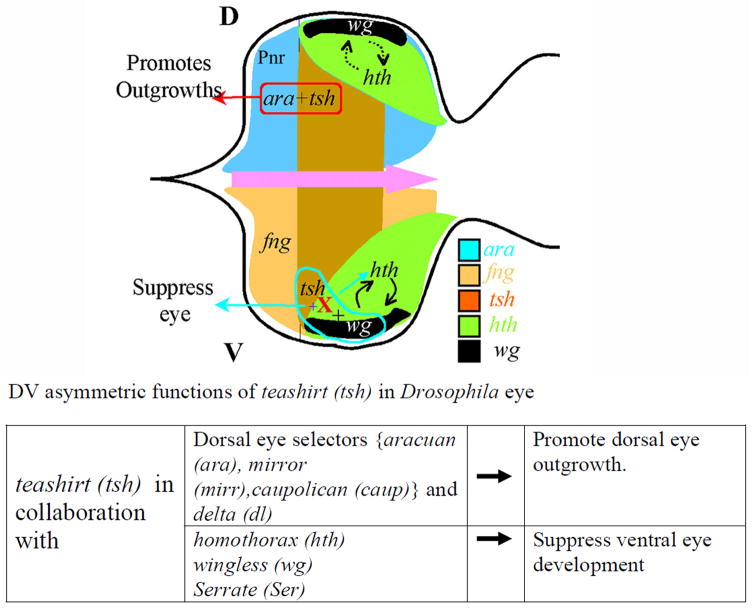
Figure 6. DV asymmetric function of homeotic gene teashirt (tsh) in the developing eye imaginal disc depends on the spatial cues provided by other genes which are expressed in DV asymmetric fashion (Singh et al., 2004).
Gain-of-function of Tsh suppresses the eye fate in the ventral eye, whereas it promotes dorsal eye enlargement. It has been shown the DV asymmetric function of Tsh in the developing eye imaginal disc depends on the domain specific cues provided by genes that are either expressed or function in a DV asymmetric fashion. Tsh in collaboration with Wg and Ser can induce Hth in the ventral eye to suppress the eye fate. The ventral eye suppression function of tsh is independent of other ventral specific genes like fng and L. In the dorsal eye, tsh is known to promote growth and eye in collaboration with Iro-C members and a N ligand in the dorsal eye, Dl (Singh et al., 2002; Singh et al., 2004; Singh et al., 2005b). [D: Dorsal, V: ventral]
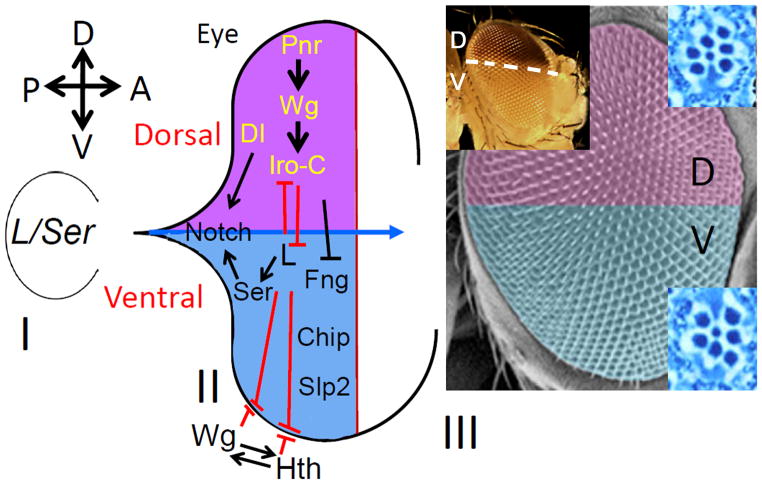
Figure 7. Genes with DV asymmetric expression or function are involved in regulating DV patterning in the Drosophila eye.
(I) Early eye imaginal disc primordium comprises of a homogeneous population of ventral eye fate cells whose growth and survival depends on L/Ser function (Singh and Choi, 2003; Singh et al., 2005b; Singh et al., 2006). (II) Later during development, DV lineage is generated by onset of expression of dorsal gene pnr (Singh and Choi, 2003). Pnr acts upstream of Wg, which in turn triggers the expression of downstream Iro-C members (Maurel-Zaffran and Treisman, 2000). Dl, an N ligand, is involved in dorsal eye development. The default ventral eye fate is maintained by function of L and Ser (Singh and Choi, 2003; Singh et al., 2005b). There are three other players involved in ventral eye development viz., fringe (fng) (Cho and Choi, 1998; Dominguez and de Celis, 1998; Papayannopoulos et al., 1998; Irvine, 1999; Cho et al., 2000; Dominguez and Casares, 2005), Chip (Roignant et al., 2010) and Sloppy paired (Slp) (Sato and Tomlinson, 2007). L is known to act antagonistically to the dorsal genes to define the boundary between the dorsal and ventral compartment of the eye (Singh et al., 2005a). L and Ser also interact antagonistically with hth on the ventral eye margin to define the ventral eye margin boundary (Singh et al., 2005b). There is a positive feedback loop between Wg and Hth on the ventral eye margin (Pichaud and Casares, 2000; Singh et al., 2004; Dominguez and Casares, 2005; Singh et al., 2005b). Chip acts independently of hth (Roignant et al., 2010). Sloppy paired (Slp) is involved in ventral eye development by repressing Iro-C at the DV midline (Sato and Tomlinson, 2007). These DV patterning genes work together and contribute towards sculpting the final shape and size of the adult eye. (III) The ommatidial clusters within an adult eye are organized into two mirror symmetric orientations that are polarized in the opposite directions in the dorsal and the ventral half. The dorsal selectors exhibit a dorsal specific expression of mini-white reporter gene in the adult eye.
Acknowledgments
The authors thank members of Singh and Kango-Singh lab for their help and comments on the manuscript. We apologize to all authors whose work could not be cited due to space limitations. AS is supported by a NIH grant (1R15 HD064557-01), start-up support from the University of Dayton. MKS is supported by start-up funds from the University of Dayton.
References
- Anderson DT. The development of hemimetabolous insects. In: Counce S, Waddington CH, editors. Developmental Systems: Insects. New York: Academic Press; 1972a. pp. 165–242. [Google Scholar]
- Anderson DT. The development of hemimetabolous insects. In: Counce S, Waddington CH, editors. Developmental Systems: Insects. New York: Academic Press; 1972b. pp. 96–163. [Google Scholar]
- Atkins M, Mardon G. Signaling in the third dimension: the peripodial epithelium in eye disc development. Dev Dyn. 2009;238:2139–2148. doi: 10.1002/dvdy.22034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NE. Embryonic and imaginal requirements for wingless, a segment-polarity gene in Drosophila. Dev Biol. 1988;125:96–108. doi: 10.1016/0012-1606(88)90062-0. [DOI] [PubMed] [Google Scholar]
- Baker WK. A clonal analysis reveals early developmental restrictions in the Drosophila head. Dev Biol. 1978;62:447–463. doi: 10.1016/0012-1606(78)90227-0. [DOI] [PubMed] [Google Scholar]
- Baonza A, Garcia-Bellido A. Notch signaling directly controls cell proliferation in the Drosophila wing disc. Proc Natl Acad Sci U S A. 2000;97:2609–2614. doi: 10.1073/pnas.040576497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, O’Kane CJ, Wilson C, Grossniklaus U, Pearson RK, Gehring WJ. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- Bessa J, Gebelein B, Pichaud F, Casares F, Mann RS. Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes Dev. 2002;16:2415–2427. doi: 10.1101/gad.1009002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa J, Tavares MJ, Santos J, Kikuta H, Laplante M, Becker TS, Gomez-Skarmeta JL, Casares F. meis1 regulates cyclin D1 and c-myc expression, and controls the proliferation of the multipotent cells in the early developing zebrafish eye. Development. 2008;135:799–803. doi: 10.1242/dev.011932. [DOI] [PubMed] [Google Scholar]
- Bhojwani J, Singh A, Misquitta L, Mishra A, Sinha P. Search for the Drosophila genes based on patterned expression of mini-white reporter gene of a P lacW vector in adult eyes. Roux’s Arch Dev Biol. 1995;205:114–121. doi: 10.1007/BF00357757. [DOI] [PubMed] [Google Scholar]
- Bier E, Vaessin H, Shepherd S, Lee K, McCall K, Barbel S, Ackerman L, Carretto R, Uemura T, Grell E, et al. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 1989;3:1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- Blair SS. Cell lineage: compartments and Capricious. Curr Biol. 2001;11:R1017–1021. doi: 10.1016/s0960-9822(01)00614-5. [DOI] [PubMed] [Google Scholar]
- Bodentstein D. The Postembryonic development of DRosophila melanogaster. In: Demerec M, editor. Biology of Drosophila. New York: Wiley; 1950. pp. 275–367. [Google Scholar]
- Brook WJ, Cohen SM. Antagonistic interactions between wingless and decapentaplegic responsible for dorsal-ventral pattern in the Drosophila Leg. Science. 1996;273:1373–1377. doi: 10.1126/science.273.5280.1373. [DOI] [PubMed] [Google Scholar]
- Brower DL. Engrailed gene expression in Drosophila imaginal discs. EMBO J. 1986;5:2649–2656. doi: 10.1002/j.1460-2075.1986.tb04547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavodeassi F, Modolell J, Campuzano S. The Iroquois homeobox genes function as dorsal selectors in the Drosophila head. Development. 2000;127:1921–1929. doi: 10.1242/dev.127.9.1921. [DOI] [PubMed] [Google Scholar]
- Chang T, Mazotta J, Dumstrei K, Dumitrescu A, Hartenstein V. Dpp and Hh signaling in the Drosophila embryonic eye field. Development. 2001;128:4691–4704. doi: 10.1242/dev.128.23.4691. [DOI] [PubMed] [Google Scholar]
- Chanut F, Heberlein U. Role of decapentaplegic in initiation and progression of the morphogenetic furrow in the developing Drosophila retina. Development. 1997;124:559–567. doi: 10.1242/dev.124.2.559. [DOI] [PubMed] [Google Scholar]
- Charlton-Perkins M, Cook TA. Building a fly eye: terminal differentiation events of the retina, corneal lens, and pigmented epithelia. Curr Top Dev Biol. 2010;93:129–173. doi: 10.1016/B978-0-12-385044-7.00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern JJ, Choi KW. Lobe mediates Notch signaling to control domain-specific growth in the Drosophila eye disc. Development. 2002;129:4005–4013. doi: 10.1242/dev.129.17.4005. [DOI] [PubMed] [Google Scholar]
- Cho KO, Chern J, Izaddoost S, Choi KW. Novel signaling from the peripodial membrane is essential for eye disc patterning in Drosophila. Cell. 2000;103:331–342. doi: 10.1016/s0092-8674(00)00124-0. [DOI] [PubMed] [Google Scholar]
- Cho KO, Choi KW. Fringe is essential for mirror symmetry and morphogenesis in the Drosophila eye. Nature. 1998;396:272–276. doi: 10.1038/24394. [DOI] [PubMed] [Google Scholar]
- Choi KW, Mozer B, Benzer S. Independent determination of symmetry and polarity in the Drosophila eye. Proc Natl Acad Sci U S A. 1996;93:5737–5741. doi: 10.1073/pnas.93.12.5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B, McGuffin ME, Pfeifle C, Segal D, Cohen SM. apterous, a gene required for imaginal disc development in Drosophila encodes a member of the LIM family of developmental regulatory proteins. Genes Dev. 1992;6:715–729. doi: 10.1101/gad.6.5.715. [DOI] [PubMed] [Google Scholar]
- Cohen B, Simcox AA, Cohen SM. Allocation of the thoracic imaginal primordia in the Drosophila embryo. Development. 1993;117:597–608. doi: 10.1242/dev.117.2.597. [DOI] [PubMed] [Google Scholar]
- Cohen SM. Imaginal Disc development. In: Bate M, Martinez-Arias A, editors. The development of Drosophila melanogaster. New York: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Crick FH, Lawrence PA. Compartments and polyclones in insect development. Science. 1975;189:340–347. doi: 10.1126/science.806966. [DOI] [PubMed] [Google Scholar]
- Curtiss J, Halder G, Mlodzik M. Selector and signalling molecules cooperate in organ patterning. Nat Cell Biol. 2002;4:E48–51. doi: 10.1038/ncb0302-e48. [DOI] [PubMed] [Google Scholar]
- Dahmann C, Oates AC, Brand M. Boundary formation and maintenance in tissue development. Nat Rev Genet. 2011;12:43–55. doi: 10.1038/nrg2902. [DOI] [PubMed] [Google Scholar]
- Datta RR, Lurye JM, Kumar JP. Restriction of ectopic eye formation by Drosophila teashirt and tiptop to the developing antenna. Dev Dyn. 2009;238:2202–2210. doi: 10.1002/dvdy.21927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis JF, Garcia-Bellido A, Bray SJ. Activation and function of Notch at the dorsal-ventral boundary of thewing imaginal disc. Development. 1996;122:359–369. doi: 10.1242/dev.122.1.359. [DOI] [PubMed] [Google Scholar]
- Dietrich W. Die Facettenaugen der Dipteran. Z Wiss Zool. 1909;92:465–539. [Google Scholar]
- Dominguez M, Casares F. Organ specification-growth control connection: new in-sights from the Drosophila eye-antennal disc. DevDyn. 2005;232:673–684. doi: 10.1002/dvdy.20311. [DOI] [PubMed] [Google Scholar]
- Dominguez M, de Celis JF. A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature. 1998;396:276–278. doi: 10.1038/24402. [DOI] [PubMed] [Google Scholar]
- Erclik T, Hartenstein V, McInnes RR, Lipshitz HD. Eye evolution at high resolution: the neuron as a unit of homology. Dev Biol. 2009;332:70–79. doi: 10.1016/j.ydbio.2009.05.565. [DOI] [PubMed] [Google Scholar]
- Fasano L, Roder L, Core N, Alexandre E, Vola C, Jacq B, Kerridge S. The gene teashirt is required for the development of Drosophila embryonic trunk segments and encodes a protein with widely spaced zinc finger motifs. Cell. 1991;64:63–79. doi: 10.1016/0092-8674(91)90209-h. [DOI] [PubMed] [Google Scholar]
- Ferris GF. External morphology of the Adult. In: Demerec M, editor. Biology of Drosophila. New York: Wiley; 1950. pp. 368–419. [Google Scholar]
- Fleming RJ, Gu Y, Hukriede NA. Serrate-mediated activation of Notch is specifically blocked by the product of the gene fringe in the dorsal compartment of the Drosophila wing imaginal disc. Development. 1997;124:2973–2981. doi: 10.1242/dev.124.15.2973. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido A, Merriam JR. Cell lineage of the imaginal discs in Drosophila gynandromorphs. J Exp Zool. 1969;170:61–75. doi: 10.1002/jez.1401700106. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido A, Ripoll P, Morata G. Developmental compartmentalisation of the wing disk of Drosophila. Nat New Biol. 1973;245:251–253. doi: 10.1038/newbio245251a0. [DOI] [PubMed] [Google Scholar]
- Gaul U. The establishment of retinal connectivity. In: Moses K, editor. Drosophila Eye Development. Berlin: Springer-Verlag; 2002. pp. 205–216. [Google Scholar]
- Gehring WJ. New perspectives on eye development and the evolution of eyes and photoreceptors. J Hered. 2005;96:171–184. doi: 10.1093/jhered/esi027. [DOI] [PubMed] [Google Scholar]
- Gibson MC, Schubiger G. Drosophila peripodial cells, more than meets the eye? Bioessays. 2001;23:691–697. doi: 10.1002/bies.1098. [DOI] [PubMed] [Google Scholar]
- Glazov EA, Pheasant M, McGraw EA, Bejerano G, Mattick JS. Ultraconserved elements in insect genomes: a highly conserved intronic sequence implicated in the control of homothorax mRNA splicing. Genome Res. 2005;15:800–808. doi: 10.1101/gr.3545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go MJ, Eastman DS, Artavanis-Tsakonas S. Cell proliferation control by Notch signaling in Drosophila development. Development. 1998;125:2031–2040. doi: 10.1242/dev.125.11.2031. [DOI] [PubMed] [Google Scholar]
- Gomez-Skarmeta JL, Modolell J. araucan and caupolican provide a link between compartment subdivisions and patterning of sensory organs and veins in the Drosophila wing. Genes Dev. 1996;10:2935–2945. doi: 10.1101/gad.10.22.2935. [DOI] [PubMed] [Google Scholar]
- Gomez-Skarmeta JL, Modolell J. Iroquois genes: genomic organization and function in vertebrate neural development. Curr Opin Genet Dev. 2002;12:403–408. doi: 10.1016/s0959-437x(02)00317-9. [DOI] [PubMed] [Google Scholar]
- Grillenzoni N, van Helden J, Dambly-Chaudiere C, Ghysen A. The iroquois complex controls the somatotopy of Drosophila notum mechanosensory projections. Development. 1998;125:3563–3569. doi: 10.1242/dev.125.18.3563. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Pearson RK, Gehring WJ. The Drosophila sloppy paired locus encodes two proteins involved in segmentation that show homology to mammalian transcription factors. Genes Dev. 1992;6:1030–1051. doi: 10.1101/gad.6.6.1030. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Reh TA. Homologies between vertebrate and invetebrate eyes. In: Moses K, editor. Drosophila Eye Development. Berlin: Springer-Verlag; 2002. pp. 219–251. [Google Scholar]
- Haynie JL, Bryant PJ. Development of the eye-antenna imaginal disc and morphogenesis of the adult head in Drosophila melanogaster. J Exp Zool. 1986;237:293–308. doi: 10.1002/jez.1402370302. [DOI] [PubMed] [Google Scholar]
- Hazelett DJ, Bourouis M, Walldorf U, Treisman JE. decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development. 1998;125:3741–3751. doi: 10.1242/dev.125.18.3741. [DOI] [PubMed] [Google Scholar]
- Heberlein U, Borod ER, Chanut FA. Dorsoventral patterning in the Drosophila retina by wingless. Development. 1998;125:567–577. doi: 10.1242/dev.125.4.567. [DOI] [PubMed] [Google Scholar]
- Held LIJ. The eye disc. In: Held LI, editor. Imaginal Disc. Cambridge University Press; 2002. pp. 197–236. [Google Scholar]
- Hidalgo A. Growth and patterning from the engrailed interface. Int J Dev Biol. 1998;42:317–324. [PubMed] [Google Scholar]
- Irvine KD. Fringe, Notch, and making developmental boundaries. Curr Opin Genet Dev. 1999;9:434–441. doi: 10.1016/S0959-437X(99)80066-5. [DOI] [PubMed] [Google Scholar]
- Irvine KD, Wieschaus E. fringe, a Boundary-specific signaling molecule, mediates interactions between dorsal and ventral cells during Drosophila wing development. Cell. 1994;79:595–606. doi: 10.1016/0092-8674(94)90545-2. [DOI] [PubMed] [Google Scholar]
- Jaw TJ, You LR, Knoepfler PS, Yao LC, Pai CY, Tang CY, Chang LP, Berthelsen J, Blasi F, Kamps MP, Sun YH. Direct interaction of two homeoproteins, homothorax and extradenticle, is essential for EXD nuclear localization and function. Mech Dev. 2000;91:279–291. doi: 10.1016/s0925-4773(99)00316-0. [DOI] [PubMed] [Google Scholar]
- Jordan KC, Clegg NJ, Blasi JA, Morimoto AM, Sen J, Stein D, McNeill H, Deng WM, Tworoger M, Ruohola-Baker H. The homeobox gene mirror links EGF signalling to embryonic dorso-ventral axis formation through notch activation. Nat Genet. 2000;24:429–433. doi: 10.1038/74294. [DOI] [PubMed] [Google Scholar]
- Ju BG, Jeong S, Bae E, Hyun S, Carroll SB, Yim J, Kim J. Fringe forms a complex with Notch. Nature. 2000;405:191–195. doi: 10.1038/35012090. [DOI] [PubMed] [Google Scholar]
- Jurgens J, Hartenstein V. The terminal regions of body pattern. In: Martinez-Arias MBaA., editor. The Development of Drosophila melanogaster. Cold-Spring Harbor: Cold Spring Harbor Laboratory Press; 1993. pp. 687–746. [Google Scholar]
- Kehl BT, Cho KO, Choi KW. mirror, a Drosophila homeobox gene in the Iroquois complex, is required for sensory organ and alula formation. Development. 1998;125:1217–1227. doi: 10.1242/dev.125.7.1217. [DOI] [PubMed] [Google Scholar]
- Kenyon KL, Ranade SS, Curtiss J, Mlodzik M, Pignoni F. Coordinating proliferation and tissue specification to promote regional identity in the Drosophila head. Dev Cell. 2003;5:403–414. doi: 10.1016/s1534-5807(03)00243-0. [DOI] [PubMed] [Google Scholar]
- Kim BJ, Fulton AB. The genetics and ocular findings of Alagille syndrome. Semin Ophthalmol. 2007;22:205–210. doi: 10.1080/08820530701745108. [DOI] [PubMed] [Google Scholar]
- Kim J, Irvine KD, Carroll SB. Cell recognition, signal induction, and symmetrical gene activation at the dorsal-ventral boundary of the developing Drosophila wing. Cell. 1995;82:795–802. doi: 10.1016/0092-8674(95)90476-x. [DOI] [PubMed] [Google Scholar]
- Klueg KM, Muskavitch MA. Ligand-receptor interactions and trans-endocytosis of Delta, Serrate and Notch: members of the Notch signalling pathway in Drosophila. J Cell Sci. 1999;112 ( Pt 19):3289–3297. doi: 10.1242/jcs.112.19.3289. [DOI] [PubMed] [Google Scholar]
- Koshiba-Takeuchi K, Takeuchi JK, Matsumoto K, Momose T, Uno K, Hoepker V, Ogura K, Takahashi N, Nakamura H, Yasuda K, Ogura T. Tbx5 and the retinotectum projection. Science. 2000;287:134–137. doi: 10.1126/science.287.5450.134. [DOI] [PubMed] [Google Scholar]
- Kumar JP. The molecular circuitry governing retinal determination. Biochim Biophys Acta. 2009;1789:306–314. doi: 10.1016/j.bbagrm.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP. Retinal determination the beginning of eye development. Curr Top Dev Biol. 2010;93:1–28. doi: 10.1016/B978-0-12-385044-7.00001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP. My what big eyes you have: How the Drosophila retina grows. Dev Neurobiol. 2011 doi: 10.1002/dneu.20921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP, Moses K. EGF receptor and Notch signaling act upstream of Eyeless/Pax6 to control eye specification. Cell. 2001;104:687–697. doi: 10.1016/s0092-8674(01)00265-3. [DOI] [PubMed] [Google Scholar]
- Kurant E, Pai CY, Sharf R, Halachmi N, Sun YH, Salzberg A. Dorsotonals/homothorax, the Drosophila homologue of meis1, interacts with extradenticle in patterning of the embryonic PNS. Development. 1998;125:1037–1048. doi: 10.1242/dev.125.6.1037. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Morata G. Compartments in the wing of Drosophila: a study of the engrailed gene. Dev Biol. 1976;50:321–337. doi: 10.1016/0012-1606(76)90155-x. [DOI] [PubMed] [Google Scholar]
- Leyns L, Gomez-Skarmeta JL, Dambly-Chaudiere C. iroquois: a prepattern gene that controls the formation of bristles on the thorax of Drosophila. Mech Dev. 1996;59:63–72. doi: 10.1016/0925-4773(96)00577-1. [DOI] [PubMed] [Google Scholar]
- Ma C, Moses K. Wingless and patched are negative regulators of the morphogenetic furrow and can affect tissue polarity in the developing Drosophila compound eye. Development. 1995;121:2279–2289. doi: 10.1242/dev.121.8.2279. [DOI] [PubMed] [Google Scholar]
- Maurel-Zaffran C, Treisman JE. pannier acts upstream of wingless to direct dorsal eye disc development in Drosophila. Development. 2000;127:1007–1016. doi: 10.1242/dev.127.5.1007. [DOI] [PubMed] [Google Scholar]
- McClure KD, Schubiger G. Developmental analysis and squamous morphogenesis of the peripodial epithelium in Drosophila imaginal discs. Development. 2005;132:5033–5042. doi: 10.1242/dev.02092. [DOI] [PubMed] [Google Scholar]
- McLaughlin T, Hindges R, O’Leary DD. Regulation of axial patterning of the retina and its topographic mapping in the brain. Curr Opin Neurobiol. 2003;13:57–69. doi: 10.1016/s0959-4388(03)00014-x. [DOI] [PubMed] [Google Scholar]
- McNeill H, Yang CH, Brodsky M, Ungos J, Simon MA. mirror encodes a novel PBX-class homeoprotein that functions in the definition of the dorsal-ventral border in the Drosophila eye. Genes Dev. 1997;11:1073–1082. doi: 10.1101/gad.11.8.1073. [DOI] [PubMed] [Google Scholar]
- Meinhardt H. Cell determination boundaries as organizing regions for secondary embryonic fields. Dev Biol. 1983:375–385. doi: 10.1016/0012-1606(83)90175-6. [DOI] [PubMed] [Google Scholar]
- Milner M, Bleasby A, Pyott A. The role of the peripodial membrane in the morphogenesis of the eye antennal disc of Drosophila melanogaster. Roux’s Arch Dev Biol. 1983;192:164–170. doi: 10.1007/BF00848686. [DOI] [PubMed] [Google Scholar]
- Morgan TH, Bridges CB, Strutevant AH. The genetics of Drosophila. Bibliog Genet. 1925;2:1–262. [Google Scholar]
- Morrison CM, Halder G. Characterization of a dorsal-eye Gal4 Line in Drosophila. Genesis. 2010;48:3–7. doi: 10.1002/dvg.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskow JJ, Bullrich F, Huebner K, Daar IO, Buchberg AM. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mui SH, Hindges R, O’Leary DD, Lemke G, Bertuzzi S. The homeodomain protein Vax2 patterns the dorsoventral and nasotemporal axes of the eye. Development. 2002;129:797–804. doi: 10.1242/dev.129.3.797. [DOI] [PubMed] [Google Scholar]
- Murali D, Yoshikawa S, Corrigan RR, Plas DJ, Crair MC, Oliver G, Lyons KM, Mishina Y, Furuta Y. Distinct developmental programs require different levels of Bmp signaling during mouse retinal development. Development. 2005;132:913–923. doi: 10.1242/dev.01673. [DOI] [PubMed] [Google Scholar]
- Nellen D, Burke R, Struhl G, Basler K. Direct and long-range action of a DPP morphogen gradient. Cell. 1996;85:357–368. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- Netter S, Fauvarque MO, Diez del Corral R, Dura JM, Coen D. white+ transgene insertions presenting a dorsal/ventral pattern define a single cluster of homeobox genes that is silenced by the polycomb-group proteins in Drosophila melanogaster. Genetics. 1998;149:257–275. doi: 10.1093/genetics/149.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann CJ, Nuesslein-Volhard C. Patterning of the zebrafish retina by a wave of sonic hedgehog activity. Science. 2000;289:2137–2139. doi: 10.1126/science.289.5487.2137. [DOI] [PubMed] [Google Scholar]
- Noro B, Culi J, McKay DJ, Zhang W, Mann RS. Distinct functions of homeodomain-containing and homeodomain-less isoforms encoded by homothorax. Genes Dev. 2006;20:1636–1650. doi: 10.1101/gad.1412606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Elkahloun AG, Pike BL, Okajima K, Krantz ID, Genin A, Piccoli DA, Meltzer PS, Spinner NB, Collins FS, Chandrasekharappa SC. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- Okajima T, Irvine KD. Regulation of notch signaling by o-linked fucose. Cell. 2002;111:893–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- Oros SM, Tare M, Kango-Singh M, Singh A. Dorsal eye selector pannier (pnr) suppresses the eye fate to define dorsal margin of the Drosophila eye. Dev Biol. 2010;346:258–271. doi: 10.1016/j.ydbio.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiro N, Takahashi R, Yoshino K, Tanimura K, Nakashima A, Eguchi S, Miyamoto T, Hara K, Takehana K, Avruch J, Kikkawa U, Yonezawa K. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J Biol Chem. 2007;282:20329–20339. doi: 10.1074/jbc.M702636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CY, Kuo TS, Jaw TJ, Kurant E, Chen CT, Bessarab DA, Salzberg A, Sun YH. The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, extradenticle, and suppresses eye development in Drosophila. Genes Dev. 1998;12:435–446. doi: 10.1101/gad.12.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Rubin GM. Targeted expression of teashirt induces ectopic eyes in Drosophila. Proc Natl Acad Sci U S A. 1998;95:15508–15512. doi: 10.1073/pnas.95.26.15508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papalopulu N, Kintner C. A Xenopus gene, Xbr-1, defines a novel class of homeobox genes and is expressed in the dorsal ciliary margin of the eye. Dev Biol. 1996;174:104–114. doi: 10.1006/dbio.1996.0055. [DOI] [PubMed] [Google Scholar]
- Papayannopoulos V, Tomlinson A, Panin VM, Rauskolb C, Irvine KD. Dorsal-ventral signaling in the Drosophila eye. Science. 1998;281:2031–2034. doi: 10.1126/science.281.5385.2031. [DOI] [PubMed] [Google Scholar]
- Penton A, Hoffmann FM. Decapentaplegic restricts the domain of wingless during Drosophila limb patterning. Nature. 1996;382:162–164. doi: 10.1038/382162a0. [DOI] [PubMed] [Google Scholar]
- Peters MA. Patterning the neural retina. Curr Opin Neurobiol. 2002;12:43–48. doi: 10.1016/s0959-4388(02)00288-x. [DOI] [PubMed] [Google Scholar]
- Peters MA, Cepko CL. The dorsal-ventral axis of the neural retina is divided into multiple domains of restricted gene expression which exhibit features of lineage compartments. Dev Biol. 2002;251:59–73. doi: 10.1006/dbio.2002.0791. [DOI] [PubMed] [Google Scholar]
- Pichaud F, Casares F. homothorax and iroquois-C genes are required for the establishment of territories within the developing eye disc. Mech Dev. 2000;96:15–25. doi: 10.1016/s0925-4773(00)00372-5. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Zipursky SL. Induction of Drosophila eye development by decapentaplegic. Development. 1997;124:271–278. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- Poulson DF. Histogenesis, oogenesis, and differentiation in the embryo of Drosophila melanogaster meigen. In: Demerec M, editor. Biology of Drosophila. New York: Wiley; 1950. pp. 168–274. [Google Scholar]
- Ramain P, Heitzler P, Haenlin M, Simpson P. pannier, a negative regulator of achaete and scute in Drosophila, encodes a zinc finger protein with homology to the vertebrate transcription factor GATA-1. Development. 1993;119:1277–1291. doi: 10.1242/dev.119.4.1277. [DOI] [PubMed] [Google Scholar]
- Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- Roignant JY, Legent K, Janody F, Treisman JE. The transcriptional co-factor Chip acts with LIM-homeodomain proteins to set the boundary of the eye field in Drosophila. Development. 2010;137:273–281. doi: 10.1242/dev.041244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant JY, Treisman JE. Pattern formation in the Drosophila eye disc. Int J Dev Biol. 2009;53:795–804. doi: 10.1387/ijdb.072483jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo HD, Marty T, Casares F, Affolter M, Mann RS. Regulation of Hox target genes by a DNA bound Homothorax/Hox/Extradenticle complex. Development. 1999;126:5137–5148. doi: 10.1242/dev.126.22.5137. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Zipursky SL. Design principles of insect and vertebrate visual systems. Neuron. 2010;66:15–36. doi: 10.1016/j.neuron.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Tomlinson A. Dorsal-ventral midline signaling in the developing Drosophila eye. Development. 2007;134:659–667. doi: 10.1242/dev.02786. [DOI] [PubMed] [Google Scholar]
- Singh A, Chan J, Chern JJ, Choi KW. Genetic interaction of Lobe with its modifiers in dorsoventral patterning and growth of the Drosophila eye. Genetics. 2005a;171:169–183. doi: 10.1534/genetics.105.044180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Choi KW. Initial state of the Drosophila eye before dorsoventral specification is equivalent to ventral. Development. 2003;130:6351–6360. doi: 10.1242/dev.00864. [DOI] [PubMed] [Google Scholar]
- Singh A, Kango-Singh M, Choi KW, Sun YH. Dorso-ventral asymmetric functions of teashirt in Drosophila eye development depend on spatial cues provided by early DV patterning genes. Mechanisms of Development. 2004;121:365–370. doi: 10.1016/j.mod.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Singh A, Kango-Singh M, Sun YH. Eye suppression, a novel function of teashirt, requires Wingless signaling. Development. 2002a;129:4271–4280. doi: 10.1242/dev.129.18.4271. [DOI] [PubMed] [Google Scholar]
- Singh A, Kango-Singh M, Sun YH. Eye suppression, a novel function of teashirt, requires Wingless signaling. Development. 2002b;129:4271–4280. doi: 10.1242/dev.129.18.4271. [DOI] [PubMed] [Google Scholar]
- Singh A, Lim J, Choi K-W. Dorso-ventral boundary is required for organizing growth and planar polarity in the Drosophila eye. In: Mlodzik M, editor. Planar Cell Polarization during Development: Advances in Developmental Biology and Biochemistry. Elsevier Science & Technology Books; 2005b. pp. 59–91. [Google Scholar]
- Singh A, Shi X, Choi KW. Lobe and Serrate are required for cell survival during early eye development in Drosophila. Development. 2006;133:4771–4781. doi: 10.1242/dev.02686. [DOI] [PubMed] [Google Scholar]
- Speicher SA, Thomas U, Hinz U, Knust E. The Serrate locus of Drosophila and its role in morphogenesis of the wing imaginal discs: control of cell proliferation. Development. 1994;120:535–544. doi: 10.1242/dev.120.3.535. [DOI] [PubMed] [Google Scholar]
- Stevens KE, Mann RS. A balance between two nuclear localization sequences and a nuclear export sequence governs extradenticle subcellular localization. Genetics. 2007;175:1625–1636. doi: 10.1534/genetics.106.066449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G. A blastoderm fate map of compartments and segments of the Drosophila head. Dev Biol. 1981;84:386–396. doi: 10.1016/0012-1606(81)90407-3. [DOI] [PubMed] [Google Scholar]
- Sun X, Artavanis-Tsakonas S. The intracellular deletions of Delta and Serrate define dominant negative forms of the Drosophila Notch ligands. Development. 1996;122:2465–2474. doi: 10.1242/dev.122.8.2465. [DOI] [PubMed] [Google Scholar]
- Sun YH, Tsai CJ, Green MM, Chao JL, Yu CT, Jaw TJ, Yeh JY, Bolshakov VN. White as a reporter gene to detect transcriptional silencers specifying position-specific gene expression during Drosophila melanogaster eye development. Genetics. 1995;141:1075–1086. doi: 10.1093/genetics/141.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen H, Haerry TE, O’Connor MB, Marsh JL. Developmental territories created by mutual antagonism between Wingless and Decapentaplegic. Development. 1996;122:3939–3948. doi: 10.1242/dev.122.12.3939. [DOI] [PubMed] [Google Scholar]
- Treisman JE, Rubin GM. wingless inhibits morphogenetic furrow movement in the Drosophila eye disc. Development. 1995;121:3519–3527. doi: 10.1242/dev.121.11.3519. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Segment polarity and DV patterning gene expression reveals segmental organization of the Drosophila brain. Development. 2003;130:3607–3620. doi: 10.1242/dev.00532. [DOI] [PubMed] [Google Scholar]
- Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Wang YH, Huang ML. Reduction of Lobe leads to TORC1 hypoactivation that induces ectopic Jak/STAT signaling to impair Drosophila eye development. Mech Dev. 2009;126:781–790. doi: 10.1016/j.mod.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Wawersik S, Maas RL. Vertebrate eye development as modeled in Drosophila. Hum Mol Genet. 2000;9:917–925. doi: 10.1093/hmg/9.6.917. [DOI] [PubMed] [Google Scholar]
- Wilson C, Pearson RK, Bellen HJ, O’Kane CJ, Grossniklaus U, Gehring WJ. P-element-mediated enhancer detection: an efficient method for isolating and characterizing developmentally regulated genes in Drosophila. Genes Dev. 1989;3:1301–1313. doi: 10.1101/gad.3.9.1301. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. Pattern formation in the Drosophila retina. In: Martinez-Arias MBaA., editor. The Development of Drosophila melanogaster. Cold-Spring Harbor: Cold Spring Harbor Laboratory Press; 1993. pp. 1277–1325. [Google Scholar]
- Xue Y, Gao X, Lindsell CE, Norton CR, Chang B, Hicks C, Gendron-Maguire M, Rand EB, Weinmaster G, Gridley T. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum Mol Genet. 1999;8:723–730. doi: 10.1093/hmg/8.5.723. [DOI] [PubMed] [Google Scholar]
- Yamamoto D. Architecture of the Adult Compound eye and the Developing eye Disks. In: Yamamoto D, editor. Molecular Dynamics in the developing drosophila eye. Austin, Texas: Chapman and Hall; 1996. p. 169. [Google Scholar]
- Yang CH, Simon MA, McNeill H. mirror controls planar polarity and equator formation through repression of fringe expression and through control of cell affinities. Development. 1999;126:5857–5866. doi: 10.1242/dev.126.24.5857. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Hartenstein V. The role of the tracheae and musculature during pathfinding of Drosophila embryonic sensory axons. Dev Biol. 1993;158:430–447. doi: 10.1006/dbio.1993.1201. [DOI] [PubMed] [Google Scholar]