Abstract
Quorum sensing (QS) systems play global regulatory roles in bacterial virulence. They synchronize the expression of multiple virulence factors and they control and modulate bacterial antibiotic tolerance systems and host defense mechanisms. Therefore, it is important to obtain knowledge about QS modes of action and to test putative therapeutics that may interrupt QS actions in the context of infections. This chapter describes methods to study bacterial pathogenesis in murine acute and persistent/relapsing infection models, using the Gram-negative bacterial pathogen Pseudomonas aeruginosa as an example. These infection models can be used to probe bacterial virulence functions and in mechanistic studies, as well as for the assessment of the therapeutic potential of antibacterials, including anti-virulence agents.
Keywords: Lung infection, Abdominal burn infection, Open wound infection, Back burn infection, Antibiotic tolerance, Persistence, Pseudomonas aeruginosa
1. Introduction
Pseudomonas aeruginosa is a widespread opportunistic human pathogen responsible for acute and chronic/persistent infections, primarily in patients with immuno-compromising conditions (e.g., HIV infection, cancer, and diabetes), cystic fibrosis (CF), and trauma [1, 2]. Furthermore, P. aeruginosa infections have been exhibiting increasing recalcitrance to all available antibiotics [3].
Clinically relevant model systems that allow the assessment of virulence factors, their actions, and new therapeutics are needed. Notably, there are two distinct clinical syndromes of P. aeruginosa lung infection that require different models. In ventilated patients, P. aeruginosa infection causes an acute pneumonia with a high mortality rate [4]. Meanwhile, in patients with CF, P. aeruginosa infection is the primary cause of chronic inflammation, which is a key factor in the progression of CF lung disease [5]. In addition, serious burn injuries result in immunosuppression that predisposes affected patients to opportunistic nosocomial infections. In this context, P. aeruginosa infection, in particular, is feared due to its high mortality and pervasiveness worldwide [6–8]. Indeed, most deaths in severely burn-injured patients are due to burn wound sepsis. Immunocompromised patients, including burn patients, are also at risk for developing sepsis secondary to pneumonia and catheter-related infections [9].
P. aeruginosa infections are facilitated by a wide array of virulence factors that impact various stages of the infection process, host defenses, and host metabolic systems. Many of these factors are regulated by three major quorum sensing (QS) regulators, namely LasR, RhlR, and MvfR [10–13]. Accordingly, QS has been the focus of extensive mechanistic and therapeutic studies over the past 20 years [10, 14–16]. Several animal models have been developed and used in order to evaluate these findings in vivo in the context of mammalian infections [17, 18].
In this chapter, we describe five clinically relevant murine infection models that are used to assess the role of biological pathways in acute or persistent P. aeruginosa infections. These models provide a means of evaluating antibacterial, anti-virulence, or anti-persister drugs in vivo, a prerequisite to move forward in the discovery of drugs for the treatment of multidrug resistant P. aeruginosa infections, which are currently lacking.
The first model simulates a soft-tissue invasive wound infection [19]. It consists of an abdominal full-skin thickness burn generated with heated brass plugs, wherein the underlying rectus abdominus muscle is left uninjured, followed by local P. aeruginosa inoculation at the burn eschar site. This kind of burn injury disrupts the skin barrier and skin vascularization, dampens re-epithelization of the basal dermal tissue, and promotes systemic disturbances that lead to immune suppression [20, 21]. The risk of subsequent burn wound infection and systemic infection may correlate with the size of the burn injury [22, 23]. This full-skin thickness burn injury model has been used extensively to assess the role of the MvfR QS system in P. aeruginosa virulence as well as the therapeutic potential of anti-QS inhibitors [11, 24–26].
Recently, we adapted the aforementioned abdominal burn and infection model for studies of bacterial antibiotic tolerance and persistence [25]. Antibiotic tolerance—defined as the ability of a fraction of an antibiotic-susceptible bacterial population to survive exposure to normally lethal concentrations of bactericidal antibiotics—was demonstrated to be regulated by QS [12, 25, 27, 28]. The clinical importance of bacterial antibiotic tolerance is reflected by many cases in which antibiotics fail to clear infections despite the absence of resistant bacteria. Furthermore, clinical reports suggest that the contribution of bacterium tolerance to treatment failure and mortality in some patients with infections can be as significant as that of antibiotic resistance. The murine persistent/relapsing full-skin thickness burn injury model utilizes a short-term antibiotic treatment postinfection to allow assessment of antibiotic tolerant cells that survive antibiotic killing, repopulate the infected tissues, and thus resume infection following antibiotic cessation. Recently, this model was used to examine the therapeutic potential of P. aeruginosa antibiotic tolerance inhibitors [25].
The third model simulates an invasive infection of large-area burn wounds [30% total body surface area, (TBSA)]. In this model, animals receive a burn injury on the back followed by intradermal injection of the bacterial inoculum. This model has been used widely in burn infection studies, including bacterial translocation [29], gene therapy, and antibiotic efficacy studies [9].
The fourth model discussed herein models acute lung infection in a manner considered to be clinically relevant to pneumonia and potentially CF. Typically, pathophysiological changes in the lung due to P. aeruginosa infection include micro-abscesses with focal hemorrhage and the formation of bacteria filled necrotic foci throughout the lung parenchyma [30, 31]. In this lung infection mouse model, bacterial inoculum is administrated via a simple-to-administer intranasal route. Consequently, it has been used extensively in studies of acute pneumonia examining the biological pathways of various pathogens, as well as the therapeutic potential of antibacterial agents [25, 32–34].
Finally, the fifth model mimics P. aeruginosa open wound infection. It is highly clinically important given that P. aeruginosa can be found in about half of all human chronic wounds [35]. In these wounds, pathogens persist in adhesive, polymeric matrix biofilm communities, which induce chronic inflammation that delays healing and increases antimicrobial tolerance [36]. In this model, mice receive a full thickness excisional wound on the back, into the center of which bacterial cells are applied. The wound is then covered with Tegaderm film to prevent secondary infections. This model is used to investigate the virulence and QS properties of pathogens as well as the efficacy of combined topical and systemic antibiotic prophylaxis in experimental wound infection studies [17, 37–39].
2. Materials
2.1. Bacterial Inoculum Preparation
LB-Lennox liquid medium: 10 g/l tryptone, 5 g/l yeast extract, 5 g/l NaCl. Sterilize by filtration.
LB-Lennox agar plates: LB-Lennox liquid medium plus 15 g/l agar. Sterilize by autoclaving.
Incubator set at 37 °C for incubation of agar plates.
Shaker incubator set at 37 °C 200 rpm for bacterial culture growth.
1.5 and 15 ml bacterial culture glass tubes.
Spectrophotometer to measure OD600nm of bacterial cultures.
Centrifuge with a rotor for 1.5 ml tubes.
Filter-sterilized 10 mM MgSO4 solution.
2.2. Animal Infection
2.2.1. Core Material Required for Every Infection Model
6-week-old CD1 male mice weighing 22–25 g (Charles River Laboratories).
Anesthesia solution: 6.25 mg/ml ketamine and 0.625 mg/ml xylazine in sterile saline.
0.5 ml insulin syringes.
Standard dissection tools and sterile alcohol pads.
5 ml plastic tubes for tissue collection.
Tissue homogenizer (Brinkman Polytron PT3000).
5 ml round bottom polystyrene tubes.
LB-Lennox agar plates containing 100 μg/ml rifampicin.
2.2.2. Additional Material Required for the Burn-Infection Model
Electric hair clipper.
Hair remover cream.
Heating plate, metal bowl, and brass plugs (see Fig. 1a, c).
Fig. 1.
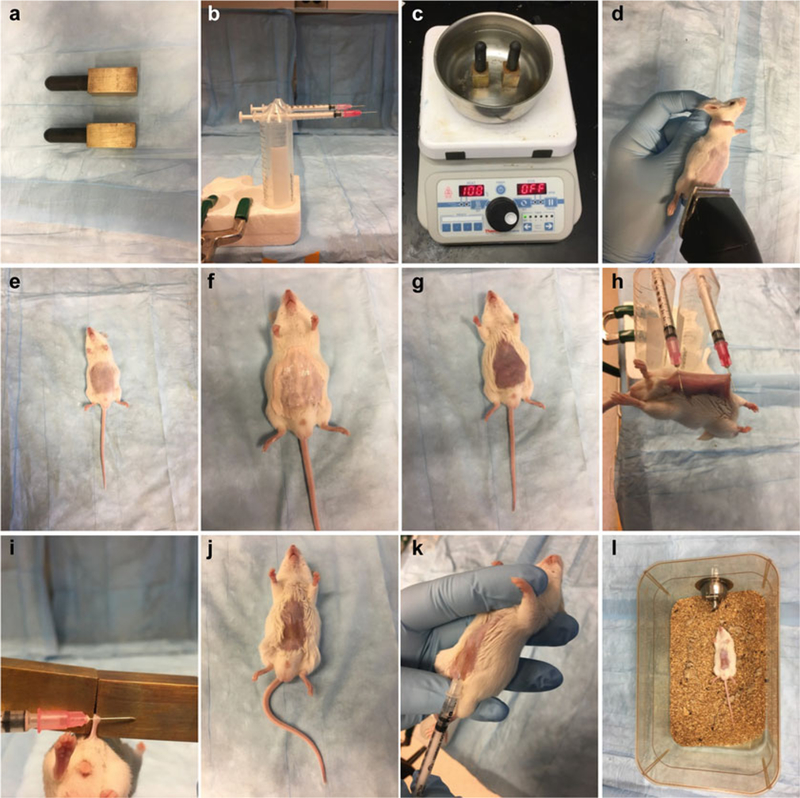
Burn and infection model. Materials used in this model (a–c). Mice are shaved (d, e), depilated (f, g), given a full thickness skin burn injury (h–j), infected (k), and put back in their cages (l)
2.2.3. Additional Material Required for the Antibiotic Tolerance/Relapsing Model
Ciprofloxacin solution, 10 mg/ml.
Ciprofloxacin E-test.
2.2.4. Additional Material Required for the Back Burn Infection Model
Electric hair clipper.
Hair remover cream.
Heating plate and clear water container (Fig. 2g).
Foam template (Fig. 2d).
Fig. 2.

Back burn and infection model. Mice are shaved (a), depilated (b, c), injected with saline (d), burned (e–h), infected (i), and put back in their cages (j)
2.2.5. Additional Material Required for the Lung Infection Model
Metal rack (Fig. 3b).
Fig. 3.
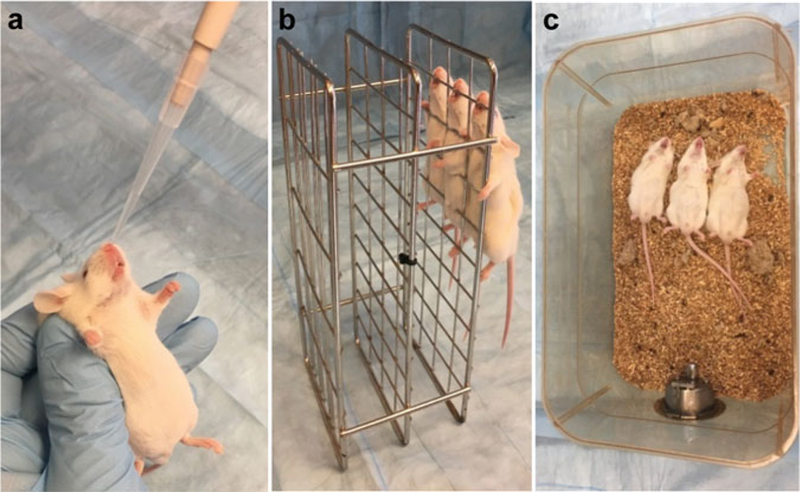
Lung infection model. After intranasal infection (a), place mice upright for a few minutes to facilitate drainage from the nostrils into the lower respiratory system (b), and then put them back in their cages (c)
2.2.6. Additional Material Required for the Open Wound Infection Model
Electric hair clipper.
Hair remover cream.
Betadine Antiseptic Pads (Fig. 4c).
Acu-Punch 12 mm skin biopsy punch (Fig. 4a).
Skin covering film (Tegaderm™) (Fig. 4b).
Fig. 4.
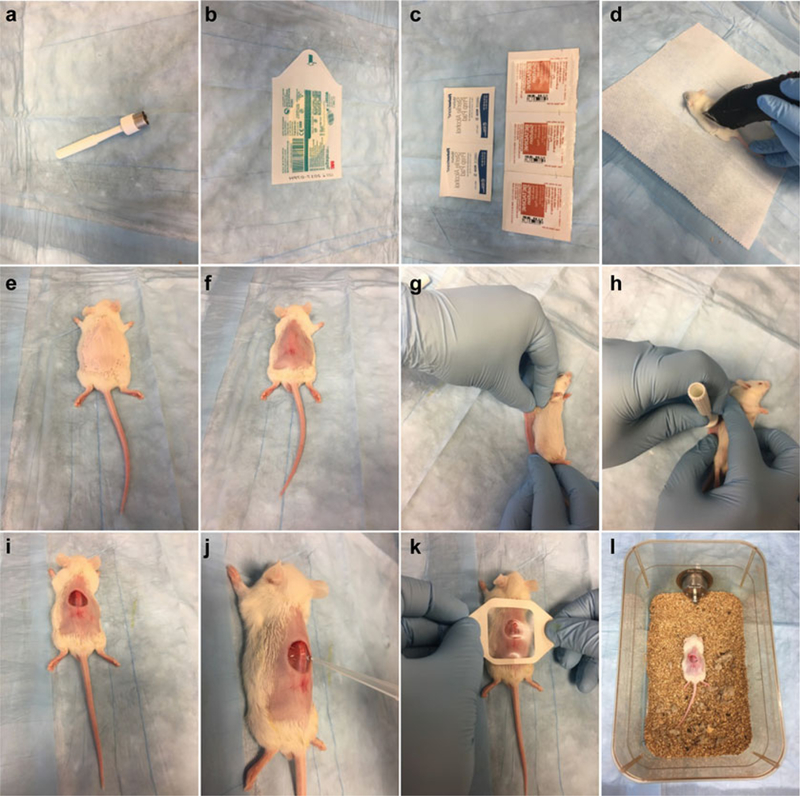
Open wound and infection model. Materials used in this model (a–c). Mice are shaved (d), depilated (e, f), wounded (g–i), infected (j), covered (k), and put back in their cages (l)
3. Methods
3.1. Burn and Infection Model
3.1.1. Bacterial Inoculum Preparation
Streak P. aeruginosa PA14 cells from a –80 °C stock onto LB agar plate and incubate the plates at 37 °C overnight.
The next day, inoculate one bacterial culture tube containing 5 ml of LB-Lennox liquid medium with one isolated colony and incubate at 37 °C, rotating at 200 rpm, until an OD600nm = 1.0 is reached.
Dilute the bacterial culture 1/1000 in LB-Lennox, then dilute 1/3 serially 20 times in 5 ml LB-Lennox and incubate the tubes at 37 °C, rotating at 200 rpm, overnight.
The next morning, select a culture tube containing cells at exactly OD600nm = 3.0 (see Note 1).
Spin down 1 ml of bacterial culture solution for 5 min at 12,000 × g, remove the supernatant, and resuspend the pellet in 10 mM MgSO4.
Dilute cells 1/20,000 in 10 mM MgSO4 to obtain 2.5 × 104 colony forming units (CFU) in 100 μl (see Note 2).
Keep the prepared bacterial cells on ice until they are used for animal infection (see Note 3).
3.1.2. Animal Infection
Perform intraperitoneal (i.p.) injection of 500 μl of a 6.25 mg/ml ketamine and 0.625 mg/ml xylazine anesthesia solution per animal (final concentration: 125 mg/kg ketamine 12.5 mg/kg xylazine). Keep anesthetized animals on a heating pad.
Shave each animal’s abdomen with an electric hair clipper (Fig. 1d, e)
Eliminate remaining fur from the abdomen by applying a coat of hair remover cream for 1 min (Fig. 1f); wipe the abdomen clean with soft tissue paper, removing any trace of the cream (Fig. 1g; see Note 4).
Place brass plugs in water and bring the water to a boil (Fig. 1c).
Lift the mouse abdominal skin and hang the animal by the skin by two syringe needles mounted parallel to the table surface (Fig. 1h; see Note 5).
Apply the brass plugs for 10 s on the mouse’s abdominal skin to produce a full thickness burn (5–8% TBSA) (Fig. 1i, j).
Following burn injury, deliver a 500 μl i.p. injection of saline for animal resuscitation.
Inject 100 μl of bacterial suspension intradermally in the burn eschar. The fold of the eschar will provide easy access (Fig. 1k).
Place the animals back in their cages lying in a supine position until they recover from the anesthesia (Fig. 1l).
For inoculum CFU assessment, serially dilute the bacterial suspension in LB and plate 100 μl of each dilution on LB agar plates. Count the CFUs after an overnight incubation at 37 °C to determine the bacterial inoculum concentration.
Assess animal survival over time (see Note 6).
At the experimentally determined assessment time points, sacrifice subgroups of animals. Dissect the rectus abdominis and pectoralis major muscles and place muscle samples into 5 ml plastic tubes for bacterial load quantification (see Note 7).
Blood samples by submandibular or cardiac puncture can be used to assess bacterial systemic dissemination serially or at the time or euthanasia. Kidney, liver, or spleen samples can be also obtained to assess systemic dissemination.
Cut the tissue samples into small pieces with dissection scissors to facilitate homogenization.
Weigh the tissue samples and add 1 ml of sterile phosphate buffered saline pH 7.4 to each sample. Keep samples in 5 ml round bottom polystyrene tubes. For blood samples, move directly to step 16.
Homogenize each sample using a Polytron blender at maximum speed for 10–15 s or until all tissue is homogenized and not tissue pieces are present in the tip of the blade.
Serially dilute the sample homogenates in phosphate buffered saline and plate 100 μl of the dilutions on LB agar plates containing 100 μg/ml rifampicin. Count the CFUs after an overnight incubation at 37 °C to determine the bacterial concentration in each sample (see Notes 8 and 9).
3.1.3. Adaptation of the Burn and Infection Model for Assessment of Antibiotic Tolerance
Infect animals with a bacterial inoculum of 8 × 103 CFUs (rather than 2.5 × 104 CFUs) to avoid animal mortality and permit long-term infection assessment following the procedures described in Subheadings 3.1.1 and 3.1.2. A 1/62,500 dilution of bacterial cells is required instead of 1/20,000 as described in Subheading 3.1.1, step 6.
Separate infected mice into two groups: one in which mice are given 10 mg/kg ciprofloxacin twice a day, and the other to serve as an untreated control group. These groups can be subdivided to assess the efficacy of antibiotic tolerance inhibitors [25].
At 6, 24, and 32 h postinfection, inject 10 mg/kg (50 μl of 5 mg/ml) ciprofloxacin into the tail vein.
At 48 h postinfection, sacrifice five animals per group and collect rectus abdominis and pectoralis major muscle samples.
Process the samples as described in Subheading 3.1.2, steps 11–16.
At 48 and 56 h postinfection, inject 10 mg/kg ciprofloxacin as above.
Continue ciprofloxacin injections twice a day and muscle sampling once a day to assess CFU as described above until no bacterial cells are detected in the muscle samples (see Note 10).
Stop antibiotic treatment as soon as PA14 cells are no longer detectable in the muscle samples.
48 h post-ciprofloxacin treatment arrest and every 2 days there-after, collect muscle samples and quantitate PA14 cells as described above.
Check the ciprofloxacin minimal antibiotic concentration (MIC) on bacterial colonies that emerge after ciprofloxacin treatment arrest using E-test to confirm that cells are antibiotic tolerant (unchanged MIC) rather than antibiotic resistant (increased MIC).
A representative example of PA14 cells killing by ciprofloxacin and relapsing infection caused by antibiotic tolerant PA14 cells can be seen in [25].
3.2. Back Burn Infection Model
3.2.1. Bacterial Inoculum Preparation
For the back burn infection model, prepare the bacterial inoculum using the procedure described in Subheading 3.1.1, except with a dose of 2.5 × 105 CFUs per 100 μl. Accordingly, a 1/2500 dilution of the OD600nm = 3 bacterial cells will be used.
3.2.2. Animal Infection
Anesthetize the animals as described in Subheading 3.1.2, step 1.
Administer the analgesic buprenorphine via i.p. injection (0.05–0.10 mg/kg) while the animals are anesthetized.
Shave an area of skin on each mouse’s back with mouse fur clippers (Fig. 2a).
Depilate with depilatory cream for 1 min (Fig. 2b), then wipe the abdomen clean with soft tissue paper to remove any trace of cream (Fig. 2c).
Calculate TBSAusing Meehs formula (A = k × W(2/3), where A = surface area in cm2; k = proportionality constant 12.3; W = weight in gram).
Inject 0.5 ml of saline [0.9% (wt/vol) NaCl in distilled water] subcutaneously beside the designated burn site before inflicting the back burn to facilitate resuscitation and protect the spinal cord from burn damage (Fig. 2d).
Lay each animal in a supine position on a foam template of an appropriate size, such that the skin on the animal’s back and sides that protrudes from the template equals 30% TBSA based on the calculation (Fig. 2e, f).
Immerse the back of the animal in hot water (90 °C) for 8 s while being careful not to expose its head or limbs to the hot water; then dry the burned area gently with paper towels (Fig. 2g; see Note 11).
Resuscitate each mouse with another 0.5 ml of saline, injected i.p. while the animal is still under anesthesia.
Inject 100 μl of bacterial suspension intradermally immediately underneath the burn eschar (Fig. 2i).
Place the animals back into their cages, making sure to lay them on their abdomens, while they recover from the anesthesia (Fig. 2j).
Assess the bacterial inoculum concentration as described in Subheading 3.1.2, step 8.
Record animal survival over time (see Note 12).
At the experimentally determined time points, sacrifice subgroups of animals and collect muscle from the wound for bacterial load quantification.
Process the animal tissue samples as described in Subheading 3.1.2, steps 11–16.
3.3. Lung Infection Model
3.3.1. Bacterial Inoculum Preparation
For the lung infection model, prepare the bacterial inoculum according to the procedure in Subheading 3.1.1, except with an inoculum dose of 5 × 106 CFUs per 20 μl. Accordingly, a 1/20 dilution of the OD600nm = 3 bacterial cells is required.
3.3.2. Animal Infection
Anesthetize the animals as described in Subheading 3.1.2, step 1.
Administer 20 μl of bacterial suspension slowly into the mouse nose with a micropipette, ~10 μl in each nostril (Fig. 3a; see Note 13).
Hold the animals upright for 2 min by placing them vertically on a metal rack to allow the inoculum to pass the sinuses and reach the lungs (Fig. 3b).
Place the animals back into their cages, lying on their backs, and allow them to recover from the anesthesia (Fig. 3c).
Assess the bacterial inoculum concentration as described in Subheading 3.1.2, step 8.
Record animal survival over time (see Note 14).
At experimentally determined time points, sacrifice subgroups of animals. Collect the lungs, and any other tissues of interest, for bacterial load quantification.
Process the animal tissue samples as described in Subheading 3.1.2, steps 11–16. Both lungs from each animal should be pooled and homogenized together.
3.4. Open Wound Infection Model
3.4.1. Bacterial Inoculum Preparation
Prepare the bacterial inoculum according to the procedure in Subheading 3.1.1, except with a dose of 2.5 × 106 CFUs per 10 μl. Use a 1/20 dilution of bacterial cells for the open wound infection model.
3.4.2. Animal Infection
Anesthetize the animals as described in Subheading 3.1.2, step 1.
Shave target area of skin on the back with mouse fur clippers (Fig. 4d).
Depilate with depilatory cream for 1 min (Fig. 4e), then wipe the abdomen clean with soft tissue paper to remove any trace of cream (Fig. 4f).
Administer 0.1 mg/kg buprenorphine by i.p. injection right before the wounding procedure and once daily for up to 3 days.
Sterilize the skin with iodine and alcohol swabs.
Lay the mouse on its side and lift skin from the back, pushing it down onto the working surface (Fig. 4g).
Make an excision on the mouse’s back with a skin biopsy punch (12 mm diameter) on top of a sterile sheet (Fig. 4h, i; see Note 15).
Inoculate 10 μl of bacterial suspension into the center of the wound (Fig. 4j).
Allow the suspension to be absorbed and cover the wound with a 4 cm × 4 cm square Tegaderm film (Fig. 4k).
Place the animals back in their cages, making sure to lay them on their abdomens, and allow them to recover from the anesthesia (Fig. 4l).
Assess the bacterial inoculum concentration as described in Subheading 3.1.2, step 8.
Record animal survival over time (see Note 16).
At experimentally determined time points, sacrifice subgroups of animals and collect muscle tissue from the wound site, blood through cardiac puncture, and, if indicated experimentally, other tissues for bacterial load quantification.
Process the animal tissue samples as described in Subheading 3.1.2, steps 11–16.
4. Notes
The OD600nm of the bacterial culture used to prepare the infection inoculum is critical because it ensures that an appropriate number of bacterial cells are present at a defined cell metabolic stage, which is optimal for the infection process. For these models, it is very important that cultures with an OD600nm = 3.0 are used. Assess the OD600nm by diluting culture (usually 1:10) to accurately measure cell density within the spectrophotometer’s reading limit.
For the PA14 strain, an OD600nm = 3.0 culture grown in LB-Lennox at 37 °C rotating at 200 rpm contains 5 × 109 CFU/ml. For other strains, the CFU quantity at OD600nm = 3.0 may be slightly different and needs to be taken into consideration in the inoculum preparation.
Perform all animal infections within 2 h of cell preparation.
Make sure to wipe each mouse’s abdomen thoroughly first with a soft dry tissue and then with a wet one to remove any trace of cream, which can interfere with the burn step and inflict a chemical burn.
When lifting the skin, be sure to lift only the skin to ensure that the underlying muscle and internal organs are not punctured by the needles or burned in the following step.
In the burn infection model, an inoculum of 2.5 × 104 CFU induces ~50% mortality. Most animals succumb to infection between 32 and 56 h. Moribund animals should be euthanized. The moribundity is defined as when the animal shows (1) difficulty with ambulation or reluctant to move when given stimuli, or at least three of the following five symptoms known to be associated with infection morbidities: (2) abnormal posture, (3) ruffled hair coat, (4) head-tucked into abdomen, (5) exudate around eyes or closed eyes, and (6) abnormal breathing.
Samples obtained from the rectus abdominis muscle area underlying the burn site allow assessment of bacterial invasion at the most proximal site ofinfection, whereas samples from the pectoralis major muscle adjacent to the infection site enable assessment of bacterial dissemination from the inoculation site via the bloodstream to other tissues, including kidney, spleen, and liver.
LB agar plates containing antibiotic (i.e., rifampicin for PA14) are required to determine the exact number of PA14 cells present in each sample and to prevent the growth of other bacteria. The particular antibiotic used should be selected based on the strain used. Pseudomonas isolation agar can be used instead of LB agar plates containing antibiotic.
Express bacterial cell concentrations in CFU per gram of tissue to take into account the variation in the amount of tissue in each sample.
Usually, it takes 4–5 days to clear antibiotic sensitive bacterial cells from the animals.
Apply EMLA cream to the margin of burn wounds in burn-infection model animals at least once after the procedure.
In the back burn infection model, an inoculum of 2.5 × 105 CFU induces ~50% mortality. Most animals succumb to the infection within 24–72 h. Moribund animals should be euthanized.
Make sure to administer the bacterial suspension very slowly and gingerly to avoid coughing or suffocation of the animal. Note that this is a very delicate step.
In this lung acute infection model, an inoculum of 5 × 106 CFU induces ~50% mortality. Most animals start succumbing infection by 24 h postinfection and continue up to 48 h. Moribund animals should be euthanized.
When inflicting the wound, take care to avoid injuring the panniculus carnosus. The excision will measure roughly 1 cm2. Give each mouse a single wound.
In the open wound infection model, an inoculum of 2.5 × 106 CFU induces ~50% mortality. Most animals succumb the infection between 48 and 96 h. Moribund animals should be euthanized.
References
- 1.Gellatly SL, Hancock RE (2013) Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis 67:159–173 [DOI] [PubMed] [Google Scholar]
- 2.Kerr KG, Snelling AM (2009) Pseudomonas aeruginosa: a formidable and ever-present adversary. J Hosp Infect 73:338–344 [DOI] [PubMed] [Google Scholar]
- 3.Lister PD, Wolter DJ, Hanson ND (2009) Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crouch Brewer S, Wunderink RG, Jones CB, Leeper KV Jr (1996) Ventilator-associated pneumonia due to Pseudomonas aeruginosa. Chest 109:1019–1029 [DOI] [PubMed] [Google Scholar]
- 5.Pier GB, Ramphal R (2004) Pseudomonas aeruginosa In: Mandell GL, Bennet JE, Dolin R (eds) Principles and practise of infectious diseases, vol 6 Elsevier, Amsterdam, pp 2587–2615 [Google Scholar]
- 6.Wibbenmeyer L, Danks R, Faucher L, Amelon M, Latenser B, Kealey GP et al. (2006) Prospective analysis of nosocomial infection rates, antibiotic use, and patterns of resistance in a burn population. J Burn Care Res 27:152–160 [DOI] [PubMed] [Google Scholar]
- 7.Azzopardi EA, Azzopardi E, Camilleri L, Villapalos J, Boyce DE, Dziewulski P et al. (2014) Gram negative wound infection in hospitalised adult burn patients-systematic review and met-analysis. PLoS One 9:e95042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lipovy B, Rihova H, Hanslianova M, Gregorova N, Suchanek I, Brychta P (2010) Prevalence and resistance of Pseudomonas aeruginosa in severely burned patients: a 10-year retrospective study. Acta Chir Plast 52:39–43 [PubMed] [Google Scholar]
- 9.Church D, Elsayed S, Reid O, Winston B, Lindsay R (2006) Burn wound infections. Clin Microbiol Rev 19:403–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venturi V (2006) Regulation of quorum sensing in Pseudomonas. FEMS Microbiol Rev 30:274–291 [DOI] [PubMed] [Google Scholar]
- 11.Deziel E, Gopalan S, Tampakaki AP, Lepine F, Padfield KE, Saucier M et al. (2005) The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol Microbiol 55:998–1014 [DOI] [PubMed] [Google Scholar]
- 12.Maura D, Hazan R, Kitao T, Ballok AE, Rahme LG (2016) Evidence for direct control of virulence and defense dene circuits by the Pseudomonas aeruginosa quorum sensing regulator, MvfR. Sci Rep 6:34083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert KB, Kim TH, Gupta R, Greenberg EP, Schuster M (2009) Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol Microbiol 73:1072–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heeb S, Fletcher MP, Chhabra SR, Diggle SP, Williams P, Camara M (2011) Quinolones: from antibiotics to autoinducers. FEMS Microbiol Rev 35:247–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maura D, Ballok AE, Rahme LG (2016) Considerations and caveats in anti-virulence drug development. Curr Opin Microbiol 33:41–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner S, Sommer R, Hinsberger S, Lu C, Hartmann RW, Empting M et al. (2016) Novel strategies for the treatment of Pseudomonas aeruginosa Infections. J Med Chem 59:5929–5969 [DOI] [PubMed] [Google Scholar]
- 17.Papaioannou E, Utari PD, Quax WJ (2013) Choosing an appropriate infection model to study quorum sensing inhibition in Pseudomonas infections. Int J Mol Sci 14:19309–19340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kukavica-Ibrulj I, Levesque RC (2008) Animal models of chronic lung infection with Pseudomonas aeruginosa: useful tools for cystic fibrosis studies. Lab Anim 42:389–412 [DOI] [PubMed] [Google Scholar]
- 19.Stevens EJ, Ryan CM, Friedberg JS, Barnhill RL, Yarmush ML, Tompkins RG (1994) A quantitative model of invasive Pseudomonas infection in burn injury. J Burn Care Rehabil 15:232–235 [DOI] [PubMed] [Google Scholar]
- 20.Murray CK (2007) Infections in burns. J Trauma 62:S73. [DOI] [PubMed] [Google Scholar]
- 21.Pruitt BA Jr, McManus AT, Kim SH, Goodwin CW (1998) Burn wound infections: current status. World J Surg 22:135–145 [DOI] [PubMed] [Google Scholar]
- 22.Coban YK (2012) Infection control in severely burned patients. World J Crit Care Med 1:94–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cochran A, Morris SE, Edelman LS, Saffle JR (2002) Systemic Candida infection in burn patients: a case-control study of management patterns and outcomes. Surg Infect (Larchmt) 3:367–374 [DOI] [PubMed] [Google Scholar]
- 24.Lesic B, Lepine F, Deziel E, Zhang J, Zhang Q, Padfield K et al. (2007) Inhibitors of pathogen intercellular signals as selective anti-infective compounds. PLoS Pathog 3:1229–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Starkey M, Lepine F, Maura D, Bandyopadhaya A, Lesic B, He J et al. (2014) Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity. PLoS Pathog 10:e1004321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bandyopadhaya A, Tsurumi A, Maura D, Jeffrey KL, Rahme LG (2016) A quorum-sensing signal promotes host tolerance training through HDAC1-mediated epigenetic reprogramming. Nat Microbiol 1:16174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Que YA, Hazan R, Strobel B, Maura D, He J, Kesarwani M et al. (2013) A quorum sensing small volatile molecule promotes antibiotic tolerance in bacteria. PLoS One 8:e80140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kayama S, Murakami K, Ono T, Ushimaru M, Yamamoto A, Hirota K et al. (2009) The role of rpoS gene and quorum-sensing system in oflox-acin tolerance in Pseudomonas aeruginosa. FEMS Microbiol Lett 298:184–192 [DOI] [PubMed] [Google Scholar]
- 29.Earley ZM, Akhtar S, Green SJ, Naqib A, Khan O, Cannon AR et al. (2015) Burn injury alters the intestinal microbiome and increases gut permeability and bacterial translocation. PLoS One 10:e0129996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang HB, DiMango E, Bryan R, Gambello M, Iglewski BH, Goldberg JB et al. (1996) Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis ofpneumonia in a neonatal mouse model of infection. Infect Immun 64:37–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Heeckeren AM, Schluchter MD, Xue W, Davis PB (2006) Response to acute lung infection with mucoid Pseudomonas aeruginosa in cystic fibrosis mice. Am J Respir Crit Care Med 173:288–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Debarbieux L, Leduc D, Maura D, Morello E, Criscuolo A, Grossi O et al. (2010) Bacteriophages can treat and prevent Pseudomonas aeruginosa lung infections. J Infect Dis 201:1096–1104 [DOI] [PubMed] [Google Scholar]
- 33.Dufour N, Clermont O, La Combe B, Messika J, Dion S, Khanna V et al. (2016) Bacteriophage LM33_P1, a fast-acting weapon against the pandemic ST131-O25b:H4 Escherichia coli clonal complex. J Antimicrob Chemother 71:3072–3080 [DOI] [PubMed] [Google Scholar]
- 34.Harris G, Kuo Lee R, Lam CK, Kanzaki G, Patel GB et al. (2013) A mouse model of Acinetobacter baumannii-associated pneumonia using a clinically isolated hypervirulent strain. Antimicrob Agents Chemother 57:3601–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gjodsbol K, Christensen JJ, Karlsmark T, Jorgensen B, Klein BM, Krogfelt KA (2006) Multiple bacterial species reside in chronic wounds: a longitudinal study. Int Wound J 3:225–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhoads DD, Wolcott RD, Percival SL (2008) Biofilms in wounds: management strategies. J Wound Care 17:502–508 [DOI] [PubMed] [Google Scholar]
- 37.Bergamini TM, Lamont PM, Cheadle WG, Polk HC Jr (1984) Combined topical and systemic antibiotic prophylaxis in experimental wound infection. Am J Surg 147:753–756 [DOI] [PubMed] [Google Scholar]
- 38.McHugh SM, Collins CJ, Corrigan MA, Hill AD, Humphreys H (2011) The role of topical antibiotics used as prophylaxis in surgical site infection prevention. J Antimicrob Chemother 66:693–701 [DOI] [PubMed] [Google Scholar]
- 39.Bandyopadhaya A, Kesarwani M, Que YA, He J, Padfield K, Tompkins R et al. (2012) The quorum sensing volatile molecule 2-amino acetophenon modulates host immune responses in a manner that promotes life with unwanted guests. PLoS Pathog 8:e1003024. [DOI] [PMC free article] [PubMed] [Google Scholar]


