Abstract
Oxylipins, or oxygenated lipids, are universal signaling molecules across all kingdoms of life. These molecules, either produced by microbial pathogens or their mammalian host, regulate inflammation during microbial infection. In this review, we summarize current literature on the biosynthesis pathways of microbial oxylipins and their biological activity towards mammalian cells. Collectively, these studies have illustrated how microbial pathogens can modulate immune response and disease outcome via oxylipin-mediated mechanisms.
1 |. INTRODUCTION
Microbes communicate with their hosts using small molecules, some unique to the microbe and some similar or even identical to host metabolites. Of the latter, oxygenated lipids called oxylipins are increasingly understood as key chemical signals that are produced and perceived by both microbe and host. These bioactive signaling molecules mediate many important biological processes, including immune homeostasis in plants and animals as well as microbial development. Despite that both microbial pathogen and the host can produce bioactive oxylipins, the role of microbial oxylipins in pathogenesis have been underappreciated. Herein we provide an overview of biosynthesis of oxylipins derived from prokaryotic and eukaryotic microorganisms that cause human infections, and the biological functions of microbial oxylipins in shaping the pathogenic relationships.
2 |. MICROBIAL OXYLIPINS: ENZYMES AND PRODUCTS
Oxylipins are oxygenated derivatives of long chain mono- or poly-unsaturated fatty acids (PUFAs). Host oxylipins are mainly synthesized with 20:4 (n-6) arachidonic acid (AA), 20:5 (n-3) eicosapentaenoic acid (EPA), and 22:6(n-3) docosahexaenoic acid (DHA). Microbial pathogens can synthesize oxylipins using these PUFAs as well as other substrates such as 18:1 (n-9) oleic acid, 18:2 (n-6) linoleic acid (LA), and 18:3 (n-6) linolenic acid. Table 1 shows the enzymes and oxylipins produced by the pathogenic microbes discussed in this review. Microorganisms possess the same classes of eicosanoids biosynthesis enzymes as mammals including cyclooxygenases, lipoxygenases, and cytochrome P450s as well as microbial specific enzymes (Figure 1). Other microbial proteins, such as phospholipases and epoxide hydrolases, don’t directly catalyze oxylipin biosynthesis but alter PUFA substrate availability and modify host oxylipins during host-microbe encounters. Although some oxylipins can be non-enzymatically derived, this review will focus on microbial enzymatic sources of these lipids.
TABLE 1.
List of enzyme-derived microbial oxylipins associated with pathogenesis and their microbial origin
| PUFA | Oxylipin name | Chemical structure | Producer species and biosynthetic enzymes | References |
|---|---|---|---|---|
| Arachidonic Acid | Prostaglandin D2 (PGD2) |
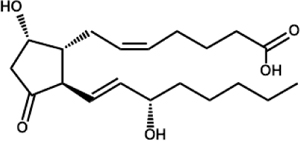 |
C. albicans C. parapsilosis (CPAR2_603600, CPAR2_800020, CPAR2_807710) T. brucei P. falciparum |
Chakraborty et al., 2018; Kubata et al., 2000; Kubata et al., 1998; Noverr et al., 2001; |
| Prostaglandin E2 (PGE2) |
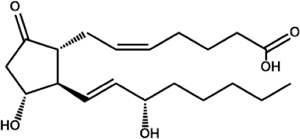 |
C. albicans (Ole2, Fet3) C. neoformans (Lac1) C. parapsilosis (CPAR2_603600, CPAR2_800020) P. brasiliensis A. fumigatus (Ppo’s) E. histolytica T. cruzi T. brucei L. mexicana P. falciparum |
Chakraborty et al., 2018; Colas et al., 2018; Dey et al., 2003; Erb-Downward et al., 2008 Erb-Downward and Noverr, 2007; Estrada-Figueroa et al., 2018; Kubata et al., 2000; Kubata et al., 1998; Noverr et al., 2001; Tsitsigiannis et al., 2005 |
|
| Prostaglandin F2α (PGF2α) | 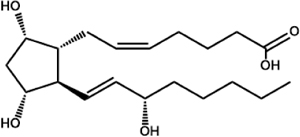 |
C. albicans
C. neoformans T. cruzi (TcOYE) T. brucei (TbPGFS) L. major, L. donovani, L. tropica L.i. chagasi P. falciparum |
Araújo-Santos et al., 2014; Ashton et al., 2007; Colas et al., 2018; Kabututu et al., 2003; Kubata et al., 2000; Kubata et al., 1998; Noverr et al., 2001; |
|
| Thromboxane A2 (TXA2) |
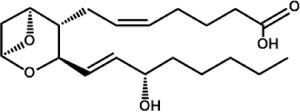 |
C. albicans T. cruzi |
Ashton et al., 2007; Noverr et al., 2001 |
|
| 15S-hydroxyeicosatetraenoic acid (15-HETE) | 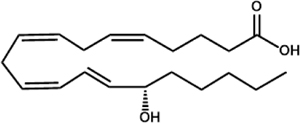 |
P. aeruginosa (LoxA) T. gondii (STAg) |
Bannenberg et al., 2004; Vance et al., 2004 |
|
| 3S-hydroxy eicosatetraenoic acid (3S-HETE) | 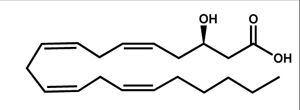 |
C. albicans | Ciccoli et al., 2005 | |
| Leukotriene B4 (LTB4) |
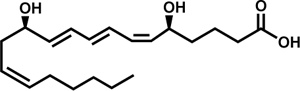 |
P. brasiliensis | Bordon et al., 2007 | |
| Lipoxin A4 (LXA4) |
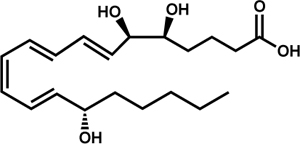 |
T. gondii | Bannenberg et al., 2004 | |
| Linoleic Acid | 13-hydroxyoctadecadienoic acid (13-HODE) | 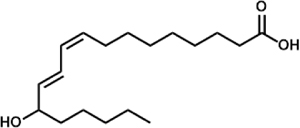 |
P. aeruginosa (LoxA) A. fumigatus (LoxB) |
Banthiya et al., 2015; Fischer et al., 2017; |
| 10R-hydroxyoctadecadienoic acid (10R-HODE) | 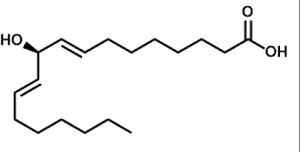 |
A. fumigatus (PpoC) A. nidulans (PpoC) A. terreus (ATEG_03992) |
Garscha et al., 2007; Hoffmann et al., 2013 | |
| 8R-hydroxyoctadecadienoic acid (8R-HODE) | 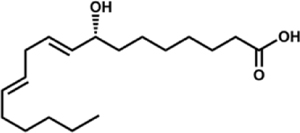 |
A. nidulans (PpoA) A. fumigatus (PpoA) C. immitis (8R-DOX-AOS) |
Garscha et al., 2007; Oliw et al., 2016; Tsitsigiannis, Zarnowski, & Keller, 2004 | |
| 5S,8R-dihydroxyoctadecadienoic acid (5S,8R-diHODE) | 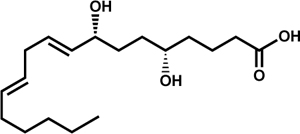 |
A. fumigatus (PpoA) | Garscha et al., 2007 | |
| Eicosapentaenoic Acid | Resolvin E1 (RvE1) | 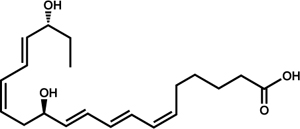 |
T. cruzi C. albicans | Colas et al., 2018; Hass-Stapleton et al., 2007 |
| Resolvin E2 (RvE2) | 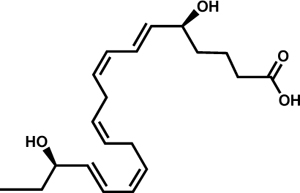 |
T. cruzi | Colas et al., 2018 | |
| Docosahexaenoic Acid | Resolvin D1 (RvD1) | 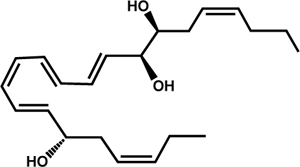 |
T. cruzi | Colas et al., 2018 |
FIGURE 1.
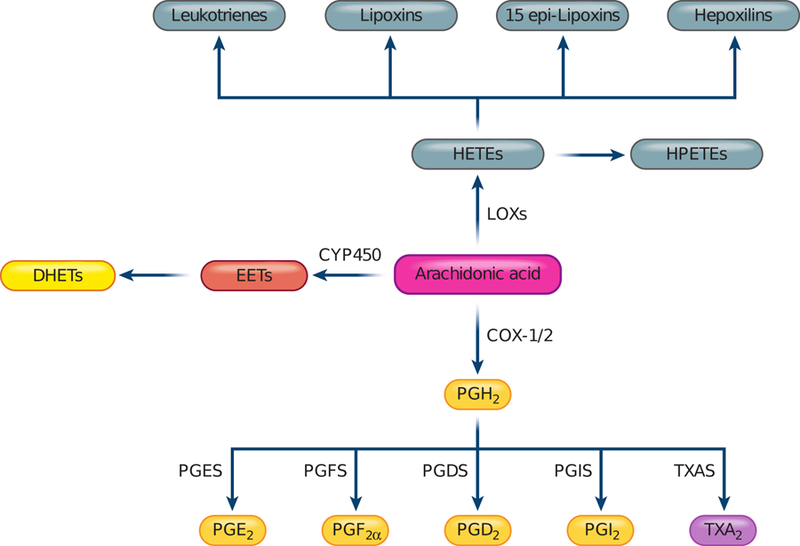
Eicosanoid biosynthesis pathways in animals. Arachidonic acid-derived eicosanoids are biosynthesized through cyclooxygenase (COX), lipoxygenase (LOX), and cytochrome P450 (CYP) pathways. Cyclooxygenases, including COX-1 and COX-2, produce prostaglandin H2 (PGH2) that is further converted by prostaglandin synthases (PGES, PGFS, PGDS), prostacyclin synthase (PGIS) and thromboxane synthase (TXAS) to prostaglandins, prostacyclin, and thromboxane respectively. Lipoxygenases, including Alox5, Alox15, Alox12, and Alox8 synthesize hydroperoxyeicosatetraenoic acids (HPETEs) that are further converted to various hydroxylated and epoxidized eicosanoids. Cytochrome P450 (CYP 450) enzymes catalyze formation of various epoxyeicosatrienoic acids (EETs), which are short-lived and rapidly converted to dihydroxyeicosatrienoic acid (DHETs). HETEs, hydroxyeicosatetraenoic acids; PGH2, prostaglandin H2; PGE2, prostaglandin E2; PGF2α, prostaglandin F2α; PGD2, prostaglandin D2; PGI2, prostacyclin; TXA2, thromboxane A2; PGES, PGE synthase; PGFS, PGF synthase; PGDS, PGD synthase; PGIS, PGI synthase; TXA, TXA synthase
2.1 |. Cyclooxygenase (COX)-like enzymes
Filamentous and dimorphic fungi contain upwards of four enzymes with significant homologies to mammalian cyclooxygenases COX-1 and COX-2 that synthesize prostaglandin H2 (PGH2), the substrate for further enzymatic steps (Figure 1). The fungal COX-like oxygenases, termed Ppo proteins, have been best studied in Aspergillus spp. including the opportunistic pathogens Aspergillus nidulans, Aspergillus flavus, Aspergillus fumigatus, and Aspergillus terreus (Garscha et al., 2007; Hoffmann, Jernerén, & Oliw, 2013; Tsitsigiannis, Zarnowski, & Keller, 2004; Table 1). Ppo enzymes produce mono- or di-hydroxyl oxylipins derived from oleic acid and linoleic acid, and PpoA and PpoC also contribute to AA-derived prostaglandin formation (Kupfahl et al., 2012; Tsitsigiannis et al., 2005; Noverr, Toews, & Huffnagle, 2002). Ppo oxygenases contain a heme-containing oxygenase domain (similar to the vertebrate COX) and a cytochrome P450 domain, thus are also termed linoleate dioxygenase-cytochrome P450. One of these enzymes has also been biochemically characterized in the dimorphic valley fever causing fungus Coccidioides immitis (Oliw et al., 2016). Other fungal pathogens, including the thermal dimorphic fungus Paracocciodiodes brasiliensis and pathogenic yeasts Candida albicans, Candida parapsilosis and Cryptococcus neoformans, can produce PGE2 and other prostaglandins, yet the responsible COX has not been identified (Bordon et al., 2007; Erb-Downward and Noverr, 2007; Erb-Downward et al., 2008; Fischer and Keller, 2016).
The intestinal amebiasis-causing agent Entameoba histolytica produces PGE2 via a nuclear COX-like protein and this production is sensitive to the non-selective COX inhibitor aspirin (Dey et al., 2003). COX activity of the protease Gp63 in the Leishmania causing Leishmania mexicana was reported, using a chemiluminescence-based kit for COX activity (Estrada-Figueroa et al., 2018). Phylogenetic analysis revealed that the protozoan COX-like proteins, including Gp63 proteins from L. mexicana and the Chaga’s disease agent Trypanosoma cruzi and the Cox-like protein of E. histolytica, are evolutionary distant from mammalian COXs (Estrada-Figueroa et al., 2018).
2.2 |. Prostaglandin synthases and thromboxane synthase
In humans, the COX product PGH2 serves as substrate for synthesizing various structurally similar oxylipins, including other prostaglandins PGD2, PGE2 and PGF2α, prostacyclin PGI2, and thromboxane TXA2 (Figure 1). Prostaglandin and thromboxane synthases were identified in microbial pathogens based on their biosynthetic property rather than homology to mammalian enzymes.
The African trypanosomiasis agent Trypanosoma brucei, the Leishmaniasis pathogen Leishmania spp., and malaria pathogen Plasmodium falciparum can produce PGD2, PGE2, and PGF2α when supplemented with AA (Araújo-Santos et al., 2014; Kubata et al., 1998; Kubata et al., 2000; Kabututu et al., 2003). Unlike the E. histolytica COX activity, biosynthesis of these prostaglandins in T. brucei is not sensitive to COX inhibitors (Kubata et al., 2000). Trypanosoma cruzi produces TXA2 as its major eicosanoid with minor amount of PGF2α (Ashton et al., 2007). PGF2α synthases were identified in T. brucei (TbPGFS), T. cruzi (TcOYE), and Leishmania spp. and reviewed in Kubata et al., (2007).
Several enzymes were found to contribute to prostaglandin production in pathogenic yeasts. C. neoformans laccase Lac1, a multicopper oxidoreductase, catalyzes conversion of PGG2 to PGE2 (Erb-Downward et al., 2008). In C. albicans, the oleate ∆9 fatty acid stearyl-coenzyme A desaturase Ole2 and the laccase homolog ferroxidase Fet3 contribute to PGE2 production (Erb-Downward and Noverr, 2007). However, Ole2 of C. parapsilosis doesn’t contribute to PGE2 biosynthesis (Grózer et al., 2015). Three C. parapsilosis genes highly induced by AA were identified through RNA sequencing: CPAR2_603600 (Fet3 homolog), CPAR2_800020 (a putative thiolase) and CPAR2_807710 (a putative Acyl-CoA dehydrogenase). The CPAR2_603600 and CPAR2_800020 proteins contribute to PGE2 and PGD2 production while the CPAR2_807710 protein contributes to PGE2 and 15-keto-PGE2 production (Chakraborty et al., 2018).
2.3 |. Lipoxygenases (LOXs)
A. fumigatus has two lipoxygenase homologs of human Alox15 and Alox5, denoted LoxA and LoxB (Heshof et al., 2014; Fischer et al., 2017). LoxB is secreted extracellularly and produces 13-hydroxyoctadecadienoic acid (13-HODE). A single Lox, the LoxB homolog is present in the carcinogenic fungus Aspergillus flavus (Horowitz Brown, 2008). Lipoxygenase activity with subsequent synthesis of leukotriene B4 (LTB4) has been reported in P. brasiliensis although no associated enzyme has been characterized (Biondo et al., 2012).
Prokaryotic LOX-encoding genes are evolutionarily distant from human LOX and have been found predominantly in Gram-negative environmental species (Hansen et al., 2013). The first bacterial LOX to be identified was LoxA in the opportunistic bacterial pathogen P. aeruginosa (Vance et al., 2004). LoxA has multiple PUFA substrates and can convert AA and LA to 15S-hydroxyeicosatetraenoic acid (15-HETE) and 13-HODE. LoxA lacks the leukotriene synthase activity and whether it synthesize lipoxin is still debatable (Banthiya et al., 2016; Deschamps et al., 2016; Vance et al., 2004). The Toxoplasmosis causing parasite Toxoplasma gondii produces the anti-inflammatory lipoxin LTA4 and the extracted soluble tachyzoites antigen (STAg) converts AA to 15-HETE (Bannenberg et al., 2004).
Biosynthesis of specialized pro-resolving mediators, such as lipoxins and resolvins, often requires multiple enzymes, sometimes in different classes (Basil and Levy, 2016). A survey of T. cruzi oxylipins revealed production of AA derived prostaglandins and HETE, EPA derived RvE2, and DHA derived RvD1 and RvD5 from lysates of free trypomastigotes, suggesting presence of enzymes required for biosynthesis (Colas et al., 2018). RvE1 can be produced by C. albicans when supplemented with EPA, suggesting that cytochrome P450 and LOX activities are present (Hass-Stapleton et al., 2007).
2.4 |. Other enzymes in oxylipin-mediated crosstalk: phospholipase and epoxide hydrolase
Human cytosolic phospholipase A2 releases AA from plasma membrane to initiate inflammation (Dennis and Norris, 2015). Microbial phospholipases are widely present, and some can cleave host AA to mediate a host response. These studies are reviewed extensively in Ghannoum (2000) and Istivan and Coloe (2006). The most extensively characterized PLA is ExoU in P. aeruginosa. ExoU, a patatin-like phospholipase and cytotoxin, is injected through the Type III secretion system to the host cell (Anderson et al., 2011; Sato et al., 2003; Sawa et al., 2016). ExoU liberates AA from eukaryotic cell membrane, an activity required for its toxicity (Saliba et al., 2005; Rabin and Hauser, 2005). Other bacterial phospholipases implicated in host interaction include Streptococcus pyogenes phospholipase A2 SlaA (Oda et al., 2017) and the ExoU homolog VipD in Legionella pneumophila (Gasper and Machner, 2014; Ku et al., 2012).
Epoxide hydrolase hydrolyzes epoxides to corresponding diols. P. aeruginosa secretes an epoxide hydrolase termed the CFTR inhibitory factor or Cif. Cif hydrolyzes 14,15-epoxyeicosatrienoic acid (14,15-EET) produced by epithelial cells to 14,15-dihydroxyeicosatrienoic acid (DHET; Flitter et al., 2016). Two epoxide hydrolases in P. falciparum, PfEH1 and PfEH2, hydrolyze epoxyeicosatrienoic acids (EETS) and epoxyoctadecenoic acids (EpOMEs) (Spillman et al., 2016). These microbial epoxide hydrolases have non-canonical active sites compared to human counterparts (Bahl et al., 2010; Spillman et al., 2016).
3 |. MICROBIAL OXYLIPINS: EFFECT ON INFLAMMATION AND VIRULENCE
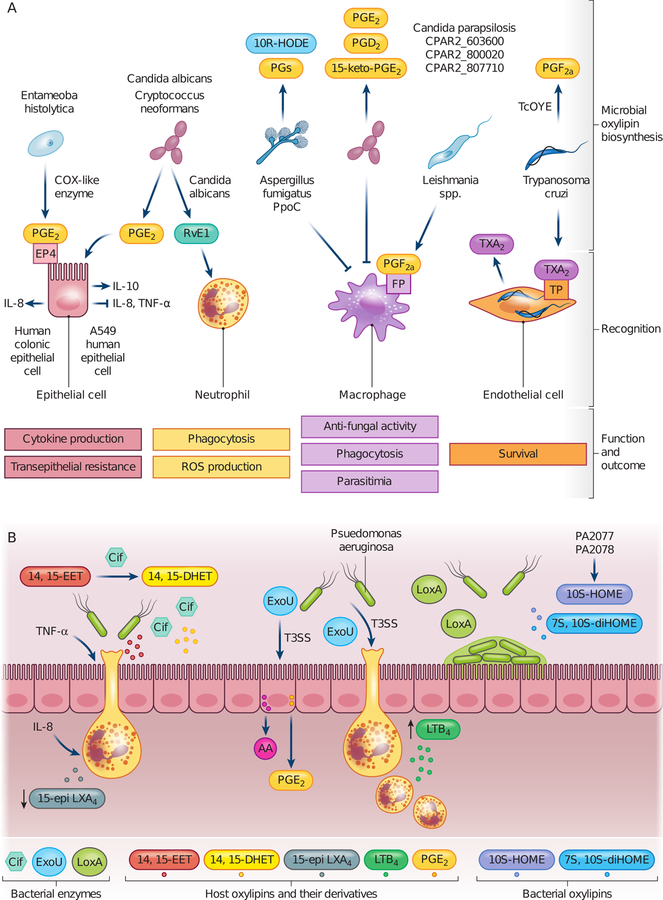
Pathology of microbial infections is dependent on the immunological state of the host as well as the metabolism, persistence, and growth of the pathogen. The host initiates and resolves infection through pro-inflammatory and anti-inflammatory responses mediated by AA, EPA, and DHA- derived lipid mediators and their cognate receptors (Basil and Levy, 2016; Dennis and NForris, 2015). Pro-inflammatory lipid mediators induce inflammatory cell infiltration and elevate production of chemokines and pro-inflammatory cytokines. Anti-inflammatory lipid mediators inhibit infiltration of inflammatory cells and induce anti-inflammatory cytokine production, which induces apoptosis of immune cells and resolves inflammation. Since many microbial pathogens can produce eicosanoids and other potentially immuno-modulatory oxylipins, they can significantly steer the immunological response and influence disease outcomes. Many studies – primarily with fungi - have illustrated that microbial oxylipins are also important for development and cell morphogenesis (Fischer and Keller, 2016; Deschampes et al., 2016). These effects may alter disease outcome; here we specifically focus on roles of microbial oxylipins as immune-modulators during host confrontation. Figure 2 depicts various interactions between microbial pathogens and host cells mediated by microbial oxylipins and associated enzymes.
FIGURE 2.
Host-pathogen interactions mediated by microbial oxylipins and enzymes. (a) Eukaryotic microbial pathogens, predominantly pathogenic fungi and protozoan parasites, produce the same oxylipins as the host eicosanoids and those unique to microbes. Host cells in close contact with pathogens through either infection (endothelial cell and macrophage) or immune response (macrophage, neutrophil, epithelial cell) can recognize microbe-derived oxylipins and respond with altered functionality. (b) Bacterial pathogens, such as Pseudomonas aeruginosa, produce enzymes that influence the type and abundance of host-derived oxylipins, leading to skewed inflammatory state. These include the epoxide hydrolase Cif and the phospholipase A2 ExoU. LoxA in P. aeruginosa also promotes biofilm formation on epithelial surface. IL-8, interleukin 8; IL-10, interleukin 10; TNF-α, tumor necrosis factor alpha; PGs, prostaglandins; PGE2, prostaglandin E2; PGD2, prostaglandin D2; PGF2α, prostaglandin F2α; 15-keto-PGE2, 15-keto-prostaglandin E2; RVE1, resolvin E1; 10R-HODE, 10R-hydroxyoctadecadienoic acid; TXA2, thromboxane A2; EP4, PGE receptor; FP, PGF receptor; TP, thromboxane receptor; TcOYE, Trypanosoma cruzi Old Yellow Enzyme; PpoC, Psi-producing oxygenase C; 14,15-EET, 14,15-epoxyeicosatrienoic acid; 14,15-DHET, 14,15-dihydroxyeicosatrienoic acid; 15-epi-LXA4, 15-epi-lipoxin A4; AA, arachidonic acid; LTB4, leukotriene B4; T3SS, Type III Secretion System; Cif, conductance regulator inhibitory factor
3.1 |. Fungal pathogens
Oxylipin mediated host crosstalk have been studied most predominantly in Aspergillus, Candida, and Cryptococcus spp., the three species that contribute to most of the fungal attributed mortality (Brown, 2012). These fungi can produce the same oxylipins present in the host and those unique to the microbe.
It is currently not known if Aspergillus LA- derived oxylipins are synthesized de novo in animals but the A. fumigatus LoxB LA-metabolite 13-HODE is also made by humans and is implicated in airway hyper-responsiveness (Mabaliraian et al., 2013). A. fumigatus PpoC contributes to resisting phagocytosis and killing by murine bone marrow derived macrophages (Dagenais et al., 2008). An RNAi mutant with reduced expression of ppoA, ppoB, and ppoC shows lower prostaglandin production in vitro and increased virulence in a chemotherapeutic murine model of invasive aspergillosis (Tsitsigiannis et al., 2005). The mechanism of how AA and LA-derived oxylipins catalyzed by Ppo proteins mediate macrophage interaction and virulence remains to be elucidated.
Purified C. albicans and C. neoformans PGE2 has an anti-inflammatory effect: it reduces chemokine IL-8 and inflammatory cytokine TNF-α production but increases anti-inflammatory IL-10 production in A549 epithelial cells (Noverr et al., 2001). Further study is required to assess whether the identified oxylipin producing enzymes in these pathogens (Table 1) mediate pathogenesis through altered PGE2 production. Deletion mutants of the three C. parapsilosis enzymes mentioned earlier (CPAR2_603600, CPAR2_800020, and CPAR2_807710) are more susceptible to phagocytosis and killing by human peripheral blood monocyte derived macrophages (PBMC-DM; Chakraborty et al., 2018). PBMC-DM infected by the CPAR2_807710∆/∆ strain produced higher levels of IL-6, TNF-α and IL-8. All three mutants showed reduced fungal burden in various organs in a murine model of intravenous infection (Chakraborty et al., 2018).
3-HETE produced by C. albicans can serve as substrate for human COX-2 to synthesize 3-OH-PGE2 (Ciccoli et al., 2005). 3-OH-PGE2 triggers increased IL-6 expression in A549 epithelial cells and cAMP release in Jurkat T cells, mediated by the PGE2 cognate receptors EP3 and EP4, respectively. As C. albicans produces RvE1 that’s structurally identical to human RvE1, synthetic RvE1 was used to study neutrophil phagocytosis and pathogenesis of C. albicans. Treatment of low concentrations of RvE1 enhances neutrophil phagocytosis and killing of C. albicans and intravenous injection of RvE1 reduces C. albicans replication in a murine model of intravenous C. albicans infection (Hass-Stapleton et al., 2007). However, it’s unclear whether these findings are physiologically relevant and whether RvE1 itself inhibits C. albicans growth.
3.2 |. Bacterial pathogens
While investigations on bacteria derived oxylipins are scarce, studies on P. aeruginosa enzymes, including the epoxide hydrolase Cif, the phospholipase ExoU, and the lipoxygenase LoxA, have shed light on mechanisms of oxylipin-mediated host crosstalk. These studies present a view that bacterial pathogens may have evolved to manipulate host immunity in environments where oxylipins and their substrates are abundant.
Pseudomonas aeruginosa colonizes immuno-compromised individuals, such as cystic fibrosis (CF) patients. Cif, a virulence factor in P. aeruginosa colonization of CF lungs, degrades the potent anti-inflammatory 14,15-EET from airway epithelial cells to less potent 14,15-DHET. This leads to reduced 15-epi LXA4, a pro-resolving product of neutrophils, and elevated inflammation (Bahl et al., 2011; Flitter et al., 2017). Furthermore, Cif activity in CF bronchial lavage fluid is positively associated with IL-8 level, and negatively associated with lung functions and 15-epi-LXA4 abundance (Flitter et al., 2017). Cif also inhibits mucocilliary beating and hinders mechanical clearance of P. aeruginosa during infection of reconstituted human bronchial epithelium and in a murine model of acute pneumonia (Hvorecny et al.,2018).
ExoU is a well-recognized virulence factor, cytotoxin, and phospholipase (Sawa et al., 2016). ExoU releases AA and increases PGE2 and PGI2 concentrations in culture supernatant of infected cells (Saliba et al., 2005; Plotkowski et al., 2008). ExoU expressing strains induce neutrophil production of LTB4 and neutrophil trans-epithelial migration in vitro, and increase PGE2 and LTB4 production, innate immune cell infiltration, and myeloperoxidase activity during acute pulmonary infection in mice (Pazos et al., 2017; Saliba et al., 2005). A549 epithelial cells infected with ExoU-expressing P. aeruginosa have impaired antioxidant detoxification machinery and higher levels of oxidative metabolites, including lipid hydroperoxides, 8-isoprostanes, ROS, and NO (Da Cunha et al., 2015). The L. pneumophila homolog of ExoU, VipD disrupts endosomal membrane lipids and facilitates L. pneumophila escape from endosomal fusion in the Cos-1 cells (Gasper and Machner, 2014). Streptococcus pyogenes phospholipase A2 SlaA increases monocyte adhesion to endothelial cells in vitro (Oda et al., 2017).
Recombinant P. aeruginosa LoxA modifies membrane phospholipids by forming 15-HETE and 13-HODE, which is associated with induction of red blood cell hemolysis (Banthiya et al., 2015). LoxA is highly expressed in P. aeruginosa isolated from CF lungs and is implicated in biofilm formation on biotic surfaces, such as epithelium (Deschamps et al., 2016; Starkey et al., 2009).
3.3 |. Protozoan parasites
So far, all of the reported parasite derived oxylipins are present in mammals. As many of these organisms cannot grow in axenic culture, several studies have revealed parasite-host oxylipin signaling pathways by exploiting a mammalian receptor mutant. This tactic offers the additional value of insights into pharmacological control of parasite infections.
Trypanosoma cruzi produced TXA2 and PGF2α, are important in maintaining long-term infection. TXB2 derived from T. cruzi, the stable derivative of TXA2, consists most of the circulating TXB2 in infected mice during later stage of infection (Ashton et al, 2007). Deletion of the TXA2 receptor TP in mice results in increased inflammatory cell infiltration, increased parasitic load in myocardium, and increased host mortality (Ashton et al, 2007). These findings suggest that T. cruzi exploits the TXA2-TP signaling to prevent host from early death and maintain infection longevity. PGF2 synthase TcOYE is important for completion and progression of its infective cycle (Díaz-Viraqué et al., 2018). The TcOYE overexpression mutant displays more aggressive parasitemia and increased parasitic load in cardiac tissue.
The Leishmania spp. protein Gp63 and its product PGF2α are implicated in mediating host responses to the parasite. Gp63 is a well-recognized virulence factor and was extensively reviewed prior to the knowledge that it possessed COX-like activity (Olivier et al., 2012). Comparison of Leishmania amazonensis gp63 RNAi mutants expressing differential levels of Gp63 revealed that Gp63 promotes parasite binding to J774 macrophages and intracellular survival (Chen et al., 2000). Future studies on Gp63 should consider its newly identified COX activity in interpreting past studies of such mutants. The PGF2α receptor FP of bone marrow derived macrophages localizes to the parasitophorous vacuoles when infected with Leishmania infantum chagasi (Araújo-Santos et al., 2014). Treatment with a FP inhibitor decreases the infection index of macrophage by various stages of L. i. chagasi (Araújo-Santos et al., 2014). Activation of PGF2α-FP signaling allows L. i. chagasi to evade the macrophage anti-parasitic activity. Therefore, blockade of this pathway may present a therapeutic avenue to manage parasitic load and treat Leishmaniasis.
Invasive infection of E. histolytica induces robust inflammation and tissue damage. E. histolytica or its secreted components induces IL-8 production by infected Caco-2 epithelial cells (Dey et al., 2008). This induction is absent using de-lipidized soluble components, aspirin treated E. histolytica, or Caco-2 cells with inhibited PGE2 receptor EP4. The same group later found that the secreted components also decreases trans-epithelial resistance of the human colonic epithelial monolayer through dissociating the tight junction protein Claudin-4, which is abolished through aspirin pretreatment of E. histolytica or inhibition of EP4 (Lejeune et al., 2011). The role of the previously identified COX-like enzyme in these interactions is worth further investigation.
The aminopeptidase activity of PfEH2 and the epoxide hydrolase activity of PfEH1 and PfEH2 in P. falciparum were independently reported (Spillman et al., 2016; da Silva et al., 2016). The PfEH2 deletion mutant is more easily filtered outside of the infected erythrocytes, and the PfEH1 and PfEH2 overexpression mutants decreases abundance of several EETs and EpOMEs in infected erythrocytes.
4 |. CONCLUSIONS
The critical nature of oxylipins in inflammation and infection is clear from myriad studies as reflected in the moniker ‘eicosanoid storm’ (Dennis and Norris, 2015). Yet the majority of these studies have focused solely on host oxylipins. Collectively, the studies in this review support an important but underappreciated role of microbial oxylipins in human infectious disease.
Where might the future directions microbial oxylipin research head? An increasing body of literature has reported that microbial extracellular vesicles modulate host immunological response (Joffe et al., 2016; Kuipers, et al., 2018). In mammalian systems, extracellular vesicles facilitate intercellular transport of eicosanoids and their biosynthetic machinery, mediating paracrine signaling (Boilard et al., 2018; Sagini et al., 2018). Proteins that mediate oxylipin biosynthesis and signaling, such as Cif from P. aeruginosa and the Leishmania COX Gp63 protein, are located in extracellular vesicles (Ghosh et al., 2013). Purified EVs from C. albicans and C. neoformans are internalized by macrophage and dendritic cells and stimulate production of nitric oxide and proinflammatory cytokine (Oliveira et al., 2010; Vargas et al., 2015). Extracellular vesicles thus can serve as a site of oxylipin biosynthesis and a vehicle in shuffling the enzymes, substrates and products between the pathogen and the host. Additionally, roles for microbial oxylipins in microbial confrontations should also be explored more thoroughly in the context of the human microbiome (Fourie et al., 2017; Krause et al., 2015).
Finally, we note that one emerging theme from the research summarized in this review is that whereas some microbial oxylipins are identical to mammalian products and impact host processes, other microbial oxylipin products and/or their synthesizing enzymes are evolutionarily distant from those in mammals (Estrada-Figueroa et al., 2018; Hansen et al., 2013). Not only is it important to investigate the potential impacts of these structurally distinct microbial oxylipins on host immunological processes but the microbial chemical signatures may be useful for molecular diagnosis of the pathogen. Furthermore, biochemical studies on microbial specific enzymes could potentially pave a way to identifying new targets for future therapies.
Acknowledgments
Funding information
National Institute of Health R01, Grant Number: AI065728–01
REFERENCES
- Anderson DM, Schmalzer KM, Sato H, Casey M, Terhune SS, Haas AL, … Frank DW (2011). Ubiquitin and ubiquitin-modified proteins activate the Pseudomonas aeruginosa T3SS cytotoxin, ExoU: Ubiquitin activation of ExoU. Molecular Microbiology, 82(6), 1454–1467. 10.1111/j.1365-2958.2011.07904.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo-Santos T, Rodríguez NE, Moura-Pontes S, Dixt UG, Abánades DR, Bozza PT, … Borges VM (2014). Role of prostaglandin F2α production in lipid bodies from Leishmania infantum chagasi: Insights on virulence. The Journal of Infectious Diseases, 210(12), 1951–1961. 10.1093/infdis/jiu299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton AW, Mukherjee S, Nagajyothi F, Huang H, Braunstein VL, Desruisseaux MS, … Tanowitz HB (2007). Thromboxane A 2 is a key regulator of pathogenesis during Trypanosoma cruzi infection. The Journal of Experimental Medicine, 204(4), 929–940. 10.1084/jem.20062432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl CD, Morisseau C, Bomberger JM, Stanton BA, Hammock BD, O’Toole GA, & Madden DR (2010). Crystal structure of the cystic fibrosis transmembrane conductance regulator inhibitory factor Cif reveals novel active-site features of an epoxide hydrolase virulence factor. Journal of Bacteriology, 192(7), 1785–1795. 10.1128/JB.01348-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannenberg GL, Aliberti J, Hong S, Sher A, & Serhan C (2004). Exogenous pathogen and plant 15-lipoxygenase initiate endogenous lipoxin A 4 biosynthesis. The Journal of Experimental Medicine, 199(4), 515–523. 10.1084/jem.20031325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banthiya S, Kalms J, Galemou Yoga E, Ivanov I, Carpena X, Hamberg M, … Scheerer P (2016). Structural and functional basis of phospholipid oxygenase activity of bacterial lipoxygenase from Pseudomonas aeruginosa. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids, 1861(11), 1681–1692. 10.1016/j.bbalip.2016.08.002 [DOI] [PubMed] [Google Scholar]
- Banthiya S, Pekárová M, Kuhn H, & Heydeck D (2015). Secreted lipoxygenase from Pseudomonas aeruginosa exhibits biomembrane oxygenase activity and induces hemolysis in human red blood cells. Archives of Biochemistry and Biophysics, 584, 116–124. 10.1016/j.abb.2015.09.003 [DOI] [PubMed] [Google Scholar]
- Basil MC, & Levy BD (2016). Specialized pro-resolving mediators: endogenous regulators of infection and inflammation. Nature Reviews Immunology, 16(1), 51–67. 10.1038/nri.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondo GA, Dias-Melicio LA, Bordon-Graciani AP, Kurokawa CS, & Soares AMV (2012). Production of leukotriene B4 by Paracoccidioides brasiliensis: Eicosanoid production by Paracoccidioides brasiliensis. Yeast, 29(6), 201–208. 10.1002/yea.2900 [DOI] [PubMed] [Google Scholar]
- Boilard E (2018). Extracellular vesicles and their content in bioactive lipid mediators: more than a sack of microRNA. Journal of Lipid Research, jlr – R084640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordon AP, Dias-Melicio LA, Acorci MJ, Biondo GA, Fecchio D, Peraçoli MTS, & de Soares ÂMVC (2007). Prostaglandin E2 production by high and low virulent strains of Paracoccidioides brasiliensis. Mycopathologia, 163(3), 129–135. 10.1007/s11046-007-0098-1 [DOI] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, & White TC (2012). Hidden killers: Human fungal infections. Science Translational Medicine, 4(165), 165rv13–rv165rv13. 10.1126/scitranslmed.3004404 [DOI] [PubMed] [Google Scholar]
- Chakraborty T, Thuer E, Heijink M, Tóth R, Bodai L, Vágvölgyi C, … Gácser A (2018). Eicosanoid biosynthesis influences the virulence of Candida parapsilosis. Virulence, 9(1), 1019–1035. 10.1080/21505594.2018.1475797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D-Q, Kolli BK, Yadava N, Lu HG, Gilman-Sachs A, Peterson DA, & Chang K-P (2000). Episomal expression of specific sense and antisense mRNAs in Leishmania amazonensis: modulation of gp63 level in promastigotes and their infection of macrophages in vitro. Infection and Immunity, 68(1), 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccoli R, Sahi S, Singh S, Prakash H, Zafiriou M-P, Ishdorj G, … Nigam S (2005). Oxygenation by COX-2 (cyclo-oxygenase-2) of 3-HETE (3-hydroxyeicosatetraenoic acid), a fungal mimetic of arachidonic acid, produces a cascade of novel bioactive 3-hydroxyeicosanoids. Biochemical Journal, 390(3), 737–747. 10.1042/BJ20041995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas R, Ashton A, Mukherjee S, Dalli J, Akide-Ndunge O, Huang H, … Tanowitz H (2018). Trypanosoma cruzi produces the specialized proresolving mediators resolvin D1, resolvin D5, and resolvin E2. Infection and Immunity, 86 (4), e00688–17. doi: 10.1128/IAI.00688-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Cunha LG, Ferreira MF, de Moraes JA, Reis PA, Castro-Faria-Neto HC, Barja-Fidalgo C, … Saliba AM (2015). ExoU-induced redox imbalance and oxidative stress in airway epithelial cells during Pseudomonas aeruginosa pneumosepsis. Medical Microbiology and Immunology, 204(6), 673–680. 10.1007/s00430-015-0418-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagenais TRT, Chung D, Giles SS, Hull CM, Andes D, & Keller NP (2008). Defects in conidiophore development and conidium-macrophage interactions in a dioxygenase mutant of Aspergillus fumigatus. Infection and Immunity, 76(7), 3214–3220. 10.1128/IAI.00009-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva FL, Dixon MWA, Stack CM, Teuscher F, Taran E, Jones MK, … Skinner-Adams TS (2016). A Plasmodium falciparum S33 proline aminopeptidase is associated with changes in erythrocyte deformability. Experimental Parasitology, 169, 13–21. 10.1016/j.exppara.2016.06.013 [DOI] [PubMed] [Google Scholar]
- Dennis EA, & Norris PC (2015). Eicosanoid storm in infection and inflammation. Nature Reviews Immunology, 15(8), 511–523. 10.1038/nri3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps JD, Ogunsola AF, Jameson JB, Yasgar A, Flitter BA, Freedman CJ, … Holman TR (2016). Biochemical and cellular characterization and Inhibitor discovery of Pseudomonas aeruginosa 15-Lipoxygenase. Biochemistry, 55(23), 3329–3340. 10.1021/acs.biochem.6b00338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey I, & Chadee K (2008). Prostaglandin E2 produced by Entamoeba histolytica binds to EP4 receptors and stimulates interleukin-8 production in human colonic cells. Infection and Immunity, 76(11), 5158–5163. 10.1128/IAI.00645-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey I, Keller K, Belley A, & Chadee K (2003). Identification and characterization of a cyclooxygenase-like enzyme from Entamoeba histolytica. Proceedings of the National Academy of Sciences, 100(23), 13561–13566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Viraqué F, Chiribao ML, Trochine A, González-Herrera F, Castillo C, Liempi A, … Robello C (2018). Old Yellow Enzyme from Trypanosoma cruzi exhibits in vivo prostaglandin F2α synthase activity and has a key role in parasite infection and drug susceptibility. Frontiers in Immunology, 9 10.3389/fimmu.2018.00456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb-Downward JR, Noggle RM, Williamson PR, & Huffnagle GB (2008). The role of laccase in prostaglandin production by Cryptococcus neoformans. Molecular Microbiology, 68(6), 1428–1437. 10.1111/j.1365-2958.2008.06245.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb-Downward JR, & Noverr MC (2007). Characterization of prostaglandin E2 production by Candida albicans. Infection and Immunity, 75(7), 3498–3505. 10.1128/IAI.00232-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-Figueroa LA, Díaz-Gandarilla JA, Hernández-Ramírez VI, Arrieta-González MM, Osorio-Trujillo C, Rosales-Encina JL, … Talamás-Rohana P (2018). Leishmania mexicana gp63 is the enzyme responsible for cyclooxygenase (COX) activity in this parasitic protozoa. Biochimie, 151, 73–84. 10.1016/j.biochi.2018.05.016 [DOI] [PubMed] [Google Scholar]
- Fischer GJ, Bacon W, Yang J, Palmer JM, Dagenais T, Hammock BD, & Keller NP (2017). Lipoxygenase activity accelerates programmed spore germination in Aspergillus fumigatus. Frontiers in Microbiology, 8 10.3389/fmicb.2017.00831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer GJ, & Keller NP (2016). Production of cross-kingdom oxylipins by pathogenic fungi: An update on their role in development and pathogenicity. Journal of Microbiology, 54(3), 254–264. 10.1007/s12275-016-5620-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flitter BA, Hvorecny KL, Ono E, Eddens T, Yang J, Kwak DH, … Bomberger JM (2017). Pseudomonas aeruginosa sabotages the generation of host proresolving lipid mediators. Proceedings of the National Academy of Sciences, 114(1), 136–141. 10.1073/pnas.1610242114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourie R, Ells R, Kemp G, Sebolai OM, Albertyn J, & Pohl CH (2017). Pseudomonas aeruginosa produces aspirin insensitive eicosanoids and contributes to the eicosanoid profile of polymicrobial biofilms with Candida albicans. Prostaglandins, Leukotrienes, and Essential Fatty Acids, 117, 36–46. 10.1016/j.plefa.2017.01.008 [DOI] [PubMed] [Google Scholar]
- Garscha U, Jernerén F, Chung D, Keller NP, Hamberg M, & Oliw EH (2007). Identification of dioxygenases required for Aspergillus development: Studies of products. Stereochemistry, and the reaction mechanism. Journal of Biological Chemistry, 282(48), 34707–34718. 10.1074/jbc.M705366200 [DOI] [PubMed] [Google Scholar]
- Gaspar AH, & Machner MP (2014). VipD is a Rab5-activated phospholipase A1 that protects Legionella pneumophila from endosomal fusion. Proceedings of the National Academy of Sciences, 111(12), 4560–4565. 10.1073/pnas.1316376111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum MA (2000). Potential role of phospholipases in virulence and fungal pathogenesis. Clinical Microbiology Reviews, 13(1), 122–143. 10.1128/CMR.13.1.122-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh J, Bose M, Roy S, & Bhattacharyya SN (2013). Leishmania donovani targets Dicer1 to downregulate miR-122, lower serum cholesterol, and facilitate murine liver infection. Cell Host & Microbe, 13(3), 277–288. 10.1016/j.chom.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grózer Z, Tóth A, Tóth R, Kecskeméti A, Vágvölgyi C, Nosanchuk JD, … Gácser A (2015). Candida parapsilosis produces prostaglandins from exogenous arachidonic acid and OLE2 is not required for their synthesis. Virulence, 6(1), 85–92. https://doi.org/10.416½1505594.2014.988097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas-Stapleton EJ, Lu Y, Hong S, Arita M, Favoreto S, Nigam S, … Agabian N (2007). Candida albicans modulates host defense by biosynthesizing the pro-sesolving mediator resolvin E1. PLoS ONE, 2(12), e1316 10.1371/journal.pone.0001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J, Garreta A, Benincasa M, Fusté MC, Busquets M, & Manresa A (2013). Bacterial lipoxygenases, a new subfamily of enzymes? A phylogenetic approach. Applied Microbiology and Biotechnology, 97(11), 4737–4747. 10.1007/s00253-013-4887-9 [DOI] [PubMed] [Google Scholar]
- Heshof R, Jylhä S, Haarmann T, Jørgensen ALW, Dalsgaard TK, & de Graaff LH (2014). A novel class of fungal lipoxygenases. Applied Microbiology and Biotechnology, 98(3), 1261–1270. 10.1007/s00253-013-5392-x [DOI] [PubMed] [Google Scholar]
- Hoffmann I, Jernerén F, & Oliw EH (2013). Expression of fusion proteins of Aspergillus terreus reveals a novel allene oxide synthase. Journal of Biological Chemistry, 288(16), 11459–11469. 10.1074/jbc.M113.458257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz Brown S, Zarnowski R, Sharpee WC, & Keller NP (2008). Morphological transitions governed by density dependence and lipoxygenase activity in Aspergillus flavus. Applied and Environmental Microbiology, 74(18), 5674–5685. 10.1128/AEM.00565-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvorecny KL, Dolben E, Moreau-Marquis S, Hampton TH, Shabaneh TB, Flitter BA, … Madden DR (2018). An epoxide hydrolase secreted by Pseudomonas aeruginosa decreases mucociliary transport and hinders bacterial clearance from the lung. American Journal of Physiology-Lung Cellular and Molecular Physiology, 314(1), L150–L156. 10.1152/ajplung.00383.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istivan TS, & Coloe P (2006). Phospholipase A in Gram-negative bacteria and its role in pathogenesis. Microbiology, 152(5), 1263–1274. 10.1099/mic.0.28609-0 [DOI] [PubMed] [Google Scholar]
- Joffe LS, Nimrichter L, Rodrigues ML, & Del Poeta M (2016). Potential roles of fungal extracellular vesicles during infection. mSphere, 1(4). 10.1128/mSphere.00099-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabututu Z, Martin SK, Nozaki T, Kawazu S, Okada T, Munday CJ, … Kubata BK (2003). Prostaglandin production from arachidonic acid and evidence for a 9,11-endoperoxide prostaglandin H2 reductase in Leishmania. International Journal for Parasitology, 33(2), 221–228. 10.1016/S0020-7519(02)00254-0 [DOI] [PubMed] [Google Scholar]
- Krause J, Geginat G, & Tammer I (2015). Prostaglandin E2 from Candida albicans stimulates the growth of Staphylococcus aureus in mixed biofilms. PloS One, 10(8), e0135404 10.1371/journal.pone.0135404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubata BK, Duszenko M, Kabututu Z, Rawer M, Szallies A, Fujimori K, … Hayaishi O (2000). Identification of a novel prostaglandin F2α synthase in Trypanosoma brucei. The Journal of Experimental Medicine, 192(9), 1327–1338. 10.1084/jem.192.9.1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubata BK, Duszenko M, Martin KS, & Urade Y (2007). Molecular basis for prostaglandin production in hosts and parasites. Trends in Parasitology, 23(7), 325–331. 10.1016/j.pt.2007.05.005 [DOI] [PubMed] [Google Scholar]
- Kubata BK, Eguchi N, Urade Y, Yamashita K, Mitamura T, Tai K, … Horii T (1998). Plasmodium falciparum produces prostaglandins that are pyrogenic, somnogenic, and immunosuppressive substances in humans. The Journal of Experimental Medicine, 188(6), 1197–1202. 10.1084/jem.188.6.1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubata BK, Kabututu Z, Nozaki T, Munday CJ, Fukuzumi S, Ohkubo K, … Urade Y (2002). A key role for Old Yellow Enzyme in the metabolism of drugs by Trypanosoma cruzi. The Journal of Experimental Medicine, 196(9), 1241–1252. 10.1084/jem.20020885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku B, Lee K-H, Park WS, Yang C-S, Ge J, Lee S-G, … Oh B-H (2012). VipD of Legionella pneumophila targets activated Rab5 and Rab22 to interfere with endosomal trafficking in macrophages. PLoS Pathogens, 8(12), e1003082 10.1371/journal.ppat.1003082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers ME, Hokke CH, Smits HH, & Nolte-’t Hoen ENM (2018). Pathogen-derived extracellular vesicle-associated molecules that affect the host immune system: An overview. Frontiers in Microbiology, 9 10.3389/fmicb.2018.02182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfahl C, Tsikas D, Niemann J, Geginat G, & Hof H (2012). Production of prostaglandins, isoprostanes and thromboxane by Aspergillus fumigatus: Identification by gas chromatography–tandem mass spectrometry and quantification by enzyme immunoassay. Molecular Immunology, 49(4), 621–627. 10.1016/j.molimm.2011.10.010 [DOI] [PubMed] [Google Scholar]
- Lejeune M, Moreau F, & Chadee K (2011). Prostaglandin E2 produced by Entamoeba histolytica signals via EP4 receptor and alters Claudin-4 to increase ion permeability of tight junctions. The American Journal of Pathology, 179(2), 807–818. 10.1016/j.ajpath.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabalirajan U, Rehman R, Ahmad T, Kumar S, Singh S, Leishangthem GD, … Ghosh B (2013). Linoleic acid metabolite drives severe asthma by causing airway epithelial injury. Scientific Reports, 3(1). 10.1038/srep01349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noverr MC, Toews GB, Huffnagle GB (2002). Production of prostaglandins and leukotrienes by pathogenic fungi. Infection and Immunity, 70(1), 400–402. 10.1128/IAI.70.1.400-402.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noverr MC, Phare SM, Toews GB, Coffey MJ, & Huffnagle GB (2001). Pathogenic yeasts Cryptococcus neoformans and Candida albicans produce immunomodulatory prostaglandins. Infection and Immunity, 69(5), 2957–2963. 10.1128/IAI.69.5.2957-2963.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda M, Domon H, Kurosawa M, Isono T, Maekawa T, Yamaguchi M, … Terao Y (2017). Streptococcus pyogenes phospholipase A2 induces the expression of adhesion molecules on human umbilical vein endothelial cells and aorta of mice. Frontiers in Cellular and Infection Microbiology, 7 10.3389/fcimb.2017.00300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M, Atayde VD, Isnard A, Hassani K, & Shio MT (2012). Leishmania virulence factors: Focus on the metalloprotease GP63. Microbes and Infection, 14(15), 1377–1389. 10.1016/j.micinf.2012.05.014 [DOI] [PubMed] [Google Scholar]
- Oliveira DL, Freire-de-Lima CG, Nosanchuk JD, Casadevall A, Rodrigues ML, & Nimrichter L (2010). Extracellular vesicles from Cryptococcus neoformans modulate macrophage functions. Infection and Immunity, 78(4), 1601–1609. 10.1128/IAI.01171-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliw EH, Aragó M, Chen Y, & Jernerén F (2016). A new class of fatty acid allene oxide formed by the DOX-P450 fusion proteins of human and plant pathogenic fungi, C. immitis and Z. tritici. Journal of Lipid Research, 57(8), 1518–1528. 10.1194/jlr.M068981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos MA, Lanter BB, Yonker LM, Eaton AD, Pirzai W, Gronert K, … Hurley BP (2017). Pseudomonas aeruginosa ExoU augments neutrophil transepithelial migration. PLoS Pathogens, 13(8), e1006548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkowski M-C, Brandão BA, de Assis M-C, Feliciano L-FP, Raymond B, Freitas C, … Bozza PT (2008). Lipid body mobilization in the ExoU-induced release of inflammatory mediators by airway epithelial cells. Microbial Pathogenesis, 45(1), 30–37. 10.1016/j.micpath.2008.01.008 [DOI] [PubMed] [Google Scholar]
- Rabin SDP, & Hauser AR (2005). Functional regions of the Pseudomonas aeruginosa cytotoxin ExoU. Infection and Immunity, 73(1), 573–582. 10.1128/IAI.73.1.573-582.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagini K, Costanzi E, Emiliani C, Buratta S, & Urbanelli L (2018). Extracellular vesicles as conveyors of membrane-derived bioactive lipids in immune system. International Journal of Molecular Sciences, 19(4), 1227 10.3390/ijms19041227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba AM, Nascimento DO, Silva MCA, Assis MC, Gayer CRM, Raymond B, … Plotkowski MC (2005). Eicosanoid-mediated proinflammatory activity of Pseudomonas aeruginosa ExoU. Cellular Microbiology, 7(12), 1811–1822. 10.1111/j.1462-5822.2005.00635.x [DOI] [PubMed] [Google Scholar]
- Sato H, Frank DW, Hillard CJ, Feix JB, Pankhaniya RR, Moriyama K, … Long RM (2003). The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. The EMBO Journal, 22(12), 2959–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa T, Hamaoka S, Kinoshita M, Kainuma A, Naito Y, Akiyama K, & Kato H (2016). Pseudomonas aeruginosa type III secretory toxin ExoU and its predicted homologs. Toxins, 8(11), 307 10.3390/toxins8110307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillman NJ, Dalmia VK, & Goldberg DE (2016). Exported epoxide hydrolases modulate erythrocyte vasoactive lipids during Plasmodium falciparum infection. mBio, 7(5). 10.1128/mBio.01538-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkey M, Hickman JH, Ma L, Zhang N, De Long S, Hinz A, … Parsek MR (2009). Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. Journal of Bacteriology, 191(11), 3492–3503. 10.1128/JB.00119-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsigiannis DI, Zarnowski R, & Keller NP (2004). The lipid body protein, PpoA, coordinates sexual and asexual sporulation in Aspergillus nidulans. Journal of Biological Chemistry, 279(12), 11344–11353. 10.1074/jbc.M310840200 [DOI] [PubMed] [Google Scholar]
- Tsitsigiannis I, Bok J, Andes D, Nielson K, Frisvad J, Keller N (2005). Aspergillus cyclooxygenase-like enzymes are associated with prostaglandin production and virulence. Infection and Immunity 73 (8), 4548–4559. https://doi:10.1128/IAI.73.8.4548–4559.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance RE, Hong S, Gronert K, Serhan CN, & Mekalanos JJ (2004). The opportunistic pathogen Pseudomonas aeruginosa carries a secretable arachidonate 15-lipoxygenase. Proceedings of the National Academy of Sciences, 101(7), 2135–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas G, Rocha JDB, Oliveira DL, Albuquerque PC, Frases S, Santos SS, … Nimrichter L (2015). Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans: Extracellular vesicles from Candida albicans. Cellular Microbiology, 17(3), 389–407. 10.1111/cmi.12374 [DOI] [PubMed] [Google Scholar]