Abstract
Environmental circadian disruption (ECD), characterized by repeated or long-term disruption in environmental timing cues which require the internal circadian clock to change its phase to resynchronize with the environment, is associated with numerous serious health issues in humans. While animal and isolated cell models exist to study the effects of destabilizing the relationship between the circadian system and the environment, neither approach provides an ideal solution. Here, we developed an in vitro model which incorporates both elements of a reductionist cellular model and disruption of the clock/environment relationship using temperature as an environmental cue, as occurs in vivo. Using this approach, we have demonstrated that some effects of in vivo ECD can be reproduced using only isolated peripheral oscillators. Specifically, we report exaggerated inflammatory responses to endotoxin following repeated environmental circadian disruption in explanted spleens. This effect requires a functional circadian clock but not the master brain clock, the suprachiasmatic nucleus (SCN). Further, we report that this is a result of cumulative, rather than acute, circadian disruption as has been previously observed in vivo. Finally, such effects appear to be tissue specific as it does not occur in lung, which is less sensitive to the temperature cycles employed to induce ECD. Taken together, the present study suggests that this model could be a valuable tool for dissecting the causes and effects of circadian disruption both in isolated components of physiological systems as well as the aggregated interactions of these systems that occur in vivo.
Introduction
The circadian system is a highly conserved biological timing system in organisms from single cell bacteria to humans. In mammals, cell-autonomous clocks are present in most cells but are synchronized by a central master clock, the suprachiasmatic nucleus (SCN) of the anterior hypothalamus. Most physiological and behavioral processes are subject to circadian regulation which temporally optimizes the internal environment and allows anticipation of recurrent events in the external environment. Disruption of the circadian system in humans, as occurs during chronic shift work or multiple time-zone travel, has been linked to increased incidence and severity of several cancers [1–3], autoimmune disorders [4], obesity [5], diabetes [6], and stroke [7]. Animal models of shiftwork have also been employed to demonstrate the deleterious effects of circadian disruption [8, 9]. Though the mechanism(s) mediating the development of so called “shift work diseases” is not presently known, one potential explanation is that immune function is directly impacted by misalignment of circadian clocks with the external environment.
There is ample evidence of the importance of circadian regulation in the immune system. In mice, inflammation triggered by bacterial endotoxin (lipopolysaccharide (LPS)) fluctuates in a circadian manner [10], as does mortality [11]. The inflammatory response to intraperitoneally administered LPS is primarily mediated by macrophages, which have been shown to have a strong circadian expression of core clock genes [12, 13]. Specific deletion of clock genes in myeloid cells has been shown to have a significant impact on immune specific processing in these cells, either eliminating the rhythm in LPS response entirely [13] or substantially altering it [13–15]. Further, other aspects of immune cell function such as cell trafficking have been demonstrated to be regulated by circadian rhythms [16]. This evidence suggests an important role of immune dysregulation in mediating the significant health impacts of circadian disruption.
The study of the effects of circadian disruption is, however, complicated by several important factors. Inability to control for immunologically relevant behavioral and environmental factors make conducting these studies in humans very challenging, though some studies have been able to demonstrate immune consequences of environmental circadian disruption directly on the immune system, or through other physiological processes that indirectly affect inflammation [17–22]. Mammalian animal models provide better environmental control [8, 9], however the hierarchical organization of the circadian system, in which a master pacemaker resets other brain clocks which in turn reset clocks throughout the body, as well as the diffuse nature and inter-connectedness of the immune system represent significant obstacles to identifying a mechanism of circadian disruption. Cell lines provide an excellent reductionist approach to this problem, allowing for direct alteration of the genetic components of the clock and subsequent effect on cellular function in a precise, cell-type specific fashion. However, deletion of these genes breaks the clock rather than alters its relationship with the environment (as in vivo). Furthermore, disruption of clock genes, both in vivo and in vitro, also modifies immunological function in ways which may be distinct from the roles these genes play in rhythmicity. For example, core clock genes clock and both cryptochrome 1 and cryptochrome 2 may directly regulate expression of NF-κB [15, 23]. In such cases, clock specific effects on immune function are obscured by direct immune specific regulation by these genes. Others have previously used primary culture to demonstrate both rhythmicity in vitro as well as temperature sensitivity in some mammalian tissues [24–26], suggesting that a reductionist model of environmental control is entirely feasible.
Here, we a developed such an in vitro model of environmental circadian disruption (ECD) in which explanted spleen and lung were cultured in circadian high/low temperature cycles which were either kept constant or repeatedly advanced to investigate the consequences of repeated phase shifting on peripheral tissue function. This approach provides a reductionist application in which the disruption which takes place is of the environment rather than of the circadian clock, an important distinction. Further, organotypic, rather than cellular, cultures retain multi-oscillatory connectivity of diverse cell-types which is critical for normal circadian rhythms in vivo. In combination, these factors may represent a model of ECD which is more closely analogous to in vivo circadian disruption and results in immune dysregulation similar that what has been previously reported in animal models of ECD. Here, we have used this model to demonstrate that the immune-specific effects of ECD previously reported in animal models is a local phenomenon which does not require input from the master pacemaker in the SCN. Specifically, explanted spleen tissue subjected to at least four consecutive 6-hour phase advances and challenged with LPS undergo significantly increased transcription and secretion of the pro-inflammatory cytokine IL-6, as we have previously reported using in vivo ECD [8]. Importantly, this effect was abolished in BMAL1KO mouse spleen explants which do not possess a functional circadian clock. Exposure to fewer phase advances (less disruption) also abolished exaggerated pro-inflammatory signaling. Interestingly, advancing temperature cycles did not impact the circadian clock, nor cause a differential response to immune challenge by LPS in lung explants. Thus, the immune consequences of environmental circadian disruption can be elicited by disrupting peripheral rhythms in a reductionist model amenable to mechanistic dissection.
Materials and methods
Animals
Male Per2Luc knock-in mice on a C57BL/6 background [27] or B6.129-Arntltm1Bra/J (Jax 009100), 6–8 weeks old were used. Per2Luc mice were obtained from the Menaker lab at UVA in 2006 and maintained as an inbred colony at Morehouse School of Medicine. Heterozygous B6.129-Arntltm1Bra/J male and female mice were purchased from Jackson labs and bred to obtain homozygous BMAL1 KO mice, confirmed by genotyping (www.MouseGenotype.com), for the present experiments. Mice were group housed in polycarbonate cages (19.56 x 30.91 x 12.34 cm) with corncob bedding under a 12:12 LD cycle and cared for by Morehouse School of Medicine Animal Care staff. Prior to organ collection, mice were euthanized by CO2 asphyxiation followed by secondary cervical dislocation. All procedures and protocols were approved by the Morehouse School of Medicine Institutional Animal Care and Use Committee.
Temperature entrainment of endogenous period
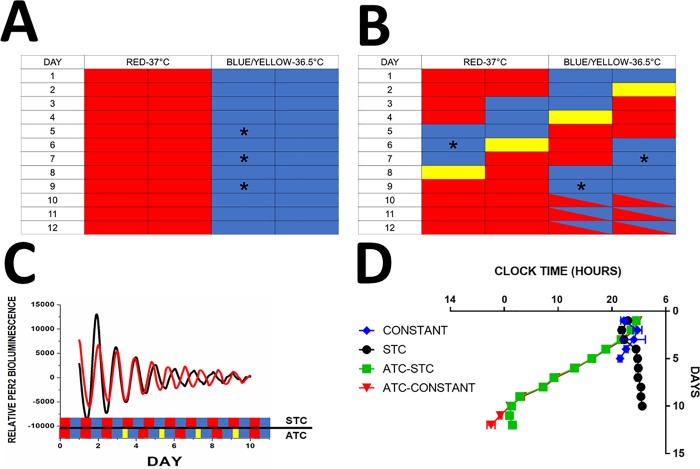
Isolated explants were maintained in temperature cycles consisting of 12 hours of a “high” (37°C) and 12 hours of “low” (36.5°C) temperature phase in temp-cycle incubators (Digitherm Incubators, Tritech Research), analogous to light:dark cycles to which animals are exposed in vivo. This amplitude of temperature cycle was selected because it was the smallest temperature oscillation which could entrain the tissue as well as produce type 1 phase resetting in response to shifts in the temperature cycle. Explants were then randomly assigned to either remain in the original (“static”) temperature cycle or undergo a phase advancing schedule. In order to achieve phase advances, the low portion of the temperature cycle was shortened by 6 hours every 42.5 hours for four consecutive advances (Fig 1A). Period, the length of time between adjacent daily Per2 expression peaks at least two peaks, was calculated by monitoring the expression of Per2Luc under photo-multiplier tubes (PMT Detector Assembly, Hamamatsu) inside of a temperature cycle incubator under a constant (fixed 37°C temperature; n = 6), a static temperature cycle (time of high/low temperature transitions never changed, STC; n = 6), an advancing temperature cycle (ATC) followed by placement into a STC (n = 6), and an ATC followed by release into constant temperature (n = 6; Fig 1B and 1C). Type 1 phase resetting is demonstrated in Fig 1C by mPer2 peak times on the circadian cycle following a phase advance which are intermediate between the old and new temperature cycle.
Fig 1. Temperature control of the splenic circadian clock.
A: Schematic illustrating static temperature cycle. Each block represents 6 hours, red blocks are “high” temperature (37°C) while blue bars are “low” temperature (36.5°C). In experiments where immune challenge was performed, LPS was added 39 hours following the second, third, or fourth phase advance (ZT 3; Asterisks). B: Schematic illustrating advancing temperature cycle. Each block represents 6 hours, red blocks are “high” temperature (37°C) while blue bars are “low” temperature (36.5°C). Phase advances in the temperature cycle were accomplished by shortening the low temperature phase (yellow bars), advancing the subsequent cycle by 6 hours. Tissue was subjected to four consecutive 6-hour phase advances. In experiments where immune challenge was performed, LPS was added 39 hours following the second, third, or fourth phase advance (ZT3; Asterisks). For days 10, 11, and 12 boxes crossed with red and blue indicate where explants were either maintained in a STC or were released into constant temperature. C: Average Bioluminescence Traces. Traces showing relative Per2 bioluminescence traces for spleen explants under static temperature cycle (black; STC; n = 6) and advancing temperature cycle (ATC; red; n = 6). Temperature cycle indicated beneath traces; static temperature cycle by alternating red (high temp) and blue (low temp) bars, advancing with alternating red and blue bars supplemented with shorter yellow bars indicating shortened low temp period. D: Entrainment of Splenic Circadian Clocks by Temperature. Peak Per2 expression time plotted by day to demonstrate maintenance or shortening of circadian period under the following conditions: blue: kept in a constant 37°C (CONSTANT; n = 6); black: kept in a static temperature cycle (STC; n = 6) 12:12 37°C:36.5°C; green: kept in an advancing temperature cycle, followed by a static temperature cycle on day 9 after 4th shift (ATC-STC; n = 6); red: kept in an advancing temperature cycle, followed by release into constant 37°C on day 9 after 4th shift (ATC-CONSTANT; N = 6).
Tissue explants
Organs were removed and micro-dissected into roughly equally sized pieces then sealed individually in 35mm dishes with 2ml of DMEM supplemented with 10mM HEPES, 2% B27, 2.5mL pen/strep, and 42μg D-(+) Glucose [8, 28, 29]. For luminometry experiments, tissue was cultured in media containing 1mM beetle luciferin (Molecular Imaging Products). Because of the phase-resetting effects of media changes no media changes were performed in while the explants were in a temperature cycle. Explants were assigned to experimental conditions at random to control for potential differences in explant size or cellular composition of individual explants.
Lipopolysaccharide
All experiments were done as previously reported ([8, 28] purified from Escherichia coli serotype O111:B4 (Sigma Aldrich) was added to culture media for a final concentration of 10μl/mL. This dose was selected because previous publications have shown that this dose elicits immune responses from peritoneal macrophage culture [8], as well as from whole blood and culture PBMCs [30].
LPS challenge protocol
We have previously reported an increase in several pro-inflammatory cytokine markers following in vivo ECD [8], though IL-6 was the most robust. Therefore, here we chose to use IL-6 secretion as an index of pro-inflammatory signaling. To examine the effect of shift number on the expression of il6, spleens were removed from the temperature cycle incubator 39 hours (ZT3) after the 2nd (n = 6), 3rd (n = 6), or 4th (n = 6) phase advance in order to allow the tissue to complete the last shift to which they were exposed before LPS treatment. Explants were shown to complete a 6hr phase advance in just over 36 hours via analysis of bioluminescence during entrainment experiments. For all other experiments, 39 hours after the final shift at ZT 3 (or the same day/time for unshifted explants), LPS (1mM) was added to culture dishes which were then kept at 37°C for 6h for both spleen and lung explants (n = 6 for all conditions).
qPCR
Explants were frozen in Trizol at -80°C until RNA extraction. RNA was extracted grinding explants in Trizol using a sterile pestle, then adding chloroform and spinning at 12,000 RPM for 15 mins. Supernatant is then added to isopropanol and centrifuged again to obtain RNA pellet. RNA was quantified by Specrophotometry (Nanodrop, Thermo Scientific) and standardized to 1000 ng/μl. Reverse transcription was achieved using a High Capacity cDNA Reverse Transcription Kit (Applied BioSystems) using the manufacturer recommended temperature settings for amplification. For qPCR, samples were amplified in triplicate using SsoAdvanced Universal SYBR Green Supermix (Bio Rad) and run through 39 cycles of 3 mins 10 secs at 98°C, 30 seconds of 60°C in a CGX96 Real-Time System (BioRad). Il-6 (primer, F: TAGTCCTTCCTACCCCAATTTCC, R: TGGTCCTTAGCCACTCCTTC) was amplified and reported as relative expression of transcript (RE) normalized to rpl5 (F: CTCGGATAGCAGCATGAGC, R: GGAGGAAGATGAAGATGCGTA) and to unshifted, unstimulated controls by ΔΔCt with the exception of BMAL1 KO, which was normalized to the STC. Rpl5 was used because in pilot experiments its expression was not induced by LPS (Unstimuled = 23.16±0.21, 1hr = 27.13±0.36, 3hr = 26.07±0.28, 5hr = 26.09±0.22, 7hr = 26.13±0.36, 9hr = 26.07±0.23, 11hr = 26.28±0.29; F6,35 = 1.844; DF = 6; p = 0.12).
Cytokine release
39 hours after the fourth phase advance at ZT3, tissue explants were transferred to fresh 35mm dishes with new media supplemented with LPS (n = 6) or PBS (n = 6) for 24 hours at which time an aliquot of the media was collected and frozen for analysis. Secretion of IL6 was assayed using Mouse IL-6 ELISA Set (Lower detection limit: 3.8pg/mL, BD Biosciences) in triplicate and were measured using a Spectra Max M5 plate reader (Molecular Devices).
Statistics
For all experiments, n = 6 was used. Pilot experiments determined that this was a sufficient sample size to yield statistical power. Student’s t-test or two-way ANOVA were used as indicated in the figure legends. Posthoc analysis was done using Sidak’s multiple comparisons.
Results
Temperature entrainment of explanted spleens
Under constant 37°C conditions, explanted spleens have a free-running period of 23.82±0.79 hours which persisted though for only 5 days (blue, Fig 1C). In contrast, when maintained in an STC, explants retained measurable daily oscillations in mPer2 for 10 days (black), consistent with the hypothesis that temperature acts to synchronize the circadian clocks in explanted spleens. This is further supported by a lengthening of the endogenous free-running period to 24.29±1.24 under a 24-hr STC. As expected, an ATC causes the clock to run faster in explanted spleens, as measurable by the advanced peak expression time of mPer2 bioluminescence following an advance in the temperature cycle which are intermediate between the old and new temperature cycle. Under ATC conditions, explants had an average period of 21.43±0.21 hours while shifting persists which allows explants to complete a 6hr phase advance in ~36 hours. When returned to an STC following 4 phase advances, the period lengthens to 23.10±0.33 hours compared to 21.93±0.19 hours when released into constant 37°C. Importantly, both re-entrainment (green) and free-running (red) commence from the peak time of the last shift as opposed to the peak of per2 expression prior to shifting (Fig 1C), further suggesting resetting of the circadian clock rather than masking by temperature.
Effect of shift number on expression of il6
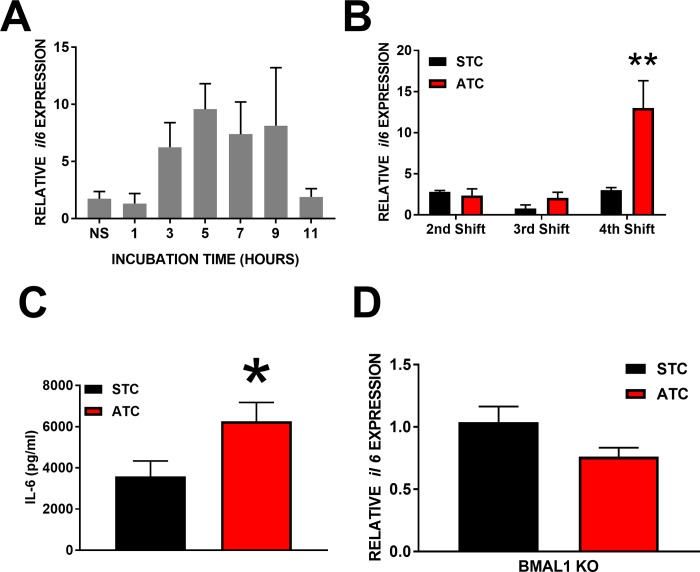
Optimal LPS incubation duration was experimentally determined as being maximal after approximately 6 hours of exposure in spleen explants (Fig 2A). After an STC, LPS drove an increase in il6 expression after the equivalent of two (2.80±0.18 RE), three (0.80±0.43 RE), or four (3.02±0.30 RE) phase advances in the ATC group, respectively. Following exposure to an ATC, LPS induced an increase in il6 expression after two (2.36±0.81 RE), three (2.08±0.67 RE) or four (12.99±3.33 RE) phase advances. Two-way ANOVA of il-6 induction following exposure to STC or ATC indicated that there was a main effect of both temperature cycle (p<0.05; f2,30 = 11.82; DF = 2) and number of shifts (p<0.05, F2,30 = 9.33, DF = 2), as well as a significant interaction (p<0.05, F1,30 = 7.47, DF = 2). Multiple comparisons revealed this was driven by increased induction of il-6 in the ATC following the 4th advance specifically (Fig 2B).
Fig 2. Immune consequences of circadian disruption in vitro.
A: Induction of il-6 by LPS exposure of increasing duration following four 6hr phase advances (in hours; n = 6 per condition, NS signifies not stimulated with LPS). B: Induction of il-6 by LPS. Following 2, 3, or 4 six-hour phase advances (ATC; red; n = 6 per condition) or the same duration of a static temperature cycle (STC; black; n = 6 per condition), spleen explants were challenged with LPS for 6-hr. Significance determined by Two-way ANOVA. C: IL-6 protein secreted by spleen explants subjected to a 6-hr LPS challenge after being maintained in a static (STC; black; n = 6) or advancing (ATC; red; n = 6) temperature cycle (4 phase advances). Significance determined by ANOVA. D: Induction of IL-6 transcript by LPS in BMAL1KO spleen explants following either a static (STC; black; n = 6) or 4 shifts in an advancing (ATC; red; n = 6) temperature cycle (4 phase advances). Significance determined by Two-Way ANOVA (B) or Student’s T-Test (C and D). *p<0.05; **p<0.01.
Secretion of IL6 after 4 phase advances
The increase in il6 was confirmed by measuring the amount of the IL6 protein in the media following the 4th shift (3590.35±744.70 pg/μl vs. 6264.85±911.62 pg/μl, respectively; p<0.05; t = 2.27, DF = 10; Fig 2C).
Effect of advancing temperature cycles with no circadian clock
Spleen explants from BMAL1KO mice exposed to an ATC did not exhibit an increase in il6 expression (1.04±0.13 RE) compared with BMAL1KO spleen explants under STC (0.76±0.07 RE; p>0.10; t = 1.92; DF = 10; Fig 2D).
Temperature entrainment of explanted lung
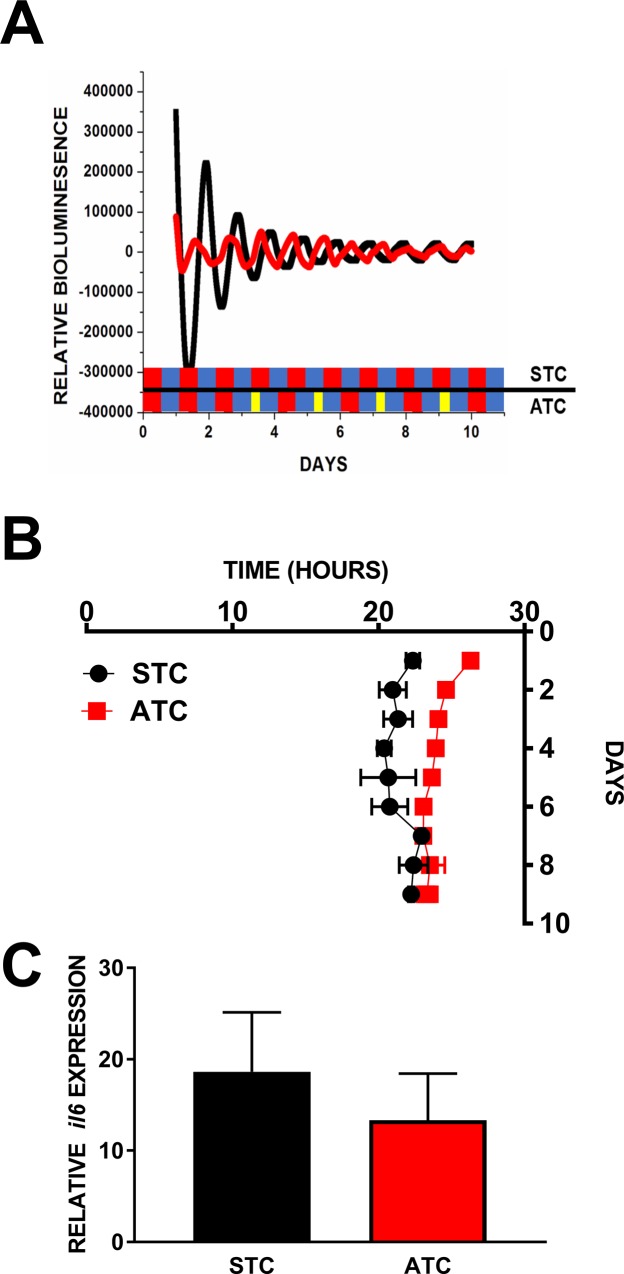
In contrast to spleen explants, explanted lung tissue cultures did not appear to entrain to temperature (24.01±0.38 hours vs 24.37±0.43 hours, STC and ATC respectively; Fig 3A and 3B).
Fig 3. Tissue specificity of ECD in immunological function.
Lung explants are not entrained by temperature. A: Average Bioluminescence Traces. Relative Per2 bioluminescence traces for lung explants under static (STC; black; n = 6) and advancing (ATC; red; n = 6) temperature cycles (4 phase advances), schedule denoted in colored bars as in Fig 1B. B: Peak Per2 expression time plotted by day to demonstrate that period in lung is not affected by a static temperature (black; STC; n = 6) or advancing temperature cycle (ATC; red; n = 6). C: Induction of IL-6 transcript by LPS in lung explants following either a static (black; n = 6) or advancing (red; n = 6) temperature cycle. Lack of significance was confirmed by Student’s T-Test.
Induction of il6 by LPS following 4 phase advances in lung
There was no difference between lung explants maintained in STC and ATC in il-6 elicited by LPS (18.63±6.51 RE vs 13.33±5.10 RE, respectively; p>0.11; t = 0.64; DF = 10; Fig 3C).
Discussion
Need for a model of environmental circadian disruption
While the deleterious effects of circadian disruption have long been known, understanding how misaligning the internal clock with the external environment, however transiently, causes these effects remains obscure. Due to the inability to control for many behavioral, environmental, and genetic components of environmental circadian disruption, investigation of these causes in humans is extremely challenging. Animal models of circadian disruption allow for control of these external factors, which are very useful in understanding the difference between disruption of the relationship of the circadian clock with the environment compared with the effects of sleep loss, for example[31]. However, attempting to identify mechanisms regulating diffuse and complex processes, such as immunity, is impeded in these models by reciprocal interaction with other systems. Further, because the circadian system is organized hierarchically, with environmental input being integrated by the suprachiasmatic nucleus (SCN) of the hypothalamus and distributed to cell autonomous clocks throughout the body, identifying which levels of this hierarchy are important for mediating the effects of circadian disruption is difficult to accomplish. Much has been accomplished by using a reductionist approach in which individual clock genes are manipulated or eliminated in specific cell types to understand the role of the clock in maintaining normal physiological function. Critically however, these models do not reflect the physiological process that occurs during circadian disruption, which involves repeated, temporarily misalignment of clocks with their proximal and distal environments. Negative health outcomes in shiftwork disorders are not caused by an absent or broken circadian clock, but rather long-term, repeated exposure of a working clock to abnormal light/dark cycles. This suggests that a critical component of circadian disruption stems from a functional clock which is repeatedly establishing synchronization with the environment. In the present work, we have attempted to combine a reductionist approach with environmental circadian disruption to more closely model this repeated, temporary misalignment of the mammalian circadian system in vivo.
Environmental circadian disruption in spleen explants
It has been previously shown that the molecular circadian clock in some peripheral tissues, including spleen, can be reset by temperature [24, 25]. Here, we show that simply subjecting spleen explants to an environmental temperature cycle of 0.5°C is sufficient to more than double the number of days in which a population Per2 rhythm is detectable, compared with a static 37°C culture environment (Fig 1C). In addition, when exposed to an advancing temperature cycle, the period in Per2::luc expression is shortened such that the peak expression time occurs early on subsequent cycles (Fig 1B). Critically, when released into a non-advancing temperature cycle or a constant temperature following four consecutive shifts, the peak expression time of Per2 1) lengthens to readjust to environmental conditions, and 2) the phase of these oscillations begin from the phase of the last phase-advance (Fig1C). Taken together this evidence suggests real circadian phase resetting by temperature rather than masking of an unperturbed endogenous rhythm by an environmental stimulus.
Immune dysregulation by environmental circadian disruption
It has previously been shown that the cell autonomous circadian clock in immune cells is important for their function. Immune cell trafficking and recruitment is strongly regulated by the circadian system[16, 32, 33]. Clock genes are expressed in a highly circadian manner in macrophages, which corresponds to a circadian pattern in cytokine release in response to LPS stimulation [12]. These patterns can be eliminated or shifted by deleting components of the molecular clock such as BMAL1 or REV-ERBα [13], Clock [14], and Cry1/Cry2 [15]. Here, rather than break the clock, we sought to misalign a functional circadian clock (or small suite of clocks within an organ) with phase-resetting cues in the environment, which is hypothesized to be the cause of shift work disorders [20, 34]. We have previously reported that mice subjected to 4 consecutive 6hr phase advances and then given an immune challenge in the form of LPS injection demonstrate a number of pro-inflammatory markers, notably IL6 [8]. Importantly, when peritoneal macrophages are obtained from mice subjected this shifting schedule, an exaggerated immune response is still elicited ex vivo [30].
Here, we demonstrate that spleens removed from unshifted mice which were subjected to ECD and LPS challenge entirely ex vivo show a similar increase in il6 transcription and IL6 secretion following 4 consecutive 6hr phase advances (Fig 2B and 2C), as occurs in vivo. In addition, we show that this is a result of cumulative, rather than acute, circadian disruption as it is not evident after 2 or even 3 phase advances. This finding is also consistent with previous reports [8] and the etiology of shift work disorders [35, 36]. The exaggerated immune response to LPS is abolished in spleens explanted from BMAL1 KO mice (Fig 2D) which lack a circadian clock[37]. Recent work suggests that BMAL1 acts as a positive regulator of IL6 in astrocytes[38], and its loss could result in less IL6 release in response to inflammatory events. Whether this holds true in peripheral tissues such as spleen and lung is not presently known, but as IL6 production in that study was reduced rather than abolished, the lack of exaggerated il6 induction following ECD in BMAL1 KO mice in unlikely to be caused by any regulatory role of BMAL on IL6, but rather to reflect that the repeated shifting of the clock is responsible for the exaggerated response. Thus, though the loss of timing information from the SCN rapidly results in arrhythmicity of most behavioral and physiological processes [39–41], true circadian disruption can be executed in a reductionist model if an appropriate environmental cue is used. Interestingly, these results suggest that the SCN is not necessary for the development of misalignment, or the consequence of that misalignment, between the environment and peripheral oscillators. Further, because it is known that the SCN does independently regulate innate immune cell reactivity to LPS [42], use of a reductionist model in conjunction with in vivo studies can tease apart the roles of local vs global contributions to circadian disruption that occurs at the organismal level. Though we acknowledge that the spleen is a heterogenous aggregation of cell types and that we have not identified specific cell-types responsible, these results provide compelling evidence that environmental circadian disruption can be successfully applied to a reduced model of immune function.
Tissue-specific susceptibility to temperature as an entrainment signal
Having demonstrated that explanted spleens are amenable to circadian disruption using temperature as an environmental cue, we next wanted to evaluate whether this was generally true of mammalian tissue. The lung has strong Per2Luc rhythms ex-vivo [24] and contains a large number of innate immune cells which are highly reactive to bacterial endotoxin [43, 44]. Here we were able to observe strong daily rhythms in Per2 from explanted lung tissue but were unable to observe any change in Per2 expression in response to an advancing temperature cycle (Fig 3A and 3B). This contrasts with previous work showing the lung capable of shifting phase in response to temperature pulses [24]. However, in that study temperature pulses of 2.5°C (36–38.5°C) were used which elicited strong phase resetting in all tissues suggesting such a large pulse acts as a very powerful entraining cue. In the present work, we found that 0.5°C temperature oscillations (37–36.5°C) were sufficient to induce phase resetting of explanted spleens such that transient peaks were observed on subsequent cycles during re-entrainment indicating perhaps a physiologically relevant environmental cue [45]. Taken together, these studies support the conclusion that while the circadian clock in lung cells can be affected by large temperature changes, it is less sensitive than the spleen to temperature as an entrainment cue. Such differential sensitivity allowed us to test whether the clock actually needs to shift in response to the environmental signal in order to produce such consequences. In contrast with spleen explants, following four 6-hr phase advances in temperature, when lung explants were challenged with bacterial endotoxin there was no difference in il-6 induction when compared with explants maintained in a non-advancing temperature cycle (Fig 3C). The lung is composed of diverse cell types with a variety of inflammatory profiles which were not identified or sorted here but could dramatically affect the LPS response, though a useful population response was obtained from randomized explants in order to minimize the impact of the lung’s physiology on the present results. This is important because it suggests that the presence of a functional circadian clock alone is not sufficient to ensure dysregulated immune function in the presence of ECD, the clock needs to be forced to shift. At present it is not known whether temperature is an important entraining cue in vivo, and additional work is required to conclusively demonstrate a potential role of differential temperature sensitivity of peripheral oscillators. If true, it would provide evidence for a complex network of peripheral oscillators which have their own milieu of entrainment signals which determine the timing of their activity.
Conclusions
In the present study we have demonstrated that at least some of the in vivo effects of circadian disruption on immune function can be recapitulated using a reductionist model of environmental circadian disruption. Given the importance of understanding how circadian disruption negatively impacts physiology, this model, particularly used in conjunction with in vivo studies, could be an important tool to identify mechanisms of circadian disruption at the cellular and organismal level. Importantly, because the organotypic culture approach in the present work did not allow for the identification of cell types or putative mechanisms involved in mediating these effects, future work should take this model even further and address these questions in a cell-type specific manner.
Data Availability
All files are available from the OSF database DOI 10.17605/OSF.IO/5ZGQ4.
Funding Statement
This work was supported by the National Institute of General Medical Sciences of the National Institute of Health (https://www.nigms.nih.gov/) grant number SC1GM112567 to Alec J Davidson. The funding agency played no role in study design, data collection or analysis, decision to publish, or preparation of this manuscript.
References
- 1.Straif K, Baan R, Grosse Y, Secretan B, El Ghissassi F, Bouvard V, et al. Carcinogenicity of shift-work, painting, and fire-fighting. The Lancet Oncology. 2007;8(12):1065–6. Epub 2007/12/01. 10.1016/S1470-2045(07)70373-X . [DOI] [PubMed] [Google Scholar]
- 2.Lahti TA, Partonen T, Kyyronen P, Kauppinen T, Pukkala E. Night-time work predisposes to non-Hodgkin lymphoma. International journal of cancer Journal international du cancer. 2008;123(9):2148–51. Epub 2008/08/13. 10.1002/ijc.23566 . [DOI] [PubMed] [Google Scholar]
- 3.Conlon M, Lightfoot N, Kreiger N. Rotating shift work and risk of prostate cancer. Epidemiology (Cambridge, Mass). 2007;18(1):182–3. Epub 2006/12/21. 10.1097/01.ede.0000249519.33978.31 . [DOI] [PubMed] [Google Scholar]
- 4.Magrini A, Pietroiusti A, Coppeta L, Babbucci A, Barnaba E, Papadia C, et al. Shift work and autoimmune thyroid disorders. International journal of immunopathology and pharmacology. 2006;19(4 Suppl):31–6. Epub 2007/02/13. . [PubMed] [Google Scholar]
- 5.Karlsson B, Knutsson A, Lindahl B. Is there an association between shift work and having a metabolic syndrome? Results from a population based study of 27,485 people. Occupational and environmental medicine. 2001;58(11):747–52. Epub 2001/10/16. 10.1136/oem.58.11.747 ; PubMed Central PMCID: PMCPmc1740071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morikawa Y, Nakagawa H, Miura K, Soyama Y, Ishizaki M, Kido T, et al. Shift work and the risk of diabetes mellitus among Japanese male factory workers. Scandinavian journal of work, environment & health. 2005;31(3):179–83. Epub 2005/07/08. . [DOI] [PubMed] [Google Scholar]
- 7.Beker MC, Caglayan B, Yalcin E, Caglayan AB, Turkseven S, Gurel B, et al. Time-of-Day Dependent Neuronal Injury After Ischemic Stroke: Implication of Circadian Clock Transcriptional Factor Bmal1 and Survival Kinase AKT. Molecular neurobiology. 2018;55(3):2565–76. Epub 2017/04/20. 10.1007/s12035-017-0524-4 . [DOI] [PubMed] [Google Scholar]
- 8.Castanon-Cervantes O, Wu M, Ehlen JC, Paul K, Gamble KL, Johnson RL, et al. Dysregulation of inflammatory responses by chronic circadian disruption. J Immunol. 2010;185(10):5796–805. 10.4049/jimmunol.1001026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guerrero-Vargas NN, Guzman-Ruiz M, Fuentes R, Garcia J, Salgado-Delgado R, Basualdo Mdel C, et al. Shift Work in Rats Results in Increased Inflammatory Response after Lipopolysaccharide Administration: A Role for Food Consumption. J Biol Rhythms. 2015;30(4):318–30. Epub 2015/05/29. 10.1177/0748730415586482 . [DOI] [PubMed] [Google Scholar]
- 10.Halberg F, Johnson EA, Brown BW, Bittner JJ. Susceptibility rhythm to E. coli endotoxin and bioassay. Proceedings of the Society for Experimental Biology and Medicine. 1960;103(1):142–4. [DOI] [PubMed] [Google Scholar]
- 11.Davidson AJ. Chronic jet-lag increases mortality in aged mice. Current Biology. 2006;16(21):R914–6. 10.1016/j.cub.2006.09.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, Volk HD, et al. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci U S A. 2009;106(50):21407–12. 10.1073/pnas.0906361106 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, Boyce SH, et al. The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci U S A. 2012;109(2):582–7. Epub 2011/12/21. 10.1073/pnas.1106750109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellet MM, Deriu E, Liu JZ, Grimaldi B, Blaschitz C, Zeller M, et al. Circadian clock regulates the host response to Salmonella. Proc Natl Acad Sci U S A. 2013;110(24):9897–902. Epub 2013/05/30. 10.1073/pnas.1120636110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narasimamurthy R, Hatori M, Nayak SK, Liu F, Panda S, Verma IM. Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines. Proc Natl Acad Sci U S A. 2012;109(31):12662–7. Epub 2012/07/11. 10.1073/pnas.1209965109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato S, Sakurai T, Ogasawara J, Takahashi M, Izawa T, Imaizumi K, et al. A circadian clock gene, Rev-erbalpha, modulates the inflammatory function of macrophages through the negative regulation of Ccl2 expression. J Immunol. 2014;192(1):407–17. Epub 2013/12/07. 10.4049/jimmunol.1301982 . [DOI] [PubMed] [Google Scholar]
- 17.Kervezee L, Cuesta M, Cermakian N, Boivin DB. Simulated night shift work induces circadian misalignment of the human peripheral blood mononuclear cell transcriptome. Proc Natl Acad Sci U S A. 2018;115(21):5540–5. Epub 2018/05/08. 10.1073/pnas.1720719115 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leproult R, Holmback U, Van Cauter E. Circadian misalignment augments markers of insulin resistance and inflammation, independently of sleep loss. Diabetes. 2014;63(6):1860–9. Epub 2014/01/25. 10.2337/db13-1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris CJ, Purvis TE, Hu K, Scheer FA. Circadian misalignment increases cardiovascular disease risk factors in humans. Proc Natl Acad Sci U S A. 2016;113(10):E1402–11. 10.1073/pnas.1516953113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris CJ, Purvis TE, Mistretta J, Hu K, Scheer F. Circadian Misalignment Increases C-Reactive Protein and Blood Pressure in Chronic Shift Workers. J Biol Rhythms. 2017;32(2):154–64. Epub 2017/03/30. 10.1177/0748730417697537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris CJ, Purvis TE, Mistretta J, Scheer FA. Effects of the Internal Circadian System and Circadian Misalignment on Glucose Tolerance in Chronic Shift Workers. The Journal of clinical endocrinology and metabolism. 2016;101(3):1066–74. Epub 2016/01/16. 10.1210/jc.2015-3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright KP, Bogan RK Jr., Wyatt JK. Shift work and the assessment and management of shift work disorder (SWD). Sleep medicine reviews. 2013;17(1):41–54. Epub 2012/05/09. 10.1016/j.smrv.2012.02.002 . [DOI] [PubMed] [Google Scholar]
- 23.Spengler ML, Kuropatwinski KK, Comas M, Gasparian AV, Fedtsova N, Gleiberman AS, et al. Core circadian protein CLOCK is a positive regulator of NF-kappaB-mediated transcription. Proc Natl Acad Sci U S A. 2012;109(37):E2457–65. Epub 2012/08/17. 10.1073/pnas.1206274109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science (New York, NY). 2010;330(6002):379–85. Epub 2010/10/16. 10.1126/science.1195262 ; PubMed Central PMCID: PMCPmc3625727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saini C, Morf J, Stratmann M, Gos P, Schibler U. Simulated body temperature rhythms reveal the phase-shifting behavior and plasticity of mammalian circadian oscillators. Genes & development. 2012;26(6):567–80. Epub 2012/03/02. 10.1101/gad.183251.111 ; PubMed Central PMCID: PMCPmc3315118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gossan N, Zeef L, Hensman J, Hughes A, Bateman JF, Rowley L, et al. The circadian clock in murine chondrocytes regulates genes controlling key aspects of cartilage homeostasis. Arthritis and rheumatism. 2013;65(9):2334–45. Epub 2013/07/31. 10.1002/art.38035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101(15):5339–46. Epub 2004/02/14. 10.1073/pnas.0308709101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davidson AJ, Castanon-Cervantes O, Leise TL, Molyneux PC, Harrington ME. Visualizing jet lag in the mouse suprachiasmatic nucleus and peripheral circadian timing system. The European journal of neuroscience. 2009;29(1):171–80. Epub 2008/11/27. 10.1111/j.1460-9568.2008.06534.x . [DOI] [PubMed] [Google Scholar]
- 29.Yamazaki S, Takahashi JS. Real-time luminescence reporting of circadian gene expression in mammals. Methods in enzymology. 2005;393:288–301. Epub 2005/04/09. 10.1016/S0076-6879(05)93012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams KL, Castanon-Cervantes O, Evans JA, Davidson AJ. Environmental circadian disruption elevates the IL-6 response to lipopolysaccharide in blood. J Biol Rhythms. 2013;28(4):272–7. 10.1177/0748730413494561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brager AJ, Ehlen JC, Castanon-Cervantes O, Natarajan D, Delisser P, Davidson AJ, et al. Sleep loss and the inflammatory response in mice under chronic environmental circadian disruption. PloS one. 2013;8(5):e63752 Epub 2013/05/23. 10.1371/journal.pone.0063752 ; PubMed Central PMCID: PMCPmc3656961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Druzd D, Matveeva O, Ince L, Harrison U, He W, Schmal C, et al. Lymphocyte Circadian Clocks Control Lymph Node Trafficking and Adaptive Immune Responses. Immunity. 2017;46(1):120–32. Epub 2017/01/15. 10.1016/j.immuni.2016.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheiermann C, Kunisaki Y, Lucas D, Chow A, Jang JE, Zhang D, et al. Adrenergic nerves govern circadian leukocyte recruitment to tissues. Immunity. 2012;37(2):290–301. Epub 2012/08/07. 10.1016/j.immuni.2012.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright KP, Drake AL Jr., Frey DJ, Fleshner M, Desouza CA, Gronfier C, et al. Influence of sleep deprivation and circadian misalignment on cortisol, inflammatory markers, and cytokine balance. Brain, behavior, and immunity. 2015;47:24–34. Epub 2015/02/03. 10.1016/j.bbi.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knutsson A, Hallquist J, Reuterwall C, Theorell T, Akerstedt T. Shiftwork and myocardial infarction: a case-control study. Occupational and environmental medicine. 1999;56(1):46–50. Epub 1999/05/26. 10.1136/oem.56.1.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan A, Schernhammer ES, Sun Q, Hu FB. Rotating night shift work and risk of type 2 diabetes: two prospective cohort studies in women. PLoS medicine. 2011;8(12):e1001141 Epub 2011/12/14. 10.1371/journal.pmed.1001141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103(7):1009–17. Epub 2001/02/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakazato R, Hotta S, Yamada D, Kou M, Nakamura S, Takahata Y, et al. The intrinsic microglial clock system regulates interleukin-6 expression. Glia. 2017;65(1):198–208. Epub 2016/10/12. 10.1002/glia.23087 . [DOI] [PubMed] [Google Scholar]
- 39.Stephan FK, Zucker I. Circadian Rhythms in Drinking Behavior and Locomotor Activity of Rats Are Eliminated by Hypothalamic Lesions. Proceedings of the National Academy of Sciences of the United States of America. 1972;69(6):1583–6. PMC426753. 10.1073/pnas.69.6.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore RY, Eichler VB. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain research. 1972;42(1):201–6. Epub 1972/07/13. . [DOI] [PubMed] [Google Scholar]
- 41.Rusak B. The role of the suprachiasmatic nuclei in the generation of circadian rhythms in the golden hamster,Mesocricetus auratus. Journal of comparative physiology. 1977;118(2):145–64. 10.1007/BF00611819 [DOI] [Google Scholar]
- 42.Guerrero-Vargas NN, Salgado-Delgado R, Basualdo Mdel C, Garcia J, Guzman-Ruiz M, Carrero JC, et al. Reciprocal interaction between the suprachiasmatic nucleus and the immune system tunes down the inflammatory response to lipopolysaccharide. Journal of neuroimmunology. 2014;273(1–2):22–30. Epub 2014/06/12. 10.1016/j.jneuroim.2014.05.012 . [DOI] [PubMed] [Google Scholar]
- 43.Morales-Nebreda L, Misharin AV, Perlman H, Budinger GR. The heterogeneity of lung macrophages in the susceptibility to disease. European respiratory review: an official journal of the European Respiratory Society. 2015;24(137):505–9. Epub 2015/09/02. 10.1183/16000617.0031-2015 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sender V, Stamme C. Lung cell-specific modulation of LPS-induced TLR4 receptor and adaptor localization. Communicative & integrative biology. 2014;7:e29053 Epub 2014/08/20. 10.4161/cib.29053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winfree AT. Biological rhythms and the behavior of populations of coupled oscillators. Journal of theoretical biology. 1967;16(1):15–42. Epub 1967/07/01. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All files are available from the OSF database DOI 10.17605/OSF.IO/5ZGQ4.