Abstract
PURPOSE:
Radiomics is the extraction of multidimensional imaging-features which when correlated with genomics is termed radiogenomics. However, radiogenomic biological validation is not sufficiently described in the literature. We seek to establish causality between differential gene expression status and MRI-extracted radiomic-features in glioblastoma.
METHODS:
Radiogenomic predictions and validation were done using the Cancer Genome Atlas and Repository of Molecular Brain Neoplasia Data glioblastoma patients (N=93) and orthotopic xenografts (OX)(N=40). Tumor phenotypes were segmented, and radiomic-features extracted using the developed radiome-sequencing pipeline. Patients and animals were dichotomized based on Periostin (POSTN) expression levels. RNA and protein levels confirmed RNAi-mediated POSTN knockdown in OX. Total RNA of tumor cells isolated from mouse brains (knockdown and control) was used for microarray-based expression profiling. Radiomic-features were utilized to predict POSTN expression status in patient, mouse, and inter-species.
RESULTS:
Our robust pipeline consists of segmentation, radiomic-feature extraction, feature normalization/selection, and predictive-modeling. The combination of skull stripping, brain-tissue focused normalization and patient-specific normalization are unique to this study, providing comparable cross-platform, cross-institution radiomic-features. POSTN expression status was not associated with qualitative or volumetric MRI parameters. Radiomic-features significantly predicted POSTN expression status in patients (AUC 76.56%, sensitivity/specificity: 73.91/78.26%) and OX (AUC 92.26%, sensitivity/specificity: 92.86%/91.67%). Furthermore, radiomic-features in OX were significantly associated with patients with similar POSTN expression levels (AUC 93.36%, sensitivity/specificity: 82.61%/95.74%; p-value 02.021E-15).
CONCLUSION:
We determined causality between radiomic texture features and POSTN expression levels in a pre-clinical model with clinical validation. Our biologically validated radiomic pipeline also showed the potential application for human-mouse matched co-clinical trials.
Keywords: radiomics, radiogenomics, validation, glioma, POSTN
INTRODUCTION
Imaging has transformed the medical field by providing a non-invasive method to interrogate the human body and underlying biological processes. Particularly in cancer patients, imaging plays a key role throughout the entire treatment paradigm ranging from diagnosis and assessing treatment response to post-care follow-up. However, despite recent exponential refinements in imaging technologies in terms of acquisition time and resolution, we have barely begun to tap the potential of imaging to characterize tissues or tumors beyond qualitative description or gross tumor size on routine imaging sequences. In fact, all current imaging assessment criteria (such as response evaluation criteria in solid tumors (RECIST), response assessment in neuro-oncology (RANO), immune related RECIST (irRECIST), and immune related response criteria (irRC)) used to evaluate tumor response in the clinical setting and in clinical trials are dependent on changes in tumor size and do not accurately capture responses to therapy [1–4]. With the development of immune and molecularly targeted therapies, alternative imaging assessment criteria and markers are needed that go beyond mere changes in tumor size. In fact, with some therapies such as immunotherapy, positive response to treatment is associated with initial decrease in tumor size in only 10% of patients, while other positive-responders fail to show such a measurable initial decline in tumor size [4]. This uncoupling of tumor metrics and response to therapy is a clinical dilemma and challenge for clinicians. Further, the specific “makeup” of a tumor’s imaging characteristics reflects underlying molecular processes, which cannot be evaluated using changes in tumor size alone. Unlocking the full breadth of data contained in routine imaging studies would enable the identification of high quality, noninvasive, clinically relevant, and actionable imaging markers.
Radiomics, an emerging field within imaging, is poised to do just that. An automated high-throughput extraction of multidimensional imaging features (on a pixel and voxel level), radiomics captures microscale information hidden within conventional imaging and beyond what is visible to the naked human eye [5–7]. Like the layer of complexity and resolution that genomics has added to tumor biology, radiomics similarly adds to conventional imaging. However, a critical need exists to understand the biological underpinnings of imaging, specifically radiomics, for informed clinical decision making. This also necessitates a proper understanding of how changes in genomics affect imaging information. With radiomics, it is now realistic to begin testing precise linkages and investigate causality between imaging and genomics. Understanding the molecular bedrocks of radiomics is important due to imaging’s inherent advantages; in contrast to genomic analyses which are often performed on only a fraction of the tumor, an advantage of imaging is that it assesses the entire three-dimensional tumor volume inclusive of spatial heterogeneity [8, 9]. Recent studies have demonstrated an association between imaging features (also termed phenotypes) and cancer histology, tumor grades, and genomics [5, 10–13]. The linkage of imaging phenotypes with genomic data is termed radiogenomics (also termed imaging genomics)[10]. Fundamental to radiogenomics is the hypothesis that expression of specific sets of genes or driver mutations impact the extractable imaging features, but this has not been directly demonstrated in a highly robust model.
Imaging-molecular connections must be established in a robust manner, specifically by establishing causality (as opposed to correlations), for its acceptance as a validated tool. To establish causality, a closed system is required for in-vivo manipulation of genomic expression patterns and linkage to radiomic features using a preclinical model system. A few studies have tried to elucidate the biological significance of imaging characteristics [14, 15]. Joo et al. demonstrated that xenograft tumors produced from patient-derived glioblastoma (GBM) cell lines were similar to their paired human parental tumors [14]. In the latter study, they found the invasiveness of the parental tumor on MRI correlated with invasive measures based on paraffin histopathology sections of the xenograft tumor. In a different study using a heterotopic colorectal cancer mouse model, Panth et al. found changes in computerized tomography (CT) imaging features after induced alterations in gene expression and radiation treatment [15]. However, to date, the linkages between imaging and genomics largely remain at a correlative stage and lack established causality in a model system relevant for human disease.
In this study, we seek to address this gap in knowledge which can be expected to change patient management. To our knowledge, this is the first study to establish causality between imaging-molecular (radiogenomic) features. In this study, we bring forward an approach for an integrated end-to-end methodology for extraction of imaging radiomic features and its validation in both a preclinical closed model system and in a clinical patient cohort. We use the most common primary malignant brain tumor characterized by extreme heterogeneity and poor patient survival [16–18], GBM, as the solid cancer prototype in this study. Periostin (POSTN), was used as the gene prototype, given its previous radiogenomic description in the first published quantitative radiogenomic study in glioma of the Cancer Genome Atlas (TCGA)[13]; its more recent surfacing as an important gene involved in GBM invasion[19], one of the most important causes of GBM recurrence; and its potential use as a GBM therapeutic target in developing clinical trials[20].
METHODS
Patient-derived imaging, clinical and genomic material were collected from TCGA, The Cancer Imaging Archive (TCIA) and the Repository of Molecular Brain Neoplasia Data (REMBRANDT); all patient data collection were HIPAA compliant and approved by MDACC’s Institutional Review Board (PA130720). Analyses were based on a total of 79 TCGA patients and 14 REMBRANDT patients for whom complete baseline MRI data and POSTN gene expression data were available. Detailed demographic information is presented in Table 1a and b.
Table 1a:
TCGA Patient Demographic Table.
| Patients with High POSTN level expression (n=39) | Patients with Low POSTN level expression (n=40) | ||
|---|---|---|---|
| Age (years) | 60 ±24 | 63±18 | |
| Sex (male/female) | 28(71.7%)/11(28.2%) | 23(57.5%)/17(42.5%) | |
| KPS (≥80/<80/Not reported) | 25(64.1%)/4(10.2%)/ 9(23 | 29(72.5%)/6(15%)/5(12,5%) |
Table 1b:
Rembrandt Patient Demographic Table *.
| Patients with High POSTN level expression (n= 7) | Patients with Low POSTN level expression (n= 7) | ||
|---|---|---|---|
| Age (years) | 59±12 | 51.5±5 | |
| Sex (male/female/not reported) | 3 (43%)/2(28.5%)/2 (28.5) | 1 (14.25%)/1(14.25)/5(71.5%) | 0.552 |
| KPS (≥80/not reported) | 4(57%)/3(43%) | 2(28.5%)/5 (71.5%) | 0.473 |
demographic information was missing for 7 out of the 14 patients used from Rembrandt database.
For the preclinical portion of the study, MD Anderson Cancer Center (MDACC) patient–derived glioma stem cells (GSC) lines were isolated in accordance with a protocol (LAB04–0001) approved by MDACC’s Institutional Review Board. Patient consent was obtained. For the cell study, the IRB issued a waiver of informed consent since data collection was retrospective. All animal experiments complied with institutional regulations and was approved by MDACC’s Institutional Animal Care and Use Committee (protocol 00001100) in accordance with the guidelines of the American Association for Laboratory Animal Science.
Image segmentation
Image analysis and software
We used 3D Slicer version 4.3.1 (www.slicer.org), an open-source image analytics platform, for tumor segmentation [21–23]. The segmented images were reviewed in consensus by two board-certified neuroradiologists with 9 (R.R.C.) and 35 (A.J.K.) years of experience. For the segmentation of the mouse MRI, we followed an approach similar to that used for the patient imaging analysis using 3D Slicer software. In brief, conventional MRI series were co-registered to bring all mouse images to same space, using a registration method identical to the method used in humans (detailed below). Then, we outlined different tumor phenotypes including edema, enhancement and necrosis on the corresponding imaging series similar to the approach used in humans (detailed below). The image analysis team was blinded to the patient cohort as well as animal groups (control vs. knockdown).
Volume selection
We used contrast-enhanced axial T1-weighted imaging (T1WI) sequences and pre-contrast axial Fluid-Attenuated Inversion Recovery (FLAIR) sequences. Contrast-enhanced T1WI was used for segmentation of the enhancing component (i.e., active enhancing tumor) and non-enhancing central tumor component (i.e., necrosis). The edema/invasion portion was segmented using the FLAIR sequence; this region was assessed based on the peritumoral hyperintensity seen on the FLAIR sequence (Fig. 1a). Hyperintensity on pre- contrast T1WI was used to assess hemorrhage. Of the 93 TCGA/TCIA/REMBRANDT patients, 1 was scanned with a 1T system (1.08%), 57 were scanned with a 1.5T system (61.3%), and 26 were scanned with a 3T system (28%). Scanner strength information was not available for 9 patients (9.7%). Standard imaging parameters (slice thickness, voxel size, and slice gap) were used for each sequence (Supplementary Tables 1a, 1b).
Figure 1: Radiome sequencing pipeline for solid tumor (glioblastoma) and tissue characterization.
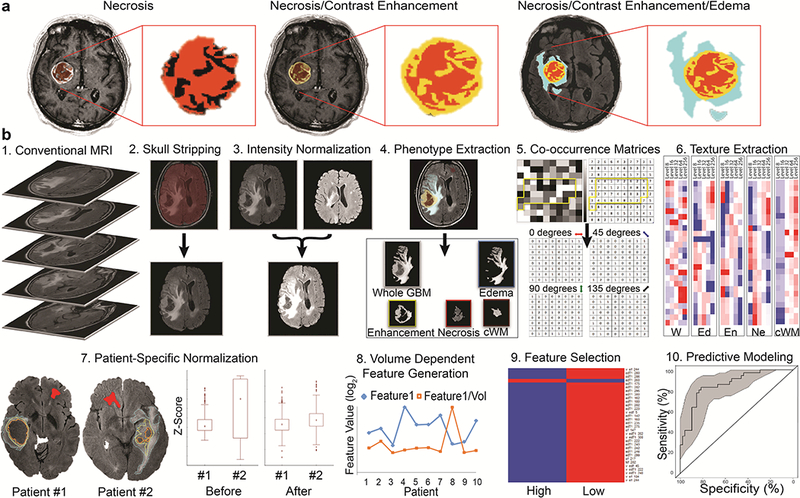
(a) Semi- automated segmentation of the three imaging phenotypes: necrosis (left), post–contrast active enhancing tumor (middle), and Fluid Attenuated Inversion Recovery (FLAIR) peritumoral edema/invasion, representing edematous tumor as well as sites of cellular invasion into brain tissue (right). (b) Automated segmentation-based radiomic feature extraction is followed by patient specific normalization and feature selection, which consists of the normalization of contralateral normal-appearing white matter as an internal control normalizer to account for various potential intra- and inter- institutional biases, a crucial step that ensures comparability. The next step is volume-dependent feature generation, which uses the necrosis, post–contrast enhancement, and FLAIR volumes for the corresponding radiomic feature sets acquired from the respective sequences, thus doubling the amount of radiomic features and creating a set of tumor volume–independent and –dependent radiomic features. Finally, this homogenous dataset can then enter feature selection and predictive modeling for any GBM trait of interest.
Image registration, segmentation, and model making
Image registration and segmentation were done as previously published by our group[24]. Prior to image segmentation, within-patient image registration was performed to align the FLAIR imaging and contrast-enhanced T1WI into the same geometric space. Images were registered using the General Registrations (BRAINS) Toolbox in 3D Slicer (affine registration, 12 degrees of freedom; Interpolation mode: Nearest Neighbor; https://www.slicer.org/wiki/Documentation/4.8/Registration/RegistrationLibrary), and the transformation matrix that maps each point of the reference image to the target’s image space was obtained. An adequate registration was designated as having an error of 2mm or less; all patient data were reviewed post-registration and if the error was more than 2mm then the patient was not included in the study. After image registration, images were segmented from the periphery (edema/invasion) to center (necrosis). Three distinct imaging phenotypes were segmented semi-automatically: edema/invasion, active enhancing tumor, and necrosis. A region of the contralateral normal-appearing white matter (opposite hemisphere and opposite lobe with respect to a line perpendicular to the midline) was also segmented for within-sequence normalization of the data. Hemorrhage appearing as hyperintensity on pre-contrast T1WI and contrast-enhancing vessels were subtracted from the FLAIR region to prevent contamination of the radiomic features obtained thereafter. The outlines of all segmented phenotypes were saved in a label volume. Finally, the inverted transformation matrix was applied to the label volume using nearest-neighbor interpolation to propagate the label volume to the original image. The volume of each phenotype was calculated by multiplying the number of voxels in a phenotype with the voxel volume.
Radiome sequencing textural analysis
Image preprocessing
An overview of the approach used for radiomic feature extraction is shown in Fig. 1b. All MRI scans were processed using the same pipeline. In the first step, we used FMRIB’s Brain Extraction Tool (BET) (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/BET) to remove non-brain tissue from the anatomical MR images (Fig. 1b) [25, 26]. Contrast-enhanced T1WI was used as an input to BET. In all runs, the BET option for “bias field correction and neck removal” was selected. The resulting brain mask from BET was applied to the FLAIR sequences, thus removing non-brain tissue from those images also. In the second step, to account for scanner differences, we applied the Nyul intensity normalization algorithm to standardize the intensity scales across MR images of the same contrast (brain-specific normalization) (Fig. 1b) [27–29]. In brief, Nyul normalization involves the matching of the histograms between a reference image and the image of each subject in the cohort. In our implementation, firstly, the MR images (FLAIR and contrast-enhanced T1WI) of one patient from the cohort were randomly selected as the reference, and the corresponding histograms were obtained; the intensities of the tumor were excluded from the calculation of the histogram, because the tumor is an abnormal environment. In the next step, for each patient in the cohort, the histograms were mapped via a piecewise linear transformation to the reference histogram; thus, the resulting transformed image has similar range of intensities. Finally, we extracted radiomic textural features from each phenotype (i.e., edema/invasion, active enhancing tumor, and necrosis, and contralateral normal-appearing white matter) as well as the whole tumor volume on the FLAIR sequences and contrast-enhanced T1WI. We followed an identical approach for processing the mouse MRI prior to radiomic feature calculation, except that we omitted the skull-stripping step.
Textural radiome sequencing
Whole radiome sequencing was performed, and the radiomic features were extracted for each phenotype and whole-tumor volume, using: a) intensity-level histogram (first-order features), and b) gray-level co-occurrence matrices (GLCMs; second-order features) [30]. The intensity-level histogram is a function showing the number of voxels that have a specific intensity in the original volume of interest; thus, the extracted features provide information about the intensity distribution within the volume of interest. From each intensity-level histogram, the following 10 features were extracted: minimum, maximum, mean, standard deviation, skewness, kurtosis and four percentiles (1%, 5%, 95%, 99%) [31, 32].
GLCM is a matrix computed from the original volume of interest, and each element of the matrix Ai,j represents the joint probability of two pixels with gray levels i and j with distance d apart and a certain angular direction θ. In our implementation, the GLCMs were calculated in four angular directions corresponding to in-plane rotations (θ=0°, 45°, 90°, 135°) and d=1 voxel (Fig. 1b). Each GLCM is normalized to the sum of co-occurrence pairs; thus, the extracted features are independent of the number of original observations. From each GLCM, the following 20 texture features were extracted: autocorrelation, contrast, correlation, cluster shade, cluster prominence, dissimilarity, energy, entropy, homogeneity, maximum probability, variance, sum average, sum variance, sum entropy, difference variance, difference entropy, information measure of correlation 1 and 2, inverse difference moment, and normalized inverse difference moment [30, 33, 34]. To obtain invariant measures of the features across different rotations, we calculated the average, range, and angular variance of each feature for different θ, thereby resulting in 60 rotation-invariant texture features for each volume of interest for every gray level.
In our implementation, the original images were discretized into N number of gray levels, where N=8, 16, 32, 64, or 256. Reducing the number of gray levels increases the signal-to-noise ratio and eliminates sparseness in the final GLCM. Intensity-level histogram-based features were calculated using images discretized at 256 gray levels and GLCM-based features were calculated using all gray levels. In summary, our analysis resulted in a total of 310 radiomic features for each phenotype per MRI sequence. Taking into account the two MRI sequences (contrast-enhanced T1WI and T2/FLAIR) along with the phenotypes (edema/invasion, enhancing active tumor, and necrosis) and whole-tumor volume, a total of 2,480 radiomic features (2,400 second-order and 80 first order features) were extracted from human GBM. An additional 620 radiomic features from the contralateral normal-appearing white matter were extracted from human MRIs and used for within- sequence normalization purposes. Similarly, in the case of mice, a total of 2,480 (2,400 second-order and 80 first order features) features were obtained.
Extracted feature generation
Out of the 310 radiomic features, a total of 300 MRI radiomic features per phenotype (enhancing active tumor, edema/invasion, necrosis, and whole GBM tumor) extracted per each imaging sequence (T1 and T2/FLAIR) were GLCM-based features as described above. This represents a total of 2,400 GLCM-based features extracted per tumor per patient (as well as per animal). To consider tumor volumes, 2,400 new variables (xij) were added by dividing each of these feature (f’ij ) by the corresponding phenotype volume. As a result, 4,880 (volume-dependent and volume-independent) features (2,400 original second-order, 2,400 volume-dependent second-order and 80 first order features) per tumor per patient (animal) were obtained.
Contralateral normalization
To account for differences in the image acquisition hardware and software used across TCGA institutions, we performed a patient-specific contralateral hemisphere normalization of the features using the features extracted from the contralateral normal-appearing white matter. Consequently, each feature (fij) was subtracted from the corresponding contralateral radiomic feature fij(wm). We assessed the performance of the patient-specific contralateral hemisphere normalization of the features across institutions, MRI scanner brands, and magnetic field strengths by plotting pre- and post- normalization feature distribution.
Biostatistical radiomic analysis: feature selection, classification, and predictive modeling
We used R software (version 3.4.0, R Foundation for Statistical Computing, Vienna, Austria) for all the statistical analysis: package xgboost (version 0.6.4.1) for the feature selection task and the Machine Learning package mlr (version 2.11) to build the binary logistic model of XGboost using “gbtree” booster for classification. Finally, Receiver Operating Characteristic (ROC) analysis was performed using pROC package (version 1.9.1). AUC, sensitivity, specificity, PPV, NPV and p-values are reported for each LOOCV and prediction outputs.
Radiomic feature selection
Given the large number of features provided by radiomics, feature selection is the first step in the pipeline. For feature selection, we used Least Absolute Shrinkage and Selection Operator (LASSO) regularization (L1 regularization) technique[35]. LASSO is a regression analysis method that typically performs variable selection and regularization to enhance the prediction accuracy and avoid overfitting of the features on the model. L1 regularization coupled with early stopping (at one third of the patient size) was used for feature selection to select the most important and relevant features required for the model building. Later, these important features were used for model building. We used the latter models in the prediction of high versus low POSTN mice and human. The criteria of groupings into high versus low POSTN gene expression levels were based on cohort (N=93) specific median expression levels.
The reliability of our radiomic pipeline was assessed by Intraclass Correlation Coefficient (ICC) values across institutions, scanner etc. before and after normalization steps for the features in the final predictive model. Additionally, the non-normalized features were tested for their ability to predict the outcome.
Patient and mice multivariable predictive modeling
Radiomic feature selection and classification model building was performed by implementing gradient boosting from the XGboost package (eXtreme Gradient Boosting) and works by implementing gradient boosting. The XGboost algorithm works by converting weak learners to strong learners using the boosting technique, in which trees are developed with the help of information from the previously grown trees and passing that information from one tree to another iteratively. Hence, slowly the trees to learn from the data and improve its prediction by minimizing the loss function or mis-classification error rate. XGboost enables parallel computation, regularization, cross validation, missing value imputation (if needed) and tree pruning [37].
For classification and model building, we used gbtree booster of XGboost and built binary logistic regression model to classify patients into high and low POSTN groups based on the selected radiomic features resulted from LASSO regularization. We performed hyperparameter tuning required for tree booster and cross validation using Leave One Out Cross Validation (LOOCV) over 100 iterations (n rounds = 100) to avoid overfitting and acquire best prediction model that has better accuracy, AUC, sensitivity, specificity and p values.
Besides LOOCV, a 50% random split was used to train and test the model for outcome. ROC analysis and analysis with a 95% Confidence Interval (CI) were implemented to measure the model accuracy. AUC, sensitivity, specificity, PPV, NPV and p-values for each LOOCV and prediction output were calculated.
Translational Causality Investigations
Two independent GSC lines upon validation of efficient short hairpin RNA (shRNA) mediated POSTN knockdown were used to develop orthotopic tumors in mouse brains. See supplemental methods for details. In brief, leveraging a controlled preclinical experimental system for mechanistic validation of imaging genomics, we illustrate the potential for radiogenomic causality beyond mere correlation. First, we screened 47 GSC lines that met three criteria: high POSTN expression, expression of stemness markers, and their ability to produce stable orthotopic xenograft tumor using primary GSC lines to most faithfully replicate human GBM (Supplementary Fig. S1). First, we used gene expression microarray and quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) to select two GSC lines (GSC11 and GSC126) which met the latter criteria. We then transduced these GSC lines with lentiviruses containing a doxycycline-inducible shRNA against POSTN, which co-expressed GFP via an IRES, thereby allowing visualization/isolation of knockdown cells both in-vitro and ex-vivo (Supplementary Fig. S2) and selected puromycin resistant stable clones with significant knockdown of POSTN at both the messenger RNA and protein levels (Supplementary Fig. S3). These clones were orthotopically implanted into nude mice (N=40). MRI was used to confirm the growth of the xenograft tumors (Fig. 3e). Ex-vivo flow-sorted cells from the control (APC+ve) and POSTN-knockdown (APC+veGFP+ve) mouse tumors (Supplementary Fig. S2) were subjected to RNA extraction and qRT-PCR for POSTN. Those cells from knockdown tumors showed a significant decrease in POSTN mRNA levels (Fig. 3a); we also performed xenograft tumor immunohistochemistry to confirm that the POSTN-knockdown tumors had significantly lower POSTN protein expression than that of control tumors (Supplementary Fig. S3).
Figure 3: Qualitative and quantitative MRI parameters are unable to distinguish genomic and histologic heterogeneity in orthotopic GBM model system.
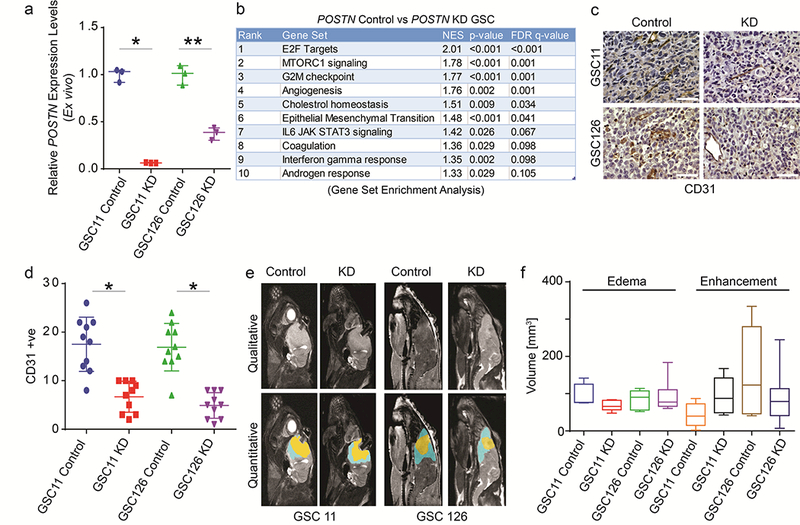
(a) Ex vivo validation of knockdown of POSTN in GSC11 and GSC126. Quantitative real time PCR analysis for determination of expression levels of POSTN gene in knockdown (KD) and control tumor cells isolated from mouse brain ex vivo. Relative expression levels of POSTN after normalization to that of GAPDH in respective samples is presented (N=3). (b) Gene set enrichment analysis of whole genome expression profiles of tumor cells (ex vivo) from control and KD mice. Top 10 gene set enriched in control tumor cells are shown in the table. Their respective rank, net enrichment score (NES), p-values, and FDR q-values are shown. (c) Immunohistochemistry staining of xenograft tumor sections from control and KD mice using anti-CD31 antibody. Nuclei are counterstained with hematoxylin. Respective cell lines giving rise to orthotopic tumors are labelled on the side of micrographs. Scale bar is 100μm. (d) Quantification of CD31 positive cells in control and KD tumors are shown in the graph. *: p-value <0.001. (e) Representative MRI scans and segmentations of GSC11 and GSC126 xenograft tumors from control (N=5 and 7, respectively) and POSTN-knockdown (KD) (N=5 and 9, respectively) animals. Tumors from POSTN-wildtype (control) and POSTN-KD GSC11 and GSC126 mice appear qualitatively (top) and quantitatively (bottom) indistinguishable on T1WI and T2WI with a 7T MRI system. (f) The box plot shows quantification of volumes of contrast enhancement and edema (T2) in control and POSTN-KD groups. p-values between groups were not significant.
RESULTS
Robust high-throughput radiomic and radiogenomic pipeline
We established a robust image analytical pipeline consisting of image segmentation, radiomic feature extraction, feature normalization and selection, and predictive model generation (Fig. 1). For an approach to be readily transferrable to the clinic, it must be robust; our analytical pipeline demonstrated high accuracy, precision, and reliability across patients, institutions, and different vendors of MRI scanners with different magnetic field strengths (Supplementary Tables 1a–b). Briefly, we obtained the outline and corresponding volume of the three GBM phenotypes (edema/invasion, enhancing active tumor and necrosis) by semi- automated consensus segmentation; consensus segmentation produces stable results compared to single- user segmentation [39] (Fig. 1a). The original MR images and corresponding segmented masks underwent image preprocessing for extraction of stable and comparable quantitative image features (Supplementary Figs. S4–S6). The inherent variability of MR signal intensities makes direct analysis of images obtained from different scanners/vendors difficult; thus, MR radiomic studies to date have mainly focused on data from single scanner and identical protocols [40, 41] and only a small number of studies have utilized data from multiple MR scanners/protocols [42, 43]. Thus, we employed a brain-tissue focused normalization consisting of two steps (i) skull stripping [25] and Nyul normalization (brain-specific normalization) [27], followed by a patient-specific contralateral hemisphere MRI normalization that allows for the extraction of stable quantitative radiomic features (Fig. 1b). Brain tissue images were first obtained by excluding extra-cerebral tissues such as skull, eyeballs, fat, and skin through skull stripping, and further used for normalization. Skull stripping is an important step in studies relying on intensity levels; however, to our knowledge, it has not been employed along with Nyul normalization before. The effectiveness of Nyul normalization compared to other image normalization techniques has been established [28]. In our study, we demonstrate its applicability in GBM. The combination of skull stripping, brain-tissue focused normalization and patient-specific normalization are unique to this study and provided comparable cross-platform, cross-institution radiomic features as demonstrated qualitatively in TCGA pre– and post– patient-specific normalization plots, Supplementary Figs. S4–S6. This novel pipeline was employed for all imaging and statistical radiomic and radiogenomic analysis in our study.
Qualitative and quantitative (volumetric) MRI is unable to classify GBM based on prototype POSTN gene expression level in patients and animal models
We evaluated POSTN expression levels in mesenchymal (MES) and proneural (PN) subgroups of GBM and found that expression of POSTN is highly correlated with MES subgroup (Fig. 2a). Gene set enrichment analysis (GSEA) of whole genome expression profiles of patient with high POSTN vs low POSTN expression (grouped according to median cutoff), revealed that high POSTN expression was significantly correlated with hallmarks such as epithelial mesenchymal transition (p-value<0.001, FDR q-value: 0), angiogenesis (p-value<0.001, FDR q-value: 0.001) etc. (Fig. 2b). As suggested by GSEA, patients with high endothelial proliferation, as extracted from Cancer digital slide archive [49] (http://cancer.digitalslidearchive.net/), (n=232) displayed significantly higher levels of POSTN expression (p-value: 0.0027) in comparison to that with low endothelial proliferation (N=152) (Fig. 2c and d). However, qualitative and quantitative volumetric analyses of MRI scans of patients with high and low POSTN expression appeared indistinguishable (Fig. 2e) and illustrated statistically insignificant variations, when various segmented volumes were quantified (Fig. 2f).
Figure 2: Qualitative and quantitative MRI parameters are unable to distinguish genomic and histologic heterogeneity in GBM patients.

Association of POSTN with distinct genomic and histologic heterogeneity in GBM. (a) POSTN expression is significantly higher in mesenchymal sub-group of GBMs in comparison to proneural sub-group. Expression levels of POSTN in mesenchymal (n=124) and proneural (n=113) sub-group is shown as box plot (p-value <0.0001). (b) Gene set enrichment analysis shows significant enrichment of key hallmark signatures in patients with high POSTN expression. Table shows top 10 enriched gene set in patients with high POSTN expression when compared to patients with low POSTN expression. Their respective rank, net enrichment score (NES), p-values, and FDR q-values are shown. (c) POSTN expression is higher in tumors with endothelial proliferation versus the tumors with undetectable endothelial proliferation. (d) Expression levels of POSTN in tumors with endothelial proliferation (n=232) and with undetectable endothelial proliferation (n=152) is shown as box plot. *: p-value 0.0027. Qualitative and quantitative MRI parameters are unremarkable in patient with high or low POSTN expression. (e)Tumors from POSTN High and POSTN Low appear qualitatively (left panels) and quantitatively (right panels) indistinguishable on representative MRI scans. (f) The box plot shows quantification and comparison of volumes of necrosis, contrast enhancement and edema (T2) in POSTN High and POSTN Low groups of patients. p-values between groups were not significant.
While in animal models, when GSEA of whole genome expression profiles of ex-vivo flow-sorted cells from the control (APC+ve) and POSTN-knockdown (APC+veGFP+ve) mouse tumors was performed, hallmarks such as angiogenesis (p-value: 0.002, FDR q-value: 0.001) and epithelial mesenchymal transition (p-value<0.001, FDR q-value: 0.041) etc. were enriched in control (i.e. high POSTN tumors) (Fig. 3b). Furthermore, as suggested by GSEA and confirmed by orthotopic xenograft tumor immunofluorescence, the control mice from both GSC11 and 126 cell lines, with high POSTN expression, displayed significantly higher levels of Ki67 and CD31+ve endothelial cells (p-values: <0.001) in contrast to tumors from the knockdown orthotopic mouse model (low POSTN) (Fig. 3c and d and Supplementary Fig. S7). Similar to GBM patients grouped according to POSTN expression levels, both GSC11 and 126 derived control and POSTN-knockdown tumors developed qualitatively similar growth and phenotype volumes and quantitative analyses revealed no significant difference in volumes of contrast enhancement or peritumoral edema/tumor invasion between the two groups of mice (Fig. 3e, f).
Radiomics predict genomic heterogeneity in GSC derived orthotopic tumors and GBM and is conserved across patient and mouse xenograft tumors
We then sought to determine whether radiomics could correctly identify patients with differential expression of POSTN and the altered POSTN expression levels between control and knockdown tumors across the two transplanted GSC lines. Our analyses of radiomic features displayed similarities between high-POSTN GBM in mice and in humans, specifically, as seen in 64 gray-level and heterogeneity histograms (Fig. 4a). We then tested the radiomic feature predictive performance by LOOCV to distinguish tumors with high POSTN (control) and low POSTN (knockdown) expression levels in two GSC lines (Fig. 4b) (AUC 92.26%, sensitivity 92.86%, specificity 91.67%; p-value 2.68E-05). Our results showed that unlike conventional qualitative and quantitative volumetric assessments, radiomic analyses (17 radiomic features) correctly distinguished tumors with differential RNAi-mediated POSTN status lines. Furthermore, we used radiomic analysis for further characterization of the mouse and human tumors. Towards this, to determine the power of radiomic features to predict targeted gene expression levels in tumors across species (mouse-to-patient), we first generated and validated a radiomic feature-based model that successfully identified tumors based on POSTN levels in four mice cohorts across two GSC lines. As shown in Fig. 4b, specific radiomic features in this model significantly distinguish control and knockdown xenograft tumors (AUC 92.26%, sensitivity 92.86%, specificity 91.67%; p-value 2.68E-05). Accordingly, radiomic features obtained using GBM patient cohort reliably predicted POSTN status in this cohort (Supplementary Table 3; 48 radiomic features). Both cross-validation (AUC 99.17%, sensitivity 97.83%, specificity 97.87%; p-value <2e-16; Supplementary Fig. S8) and prediction (AUC 76.56%%, sensitivity 73.91%, specificity 78.26 %, p-value 0.00026 Fig. 4c) in a non-overlapping validation set were highly statistically significant. As a measure of reliability across institutions, we observed a consistent decrease in ICC value for these 48 features (ranging from −7% to −104%) after normalization. This decrease was significant as the non-normalized features failed to predict POSTN levels in the patient cohort (data not shown). Furthermore, analysis with radiomic features (48 radiomic features) selected for their ability to predict POSTN levels in mouse orthotopic tumors showed significant LOOCV of POSTN levels in the patient cohort (AUC 93.36%, sensitivity 82.61%, specificity 95.74%; p-value 2.021E-15; Fig. 4d, Supplementary Table 2). Finally, radiogenomic POSTN prediction was performed in a prospective manner in individual patients; these radiogenomic maps showed gene expression status (Fig. 4e). This is summarized for the patient in a radiogenomic clinical report card.
Figure 4: Radiomics based features are conserved and are predictive of POSTN levels across xenograft and human tumors.
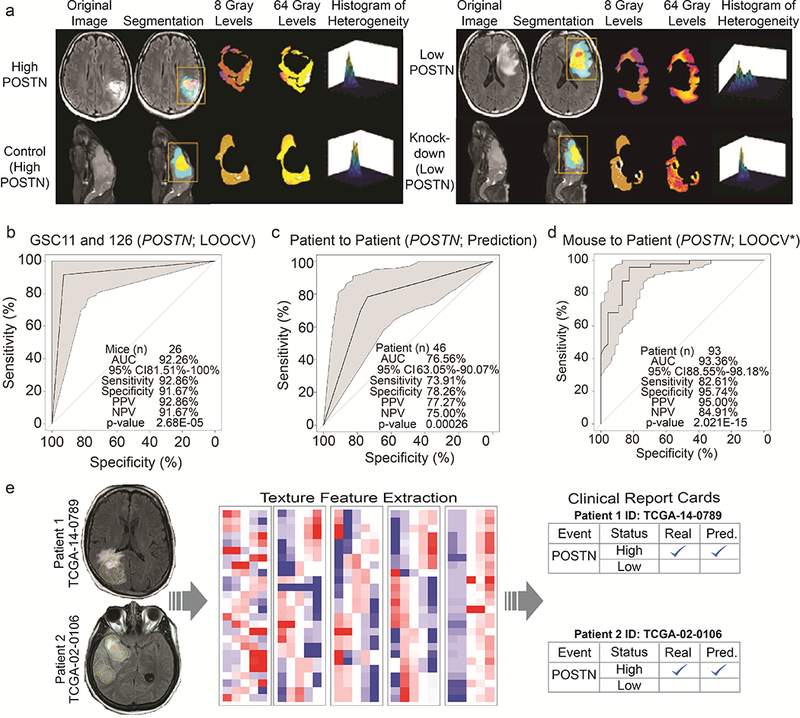
(a) High POSTN tumors from patients and orthotopic xenograft mice (Control) (left) show greater similarity of textural heterogeneity at 8 Gray and 64 Gray levels and are distinct from that of low POSTN tumors from patients and orthotopic xenograft mice (KD) (right). (b) LOOCV ROC curve depict the performance of the GSC11 and GSC126 mouse-based predictive model using 17 radiomic features to predict control and KD mice across cell lines with 92.26% accuracy and p-value 2.68E-05. 95% CI, sensitivity, specificity, PPV, NPV of models are shown within each plot. (c) ROC curve depicts the performance of TCGA training set (N=46) based predictive model using 48 radiomic features to predict POSTN expression levels in validation set (N=47) with 76.65% accuracy. (d) The LOOCV* ROC curve shows the association of 17 radiomic features derived from GSC11 and GSC126 mice from panel (b) with 93 patients with endogenous high and low POSTN expression levels. (e) Radiome sequencing in clinics. Radiogenomic probability maps and radiome sequencing clinical report cards for two representative patients. Summary of the radiomics-based prediction of the genomic events/status of GBM for two representative patients. The left panels show the segmented patient brain MRIs delineating the tumor. The right panels show the radiome sequencing clinical report cards summarizing the status of POSTN expression. A check mark indicates that the prediction was correct.
DISCUSSION
In this study, for the first time to our knowledge, we demonstrate a causal linkage between imaging radiomics and genomics. We tested our hypothesis using targeted genetic interventions. Herein, we demonstrate that gene expression, specifically POSTN expression, drives a specific imaging feature phenotype. In addition, we demonstrate that tumors with specific genomic expression profiles harbor distinct radiomic profiles. Finally, we show that patients harboring tumors with a distinct gene expression profile share significantly concordant radiomic features with their matching preclinical murine xenograft counterparts.
Fundamental to radiogenomic analysis is the presumption that expression of specific sets of genes or driver mutations impact extractable imaging features, but this presumption has not been directly demonstrated in a highly robust model. Our results suggest that imaging features, specifically radiomic features, are at least in part genetically-driven, and subtle genomic changes in tumor tissue are captured by radiomic changes on imaging. Additionally, our results show that POSTN may potentially be responsible for maintenance of a mesenchymal program and angiogenesis in GBM (a link recently illustrated in Park, S.Y. et al.) [20]. These findings support the hypothesis that the radiogenomic functional relationships can be quite robust and may contain significant causal linkages between MR imaging features and genetically-driven tumor phenotypes.
Technically underpinning this, we developed and evaluated a novel radiomics pipeline for analysis of standard of care clinical MR images that is scalable and provides widespread applicability for high-throughput cross-vendor and software platform image processing and analysis of medical imaging applied to human solid tumors and preclinical tumor models. Multiple innovative steps were required for this to occur and developed throughout this process. We established and applied two unique steps of image normalization: first, brain-specific normalization; and second, patient-specific contralateral hemisphere MRI normalization; these allowed for robust cross-platform (vendor), cross-institution patient cohort analysis and comparison. Genomic predictions were significant in the TCGA dataset, despite these data being collated from six different medical centers across the United States and patients having undergone imaging with MRI scanners from different vendors using different image acquisition protocols. This unique combination of normalization steps have not been previously used within the radiomics/tumor assessment field, although the effectiveness of Nyul normalization has been shown in image analysis of normal human MRIs and multiple sclerosis [28]. The integration of a patient-specific normalization process is new to the radiomics field and its application in conventional imaging at large has not been used in this setting. Thus, herein, we developed a novel radiomic pipeline that is agnostic to MRI scanner vendor, institution, and protocol.
Further, we developed a pipeline that can robustly extract microscale information beyond the resolution of the naked human eye. As our data demonstrate, qualitative and quantitative (volumetric) MRI assessments did not detect any differences between POSTN knockdown and wild-type xenograft tumors (Fig. 3e-f); however, radiomic analysis showed distinct radiophenotypic differences between these two groups. (See pre– and post–patient-specific normalization plots, Supplementary Figs. S4–S6). This enhanced radiomics analysis method may unlock previously inaccessible data for clinicians and investigators independent of where, when and how images were acquired. The radiomic software developed herein is scalable, potentially enabling multi-institutional collaborations, big data collation and integration into clinical care, clinical trials, and research across various arenas.
POSTN, expressed in glioma cells including glioma stem-like cells (GSCs) and secreted to extracellular matrix in tumor microenvironment, is involved in invasion as well as perivascular niche formation, metastasis and macrophage recruitment [45] [46]. More recent studies demonstrate POSTN’s role in promoting glioma invasion, tumor recurrence, and resistance to anti-angiogenic therapy [20, 47]. The peritumoral region in gliomas harbors a mixture of edema and invasive, aggressive tumor cell, a key determinant of tumor recurrence and poor patient survival [48]. Given POSTN’s role in glioma invasion as detailed above coupled with our previous imaging genomic findings, we utilized POSTN as our gene prototype. In our study, tumors with distinct POSTN expression levels harbored distinct imaging features that were not only associated with but also predictive of expression levels in both patients and murine models. Likewise, the preclinical model-derived radiomic features significantly predicted concordant gene expression in our patient GBM cohort (N=93). This is a uniquely novel finding and demonstrates that detection of selected genomic traits is conserved in appearance on imaging across species and can be expected to further facilitate integration of radiomics and radiogenomics into clinical and co-clinical trials. For a method to be translated into clinical practice robustness, generalizability and scalability of the technique are required [50]. Employing well-known robust statistical methods, we used non-overlapping discovery and validation sets to meet and achieve high protocol standards [51]. Giving further credence to the generalizability and scalability of our radiomic model is the fact that the pipeline is applicable beyond solid tumors and to other types of imaging sequences such as functional MRI [52]; in a previously published study by our group, we found that radiomics can detect and discriminate between areas of true activation in regions of eloquent cortex versus artifact [52].
The need to establish robust imaging-genomic correlations and the importance of radiomic tools to interrogate tissue is supported by multiple studies including the study by Kickingereder et al. who concluded that tumor volumes, perfusion, diffusion, and other MRI parameters are not sufficient for reliable and clinically meaningful predictions of molecular characteristics in patients [53]. A lack of established causality as to the molecular underpinnings of imaging has been a major impediment towards general clinical acceptance of radiogenomics. However, this study demonstrates that radiomic assessment can predict genomically distinct tumors with high sensitivity, specificity, accuracy and evaluate pertinent genomic alterations towards providing detailed prediction maps of the molecular GBM landscape. This method will not replace but complement conventional genomic testing, such that noninvasive genomic profiling may enhance diagnosis, help stratify patients and determine early response to targeted therapy; imaging radiomics also assess the entire three-dimensional tumor structure and may enable monitoring of genomic traits at a specific time for patients in whom biopsy or surgery cannot be performed or when cost of care limits repeated genomic profiling. Hence, comprehensive radiomics can complement genomic information obtained via biopsy or tumor resection in cancer care. It can also be anticipated that longitudinal alterations in radiomic features can be used in combination with current response assessment criteria such as RANO, RECIST1.1, irRC, etc. for a more accurate and early assessment of treatment response. Radiomics provides significantly increased predictive power and higher biomarker accuracy based on inherent tumor imaging features and pixel-based heterogeneity.
Despite this study being the pilot in establishing radiogenomic causality within the radiogenomic literature, prospective validation using spatially matched image-guided brain tumor biopsies in patients and animal models are needed to further validate the spatial distribution of the molecular underpinnings of imaging in brain cancer. Currently, topographical spatial encoding of the genomic landscape using image- guided biopsies is underway at our institution. Another limitation of the study, is that the spatial resolution (voxel size) of the human MRI is larger compared to the xenograft MRI; these differences could potentially affect the extracted radiomic features since the distance of 1-voxel used for the calculation of the GLCM matrix varies in size. Preprocessing steps that account for varying resolutions, such as up-sampling or down-sampling the volume to one resolution could be utilized, however, the goal of our study was to use the images with the resolution dictated by their respective acquisition protocols, thus no pre-processing steps were performed on xenograft models. Further, the results of our analyses (high accuracy, sensitivity and specificity) could indicate that additional preprocessing steps to account for resolution differences may not be necessary when evaluating the whole tumor compared to voxel-based radiomic analysis.
In summary, we present a clinically-applicable and -meaningful analytical imaging method for performing high- throughput image analysis that can capture key genomic events in brain cancer patients. Our radiomic analysis pipeline is a biologically validated test method. Further, the generalizability and scalability to most types of medical imaging and solid tumors can be anticipated and preliminary results underway at our institution using our pipeline are promising. It also has use beyond clinical diagnosis and prognosis, as it allows for human-mouse matched co-clinical trials, in-depth endpoint analysis, and upfront, noninvasive, high-resolution radiomics-based diagnostic, prognostic, and predictive biomarker development.
Supplementary Material
Statement of translational relevance:
Radiomics, the automated high-throughput extraction of multidimensional imaging features captures microscale information hidden within conventional imaging beyond what is visible to the naked human eye. The linkage of imaging phenotypes with genomic data is termed radiogenomics. Fundamental to radiogenomics is the hypothesis that expression of specific sets of genes or driver mutations impact the extractable imaging features, but this has not been directly demonstrated in a highly robust model; to date, the linkages between imaging and genomics remain at a correlative stage and lack established causality. In this study, we seek to address this gap in knowledge; we bring forward an approach for integrated end-to-end methodology for extraction of imaging radiomic features and validation in both a preclinical closed model system and patient cohort. Further, we demonstrate the potential of radiomics in co-clinical trials. To our knowledge, this is the first study functionally validating imaging-molecular features.
ACKNOWLEDGMENTS
We thank Joseph Munch for editorial help and V.L. Brandt for shepherding the manuscript through multiple drafts. We also thank the National Cancer Institute and the TCGA/TCIA glioma phenotype research group.
Funding:
This work was supported by John S. Dunn Sr. Distinguished chair in Diagnostic Imaging fund, MD Anderson Cancer Center startup funding. Radiological Society of North America scholar grant (RSCH11506). Cancer Prevention and Research Institute of Texas (CPRIT) grant (RP160150) (Colen). This work was also supported by NIH R25 Baylor College of Medicine, Department of Neurosurgery Research Grant and NREF (neurosurgery research and education foundation) (338703) (Zinn). Further support includes MD Anderson’s Cancer Center support grant (CA016672); national cancer institute (P50 CA127001; animal core and biospecimen core), the Elias Family Fund, and the Broach Foundation (Lang).
Footnotes
Statement of Disclosure: Authors report no conflicts of interest.
REFERENCES
- 1.Eisenhauer E, et al. , New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). European journal of cancer, 2009. 45(2): p. 228–247. [DOI] [PubMed] [Google Scholar]
- 2.Nishino M, et al. , Developing a common language for tumor response to immunotherapy: immune-related response criteria using unidimensional measurements. Clinical cancer research, 2013. 19(14): p. 3936–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wen PY, et al. , Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. Journal of Clinical Oncology, 2010. 28(11): p. 1963–1972. [DOI] [PubMed] [Google Scholar]
- 4.Wolchok JD, et al. , Guidelines for the evaluation of immune therapy activity in solid tumors: immune-related response criteria. Clin Cancer Res, 2009. 15(23): p. 7412–20. [DOI] [PubMed] [Google Scholar]
- 5.Aerts HJ, et al. , Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun, 2014. 5: p. 4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambin P, et al. , Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer, 2012. 48(4): p. 441–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, et al. , MR Imaging Radiomics Signatures for Predicting the Risk of Breast Cancer Recurrence as Given by Research Versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays. Radiology, 2016. 281(2): p. 382–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castellano G, et al. , Texture analysis of medical images. Clin Radiol, 2004. 59(12): p. 1061–9. [DOI] [PubMed] [Google Scholar]
- 9.Davnall F, et al. , Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging, 2012. 3(6): p. 573–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diehn M, et al. , Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A, 2008. 105(13): p. 5213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Preusser M, et al. , Current concepts and management of glioblastoma. Annals of Neurology, 2011. 70(1): p. 9–21. [DOI] [PubMed] [Google Scholar]
- 12.Wu W, et al. , Exploratory study to identify radiomics classifiers for lung cancer histology. Frontiers in Oncology, 2016. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zinn PO, et al. , Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PLoS One, 2011. 6(10): p. e25451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joo KM, et al. , Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell Rep, 2013. 3(1): p. 260–73. [DOI] [PubMed] [Google Scholar]
- 15.Panth KM, et al. , Is there a causal relationship between genetic changes and radiomics-based image features? An in vivo preclinical experiment with doxycycline inducible GADD34 tumor cells. Radiother Oncol, 2015. 116(3): p. 462–6. [DOI] [PubMed] [Google Scholar]
- 16.Hegi ME, et al. , MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. New England Journal of Medicine, 2005. 352(10): p. 997–1003. [DOI] [PubMed] [Google Scholar]
- 17.Stupp R, et al. , Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. New England Journal of Medicine, 2005. 352(10): p. 987–996. [DOI] [PubMed] [Google Scholar]
- 18.Verhaak RG, et al. , Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell, 2010. 17(1): p. 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou W, et al. , Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nat Cell Biol, 2015. 17(2): p. 170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park SY, et al. , Periostin (POSTN) Regulates Tumor Resistance to Antiangiogenic Therapy in Glioma Models. Mol Cancer Ther, 2016. 15(9): p. 2187–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Archip N, Jolesz FA, and Warfield SK, A Validation Framework for Brain Tumor Segmentation. Academic Radiology, 2007. 14(10): p. 1242–1251. [DOI] [PubMed] [Google Scholar]
- 22.Gering DT, et al. , An integrated visualization system for surgical planning and guidance using image fusion and an open MR. J Magn Reson Imaging, 2001. 13(6): p. 967–75. [DOI] [PubMed] [Google Scholar]
- 23.Pichon E, Tannenbaum A, and Kikinis R, A statistically based flow for image segmentation. Medical Image Analysis, 2004. 8(3): p. 267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zinn PO, et al. , Distinct Radiomic Phenotypes Define Glioblastoma TP53-PTEN-EGFR Mutational Landscape. Neurosurgery, 2017. 64(CN_suppl_1): p. 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenkinson M, Pechaud M, and Smith S, BET2: MR-based estimation of brain, skull and scalp surfaces. 2005, Eleventh Annual Meeting of the Organisation for Human Brain Mapping. [Google Scholar]
- 26.Smith SM, Fast robust automated brain extraction. Hum Brain Mapp, 2002. 17(3): p. 143–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nyul LG, Udupa JK, and Zhang X, New variants of a method of MRI scale standardization. IEEE Trans Med Imaging, 2000. 19(2): p. 143–50. [DOI] [PubMed] [Google Scholar]
- 28.Shah M, et al. , Evaluating intensity normalization on MRIs of human brain with multiple sclerosis. Med Image Anal, 2011. 15(2): p. 267–82. [DOI] [PubMed] [Google Scholar]
- 29.Shinohara RT, et al. , Statistical normalization techniques for magnetic resonance imaging. Neuroimage Clin, 2014. 6: p. 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haralick RM, Shanmugam K, and Dinstein IH, Textural Features for Image Classification. IEEE Tran Syst Man Cybern, 1973. 3: p. 610–621. [Google Scholar]
- 31.Papoulis A, Probability, random variables, and stochastic processes 3rd ed. McGraw-Hill series in electrical engineering Communications and signal processing. 1991, New York: McGraw-Hill; xvii, 666 p. [Google Scholar]
- 32.Pratt WK, Digital image processing. 2nd ed. 1991, New York: Wiley; xiv, 698 p., 4 p. of plates. [Google Scholar]
- 33.Clausi DA, An analysis of co-occurrence texture statistics as a function of grey level quantization. Canadian Journal of Remote Sensing. 28(1): p. 45–62. [Google Scholar]
- 34.Soh LK and Tsatsoulis C, Texture analysis of SAR sea ice imagery using gray level co-occurrence matrices. Geoscience and Remote Sensing, IEEE Transactions on, 1999. 37(2): p. 780–795. [Google Scholar]
- 35.Tibshirani R, Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society. Series B (Methodological), 1996: p. 267–288. [Google Scholar]
- 36. Ding C and Peng H, Minimum redundancy feature selection from microarray gene expression data. J Bioinform Comput Biol, 2005. 3(2): p. 185–205. [DOI] [PubMed] [Google Scholar]
- 37.Chen T and Guestrin C. Xgboost: A scalable tree boosting system. in Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining 2016. ACM. [Google Scholar]
- 38. Platt JC, Probabilistic Outputs for Support Vector Machines and Comparisons to Regularized Likelihood Methods ADVANCES IN LARGE MARGIN CLASSIFIERS, ed. Smola PBA, Schoelkopf B and Schuurmans D. 1999, Cambridge, MA: MIT Press. [Google Scholar]
- 39.Zinn PO, et al. , Distinct Radiomic Phenotypes Define Glioblastoma TP53-PTEN-EGFR Mutational Landscape. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu LS, et al. , Multi-Parametric MRI and Texture Analysis to Visualize Spatial Histologic Heterogeneity and Tumor Extent in Glioblastoma. PLoS One, 2015. 10(11): p. e0141506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiestler B, et al. , Multiparametric MRI-based differentiation of WHO grade II/III glioma and WHO grade IV glioblastoma. Sci Rep, 2016. 6: p. 35142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prasanna P, et al. , Radiomic features from the peritumoral brain parenchyma on treatment-naive multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: Preliminary findings. Eur Radiol, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Upadhaya T, et al. , - Prognosis classification in glioblastoma multiforme using multimodal MRI derived heterogeneity textural features: impact of pre-processing choices.
- 44. Zinn PO, et al. , Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PLoS One, 2011. 6: p. e25451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malanchi I, et al. , Interactions between cancer stem cells and their niche govern metastatic colonization. Nature, 2012. 481(7379): p. 85–89. [DOI] [PubMed] [Google Scholar]
- 46.Zhou W, et al. , Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nat Cell Biol, 2015. 17(2): p. 170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mikheev AM, et al. , Periostin is a novel therapeutic target that predicts and regulates glioma malignancy. Neuro Oncol, 2015. 17(3): p. 372–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wangaryattawanich P, et al. , Multicenter imaging outcomes study of The Cancer Genome Atlas glioblastoma patient cohort: imaging predictors of overall and progression-free survival. Neuro Oncol, 2015. 17(11): p. 1525–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gutman DA, et al. , Cancer Digital Slide Archive: an informatics resource to support integrated in silico analysis of TCGA pathology data. J Am Med Inform Assoc, 2013. 20(6): p. 1091–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar V, et al. , Radiomics: the process and the challenges. Magn Reson Imaging, 2012. 30(9): p. 1234–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bleeker SE, et al. , External validation is necessary in prediction research: a clinical example. J Clin Epidemiol, 2003. 56(9): p. 826–32. [DOI] [PubMed] [Google Scholar]
- 52.Hassan I, et al. , Radiomic Texture Analysis Mapping Predicts Areas of True Functional MRI Activity. Scientific Reports, 2016. 6: p. 25295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kickingereder P, et al. , Radiomic Profiling of Glioblastoma: Identifying an Imaging Predictor of Patient Survival with Improved Performance over Established Clinical and Radiologic Risk Models. Radiology, 2016. 280(3): p. 880–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


