Abstract
The development of anatomical structures is complex, beginning with patterning of gene expression by multiple gene regulatory networks (GRNs). These networks ultimately regulate the activity of effector molecules, which in turn alter cellular behavior during development. Together these processes biomechanically produce the three-dimensional shape that the anatomical structure adopts over time. However, the interfaces between these processes are often overlooked and also include counter-intuitive feedback mechanisms. In this review, we examine each step in this extraordinarily complex process and explore how evolutionary developmental biology model systems, such as butterfly scales, vertebrate teeth, and the Drosophila dorsal appendage offer a complementary approach to expose the multifactorial integration of genetics and morphogenesis from an alternative perspective.
Introduction
A major goal of developmental biology is to elucidate how the diverse anatomical structures throughout the organism take on their unique shapes from undefined embryonic tissues. The formation of even the simplest three-dimensional structure requires the deployment of tissue-specific gene regulatory networks (GRNs) that operate through transcriptional regulation to ultimately activate cellular effectors (Box 1). Once activated through transcriptional, post-transcriptional, or post-translational mechanisms, these effector molecules function to directly alter ubiquitously expressed proteins, such as the actin cytoskeleton (Box 1). Cumulatively, this results in a context-specific alteration to the mechanical properties of the cell, contributing to formation of the anatomical structure[1–4] (Fig. 1). However, it has been difficult to understand the interface of GRN-effector connections to understand how combinations of cellular effectors are precisely patterned to shape anatomy, and how these GRN-effector connections diversify to modify anatomical form during evolution. The goal of understanding how to connect GRNs to anatomy is complicated by mechanical or signaling influences from neighboring tissues, that are regulated by different GRNs, painting a complex and interconnected picture of multiple GRNs activated in separate tissues affecting the morphogenesis of a single structure [3,4]. While development can be studied at the level of GRNs, cellular effectors, or biophysical mechanics, it is critical that we comprehend how these distinct systems are connected and influence each other in an integrated way. Here, we will review this developmental phenomenon in epithelial tissues, highlighting several recent studies that illuminate each facet of this problem while emphasizing the novel insights they provide. Further, we propose that the study of evolutionary modifications can provide insights into how these systems interface by examining how GRN-effector connections are modified during evolution, and review up-and-coming evolutionary developmental biology (evo-devo) model systems in which these questions can be explored.
Box 1. Glossary of Terms
Gene Regulatory Network (GRN)
GRNs are composed of signaling pathways, transcription factors, and cellular effectors. Signaling pathways pattern development through cell-cell communication, often resulting in the activation of a transcription factor. Transcription factors regulate gene expression by binding to individual enhancers of downstream genes (e.g. another transcription factor, cellular effector, etc). Each enhancer requires a different set of transcription factors to bind and gene activation will only occur in cells in which the correct sets of transcription factors are present. Overall this complex set of interactions patterns development and governs the final phenotype of the cell [5,6].
Cellular Effector
Any gene that functions to non-transcriptionally activate, localize, or alter other core cellular proteins (e.g. actin, myosin, cadherin, etc.). Cellular effectors can be turned on via transcriptional regulation or can be proteins already present in the cell that are activated by other cellular effectors. Together cellular effectors function to alter the behavior of a cell by changing the mechanics within the cell or through altering mechanical connections to neighboring cells. Cellular effectors can allow a cell to express multiple phenotypes in response to different context without new transcription [3,4].
Intrinsic Mechanics
The combined effect of the cell effectors present within a cell that affects the cell’s mechanics.
Extrinsic Mechanics
The combined influence of effectors operating in neighboring cells, transferred via direct cell-cell contacts, to alter a cell’s intrinsic mechanics.
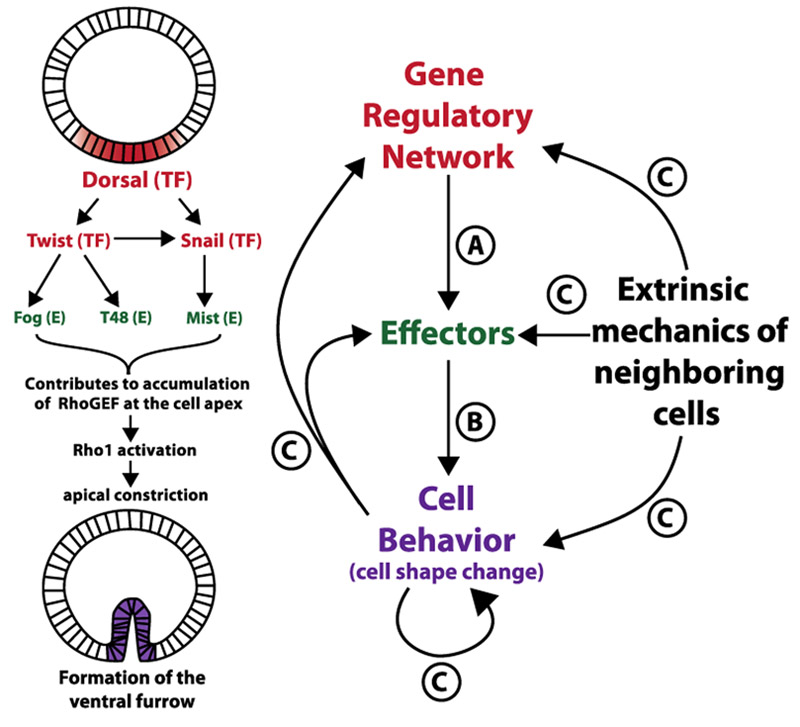
Figure 1. The influence of GRNs, cellular effectors, and neighboring mechanics on tissue morphogenesis.
Morphological structures are pre-patterned by GRNs (red) that turn on a precise set of effector molecules (green). These effectors control cell behaviors, here altering the shape of cells, together forming the final phenotype. (Left) An example of these processes occurring during ventral furrow formation where a nuclear gradient of the transcription factor Dorsal activates downstream transcription factors, which in turn activate effector molecules. These effector molecules accumulate at the apical membrane, causing the cells to apically constrict, and resulting in tissue invagination. (Right) Depiction of the interaction between gene regulatory networks, cellular effectors, and cell behaviors. While networks activate effectors (A), which drive changes in cell behaviors (B), development can also be influenced by signals and mechanical cues coming from neighboring cells in addition to feedback from different parts of the process (C).
The specification of developmental programs by gene regulatory networks
GRNs control the regulation of gene transcription in time, space, and intensity [1] to generate precise expression outputs that affect tissue morphology (Fig. 1A & Box 1) [5,6]. Recent work on the ventral furrow of Drosophila melanogaster illustrates how cell-to-cell variations in gene expression can be generated. A nuclear localized gradient of the transcription factor Dorsal establishes the dorsoventral axis of the embryo, and downstream genes Fog and T48 are expressed in a similar gradient along this axis (Fig. 1)[7]. One might hypothesize cells with the highest levels of nuclear Dorsal would transcribe fog and t48 mRNA at a higher rate. However, the gradients of fog and t48 are instead established in a progressive manner, with cells receiving the strongest Dorsal signal activating transcription earlier than cells receiving lower levels of Dorsal [8]. This causes higher levels of fog and t48 transcripts to accumulate in cells with the highest amounts of Dorsal. These dynamic differences in fog and t48 expression are vital for morphogenesis of the ventral furrow as both genes encode cellular effectors that help establish an activity gradient of non-muscle myosin II that drives apical constriction, which is essential for proper invagination of the ventral tissue (Fig. 1)[9]. Such observations emphasize the important role transcriptional dynamics can play by producing variation in gene expression, which can have fundamental mechanical consequences on morphogenesis.
At the interface between GRNs and effector molecules
As Drosophila ventral furrow formation illustrates, GRNs spatially and temporally pattern the level of expression of specific regulatory factors that impart on each cell a unique trans-regulatory environment, capable of activating a particular number of cellular effector genes, which control aspects of cellular behavior (Fig. 1B & Box 1) [3,4]. However, it is important to note that there are few universal correlations between the expression of a specific cellular effector and a certain cell behavior, because cellular context is important. Diverse cellular contexts can result from a change in the milieu of co-expressed cellular effectors, but can also result from changes in the cell's mechanical microenvironment, (e.g. strain applied to the cell), which will be discussed in the next section. As the effector repertoire of a cell is greatly influenced by its GRN, disentangling how GRNs interface with collections of cellular effectors that encode the cell’s physical responses represents a pressing need in this field.
The importance of understanding connections between GRNs and their target cellular effectors is particularly well demonstrated by the formation of morphologically diverse denticles on the larva of Drosophila. Denticles are actin rich epithelial projections that adorn the ventral surface of Drosophila larva[10]. Many signaling pathways interact to regulate the position and development of these structures, and converge to activate the transcription factor, shavenbaby (svb) [11], which is required for a cell to adopt the denticle fate. In the denticle, Svb regulates cellular effectors that promote various morphogenetic processes including actin reorganization, interaction with the extracellular matrix, and cuticle formation [10,12,13]. Interestingly, svb and several of its downstream cellular effector targets are also required for formation of other actin rich projections in Drosophila, such as the adult wing hairs, aristal laterals, and adult abdominal trichomes [11]. Although these epithelial projections all require svb, they are morphologically quite distinct, raising the possibility that svb regulates the formation of rudimentary actin rich projections, but that the final phenotype depends on the cellular context. Rizzo and Besjovec found that the transcription factor SoxNeuro (SoxN) is required to generate distinctive denticle morphologies observed on the larva of Drosophila [14]. Both svb and SoxN are required to activate shared, but also distinct sets of downstream cellular effectors with svb controlling denticle height and SoxN regulating width. The authors hypothesized that these two transcription factors respond differently to upstream signaling gradients to generate the diverse phenotypes observed across the Drosophila larva.
As this case highlights, similar GRNs can be responsible for generating generic structures whose diverse morphologies are specified by local context. Understanding how this localized context is generated at the GRN level is important, but also vital is understanding how downstream cellular effectors are precisely patterned. Essential to understanding the importance of the outcome of the GRN-effector connection is elucidating how cellular effectors exert influences on cell behaviors.
Intrinsic and extrinsic physical responses to cellular effectors
Once a cellular effector is activated, it can produce intrinsic effects by altering the function of other effectors, for instance, modulating adhesion, remodeling the cytoskeleton, changing the cell’s polarity, or targeting of cellular effectors to specific subcellular locations (Box 1) [15]. Once positioned, multiple effectors operate together to dynamically regulate the cell’s behavior, for instance, driving migration or initiating cell shape changes (Fig. 1C)[16]. Considering the interconnected nature of an epithelial tissue, these intrinsic changes can have extrinsic influences on neighboring cells and tissues. Extrinsic mechanics can influence or limit the range of shape changes the cells can adopt (Fig. 1D). Within an epithelium, these intrinsic and extrinsic mechanical processes add up to create an integrated physical response leading to distinct cell behaviors, such as cell rearrangement and shape changes. Although far more comprehensive reviews of both intrinsic and extrinsic mechanisms of epithelial cell shape change exist [16–21], we will examine recent examples that illustrate these interactions.
A case of oriented relaxation during dorsal closure in Drosophila highlights the importance of both intrinsic and extrinsic contributions to morphogenesis. Recent work has uncovered a mechanism whereby the cytohesin family member Steppke, an Arf-GEF, counteracts the assembly of actomyosin cables at the apical membrane of lateral epidermal cells [22]. Steppke allows cell-cell junctions to relax and the tissue to stretch in response to tension from neighboring tissues during the process of dorsal closure in Drosophila. Thus, Steppke operates as an intrinsic factor in lateral epidermal cells by relaxing junctions but also plays a role within the mechanically integrated dorsal closure movements as an extrinsic factor by reducing tension in the amnioserosa to aid in dorsal closure.
Intrinsic and extrinsic mechanisms also contribute to the formation of the ventral furrow during Drosophila gastrulation. As mentioned earlier, an intrinsically regulated gradient of non-muscle myosin II activity along the dorsal-ventral axis is important for invagination, however extrinsic mechanics are also vital for proper anterior-posterior orientation of actomyosin arrays within ventral furrow cells. Experimental treatments that changed the overall shape of the embryo or the dimension of the domain of gene expression in the surrounding tissue resulted in uniformly distributed actomyosin arrays in the ventral furrow [23]. This effect could be reversed through a variety of methods (e.g. laser ablations and knockdown of adhesion proteins), which restored directional tension to the ventral furrow cells and resulted in anterior-posterior actomyosin organization. This indicates that the overall shape of the embryo and the pattern of surrounding gene expression impose constraints that result in uneven tensions on the ventral furrow cells, which influences actomyosin organization in ventral furrow cells. This work highlights how intracellular force-generating and load-bearing structures might directly detect and respond to mechanical cues from the surrounding tissue to influence the final phenotype of the structure.
Finally, a striking example of the extrinsic mechanical influence of surrounding tissues on morphogenesis and gene regulation can be found in the patterning of periodic epithelial feather buds in chickens. A recent study reported that the dermal cells spontaneously aggregate below the epidermis due to their own contractility [24]. Dermal cell aggregates cause the overlying epithelial cells to bunch, resulting in the formation of a feather bud placode. Not only are epidermal cell mechanics affected by dermal cells movements, but gene regulation is also altered when β-catenin in the epidermal cells sense the dermal cell aggregation and responds by turning on a follicle GRN [24]. Overall, this suggests that the physical and regulatory state of a cell can be influenced by the mechanical movements of neighboring tissues, similar to how a signaling pathway can alter the regulatory state of neighboring cells through secretion of ligands. This and other similar cases of differentiation in response to extrinsic mechanical influences are being identified [25] and caution against focusing on intrinsic mechanical processes alone, highlighting the importance of examining the relative contributions of both intrinsic and extrinsic mechanical processes to morphogenesis.
Integrating an evolutionary perspective
So far, we have summarized recent work that is purely developmental, spanning a spectrum of morphogenetic processes that integrate GRNs, cellular effectors, and cellular mechanics. Such approaches can be complemented by evolutionary studies employing comparative methods. These studies can identify genetic variants that modify developmental processes, and have the potential to disentangle issues of cell autonomy raised by extrinsic mechanical influences and highlight which intrinsic processes were directly targeted during evolution. Below, we introduce comparative evo-devo model systems that have illuminated different aspects of morphogenesis and present new opportunities for deeper insights into the integration of GRNs, cellular effectors, mechanics, and morphogenesis.
Butterfly scales: connecting GRNs to the elaboration of single-cell appendages
Butterfly wings exhibit an enormous array of color patterns that has inspired numerous developmental, evolutionary, and ecological studies. The colors observed in butterfly wings can be formed by two mechanisms acting within the scale. The first way is through the use of different pigments, which selectively absorb certain wavelengths of light [26]. Many studies over the years have uncovered how some of these pigmentation patterns are genetically controlled. For example, Wnt signaling is responsible for regulating various pigmentation patterns across the wing [27–30].
More relevant to morphogenesis, the other way to form color is through structural changes to the butterfly scale that alters the way light is scattered [26]. The development of these complex scales begins with the projection of an epithelial cell that elongates and flattens to form the scale shape. Once scale morphogenesis is complete, the cell dies leaving both pigments and chitin, which forms the structural components of the scale [31]. The three dimensional shape of each scale is quite intricate, consisting of many chitinous substructures such as the upper lamina, which is composed of ridges and microribs, and the smooth lower lamina, both of which can contribute to structural color [32–34] (Fig. 2A). Because chitin is secreted during the development of this epithelial appendage, there are likely multiple morphogenetic processes during scale development that can be affected to alter structural color.
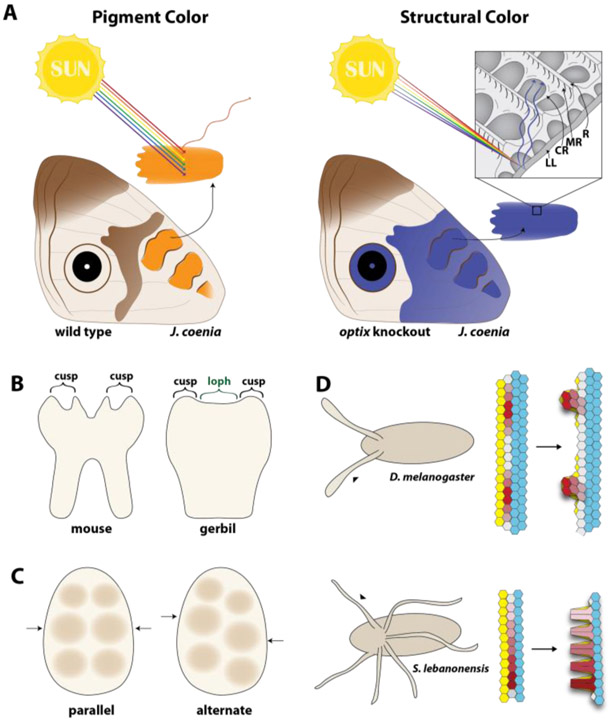
Figure 2. Evo-devo models offer many routes to explore patterning and morphogenesis.
(A) In butterflies, colors can be formed using pigments, such as the ommochrome pigment (orange), which works by absorbing all wavelengths of light except orange. In contrast, structural colors are formed by reflection of light interacting with components of the scale to selectively reflect blue wavelengths in the optix knockout. The structure of the scale is quite complex consisting of a lower lamina (LL) connected to an upper lamina composed of ridges (R), microribs (MR), and crossribs (CR). (B-C) Differences in rodent molar morphology. (B) In a cross-sectional view of the mouse molar, cusps are separated, but in gerbils cusps are connected by a ridge called a loph. (C) Occlusal view of an adult tooth with cusps represented by circles. Cusps can either be parallel or alternate in their placement on the tooth. (D) Morphology of the dorsal appendage (arrowhead) of the Drosophila eggshell differs in placement and number between species. Early morphogenesis of these homologous structures also differ, with the dorsal appendage of D. melanogaster forming through neighbor exchange events (red cells) and the dorsal appendage of S. lebanonensis (also known as S. pattersoni) forming through cell shape changes (red cells).
In the genus Bicyclus, structural violet/blue color has independently evolved twice [34]. To determine how this structural color may have evolved, researchers used artificial selection to evolve violet/blue color in a subset of scales of B. anynana, a predominately brown-pigmented species without structural color [34]. The authors detected an increase in thickness of the lower lamina of the scales that produce the structural color in both artificially selected and the naturally evolved species, suggesting that the lower lamina may be a common evolutionary target for violet/blue structural color evolution in Bicyclus.
How chitin is precisely secreted during butterfly scale development to form these architectural structures is unknown, however a previous study implicated F-actin [31]. In this study, the authors observed single bundles of actin in developing pigmented scales, with rows of chitin secreted between each actin bundle. However, in structurally colored scales of Agrulis vanillae, double bundles of actin were observed between the chitin ridges in addition to an overall increase in the amount of F-actin present during scale formation. This suggests that F-actin organization may play a key role to direct chitin secretion to form the ridges that can impart structural color. In addition, recent research has implicated pigment biosynthesis genes as contributors to chitin structure in the butterfly scale[35]. Mutations in various pigment genes can alter both pigment production and the chitin structure of the scale, which may have possible implications in limiting the potential path of evolution.
The regulatory networks controlling structural color are understudied, but recent research has pinpointed the transcription factor Optix as a repressor of blue structural color in Junonia coenia. CRISPR/Cas9 induced knockout mutations of optix in J. coenia resulted in formation blue structural color in species that normally lack it (Fig. 2A). Interestingly, this study identified two cellular effectors with known roles in F-actin filament organization that were downregulated in optix knockouts[36], correlating with the previous findings that F-actin may play an important role in determining where chitin is secreted.
Going forward, butterfly scales represent an excellent comparative model system to identify genes that regulate the morphogenesis of epidermal organs through their role in generating structural color. Of particular interest, artificially selected B. anynana strains could be used to genetically map loci that contribute variation in scale morphogenesis. Complementing this approach with descriptions of cellular effectors that are progressively activated during scale formation would illuminate the broader coupling of GRNs with effectors that operate during the formation of these complex structures.
The vertebrate tooth: elucidating evolutionarily important intrinsic and extrinsic mechanisms
Developmental and evolutionary biologists alike have long used teeth as a model system for many reasons, such as the ability to develop them ex vivo in culture and their abundant fossil record. Through the years, great progress has been made in understanding the gene network that patterns tooth development, including several signaling pathways expressed in a signaling center required to pattern tooth development (reviewed here [37–39]). Within mammals, there is substantial diversity in tooth morphology, especially in number, shape, and orientation of cusps, which are elevations on the surface of the tooth that often form a point [40]. Much of our knowledge of tooth development comes from research in mammals, fueled by extensive knowledge and tools developed for mice [38], but one very useful approach to study the evolution of tooth morphology is to leverage the extensive fossil record of mammals to infer their ancestral and derived forms. For instance, reduction of Fibroblast Growth Factor 3 (Fgf3) levels in mice and mutations in Fgf3 in humans both lead to a more ancestral tooth morphology, suggesting its involvement in more elaborate morphologies [41]. Another study found that gradual decreases in ectodysplasin (Eda) and sonic hedgehog (SHH) signaling in the mouse were able to mimic a more ancestral phenotype including a reduction in cusp number and loss of cusps on lower molars, highlighting the importance of absolute levels of growth factor signaling for tooth morphology [42]. Together these studies underscore important pathways that may be altered during tooth evolution, establishing promising systems where the connections between signaling events and specific cellular effectors that alter cell shape can be elucidated.
How cellular effectors control tooth shape is largely unexplored, but recent work has identified Rac1 and RhoA, regulators of F-actin, as important players that contribute to differences in tooth shape between gerbils and mice. Between their cusps, gerbils have ridges known as lophs that are missing in mouse (Fig. 2B)[43]. Inhibition of Rac1 or increases in RhoA in gerbils induces cell shape changes that lead to tissue invagination, eliminating lophs between the cusps to mimic the mouse phenotype. Reciprocal experiments to reduce RhoA results in loph-like ridges in mice. This study illustrates how altering expression of cellular effector can elucidate their role in controlling cell behavior and their functional influence on gross morphological difference between species. Future work can begin to connect patterning events to these important cell shape changes.
In addition to changes in signaling pathway activity and intrinsic cellular effectors, surrounding tissues can also influence the shape of teeth. Cusps can form in either parallel or alternating arrays; variations in these patterns have repeatedly evolved (Fig. 2C). Recent work has elucidated that the surrounding jaw influences the pattern of cusp formation through physical constraints [44]. Overall, given the great evolutionary diversity in tooth development, it will be interesting to determine the differences in GRNs regulating the diverse shapes between different vertebrates, in addition to understanding how changes to GRNs in the jaw can extrinsically alter the overall shape of the tooth. More generally, comparative analyses of the co-evolution of signaling networks and morphogenesis operating during tooth formation may highlight ways that epidermal organs are malleable to processes primed to select new adaptive morphologies.
Drosophila dorsal appendage: how similar structures can form by different morphogenetic processes
The dorsal appendage is a tubular structure that forms on the eggshells of Drosophilid species and is utilized for respiration during embryonic development. Its formation is a well-studied developmental process that is accompanied by a striking diversity in number and morphology across species, making it an excellent model to examine the evolutionary origins of integrated GRNs, cell signaling, and cell mechanical systems (reviewed here [45,46]). Early development of these structures begins with the projection of cells from the flat surface of the developing eggshell. Despite gross morphological similarities among species, distinct mechanisms have been found to drive the protrusion of these structures. In D. melanogaster, cell rearrangements drive protrusion of the nascent appendages, while in Scaptodrosophila lebanonensis, cell shape changes appear to be a major mechanical process in projecting the cells out (Fig. 2D) [47]. Based on the observations of these two distinct cellular mechanisms, researchers recently examined patterning systems that might account for these differences in morphogenesis [48]. The intersection of the BMP and EGF pathways regulate the formation of several eggshell structures including the dorsal appendage. The BMP signaling pathway displays similar patterning in both D. melanogaster and S. lebanonesis, but, major difference in the patterning of EGF signaling pathway is observed between the species. These changes result in one domain of expression of the transcription factor broad from which several dorsal appendages originate, as opposed to the two domains observed in D. melanogaster which each produce one dorsal appendage. This detailed knowledge of changes in patterning combined with the drastic difference in morphogenetic processes between D. melanogaster and S. lebanonensis positions the dorsal appendage for comparative analyses that can connect genetic changes in signaling pathways to downstream differences that regulate cellular effectors, providing a system to examine the importance of cell context on cellular effector function.
Conclusion
The process of developing an anatomical structure is not a simple one. It may involve many different levels that feedback upon one another and is best viewed from multiple complementary perspectives of GRNs, morphogenetic processes, and biomechanics. Studies often focus on individual steps of this process, but in order to move forward, integrative approaches must bridge these perspectives. We propose that the examination of evolutionary differences, combined with comparative morphogenetic studies can provide unique perspectives to help integrate these fields that will complement existing and upcoming developmental models. In particular, evo-devo model systems in which the contribution of individual genetic variants can be quantified (e.g. through genetic crosses) will be particularly powerful. Such systems will allow one to figure out which cells differ in cellular effector deployment and compare that information to quantitative measures of cell behavior and cell mechanics, facilitating the discrimination of intrinsic from extrinsic physical responses. Identifying these processes and how they have been altered during evolution can be a source of inspiration for engineers seeking novel methods to engineer tissues and treat disease. Above and beyond identifying genetic variants, both developmental and evolutionary model systems will require a deeper understanding of the key cellular effectors, their mechanical consequences, and how their cell-type specific influences on morphogenesis are realized.
Acknowledgements
We thank members of the Davidson and Rebeiz lab groups for helpful discussions and comments on the manuscript. The work in the Rebeiz and Davidson labs which inspired this review were supported by the NIH (HL134195, HL136566, and HD044750 to L.D. and GM107387 and GM112758 to M.R.).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Peter IS, Davidson EH: Genomic control process : development and evolution. Elsevier; 2015. [Google Scholar]
- 2.Heller E, Fuchs E: Tissue patterning and cellular mechanics. J Cell Biol 2015, 211:219–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernadskaya Y, Christiaen L: Transcriptional Control of Developmental Cell Behaviors. Annu Rev Cell Dev Biol 2016, 32:77–101. [DOI] [PubMed] [Google Scholar]
- 4.Gilmour D, Rembold M, Leptin M: From morphogen to morphogenesis and back. Nature 2017, 541:311–320. [DOI] [PubMed] [Google Scholar]
- 5.Levine M: Transcriptional enhancers in animal development and evolution. Curr Biol 2010, 20:R754–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peter IS, Davidson EH: Evolution of Gene Regulatory Networks Controlling Body Plan Development. Cell 2011, 144:970–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leptin M: Drosophila Gastrulation: From Pattern Formation to Morphogenesis. Annu Rev Cell Dev Biol 1995, 11:189–212. [DOI] [PubMed] [Google Scholar]
- 8.Lim B, Levine M, Yamazaki Y: Transcriptional Pre-patterning of Drosophila Gastrulation. Curr Biol 2017, 27:286–290. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This study illustrates the complexity of transcriptional dynamics by demonstrating how a transcription factor gradient controls the temporal expression of two downstream genes.
- 9.Heer NC, Miller PW, Chanet S, Stoop N, Dunkel J, Martin AC: Actomyosin-based tissue folding requires a multicellular myosin gradient. Development 2017, 144:1876–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price MH, Roberts DM, McCartney BM, Jezuit E, Peifer M: Cytoskeletal dynamics and cell signaling during planar polarity establishment in the Drosophila embryonic denticle. J Cell Sci 2006, 119:403–415. [DOI] [PubMed] [Google Scholar]
- 11.Delon I, Chanut-Delalande H, Payre F: The Ovo/Shavenbaby transcription factor specifies actin remodelling during epidermal differentiation in Drosophila. Mech Dev 2003, 120:747–758. [DOI] [PubMed] [Google Scholar]
- 12.Dickinson WJ, Thatcher JW: Morphogenesis of denticles and hairs in Drosophila embryos: Involvement of actin-associated proteins that also affect adult structures. Cell Motil Cytoskeleton 1997, 38:9–21. [DOI] [PubMed] [Google Scholar]
- 13.Chanut-Delalande H, Fernandes I, Roch F, Payre F, Plaza S: Shavenbaby Couples Patterning to Epidermal Cell Shape Control. PLoS Biol 2006, 4:1549–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rizzo NP, Bejsovec A: SoxNeuro and Shavenbaby act cooperatively to shape denticles in the embryonic epidermis of Drosophila. Development 2017, 144:2248–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** These authors explored how diverse denticle shapes are achieved in D. melanogaster through the characterization of SoxNeuro, a transcription factor that works with Shavenbaby to modulate overlapping aspects of denticle shape.
- 15.Nelson WJ: Adaptation of core mechanisms to generate cell polarity. Nature 2003, 422:766–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paluch E, Heisenberg C-P: Biology and physics of cell shape changes in development. Curr Biol 2009, 19:R790–799. [DOI] [PubMed] [Google Scholar]
- 17.Lecuit T, Lenne P-F: Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat Rev Mol Cell Biol 2007, 8:633–644. [DOI] [PubMed] [Google Scholar]
- 18.Heer NC, Martin AC: Tension, contraction and tissue morphogenesis. Development 2017, 144:4249–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lecuit T, Lenne P-F, Munro E: Force Generation, Transmission, and Integration during Cell and Tissue Morphogenesis. Annu Rev Cell Dev Biol 2011, 27:157–184. [DOI] [PubMed] [Google Scholar]
- 20.Devenport D: Tissue morphodynamics: Translating planar polarity cues into polarized cell behaviors. Semin Cell Dev Biol 2016, 55:99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davidson LA: Epithelial machines that shape the embryo. Trends Cell Biol 2012, 22:82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.West JJ, Zulueta-Coarasa T, Maier JA, Lee DM, Bruce AEE, Fernandez-Gonzalez R, Harris TJC: An Actomyosin-Arf-GEF Negative Feedback Loop for Tissue Elongation under Stress. Curr Biol 2017, 27:2260–2270.e5. [DOI] [PubMed] [Google Scholar]; ** This study found that cytohesins can locally antagonize actomyosin contractility in order to relax a cell junction and elongate a tissue, highlighting an important facet of active cell shape changes that are understudied.
- 23.Chanet S, Miller CJ, Vaishnav ED, Ermentrout B, Davidson LA, Martin AC: Actomyosin meshwork mechanosensing enables tissue shape to orient cell force. Nat Commun 2017, 8:15014. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This study demonstrates how cells can respond to extrinsic mechanics from surrounding tissues by orienting their cytoskeleton, emphasizing the importance of external mechanics on tissue development.
- 24.Shyer AE, Rodrigues AR, Schroeder GG, Kassianidou E, Kumar S, Harland RM: Emergent cellular self-organization and mechanosensation initiate follicle pattern in the avian skin. Science 2017, 357:811–815. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper illustrates how mechanical feedback from neighboring tissues can influence both cell shape and gene expression during feather bud development in chickens, highlighting the importance of intrinsic and extrinsic influences on development.
- 25.Chan CJ, Heisenberg C-P, Hiiragi T: Coordination of Morphogenesis and Cell-Fate Specification in Development. Curr Biol 2017, 27:R1024–R1035. [DOI] [PubMed] [Google Scholar]
- 26.Shawkey MD, Morehouse NI, Vukusic P: A protean palette: colour materials and mixing in birds and butterflies. J R Soc Interface 2009, 6:S221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazo-Vargas A, Concha C, Livraghi L, Massardo D, Wallbank RWR, Zhang L, Papador JD, Martinez-Najera D, Jiggins CD, Kronforst MR, et al. : Macroevolutionary shifts of WntA function potentiate butterfly wing-pattern diversity. Proc Natl Acad Sci 2017, 114:10701–10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin A, Papa R, Nadeau NJ, Hill RI, Counterman BA, Halder G, Jiggins CD, Kronforst MR, Long AD, McMillan WO, et al. : Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proc Natl Acad Sci U S A 2012, 109:12632–12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin A, Reed RD: Wnt signaling underlies evolution and development of the butterfly wing pattern symmetry systems. Dev Biol 2014, 395:367–378. [DOI] [PubMed] [Google Scholar]
- 30.Gallant JR, Imhoff VE, Martin A, Savage WK, Chamberlain NL, Pote BL, Peterson C, Smith GE, Evans B, Reed RD, et al. : Ancient homology underlies adaptive mimetic diversity across butterflies. Nat Commun 2014, 5:4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dinwiddie A, Null R, Pizzano M, Chuong L, Leigh Krup A, Ee Tan H, Patel NH: Dynamics of F-actin prefigure the structure of butterfly wing scales. Dev Biol 2014, 392:404–418. [DOI] [PubMed] [Google Scholar]
- 32.Vukusic P, Sambles JR, Lawrence CR, Wootton RJ: Quantified interference and diffraction in single Morpho butterfly scales. Proc R Soc London Ser B Biol Sci 1999, 266:1403–1411. [Google Scholar]
- 33.Stavenga DG, Leertouwer HL, Wilts BD: Coloration principles of nymphaline butterflies - thin films, melanin, ommochromes and wing scale stacking. J Exp Biol 2014, 217:2171–2180. [DOI] [PubMed] [Google Scholar]
- 34.Wasik BR, Liew SF, Lilien DA, Dinwiddie AJ, Noh H, Cao H, Monteiro A: Artificial selection for structural color on butterfly wings and comparison with natural evolution. Proc Natl Acad Sci U S A 2014, 111:12109–12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuoka Y, Monteiro A: Melanin Pathway Genes Regulate Color and Morphology of Butterfly Wing Scales. Cell Rep 2018, 24:56–65. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, Mazo-Vargas A, Reed RD: Single master regulatory gene coordinates the evolution and development of butterfly color and iridescence. Proc Natl Acad Sci U S A 2017, 114:10707–10712. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** The authors of this study found that the transcription factor optix represses structural color in butterfly scales. This is a fascinating example of how one genetic change can cause a switch from pigment-based to structural color.
- 37.Biggs LC, Mikkola ML: Early inductive events in ectodermal appendage morphogenesis. Semin Cell Dev Biol 2014, 25–26:11–21. [DOI] [PubMed] [Google Scholar]
- 38.Kim R, Green JBA, Klein OD: From snapshots to movies: Understanding early tooth development in four dimensions. Dev Dyn 2017, 246:442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jernvall J, Thesleff I: Tooth shape formation and tooth renewal: evolving with the same signals. Development 2012, 139:3487–97. [DOI] [PubMed] [Google Scholar]
- 40.Bergqvist LP: The role of teeth in mammal History. Brazilian J Oral Sci 2015, 2:249–257. [Google Scholar]
- 41.Charles C, Lazzari V, Tafforeau P, Schimmang T, Tekin M, Klein O, Viriot L: Modulation of Fgf3 dosage in mouse and men mirrors evolution of mammalian dentition. Proc Natl Acad Sci 2009, 106:22364–22368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harjunmaa E, Seidel K, Häkkinen T, Renvoisé E, Corfe IJ, Kallonen A, Zhang Z-Q, Evans AR, Mikkola ML, Salazar-Ciudad I, et al. : Replaying evolutionary transitions from the dental fossil record. Nature 2014, 512:44–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L, Tang Q, Nakamura T, Suh J-G, Ohshima H, Jung H-S: Fine tuning of Rac1 and RhoA alters cuspal shapes by remolding the cellular geometry. Sci Rep 2016, 6:37828. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** By altering the activity of effector genes, the authors interconverted molar cusp shapes between gerbils and mice, demonstrating the importance of individual effector genes in control of major phenotypic shapes.
- 44.Renvoisé E, Kavanagh KD, Lazzari V, Häkkinen TJ, Rice R, Pantalacci S, Salazar-Ciudad I, Jernvall J: Mechanical constraint from growing jaw facilitates mammalian dental diversity. Proc Natl Acad Sci U S A 2017, 114:9403–9408. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** These authors showed that the surrounding jaw could influence cusp pattern during tooth development. This highlights the possible importance of mechanical influence of neighboring tissue during the evolution of diverse phenotypes.
- 45.Osterfield M, Berg CA, Shvartsman SY: Epithelial Patterning, Morphogenesis, and Evolution: Drosophila Eggshell as a Model. Dev Cell 2017, 41:337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pyrowolakis G, Veikkolainen V, Yakoby N, Shvartsman SY: Gene regulation during Drosophila eggshell patterning. Proc Natl Acad Sci U S A 2017, 114:5808–5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osterfield M, Schupbach T, Wieschaus E, Shvartsman SY: Diversity of epithelial morphogenesis during eggshell formation in drosophilids. Development 2015, 142:1971–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper found that early morphogenetic events during the formation of a homologous tubular structure show dramatic differences between two species, raising the question of how genetic changes might have led to these divergent processes.
- 48.O’Hanlon KN, Dam RA, Archambeault SL, Berg CA: Two Drosophilids exhibit distinct EGF pathway patterns in oogenesis. Dev Genes Evol 2017, 228:31–48. [DOI] [PMC free article] [PubMed] [Google Scholar]