Abstract
AXL overexpression is a common resistance mechanism to anticancer therapies, including the resistance to BYL719 (Alpelisib) — the p110α isoform specific inhibitor of phosphoinositide 3-kinase (PI3K) — in esophagus squamous cell carcinoma (ESCC) and head and neck squamous cell carcinoma (HNSCC). However, the mechanisms underlying AXL overexpression in resistance to BYL719 remain elusive. Here, we demonstrate that the AP-1 transcription factors c-JUN and c-FOS regulate AXL overexpression in HNSCC and ESCC. The expression of AXL was correlated with that of c-JUN both in HNSCC patients and in HNSCC and ESCC cell lines. Silencing of c-JUN and c-FOS expression in tumor cells downregulated AXL expression and enhanced the sensitivity of human papilloma virus–positive (HPVPos) and –negative (HPVNeg) tumor cells to BYL719 in vitro. Blocking of JNK using SP600125 in combination with BYL719 showed a synergistic antiproliferative effect in vitro, which was accompanied by AXL downregulation and potent inhibition of the mTOR pathway. In vivo, the BYL719–SP600125 drug combination led to the arrest of tumor growth in cell line–derived and patient-derived xenograft models, as well as in syngeneic head and neck murine cancer models. Collectively, our data suggest that JNK inhibition, in combination with anti-PI3K therapy, is a new therapeutic strategy that should be tested in HPVPos and HPVNeg HNSCC and ESCC patients.
Keywords: Oncology, Therapeutics
Keywords: Cancer, Drug therapy, Head and neck cancer
AXL overexpression is regulated by transcription factors c-JUN and c-FOS in esophagus and head and neck squamous cell carcinoma, and JNK/c-JUN/AXL axis blockage enhances sensitivity to PI3K inhibition.
Introduction
The unsatisfactory outcomes of current therapies for patients with squamous cell carcinoma of the head and neck (HNSCC) or of the esophagus (ESCC) are underscored by the high mortality and morbidity rates of these cancers (1, 2). These outcomes, taken together with the high incidence rates of HNSCC and ESCC in many countries (3, 4), indicate an urgent need for new therapeutic approaches.
The major causes for the development of HNSCC and ESCC are alcohol consumption, smoking, and infection with human papilloma virus (HPV) (reviewed in ref. 5). Despite significant differences in the molecular landscapes of HPV-positive (HPVPos) and HPV-negative (HPVNeg) HNSCC and ESCC (6–8), both are frequently associated with alterations in the PIK3CA gene (6–8), which encodes for the p110α subunit of the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K). Activating point mutations or amplifications of the PIK3CA gene result in the hyperactivation of the PI3K/AKT/mTOR signaling pathway (reviewed in ref. 9). This pathway plays a key role in regulating cell proliferation and survival, enhancing tumor progression in PIK3CA-mutated HNSCC and ESCC. It is, thus, self-evident that new approaches for the treatment of HNSCC and ESCC should focus on blockers of the components of the PI3K/AKT/mTOR pathway, and indeed, such blockers are now under clinical development (reviewed in refs. 10–12). Among these blockers is the compound designated BYL719 (Alpelisib), which is an isoform-specific p110α inhibitor. In the first-in-human study of this compound, Juric et al. reported that 14 of 17 patients with PIK3CA-mutated HNSCC benefited from single agent administration of BYL719, although all patients eventually developed resistance to BYL719 (13). We recently showed that the emergence of resistance to BYL719 in HNSCC and ESCC involves the overexpression of AXL, which is a receptor tyrosine kinase (RTK) (14). AXL dimerizes with EGFR to activate the phospholipase Cγ–protein kinase C (PLCγ/PKC) signaling pathway, leading to the activation of mTOR in an AKT-independent manner (14). We further showed that AXL overexpression is associated with resistance to BYL719 in patients with HNSCC and that inhibition of AXL using R428 could reverse the resistance to BYL719 (14). Other studies have shown that AXL overexpression plays a key role in the resistance to many other anticancer therapies (15–19). These lines of evidence signify that treatment efficacies could be improved by blocking AXL activity, and indeed, small-molecule and antibody blockers of AXL are currently under clinical trials (ClinicalTrials.gov; NCT027929298 and NCT02988871). However, to the best of our knowledge, targeting the expression of AXL as an alternative therapeutic strategy — as will be described here — has not been explored, to date.
AXL gene transcription has been shown to be regulated by several transcription factors (TFs), such as SP1/3 (20) and MZF1 (21) in colon and cervix cancers and the AP-1 complex in melanoma, leukemia, and bladder cancer (22–24). However, the TFs that regulate AXL overexpression in ESCC and HNSCC in resistance to PI3K therapy remain uncharacterized. Here, we sought to elucidate the transcriptional machinery that regulates AXL expression and to explore whether a treatment protocol targeting AXL transcription in combination with BYL719 could serve as a therapeutic opportunity in HNSCC and ESCC.
Results
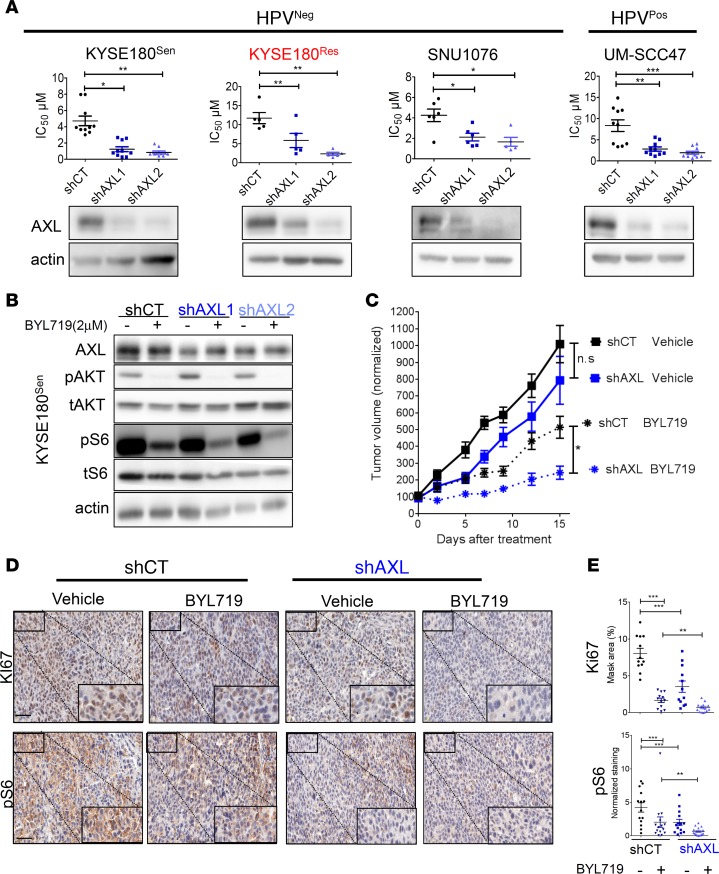
AXL expression determines sensitivity to BYL719 in HPVPos and HPVNeg cancer cell lines.
We have recently shown that AXL overexpression drives the resistance to BYL719 in HNSCC and ESCC cell lines and in HNSCC patients and that blocking of AXL with R428 sensitizes HPVNeg cells to BYL719 (14). In the current study, we first examined whether the AXL kinase activity determines the primary sensitivity to BYL719 in 2 HPVPos tumor cell lines, UM-SCC47 and UT-SCC60A. For this purpose, we tested the synergistic antitumor activity of BYL719 with R428 in these 2 HPVPos cell lines and found potent synergistic antitumor activity between the 2 agents (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.125341DS1). We next examined whether the basal expression of AXL determines the primary sensitivity to BYL719 in HPVPos and HPVNeg cell lines. As HPVNeg cell lines, we used SNU1076 (an HNSCC cell line), our previously established isogenic tumor cell line model, BYL719-sensitive KYSE180 (KYSE180Sen), and its counterpart BYL719-resistant model (KYSE180Res), which showed AXL overexpression (ESCC cell lines) (14). To this end, we knocked down the expression of AXL in HPVPos and HPVNeg HNSCC and ESCC cell lines, and we measured the half maximal inhibitory concentration (IC50) of BYL719 in vitro. Knockdown of AXL significantly reduced BYL719 IC50 values in all tumor cells (Figure 1A). A similar reduction of IC50 values was also observed for 2 additional PI3K inhibitors, GDC0941 (pan-PI3K) and GDC0032 (β-sparing) (Supplemental Figure 1B). Western blot (WB) analysis of the KYSE180Sen tumor cells following BYL719 treatment showed that AXL expression impacted the ability of BYL719 to block the mTOR pathway. Specifically, intensified inhibition of the phosphorylation of ribosomal protein S6 (pS6) was detected in tumor cells in which AXL expression had been knocked down (Figure 1B). These results are in keeping with our previous findings that AXL bypasses AKT to activate the mTOR pathway (14). Next, we confirmed the effect of AXL knockdown on the efficacy of BYL719 in vivo in a cell line–derived xenograft model (CDX) of KYSE180Sen tumor cells. In agreement with the above in vitro studies, BYL719-induced inhibition of growth was more efficient in the AXL knocked-down tumor cells (Figure 1C). This growth inhibition was associated with a reduction in mTOR pathway activation (as evaluated by pS6 IHC staining) and, consequently, with a reduction in tumor cell proliferation (as determined by Ki67 IHC staining) (Figure 1, D and E). Similar results were obtained for the HPVPos UM-SCC47 cells (Supplemental Figure 1, E and F).
Figure 1. AXL knockdown sensitizes HNSCC and ESCC cells to BYL719 in vitro and in vivo.
(A) Viability was assessed in cell lines treated with escalating doses of BYL719 for 4 days. Analysis of BYL719 IC50 values in HPVNeg and HPVPos HNSCC and ESCC cells following infection with shRNAs to silence AXL expression (shAXL1 and shAXL2) or with a nontargeting control shRNA (shCT) (n = 5–10). Lower panel: WB analysis demonstrating AXL levels in the same cells lines. (B) WB analysis showing AKT/mTOR pathway activation in KYSE180Sen cells after AXL knockdown and BYL719 treatment (2 μM, 24 hours). (C) Tumor growth of shCT or shAXL1 KYSE180Sen CDXs in mice (tumors, n = 9–11) treated daily with BYL719 25mg/kg. Presented are averaged tumor volumes ± SEM. (D) Representative images of IHC staining of the tumors showing the expression levels of the proliferation marker Ki67 and phosphorylated ribosomal S6 levels (pS6). Scale bar: 50 μm. Inset scale bar: 125 µm. (E) Analysis of expression levels in IHC staining presented in D, using the 3DHISTECH software HistoQuant. Analysis of Ki67 was performed in n = 12 different tumor tissue regions in each group, and analysis of pS6 was performed in n = 14–16 tumor tissue regions from shCT vehicle, shCT BYL719, shAXL vehicle, and shAXL BYL719 groups. All WB analysis was assessed in 2–4 independent experiments. Unpaired 2-sided t test P values are shown for in vitro studies, while in vivo 1-way ANOVA was performed. *P < 0.05; **P < 0.01; ***P < 0.001.
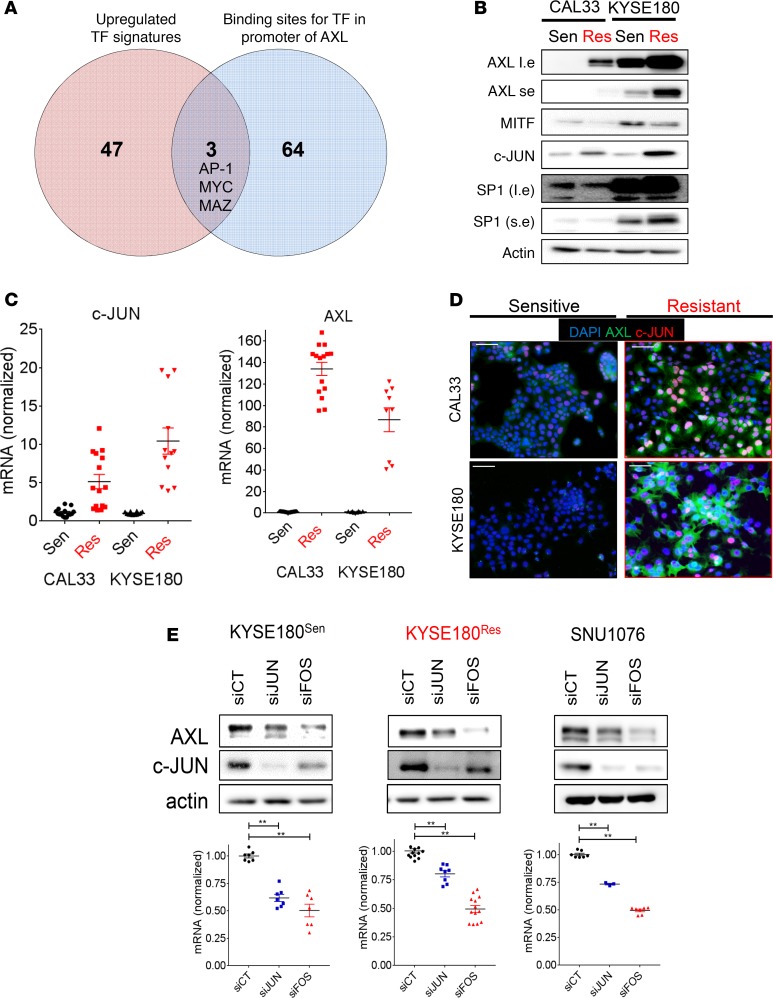
Resistance to BYL719 is associated with upregulation of the c-JUN TF.
Since we had shown that the expression level of AXL determined the response of HNSCC and ESCC cells to BYL719, we aimed to identify the molecular machinery that regulates AXL overexpression in our previously reported isogenic BYL719-sensitive and -resistant models — namely, KYSE180Sen vs. KYSE180Res and CAL33Sen vs. CAL33Res tumor cell lines (14). A gene set enrichment analysis (GSEA) of the RNA sequencing (RNA-seq) data obtained for these cells identified over 100 conserved motifs among 50 TFs that were significantly activated (Supplemental Table 1). TCF3, SP-1, AP-1, and MYC were among the most significant TF signatures that were upregulated in both resistant cell lines. Of the 50 TFs, 3 were predicted to have binding sites on the AXL promotor (QIAGEN transcription factor analysis) — namely, AP-1, MYC, and MYC-associated zinc finger protein (MAZ) (Figure 2A). Further analysis of the RNA-seq data indicated that the expression levels of MAZ and MYC were similar in BYL719-sensitive and BYL719-resistant cells, whereas the genes of the AP-1 complex — namely, FOSL1, FOS, and c-JUN — were significantly upregulated in both resistant cell models (Supplemental Figure 2A). WB and quantitative PCR (qPCR) analyses confirmed the upregulation of c-JUN in BYL719-resistant vs. -sensitive cells, with c-JUN upregulation being associated with AXL overexpression (Figure 2, B and C). The SP1 TF (previously reported to regulate AXL expression; refs. 20, 25) was upregulated in KYSE180Res cells but not in CAL33Res cells. In addition, we found an inverse correlation between AXL and the microphthalmia-associated TF (MITF) (Figure 2B), in agreement with previous reports (23, 26, 27). Increased expression of AXL and c-JUN was further confirmed in SNU1076 cells that had acquired resistance to BYL719 (Supplemental Figure 2B).
Figure 2. The AP-1 transcriptional complex regulates AXL expression in HNSCC and ESCC.
(A) RNA sequencing data obtained from isogenic BYL719 sensitive vs. acquired resistance CAL33 and KYSE180 cells (HNSCC and ESCC, respectively). Presented in the Venn diagram are TF signatures upregulated in BYL719-resistant cells and their overlap with binding sites for TF in the promoter of AXL. (B) WB analysis of CAL33 and KYSE180 BYL719-sensitive vs. -resistant cells showing the expression of TFs (l.e., long exposure; s.e., short exposure). (C) A qPCR analysis comparing the mRNA levels of AXL and c-JUN in CAL33- and KYSE180-sensitive vs.-resistant cells (n > 10). (D) IF images of CAL33- and KYSE180 BYL719–sensitive vs. –resistant cells showing c-JUN (CY-3-labeled) in red, AXL (Alexa488-labeled) in green, and DAPI in blue. Scale bar: 100 μm. (E) Upper panel: WB analysis showing AXL levels after transfection with siRNA for the silencing of c-JUN and c-FOS. Lower panel: qPCR analysis showing AXL mRNA levels in cells transfected with siRNAs for the silencing of c-JUN and c-FOS (n > 6). All WB analysis was assessed in 2–4 independent experiments. All qPCR analysis was assessed in 2–4 independent experiments. One-way ANOVA P value is shown. **P < 0.01.
To obtain further insight into AXL and c-JUN expression at the single cell level, immunofluorescent (IF) staining and imaging flow cytometry were performed. IF staining of AXL and c-JUN showed a correlation between the upregulation of AXL and that of c-JUN in BYL719-resistant cells at the single cell level (cells with high AXL displayed high c-JUN levels) (Figure 2D). Imaging flow cytometry analysis demonstrated that 97% of the KYSE180Sen cells displayed low expression levels of both c-JUN and AXL, and only a small subset of the tumor cells showed high expression levels of c-JUN that were accompanied by high AXL expression. This trend was reversed in KYSE180Res cells, most which (96.2%) showed high expression levels of both c-JUN and AXL (Supplemental Figure 2C). Given the positive correlation between AXL and c-JUN, we posited that the AP-1 transcriptional complex, which includes c-JUN and its counterpart c-FOS, regulates AXL expression. Indeed, silencing of c-JUN or c-FOS expression led to downregulation of AXL in KYSE180Sen, KYSE180Res, and SNU1076 cells, as evaluated by WB and qPCR analysis (Figure 2E and Supplemental Figure 2, D and E).
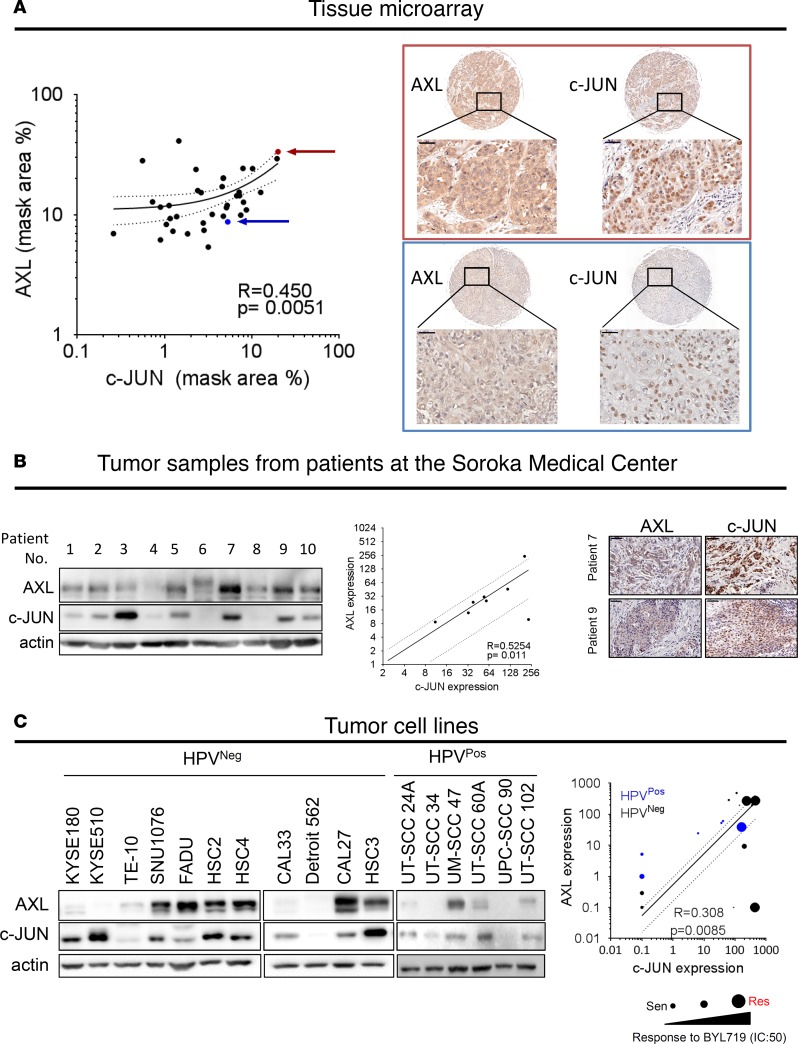
AXL and c-JUN levels are correlated in clinical samples from HNSCC patients and in HNSCC and ESCC cell lines.
To confirm the role played by c-JUN in AXL expression in HNSCC, we tested (using IHC staining) the correlation between AXL and c-JUN levels in patients with malignant HNSCC (n = 17) and in those with benign head and neck tumors (n = 15). IHC staining showed that AXL and c-JUN were positively correlated, both in tissues from the HNSCC patients (R = 0.450, P = 0.0051, Figure 3A) and in benign tissues (R = 0.6772, P < 0.0001, Supplemental Figure 3B and Supplemental Table 1). Moreover, our finding of a significant increase in both AXL and c-JUN expression in HNSCC compared with the benign tissues (Supplemental Figure 3A) is in keeping with previous reports of elevated c-JUN expression in oral SCC (28) (Supplemental Figure 3C). Validation of the expression of AXL and c-JUN was performed in an independent group of 10 freshly isolated HNSCC tumor specimens obtained from Soroka Medical Center (Beer-Sheva, Israel); the samples analyzed by WB and IHC showed a similar trend for the association of the expression levels of AXL and c-JUN (R = 0.5254, P = 0.011, Figure 3B). In addition, a positive correlation between AXL and c-JUN expression was detected in 5 HNSCC patient–derived xenografts (PDXs) (R = 0.969, P = 0.0063, Supplemental Figure 3D) and in a panel of 17 HNSCC and ESCC cell lines (R = 0.308, P = 0.0085, Figure 3C). Moreover, a positive correlation between another AP-1 member, FOSL1, was detected in 59 HNSCC and ESCC (The Cancer Cell Line Encyclopedia dataset; https://portals.broadinstitute.org/ccle) (Supplemental Figure 3E). The BYL719 IC50 values for the tested HNSCC and ESCC cell lines were also correlated to AXL and c-JUN expression, with most cell lines having high IC50 values also showing high expression levels of AXL and c-JUN (shown by the sizes of the round symbols in the plots representing each cell line in Figure 3C and in Supplemental Figure 3F). Collectively, the strong positive correlation found between AXL and c-JUN expression in HNSCC and ESCC clinical samples and in cell lines supports our premise that AXL expression is transcriptionally regulated by the AP-1 complex.
Figure 3. AXL and c-JUN levels are correlated in clinical samples of HNSCC tumors and in cell lines.
(A) IHC analysis of AXL and c-JUN expression levels in a tissue array of HNSCC tumors. The red and blue arrows indicate 2 different tumor samples that correlate with the IHC images marked by the red and blue rectangles. AXL and c-JUN expression levels were calculated using the 3DHISTECH software HistoQuant. Scale bar: 50 μm. (B) WB analysis of AXL and c-JUN expression levels in patient tumor samples from Soroka Medical Center. A densitometry analysis of expression levels of AXL and c-JUN is presented in the graph. IHC images of tumor tissues demonstrating AXL and c-JUN expression. Scale bar: 50 μm. (C) WB analysis of HPVNeg and HPVPos HNSCC and ESCC cell lines, demonstrating the expression levels of AXL and c-JUN. The graph shows quantification of the WB analysis. The size of the symbol correlates to the BYL719 IC50 value of the cell line in μM (i.e., the larger the symbol, the higher the IC50) and HPV status, represented by black or blue symbols for HPVNeg or HPVPos, respectively; IC50 values for HPVNeg cells were extracted from our previous report (14).
Knockdown of members of the AP-1 complex and inhibition of the c-JUN N-terminal kinase enhance BYL719 efficacy in vitro.
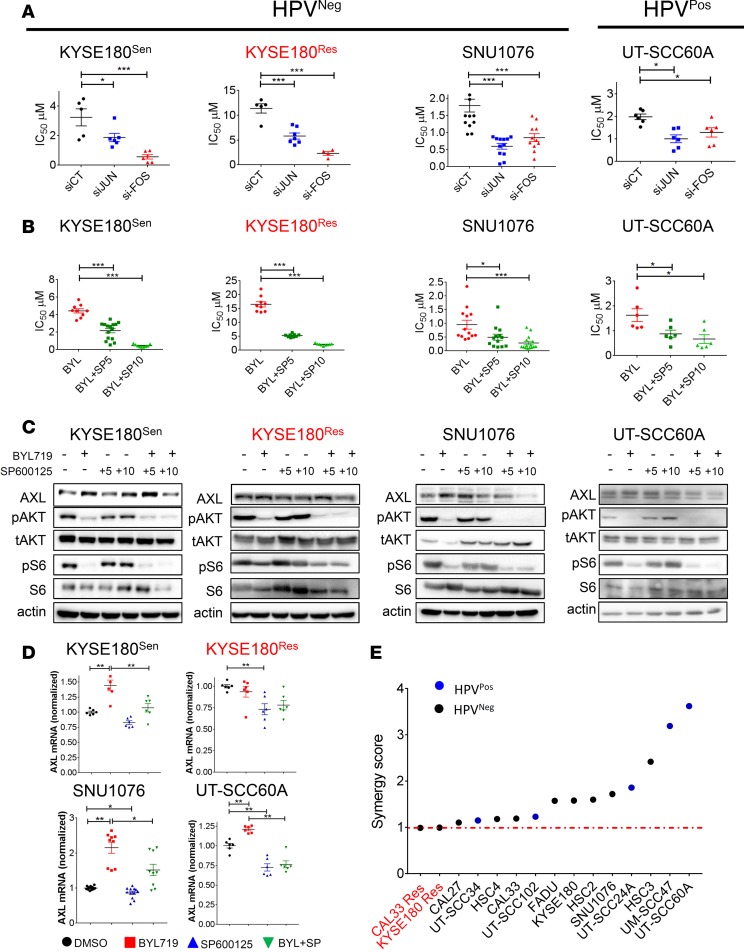
As AXL expression was shown to be regulated by the AP-1 transcription complex, we posited that knockdown of c-JUN and c-FOS would sensitize HNSCC and ESCC cells to BYL719. Indeed, we found that silencing of c-JUN and c-FOS expression in HPVNeg, KYSE180Sen, KYSE180Res, and SNU1076 cells significantly reduced the IC50 values of BYL719 (Figure 4A and Supplemental Figure 4, A and B). Similar results were obtained in the HPVPos cell lines UT-SCC60A and UM-SCC47 (Figure 4A and Supplemental Figure 4A).
Figure 4. Silencing of c-JUN and c-FOS or blocking JNK sensitizes HNSCC and ESCC cells to BYL719 in vitro.
(A) Viability was assessed in cell lines treated with escalating doses of BYL719 for 4 days. Analysis of BYL719 IC50 values in HNSCC and ESCC cells following transfection with siRNAs to silence c-JUN and c-FOS expression (n > 6). (B) Viability was assessed in cell lines treated with BYL719 and SP600125 for 4 days. Analysis of BYL719 IC50 values following JNK inhibition with SP600125 (5 and 10 μM) in HNSCC and ESCC cells (n > 6). (C) WB analysis showing AXL level and AKT/mTOR pathway activation in HNSCC and ESCC cells treated with BYL719 (2 μM), SP600125 (5 and 10 μM), and the combination therapy for 24 hours. (D) qPCR analysis of AXL mRNA levels in cells treated as in with BYL719 (2 μM), SP600125 (10 μM), and combination for 24 hours (n > 6). (E) Viability was assessed in cell lines treated with escalating doses of BYL719 and SP600125 for 4 days. Synergy test for the interaction between BYL719 and SP600125. The synergy test was analyzed using Chalice software (Horizon), and a synergy score was extracted. All WB analysis was assessed in 2–3 independent experiments. All viability experiments were assessed in 2–3 independent experiments. All qPCR experiments were assessed in 2–3 independent experiments. One-way ANOVA P values are shown. *P < 0.05; **P < 0.01; ***P < 0.001.
The direct role of c-JUN/c-FOS in AXL expression (Figure 2) and in the sensitivity to BYL719 (Figure 4A) suggests that blocking of c-JUN activity may potentiate BYL719 efficacy. As direct blockers of c-JUN are not yet available, we tested the above premise with an inhibitor of JNK, designated SP600125 (29). A combination of BYL719 and SP600125 significantly decreased the IC50 values of BYL719 in 3 HPVNeg and in 2 HPVPos HNSCC and ESCC cell lines in a dose-dependent manner (Figure 4B and Supplemental Figure 4C). Notably, the IC50 values for KYSE180Res cells treated with the BYL719–SP600125 combination were similar to those for KYSE180Sen cells, which suggests that the combined treatment delayed the acquisition of resistance. These results were validated in another isoform specific inhibitor of the PI3K, GDC0032 (Supplemental Figure 4D). A cell proliferation assay monitoring growth rates of the cells showed that the combined BYL719–SP600125 treatment had a superior antiproliferative effect compared with treatment with each of the compounds alone (Supplemental Figure 4E). WB analysis showed that, while BYL719 upregulated AXL expression, its combination with SP600125 (10 μM) resulted in decreased AXL protein levels (Figure 4C and Supplemental Figure 4F). The level of pS6 was further inhibited by the combination of BYL719 and SP600125 in KYSE180Res and SNU1076 cells (Figure 4C), when compared with the effect of BYL719. For KYSE180Sen, SNU1076, and UT-SCC60A cells, qPCR analysis showed that AXL mRNA levels increased following BYL719 treatment, but this gene upregulation was attenuated in response the combination with SP600125 (Figure 4D). In KYSE180Res cells, in which AXL transcript basal levels were stably elevated (Figure 2C), a significant downregulation of AXL mRNA levels was detected following exposure to SP600125. We confirmed the on-target effect of JNK inhibition using BI-78D3 (another JNK inhibitor) and RNAi against JNKs (Supplemental Figure 4G and Supplemental Figure 4H, respectively). Moreover, overexpression of AXL in CAL33 cells partially contrast the antitumor efficacy of the drug combination of BYL719 and SP600125, compared with CAL33 cells infected with control vector, GFP. Importantly, overexpression of AXL did not contrast the efficacy of combination of BYL719 and R428 (Supplemental Figure 4I). These rescue experiments demonstrate that AXL expression is required for the efficient treatment with of BYL719 and SP600125.
To explore whether the antiproliferative effect of BYL719 and SP600125 is additive or synergistic, a synergy test was performed in 5 PIK3CA mutant and 3 PIK3CA WT (all HPVNeg) HNSCC and ESCC cell lines, and 5 HPVPos HNSCC cell lines. Representative images of the analysis in SNU1076 cells are presented in Supplemental Figure 4J. Calculation of the synergy score using Chalice software indicated a synergistic effect (>1) in 13 of the 15 cell lines tested, whereas an additive effect was observed in the BYL719–acquired resistant cell lines KYSE180Res and CAL33Res (Figure 4E).
Superior efficacy of the combined treatment with BYL719 and SP600125 in vivo.
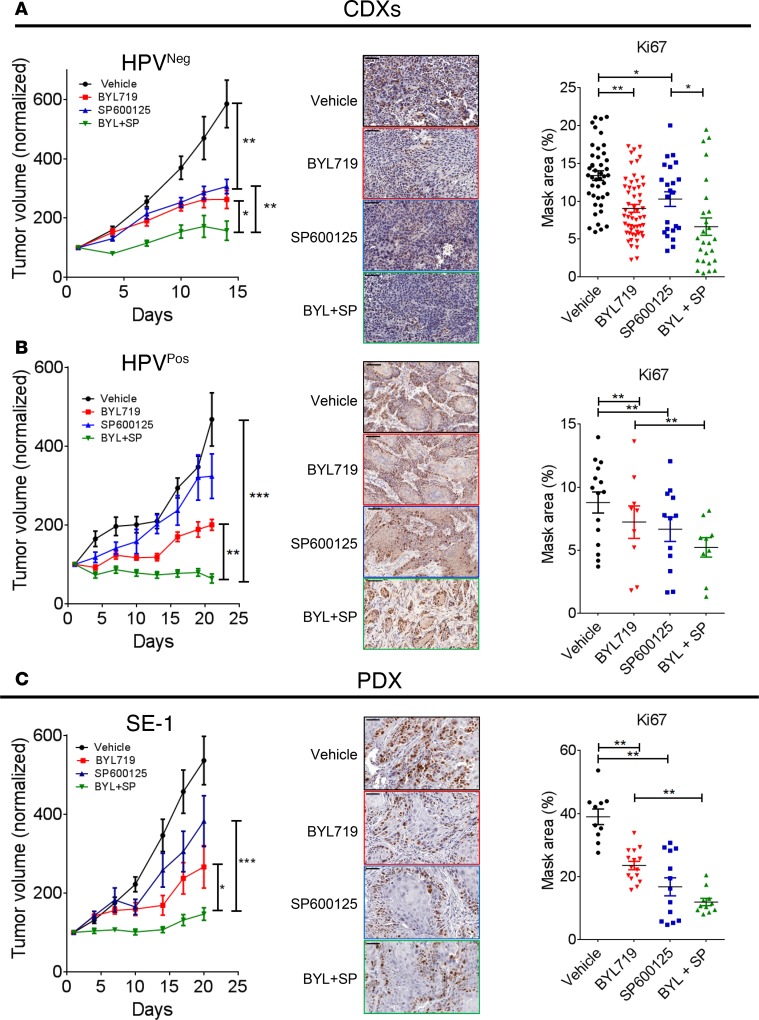
To explore the efficacy of the BYL719–SP600125 combination in vivo, CDX models of KYSE180Sen (HPVNeg) and UM-SCC47 (HPVPos) cells were generated. Daily treatments of tumor-bearing mice with BYL719 monotherapy (25 mg/kg) delayed tumor growth compared with treatment with the vehicle. Importantly, marked arrest of tumor growth (in HPVNegKYSE180Sen CDXs) and significant tumor shrinkage (in HPVPos–UM-SCC47 CDXs) were observed in mice treated with the BYL719–SP600125 combination (Figure 5, A and B, and Supplemental Figure 5, A and B). We did not detect any toxicity in that the weight of the animals remained stable (Supplemental Figure 5, A and B). IHC staining of the proliferation marker Ki67 showed decreased tumor cell proliferation in tumors treated with either BYL719 or SP600125 monotherapy. However, a more marked inhibition of cell proliferation was detected in tumors treated with the combination therapy.
Figure 5. SP600125 enhances BYL719 efficacy in vivo in CDX and PDX models.
(A–C) Tumor growth of KYSE180Sen (A), UM-SCC47 (B), and HNSCC PDX-SE1 (C) (tumor n = 8–12) following daily treatment with vehicle, BYL719 (25 mg/kg), SP600125 (15 mg/kg), or the BYL719–SP600125 combination. IHC staining of the tumors for the proliferation marker Ki67 is shown in the middle. Scale bar: 50 μm. The 3DHISTECH software HistoQuant was used analysis of IHC staining (n > 10). One-way ANOVA P values are shown. *P < 0.05; **P < 0.01; ***P < 0.001.
To further validate our observations in preclinical models, the efficacy of the drug combination was tested in 2 PDX models (SE1 and SE3). Treatment of the mice with the BYL719–SP600125 combination confirmed the superior antitumor effect of the combination, as indicated by smaller tumor volumes and lower tumor weights (Figure 5C and Supplemental Figure 5C). Ki67 staining showed a similar trend, further supporting the potency of drug combination. A reduction of AXL expression level in tumors treated with the combination of BYL719 and SP600125 was confirmed in 2 PDXs, SE1 and SE3, using WB (Supplemental Figure 5, C and D) and in UM-SCC47 xenografts using IHC (Supplemental Figure 5C).
Enhanced antitumor efficacy of the BYL719–SP600125 combination prevents tumor progression and enhances survival in syngeneic head and neck cancers.
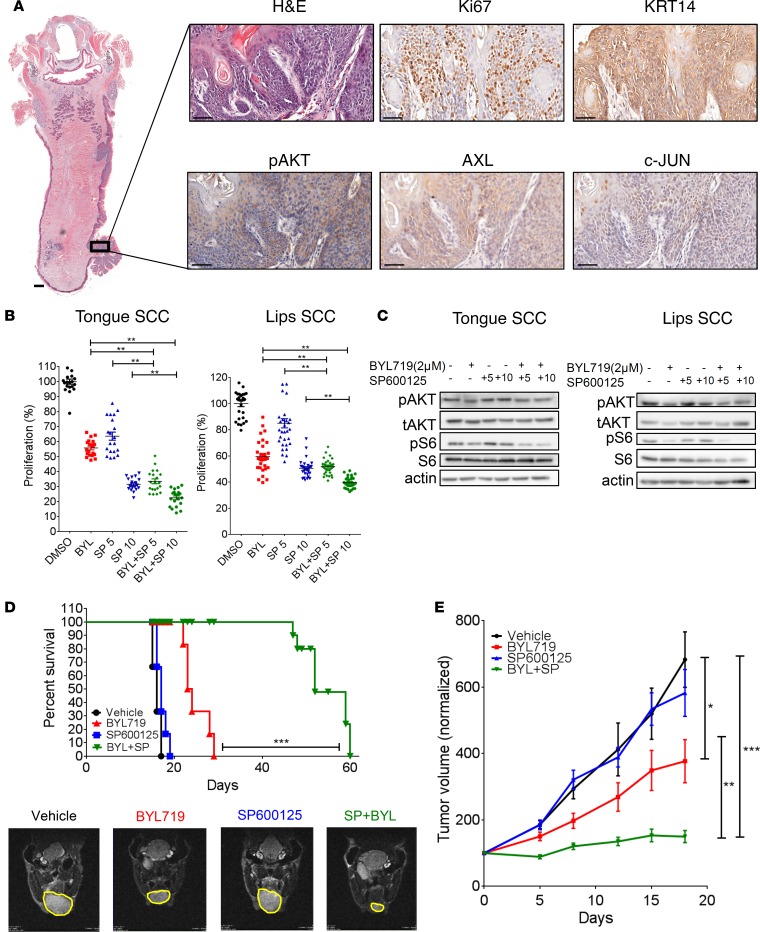
To verify the efficacy of the combination of BLY719 and SP600125 in syngeneic HNSCC tumors, we generated HPVNeg tumors in C57BL/6 mice. To this end, C57BL/6 mice were exposed to the carcinogen 4-nitroquinoline 1-oxide (4NQO) in their drinking water. Lip and tongue tumors developed following the exposure of the mice to 4NQO, as previously reported (30) (Supplemental Figure 6A). IHC staining demonstrated the activation of the AKT pathway, as well as the expression of c-JUN and AXL in the 4NQO-induced tongue malignant lesion (Ki67-positive and KRT14-positive staining; Figure 6A). Following the generation of 2 cell lines derived from the lip and tongue tumors, we studied the efficacy of BYL719, SP600125, and the combination in vitro and in vivo. In vitro, the BYL719–SP600125 combination exhibited superior antiproliferative activity, which was associated with a reduction of pS6 levels, as shown by WB analysis (Figure 6, B and C). In vivo, the efficacy of the drug combination had a significant antitumor effect in the 2 syngeneic cancer models. Measurements of the lip tumors in mice treated with the BYL719–SP600125 combination indicated a stable disease, whereas tumor progression was observed in mice treated with either BYL719 or SP600125 (Figure 6E). In an orthotropic tongue cancer model, the BYL719–SP600125 combination significantly improved the survival of the mice. The median survival for vehicle- and SP600125-treated mice was 16 and 17 days, respectively, and for BYL719-treated mice, median survival was 23 days (Supplemental Figure 6, B and C). Importantly, the median survival of mice receiving the combination therapy was more than doubled, at 52 days. MRI supported the findings; the scans showed, in particular, the inhibition of tumor progression with the drug combination (Figure 6D).
Figure 6. SP600125 increases BYL719 efficacy in vivo in syngeneic head and neck cancer models.
(A) IHC staining showing the expression of Ki67, pAKT, AXL, c-JUN, and keratin-14 (KRT14) in the 4NQO-induced tongue tumor region. Scale bars: 1000 μm (left) in the small magnification image of whole tongue and 50 μm (right) for the higher-magnification images. (B) Proliferation assay of 4NQO-induced tumor cell lines from the tongue and lips following 4 days of treatment with BYL719 (2 μM), SP600125 (5 and 10 μM), or the BYL719–SP600125 combination (n > 10). (C) WB analysis of tongue and lip 4NQO-induced tumor cell lines after 24 hours with the indicated treatments, showing activation of the AKT/mTOR pathway. (D) Survival rates of immunocompetent C57BL/6 mice (n = 5–6) in an orthotopic tongue cancer model following daily treatments with BYL719 (25 mg/kg), SP600125 (15 mg/kg), or the BYL719–SP600125 combination. Also presented are T2 weighted coronal images (obtained from the MRI) of tongues (lower panel), showing the difference in tumor progression following 10 days of treatments. Tumor margin, yellow. (E) Tumor growth (n = 8–10) of immunocompetent C57BL/6 mice bearing a syngeneic lip cancer model, treated as indicated in D. WB analysis was assessed in 2–3 independent experiments. One-way ANOVA. *P < 0.05; **P < 0.01; ***P < 0.001.
Discussion
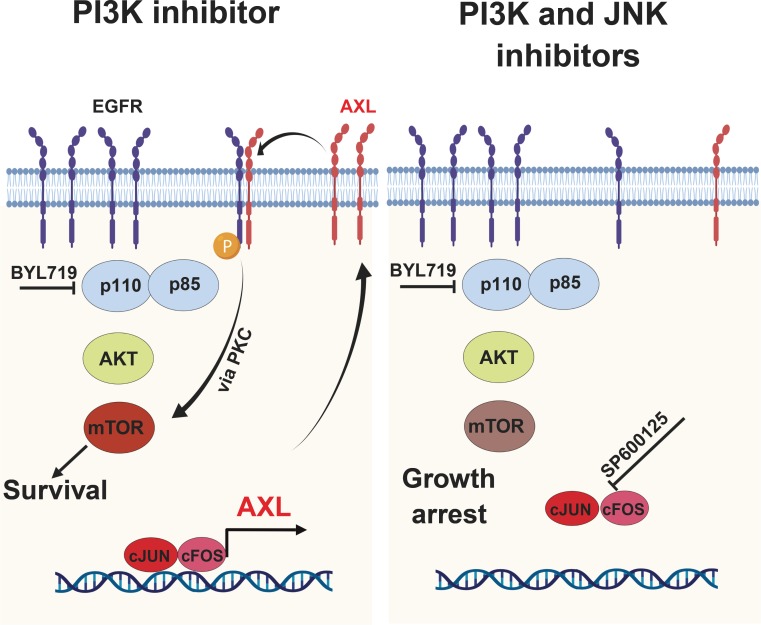
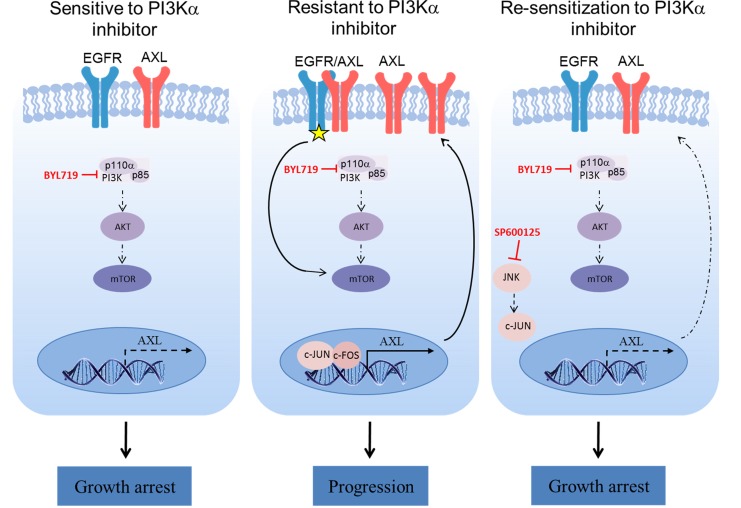
In this work, we demonstrated that the AXL expression level influences the sensitivity to BYL719 of HPVNeg and HPVPos HNSCC and ESCC cells. While we previously described that overexpression of AXL induces EGFR signaling and activation mTOR in an AKT-independent manner (14), here we identified the transcriptional machinery that induces AXL overexpression. Specifically, we identified the AP-1 transcriptional complex as a regulator of AXL expression in tumor cells, and we demonstrate that inhibition of JNK downregulated AXL mRNA and protein expression. This reduction of AXL expression enhances the antitumor efficacy of BYL719 in vitro and in vivo via further inhibition of the mTOR pathway (Figure 7).
Figure 7. Proposed mechanism of AP-1/AXL–driven resistance to PI3Kαi in HNSCC and ESCC.
In BYL719-resistant HNSCC and ESCC cell lines, overexpressed AXL dimerizes with EGFR and activates mTOR signaling in an AKT-independent manner. Upregulation of AXL is mediated by the AP-1 transcription factors c-JUN and c-FOS and induces EGFR phosphorylation (yellow star). Blocking c-JUN with the JNK inhibitor SP600125 reduces AXL expression and, hence, sensitizes the tumor cells to BYL719 via preventing mTOR pathway activation.
Our rationale for targeting the PI3K pathway with BYL719 (an isoform-specific inhibitor) is based on the encouraging results obtained from preclinical and clinical studies that tested the efficacy of the p110α inhibitor BYL719 in PIK3CA mutant cancers (13, 31–37). The major obstacle to implementing BYL719 therapy in HNSCC and ESCC lies in the primary and acquired resistance to this compound, which is driven by the overexpression of AXL (14). This overexpression of AXL in response to therapy is not unique to these malignant diseases or to PI3K-targeting therapies, as AXL pathway activation has been reported in various cancers that developed resistance to radiotherapy, chemotherapies, and EGFR inhibitors (15–19, 38–40). Indeed, small molecule inhibitors (such as BGB324, ONO-747, TP-0903, MGCD516, and BPI-9016M) or specific antibodies (CAB-AXL-ADC) that target the AXL receptor or the AXL pathway are currently under clinical evaluation in 14 clinical trials as a monotherapy or in combination with other therapies in different settings (ClinicalTrials.gov). The clinical outcomes of these studies are not yet available, but preclinical models indicate that the efficacy of a single agent targeting AXL signaling can be limited by the upregulation of other receptors, such as MERTK (41), while the potency of several combination therapies is superior to that of the monotherapies (15, 40, 42–47).
Here, we adopted an alternative approach to reducing the availability of AXL by limiting its transcription. Our initial demonstration that AXL levels determine the efficacy of treatment with BYL719 in HNSCC and ESCC cells in vitro and in vivo supported the rationale to reduce AXL expression in tumor cells as a means to enhancing BYL719 efficacy. Our results suggest that this approach could be beneficial when applied to HPVNeg, as well as to HPVPos, cells that may be intrinsically resistant to BYL719 (48). Additional confidence in our approach to reduce the AXL expression level as a therapeutic opportunity was provided by the study of Wheeler’s group showing that a reduction of AXL expression enhanced the efficacy of cetuximab in HPVPos and HPVNeg HNSCC tumors (15, 40).
A number of studies have shown that overexpression of AXL in response to therapeutic stress can evolve through a variety of mechanisms, including transcriptional regulation (15, 23, 24, 49–51), posttranslational regulation (52, 53), and expression of miRNA (54) (reviewed in ref. 55). In our isogenic tumor cell line of BYL719-sensitive and BYL719–acquired resistant cells, we observed a marked increase in the AXL transcript and a concomitant increase in members of the AP-1 complex in resistant cells. Thus, we posited that the AP-1 TF complex may be responsible for overexpression of AXL in BYL719-resistant cells. Indeed, knockdown of AP-1 complex members c-FOS and c-JUN in tumor cells resulted in significant reductions in AXL protein and mRNA expression levels in BYL719-sensitive and BYL719-resistant cells.
The mechanism by which BYL719 induces overexpression of c-JUN is not yet clear and requires further investigation. A possible explanation may involve MITF regulation of c-JUN expression. It has been shown that MITF negatively regulates c-JUN expression and is itself suppressed by proinflammatory cytokines (56). Thus, in light of recent studies demonstrating that tumor cells secrete proinflammatory cytokines, such as TNF-α following anti-PI3K treatment (57), it is tempting to suggest that BYL719 treatment induces the secretion of proinflammatory cytokines, which may downregulate MITF expression and, hence, enhance c-JUN expression.
In an attempt to target c-JUN–induced-AXL expression, we used SP600125, an inhibitor of JNK activation, as direct c-JUN inhibitors are not available (29). Supplementation of BYL719 with SP600125 improved the antiproliferative effect of BYL719 in vitro to a degree similar to that of silencing of c-JUN by using RNAi. Since the combination of BYL719 with SP600125 was shown to have a synergistic antiproliferative effect in most of the 15 cell lines tested, we used relatively low dosages of BYL719 and SP600125 (25 mg/kg and 15 mg/kg daily, respectively) in the testing of the combination in vivo. These low drug concentrations prevented tumor progression with no evidence of toxicity.
Targeting AP-1 or c-JUN signaling may have a broader effect on tumor cells than simply reducing AXL expression. Targeting the pleotropic role of AP-1 TFs in HNSCC may affect the cancer stem cells (58), epithelial-mesenchymal transition (59), and/or tumor invasiveness (60). Additionally, blocking AP-1 transcription activity may serve to influence the expression of key RTKs, such as EGFR (61, 62), and/or to reduce the expression of the immunosuppressor modulator of programmed cell death ligand 1 (PD-L1) (63) and, hence, enhance tumor elimination. The latter possibility is also supported by the findings that AXL and PD-L1 expression are correlated in different cancers (64, 65) and in the cancer genome atlas (TCGA) dataset (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) for head and neck cancer (data not shown).
In summary, we present evidence that upregulation of AXL in PI3K-resistant cells is regulated by AP-1 TFs. Knockdown of AXL or AP-1 enhanced the antitumor efficacy of BYL719. Targeting of JNK with BYL719 in combination with SP600125 provided potent antitumor activity against HPVPos and HPVNeg cells. These results provide strong grounds for investigating drug combinations of PI3K- and JNK-targeting therapies in HNSCC and ESCC.
Methods
Tumor cell lines.
All HPVNeg cell lines were purchased from commercial vendors; cell line sources and specific media are described in Supplemental Methods. BYL719-resistant KYSE180Res and CAL33Res cell lines were developed previously (14), and the SNU1076-resistant cell line was developed using the same approach as that described previously (14). All HPVPos cell lines, which were available in house, were grown in DMEM medium. For complete cell lines names, media, and source, see Supplemental Table 3. PIK3CA and PTEN status is shown in Supplemental Table 5. Two mouse HNSCC cell lines were developed in our laboratory from 4NQO-induced oral cancer. Briefly, fresh lip and tongue tumor tissues were washed with HBSS or PBS, cut into small pieces with sterile scissors, and treated with an enzyme mixture of collagenase (10 mg/ml; Thermo Fisher Scientific, catalog 17104019), hyaluronidase (1 mg/ml; MilliporeSigma, catalog H3506), and DNase (200 U/ml; Thermo Fisher Scientific, catalog18047019). The tissues were dissociated using the gentleMACS Dissociator. Cells were filtered through a 70-μM cell strainer, centrifuged (300 g, 5 minutes), and cultured in DMEM medium. The cultures were stained for the epithelial markers cytorkeratin 14 and E-cadherin for verification of epithelial origin.
All cells were maintained at 37°C in a humidified atmosphere at 5% CO2, in the relevant media supplemented with 1% l-glutamine 200 mM, 100 units each of penicillin and streptomycin, and 10% FBS. Acquired resistance cell lines were grown in the presence of BYL719 (2 μM), which was added to the medium every 3 days. Cells were routinely tested for mycoplasma infection and treated with appropriate antibiotics as needed (De-Plasma, TOKU-E, D022). HPVNeg cells were tested for cell line authentication in our previous work (14), and several cell lines (SNU1076, FADU, HSC-2, and HSC-4) were retested in this work.
WB.
Cells were washed with ice-cold PBS, harvested into lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 1% Triton X-100, 10 μM MgCl2), supplemented with phosphatase inhibitor cocktails (Biotool, B15001A/B) and protease inhibitor (MilliporeSigma, P2714-1BTL), and placed on ice for 30 minutes, followed by 3 minutes of ultrasonic cell disruption. Lysates were cleared by centrifugation (30 minutes, 20,800 g, 4°C). Supernatants were collected, protein concentrations were determined using Bio-Rad protein assay, SDS sample buffer was added, and samples were boiled for 5 minutes before being frozen at −80°C until further use. Whole cell lysates (25 μg) were separated on 10% SDS–PAGE and blotted onto PVDF membranes (Bio-Rad trans blot TurboTM transfer pack #1704157). Membranes were blocked for 1 hour in blocking solution (5% BSA [Amresco, 0332-TAM] in Tris-buffered saline [TBS] with 0.1% Tween) and then incubated with primary antibodies diluted in blocking solution. Mouse and rabbit HRP–conjugated secondary antibodies were diluted in blocking solution. Protein-antibody complexes were detected by chemiluminescence (Westar Supernova [Cyanagen, XLS3.0100] and Westar Nova 2.0 [Cyanagen, XLS071.0250]), and images were captured using the Azure C300 Chemiluminescent Western Blot Imaging System (Azure Biosystems). Details of antibodies and dilutions used are presented in Supplemental Methods (Supplemental Table 4).
Real-time PCR.
Total RNA was isolated from the cultured cells using PureLink RNA Mini Kit (Invitrogen, Thermo Fisher Scientific) according to the manufacturer’s protocol, and 1 μg RNA was converted to cDNA using qScript cDNA synthesis kit (Quanta Bioscience, 95047-100) according to the manufacturer’s protocol. Real-time PCR was performed (Roche light cycler 480 II) using PrimeTime Gene Expression Master Mix (IDT, 1055770), with matching probes from IDT: Axl gene, Hs.PT.56a.1942285; GAPH gene, Hs.PT.39a.22214836; Jun gene, Hs.PT.58.25094714.g; and Fos gene, Hs.PT.58.15540029. Analysis was performed with LightCycler 480 Software, Version 1.5.1. Fold change was calculated using the ΔΔCt method. Results were normalized to GAPDH levels and presented as a fold increase of the control cells.
GSEA.
Seven hundred and fifteen genes were upregulated in resistant cells compared with sensitive cells (cut off of >100 reads, and of 0.5 log2, Padj > 0.05). GSEA was performed using GESA of the Broad Institute (http://software.broadinstitute.org/gsea/index.jsp). The threshold for a conserved motif was defined as (Padj > 0.1 × 10-5). For the binding site of the AXL promoter, we used the Genecard and the TF binding sites on QIAGEN websites. All the data is available in Supplemental Table 1.
IHC and IF.
Tissues were fixed in a 4% paraformaldehyde (PFA) solution for a maximum of 24 hours at room temperature, dehydrated, and embedded in paraffin. The tissue sections were deparaffinized with xylene. H2O2, at 3%, was used to block the endogenous peroxidase activity for 20 minutes; thereafter, the sections were rinsed in water for 5 minutes. Antigen retrieval was performed in citrate buffer (pH 6) at 99.99°C for 5 minutes. Sections were then blocked for 1 hour at room temperature with blocking solution (PBS, 0.1% Tween [0.0125% for AXL staining], 5% BSA), followed by incubation with primary antibody (diluted in blocking solution) overnight at 4°C. The ABC kit (VECTASTAIN, VE-PK-6200) was used for detection according to the manufacturer’s protocol. Sections were counter stained with hematoxylin and mounted in mounting medium (Micromount, Leica, 380-1730).
For IF, cells were seeded on 8-well glass slides (Cellvis, C8-1.5H-N) for 48 hours. Cells were rinsed with cold PBS (4°C) and fixed in 4% PFA for 30 minutes at room temperature. Thereafter, cells were rinsed with PBS, followed by permeabilization on ice for 5 minutes in PBS with 0.05% Triton X-100 (MilliporeSigma), rerinsed with PBS, and blocked in blocking solution for 30 minutes at room temperature (5% BSA in Tween-PBS). The cells were incubated with primary antibodies overnight at 4°C, rinsed with PBS, and incubated with Cy3-conjugated anti-rabbit secondary antibody (1:250) (Jackson ImmunoResearch, 111-165-144) and Alexa Fluor 488–conjugated anti-mouse secondary antibody (1:250) (Jackson ImmunoResearch, 115-545-062) at room temperature for 1 hour. Cells were then rinsed with PBS and mounted in DAPI Fluoromount-G (Southern Biotech, 0100-20).
IHC and IF slides were digitalized using the Pannoramic Scanner (3DHISTECH) and analyzed using QuantCenter (3DHISTECH) using a single threshold parameter for all images of a specific staining in each experiment.
Tissue microarrays and HNSCC samples.
Tissue microarray slides containing 17 HNSCC and 15 benign tumors (in duplicate) were purchased from US Biomax (HNT961). The information regarding these samples and the levels of AXL and c-JUN are summarized in Supplemental Table 2. Fresh HNSCC specimens were collected from Soroka Medical Center Surgery room and stored in liquid nitrogen. Helsinki was approved by the Soroka Medical Center (0103-17-SOR and 0421-16-SOR).
IC50 and synergy assay.
Cells were seeded in 96-well plates (3000 cells per well), treated with increasing concentrations of the relevant drugs (0–10 μM), and allowed to proliferate for 4 days. Cells were then fixed with 0.6 M trichloroacetic acid (TCA) for 1 hour at 4°C, rinsed, and stained with crystal violet (1 g/l) for 10 minutes at room temperature. Following additional rinsing, the bound crystal violet was dissolved out with 10% acetic acid, and absorbance was measured at 570 nm (BioTek Epoch spectrophotometer). IC50 values were calculated using the GraphPad Software. For the synergy assays, the proliferation of the cells in the different treatment groups was presented as a percentage of control (DMSO-treated) cells, and the percent of growth inhibition was calculated. Synergy scores were analyzed using Chalice Bioinformatics Software (https://www.horizondiscovery.com/research-services/drug-combination-screening/chalice-bioinformatics-software) (66), and the synergistic effects were calculated based on the statistic Loewe Excess model; free access is available at https://synergyfinder.fimm.fi/synergy/2019040121295580052/
siRNA and shRNA.
For transient silencing of c-JUN and c-FOS, cells were transfected using GenMute siRNA Transfection Reagent (SignaGen, SL100568) according to the manufacturer’s protocol, with an siRNA nontargeting control sequence (IDT, 51-01-14-04) and a c-JUN or c-FOS gene targeting sequence (IDT; hs.Ri.JUN.13 and hs.Ri.FOS.13.1, respectively). Cells were harvested after 48 hours for WB and qPCR analysis. For BYL719 IC50 experiments, cells were treated with the relevant drugs following 24 hours of transection and allowed to proliferate for an additional 4 days.
For the production of shRNAs cell lines, we created lentiviruses by transfecting HEK293 cells with the viral plasmids psPAX2, pMD2.G, and PLKO with shRNAs — a control scrambled sequence (shCT) or 2 different sequences for the silencing of AXL expression (shAXL1 and shAXL2, MSKCC RNAi core facility identifiers TRCN0000001037 and TRCN0000194771, respectively) using PolyJet transfection reagent (SignaGen, SL100688) according to the manufacturer’s protocol. Viruses were collected after 48–72 hours and used for cell infection. Cells were seeded in a 6-well plates (150,000 cells per well) and infected with the lentiviruses in the presence of Polybrene (MilliporeSigma, 5G-H9268). Cells were selected with puromycin (Thermo Fisher Scientific, A11138-03).
Overexpression of AXL.
For the generation of CAL33 GFP and AXL cells, we created lentiviruses by transfecting HEK293 cells with the viral plasmids psPAX2, pMD2.G, and PLX302-GFP or PLX302-AXL. Viruses were collected after 48–72 hours and used for cell infection. Cells were seeded in a 6-well plates (150,000 cells per well) and infected with the lentiviruses in the presence of Polybrene (MilliporeSigma, 5G-H9268). Cells were selected with puromycin (Thermo Fisher Scientific, A11138-03).
Cell proliferation assay.
Cells were seeded in 24-well plates (10,000 cells per well) and treated as indicated in Results. The real-time cell history recorder JULI Stage was used to record cell confluence every 6 hours. Results are presented as the averaged confluence ± SEM. For the 4NQO-induced cell lines (Figure 6B), cells were treated as indicated in Figure 6B for 4 days and then fixed and stained with crystal violet. Results are presented as a percentage of the control (DMSO-treated) cells.
Establishment of tumor xenografts and studies in mice.
NOD.CB17-Prkdc-scid/NCr Hsd (Nod.Scid) and C57/BL6 mice were purchased from Envigo. NOD.Cg-Prkdc Il2rg/SzJ (NSG) mice were purchased from The Jackson Laboratory.
For CDXs, 6-week-old Nod.Scid mice were injected s.c. in the flank with 2 × 106 cells in 200 μl PBS (100 μl in each side). Tumors (60 mm3) developed after about 2 weeks.
PDXs were established from HNSCC patients treated in the Ear, Nose, and Throat Unit, Soroka Medical Center. All patients signed informed consent forms. All PDXs were first transplanted s.c. into the flanks of 6-week-old NSG mice. Upon successful tumor engraftment, tumors were expanded and retransplanted into Nod.Scid mice for drug efficacy experiments. About 2 weeks after the second transplantation, the mice were randomized into 4 groups of 6–8 mice per group.
To produce murine HNSCC, C57BL/6 mice were given 50 μg/ml 4NQO in their drinking water for 3–4 months, and tumors developed 2 months later (Supplemental Figure 6A). The lip and tougher tumor models were expended in secondary mice.
For survival experiments, an orthotropic injection of 0.5 × 106 4NQO-tongue cell line cells into the tongues of syngeneic C57BL/6 mice. At a tumor volume of 70–120 mm3 (2–4 weeks after implantation), animals were randomized into 4 groups of 10–12 mice per group.
For the in vivo experiments, animals were treated orally with vehicle (corn oil [MilliporeSigma, C8267-500ML) containing 4% DMSO [MilliporeSigma, D8418] for SP600125 administration or 0.5% carboxymethylcellulose [MilliporeSigma, 9481-1KG] for BYL719 administration), BYL719 (25 mg/kg), and/or JNK inhibitor SP600125 (15 mg/kg) daily. Tumors were measured with a digital caliper twice a week, and tumor volumes were determined according to the formula: length × width2 × (π/6). At the end of the experiment, animals were sacrificed by subjecting them to CO2 inhalation, and the tumors were harvested for investigation. Tumor volumes were normalized to initial volumes and presented as an averaged percentage of the initial volumes ± SEM.
Mice were housed in air-filtered laminar flow cabinets with a 12 hours light/dark cycle and were fed food and water ad libitum.
Drugs.
BYL719 was provided by Novartis. For in vitro experiments, BYL719 was dissolved in DMSO and, for in vivo administration, in 0.5% carboxymethylcellulose. SP600125 was purchased from Fluorochem (M02529). BI-78D3 (catalog 21183) and MPI-0479605 (catalog 22136) were purchased from Cayman Chemical. R428 (catalog S2841) was purchased from Selleckchem. For in vitro assays, SP600125 was dissolved in DMSO and for in vivo administration in corn oil with 4% DMSO.
Statistics.
Experiments were repeated at least 2–3 times, and representative data/images are shown. Statistical analysis was performed using GraphPad Prism software, and results are presented as means ± SEM. For comparisons between groups, P values were calculated. For experiment with more than 2 groups, 1-way ANOVA was calculated using Tukey’s multiple comparison test. For pathological analysis, IHC images were analyzed by Histoquant software (3DHISTECH) and 1-way ANOVA test was performed to compare control vs. treated groups. For the different experiments, P values of 0.05, 0.01, or 0.001 were considered statistically significant, as indicated by *, **, or *** in figure legends.
Study approval.
Mice were maintained and treated according to the institutional guidelines of Ben-Gurion University of the Negev. Animal experiments were approved by the IACUC (IL.80-12-2015).
Author contributions
MB, MP, LC, and ME designed research studies. MB, MP, NB, JZ, KY, BR and LC conducted the experiments and analyzed the data. JBZ, ABD, and RG provided clinical samples and cell lines. ME, LC, and MB wrote the manuscript.
Supplementary Material
Acknowledgments
This work was funded primarily by the Israel Science Foundation (ISF, 700/16) grant to ME and an INCPM (1901/17) grant to ME. Moreover, this work was also funded by the Concern Foundation (#7895) grant to ME, and the Israel Cancer Research Foundation (ICRF, 17-1693-RCDA). ME is supported by an Alon Fellowship for outstanding young researchers. MB is supported by the Planning and Budgeting Committee of the Council for Higher Education and by the national-VATA-fellowship.
Version 1. 03/12/2019
In-Press Preview
Version 2. 04/18/2019
Electronic publication
Version 3. 05/15/2019
Ben-Zion Joshua's name was corrected.
Funding Statement
To ME
to ME
to ME
Footnotes
Conflict of interest: The authors declare no conflict of interest.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(8):e125341. https://doi.org/10.1172/jci.insight.125341.
Contributor Information
Mai Badarni, Email: maibdarny@gmail.com.
Manu Prasad, Email: manupras@post.bgu.ac.il.
Noa Balaban, Email: noab151@gmail.com.
Jonathan Zorea, Email: j.zorea@gmail.com.
Anat Bahat Dinur, Email: dinura@gmail.com.
Reidar Grénman, Email: reidar.grenman@tyks.fi.
Barak Rotblat, Email: rotblat@bgu.ac.il.
Limor Cohen, Email: ayashli@post.bgu.ac.il.
Moshe Elkabets, Email: moshee@bgu.ac.il.
References
- 1.Wiegand S, Zimmermann A, Wilhelm T, Werner JA. Survival After Distant Metastasis in Head and Neck Cancer. Anticancer Res. 2015;35(10):5499–5502. [PubMed] [Google Scholar]
- 2.Ferlay J, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Curado MP, Hashibe M. Recent changes in the epidemiology of head and neck cancer. Curr Opin Oncol. 2009;21(3):194–200. doi: 10.1097/CCO.0b013e32832a68ca. [DOI] [PubMed] [Google Scholar]
- 4.Marur S, D’Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. 2010;11(8):781–789. doi: 10.1016/S1470-2045(10)70017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet. 2008;371(9625):1695–1709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seiwert TY, et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res. 2015;21(3):632–641. doi: 10.1158/1078-0432.CCR-13-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296(5573):1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 10.Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol. 2010;28(6):1075–1083. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9(8):550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 12.Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat Rev Clin Oncol. 2013;10(3):143–153. doi: 10.1038/nrclinonc.2013.10. [DOI] [PubMed] [Google Scholar]
- 13.Juric D, et al. Phosphatidylinositol 3-Kinase α-Selective Inhibition With Alpelisib (BYL719) in PIK3CA-Altered Solid Tumors: Results From the First-in-Human Study. J Clin Oncol. 2018;36(13):1291–1299. doi: 10.1200/JCO.2017.72.7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elkabets M, et al. AXL mediates resistance to PI3Kα inhibition by activating the EGFR/PKC/mTOR axis in head and neck and esophageal squamous cell carcinomas. Cancer Cell. 2015;27(4):533–546. doi: 10.1016/j.ccell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brand TM, et al. AXL mediates resistance to cetuximab therapy. Cancer Res. 2014;74(18):5152–5164. doi: 10.1158/0008-5472.CAN-14-0294. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 16.Dufies M, et al. Mechanisms of AXL overexpression and function in Imatinib-resistant chronic myeloid leukemia cells. Oncotarget. 2011;2(11):874–885. doi: 10.18632/oncotarget.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giles KM, et al. Axl mediates acquired resistance of head and neck cancer cells to the epidermal growth factor receptor inhibitor erlotinib. Mol Cancer Ther. 2013;12(11):2541–2558. doi: 10.1158/1535-7163.MCT-13-0170. [DOI] [PubMed] [Google Scholar]
- 18.Hong J, Belkhiri A. AXL mediates TRAIL resistance in esophageal adenocarcinoma. Neoplasia. 2013;15(3):296–304. doi: 10.1593/neo.122044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44(8):852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mudduluru G, Allgayer H. The human receptor tyrosine kinase Axl gene--promoter characterization and regulation of constitutive expression by Sp1, Sp3 and CpG methylation. Biosci Rep. 2008;28(3):161–176. doi: 10.1042/BSR20080046. [DOI] [PubMed] [Google Scholar]
- 21.Mudduluru G, Vajkoczy P, Allgayer H. Myeloid zinc finger 1 induces migration, invasion, and in vivo metastasis through Axl gene expression in solid cancer. Mol Cancer Res. 2010;8(2):159–169. doi: 10.1158/1541-7786.MCR-09-0326. [DOI] [PubMed] [Google Scholar]
- 22.Mudduluru G, Leupold JH, Stroebel P, Allgayer H. PMA up-regulates the transcription of Axl by AP-1 transcription factor binding to TRE sequences via the MAPK cascade in leukaemia cells. Biol Cell. 2010;103(1):21–33. doi: 10.1042/BC20100094. [DOI] [PubMed] [Google Scholar]
- 23.Maurus K, et al. The AP-1 transcription factor FOSL1 causes melanocyte reprogramming and transformation. Oncogene. 2017;36(36):5110–5121. doi: 10.1038/onc.2017.135. [DOI] [PubMed] [Google Scholar]
- 24.Sayan AE, et al. Fra-1 controls motility of bladder cancer cells via transcriptional upregulation of the receptor tyrosine kinase AXL. Oncogene. 2012;31(12):1493–1503. doi: 10.1038/onc.2011.336. [DOI] [PubMed] [Google Scholar]
- 25.Zhen Y, Lee IJ, Finkelman FD, Shao WH. Targeted inhibition of Axl receptor tyrosine kinase ameliorates anti-GBM-induced lupus-like nephritis. J Autoimmun. 2018;93:37–44. doi: 10.1016/j.jaut.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konieczkowski DJ, et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 2014;4(7):816–827. doi: 10.1158/2159-8290.CD-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Müller J, et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat Commun. 2014;5:5712. doi: 10.1038/ncomms6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu H, et al. Prognostic value from integrative analysis of transcription factors c-Jun and Fra-1 in oral squamous cell carcinoma: a multicenter cohort study. Sci Rep. 2017;7(1):7522. doi: 10.1038/s41598-017-05106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett BL, et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc Natl Acad Sci USA. 2001;98(24):13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hawkins BL, Heniford BW, Ackermann DM, Leonberger M, Martinez SA, Hendler FJ. 4NQO carcinogenesis: a mouse model of oral cavity squamous cell carcinoma. Head Neck. 1994;16(5):424–432. doi: 10.1002/hed.2880160506. [DOI] [PubMed] [Google Scholar]
- 31. Rodon J, et al. Biomarkers and clinical targeting of the PI3 Kinase and VEGF pathways. Presented at :104th Annual AACR Meeting; April 7, 2013; Washington, DC, USA. [Google Scholar]
- 32.Elkabets M, et al. mTORC1 inhibition is required for sensitivity to PI3K p110α inhibitors in PIK3CA-mutant breast cancer. Sci Transl Med. 2013;5(196):196ra99. doi: 10.1126/scitranslmed.3005747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fritsch C, et al. Characterization of the novel and specific PI3Kα inhibitor NVP-BYL719 and development of the patient stratification strategy for clinical trials. Mol Cancer Ther. 2014;13(5):1117–1129. doi: 10.1158/1535-7163.MCT-13-0865. [DOI] [PubMed] [Google Scholar]
- 34.Furet P, et al. Discovery of NVP-BYL719 a potent and selective phosphatidylinositol-3 kinase alpha inhibitor selected for clinical evaluation. Bioorg Med Chem Lett. 2013;23(13):3741–3748. doi: 10.1016/j.bmcl.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 35.Zumsteg ZS, et al. Taselisib (GDC-0032), a Potent β-Sparing Small Molecule Inhibitor of PI3K, Radiosensitizes Head and Neck Squamous Carcinomas Containing Activating PIK3CA Alterations. Clin Cancer Res. 2016;22(8):2009–2019. doi: 10.1158/1078-0432.CCR-15-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Felice F, Guerrero Urbano T. New drug development in head and neck squamous cell carcinoma: The PI3-K inhibitors. Oral Oncol. 2017;67:119–123. doi: 10.1016/j.oraloncology.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 37.Mizrachi A, et al. Tumour-specific PI3K inhibition via nanoparticle-targeted delivery in head and neck squamous cell carcinoma. Nat Commun. 2017;8:14292. doi: 10.1038/ncomms14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu L, et al. Novel mechanism of lapatinib resistance in HER2-positive breast tumor cells: activation of AXL. Cancer Res. 2009;69(17):6871–6878. doi: 10.1158/0008-5472.CAN-08-4490. [DOI] [PubMed] [Google Scholar]
- 39.Byers LA, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res. 2013;19(1):279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brand TM, et al. AXL Is a Logical Molecular Target in Head and Neck Squamous Cell Carcinoma. Clin Cancer Res. 2015;21(11):2601–2612. doi: 10.1158/1078-0432.CCR-14-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDaniel NK, et al. MERTK Mediates Intrinsic and Adaptive Resistance to AXL-targeting Agents. Mol Cancer Ther. 2018;17(11):2297–2308. doi: 10.1158/1535-7163.MCT-17-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balaji K, et al. AXL Inhibition Suppresses the DNA Damage Response and Sensitizes Cells to PARP Inhibition in Multiple Cancers. Mol Cancer Res. 2017;15(1):45–58. doi: 10.1158/1541-7786.MCR-16-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ben-Batalla I, et al. Axl Blockade by BGB324 Inhibits BCR-ABL Tyrosine Kinase Inhibitor-Sensitive and -Resistant Chronic Myeloid Leukemia. Clin Cancer Res. 2017;23(9):2289–2300. doi: 10.1158/1078-0432.CCR-16-1930. [DOI] [PubMed] [Google Scholar]
- 44.Boshuizen J, et al. Cooperative targeting of melanoma heterogeneity with an AXL antibody-drug conjugate and BRAF/MEK inhibitors. Nat Med. 2018;24(2):203–212. doi: 10.1038/nm.4472. [DOI] [PubMed] [Google Scholar]
- 45.Guo G, et al. A TNF-JNK-Axl-ERK signaling axis mediates primary resistance to EGFR inhibition in glioblastoma. Nat Neurosci. 2017;20(8):1074–1084. doi: 10.1038/nn.4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palisoul ML, et al. Inhibition of the Receptor Tyrosine Kinase AXL Restores Paclitaxel Chemosensitivity in Uterine Serous Cancer. Mol Cancer Ther. 2017;16(12):2881–2891. doi: 10.1158/1535-7163.MCT-17-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sen T, et al. Targeting AXL and mTOR Pathway Overcomes Primary and Acquired Resistance to WEE1 Inhibition in Small-Cell Lung Cancer. Clin Cancer Res. 2017;23(20):6239–6253. doi: 10.1158/1078-0432.CCR-17-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brand TM, et al. Cross-talk Signaling between HER3 and HPV16 E6 and E7 Mediates Resistance to PI3K Inhibitors in Head and Neck Cancer. Cancer Res. 2018;78(9):2383–2395. doi: 10.1158/0008-5472.CAN-17-1672. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 49.Li M, et al. Yes-associated protein 1 (YAP1) promotes human gallbladder tumor growth via activation of the AXL/MAPK pathway. Cancer Lett. 2014;355(2):201–209. doi: 10.1016/j.canlet.2014.08.036. [DOI] [PubMed] [Google Scholar]
- 50.Ghiso E, et al. YAP-Dependent AXL Overexpression Mediates Resistance to EGFR Inhibitors in NSCLC. Neoplasia. 2017;19(12):1012–1021. doi: 10.1016/j.neo.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boregowda RK, et al. The transcription factor RUNX2 regulates receptor tyrosine kinase expression in melanoma. Oncotarget. 2016;7(20):29689–29707. doi: 10.18632/oncotarget.8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bae SY, Hong JY, Lee HJ, Park HJ, Lee SK. Targeting the degradation of AXL receptor tyrosine kinase to overcome resistance in gefitinib-resistant non-small cell lung cancer. Oncotarget. 2015;6(12):10146–10160. doi: 10.18632/oncotarget.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller MA, et al. Reduced Proteolytic Shedding of Receptor Tyrosine Kinases Is a Post-Translational Mechanism of Kinase Inhibitor Resistance. Cancer Discov. 2016;6(4):382–399. doi: 10.1158/2159-8290.CD-15-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yun MR, et al. Enhancer Remodeling and MicroRNA Alterations Are Associated with Acquired Resistance to ALK Inhibitors. Cancer Res. 2018;78(12):3350–3362. doi: 10.1158/0008-5472.CAN-17-3146. [DOI] [PubMed] [Google Scholar]
- 55.Scaltriti M, Elkabets M, Baselga J. Molecular Pathways: AXL, a Membrane Receptor Mediator of Resistance to Therapy. Clin Cancer Res. 2016;22(6):1313–1317. doi: 10.1158/1078-0432.CCR-15-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Riesenberg S, et al. MITF and c-Jun antagonism interconnects melanoma dedifferentiation with pro-inflammatory cytokine responsiveness and myeloid cell recruitment. Nat Commun. 2015;6:8755. doi: 10.1038/ncomms9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Usman MW, et al. Macrophages confer resistance to PI3K inhibitor GDC-0941 in breast cancer through the activation of NF-κB signaling. Cell Death Dis. 2018;9(8):809. doi: 10.1038/s41419-018-0849-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shrivastava S, Steele R, Sowadski M, Crawford SE, Varvares M, Ray RB. Identification of molecular signature of head and neck cancer stem-like cells. Sci Rep. 2015;5:7819. doi: 10.1038/srep07819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bakiri L, et al. Fra-1/AP-1 induces EMT in mammary epithelial cells by modulating Zeb1/2 and TGFβ expression. Cell Death Differ. 2015;22(2):336–350. doi: 10.1038/cdd.2014.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong C, Ye DX, Zhang WB, Pan HY, Zhang ZY, Zhang L. Overexpression of c-fos promotes cell invasion and migration via CD44 pathway in oral squamous cell carcinoma. J Oral Pathol Med. 2015;44(5):353–360. doi: 10.1111/jop.12296. [DOI] [PubMed] [Google Scholar]
- 61.Zenz R, et al. c-Jun regulates eyelid closure and skin tumor development through EGFR signaling. Dev Cell. 2003;4(6):879–889. doi: 10.1016/S1534-5807(03)00161-8. [DOI] [PubMed] [Google Scholar]
- 62.Fang Y, et al. A new tumour suppression mechanism by p27Kip1: EGFR down-regulation mediated by JNK/c-Jun pathway inhibition. Biochem J. 2014;463(3):383–392. doi: 10.1042/BJ20140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Green MR, et al. Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: implications for targeted therapy. Clin Cancer Res. 2012;18(6):1611–1618. doi: 10.1158/1078-0432.CCR-11-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skinner HD, et al. Integrative Analysis Identifies a Novel AXL-PI3 Kinase-PD-L1 Signaling Axis Associated with Radiation Resistance in Head and Neck Cancer. Clin Cancer Res. 2017;23(11):2713–2722. doi: 10.1158/1078-0432.CCR-16-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aguilera TA, et al. Reprogramming the immunological microenvironment through radiation and targeting Axl. Nat Commun. 2016;7:13898. doi: 10.1038/ncomms13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hart LS, et al. Preclinical Therapeutic Synergy of MEK1/2 and CDK4/6 Inhibition in Neuroblastoma. Clin Cancer Res. 2017;23(7):1785–1796. doi: 10.1158/1078-0432.CCR-16-1131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.