Abstract
Conjugated bile acids (CBAs), such as tauroursodeoxycholic acid (TUDCA), are known to resolve the inflammatory and unfolded protein response (UPR) in inflammatory diseases, such as asthma. Whether CBAs exert their beneficial effects on allergic airway responses via 1 arm or several arms of the UPR, or alternatively through the signaling pathways for conserved bile acid receptor, remains largely unknown. We used a house dust mite–induced (HDM-induced) murine model of asthma to evaluate and compare the effects of 5 CBAs and 1 unconjugated bile acid in attenuating allergen-induced UPR and airway responses. Expression of UPR-associated transcripts was assessed in airway brushings from human patients with asthma and healthy subjects. Here we show that CBAs, such as alanyl β-muricholic acid (AβM) and TUDCA, significantly decreased inflammatory, immune, and cytokine responses; mucus metaplasia; and airway hyperresponsiveness, as compared with other CBAs in a model of allergic airway disease. CBAs predominantly bind to activating transcription factor 6α (ATF6α) compared with the other canonical transducers of the UPR, subsequently decreasing allergen-induced UPR activation and resolving allergic airway disease, without significant activation of the bile acid receptors. TUDCA and AβM also attenuated other HDM-induced ER stress markers in the lungs of allergic mice. Quantitative mRNA analysis of airway epithelial brushings from human subjects demonstrated that several ATF6α-related transcripts were significantly upregulated in patients with asthma compared with healthy subjects. Collectively, these results demonstrate that CBA-based therapy potently inhibits the allergen-induced UPR and allergic airway disease in mice via preferential binding of the canonical transducer of the UPR, ATF6α. These results potentially suggest a novel avenue to treat allergic asthma using select CBAs.
Keywords: Inflammation, Pulmonology
Keywords: Allergy, Asthma, Protein misfolding
Conjugated bile acid–based therapy potently inhibits the allergen-induced unfolded protein response and allergic airway disease in mice.
Introduction
Allergic asthma typically exhibits a T helper cell type 2 (Th2) inflammatory response and airway hyperresponsiveness (AHR) (1). A common aeroallergen, house dust mite (HDM), affects approximately 50%–85% of patients with asthma (2–4). Because of its complex nature, HDM likely activates several pattern recognition receptors and elicits a mixed inflammatory response, mucus metaplasia, and AHR (4). We have demonstrated that HDM challenge induces ER stress in the lung, thereby activating the unfolded protein response (UPR), which is associated with many hallmarks of allergic airway disease in humans and rodents (5–8). The UPR is an adaptive response consisting of the activation of transducers protein kinase R–like ER kinase (PERK), inositol-requiring enzyme 1α (IRE1α), and activating transcription factor 6α (ATF6α) (9). These pathways coordinate to suppress protein production, boost the ER-associated degradation machinery, and increase chaperones, members of the protein disulfide isomerase (PDI) family, and antioxidant enzymes that ensure proper protein folding (10, 11). These combined responses help maintain ER homeostasis and therefore cell survival. However, prolonged and chronic activation of the UPR may induce cellular apoptosis and has been implicated in the pathogenesis of inflammatory and fibrotic diseases (12–15).
The crucial role that the UPR plays in chronic airway disorders highlights its potential as a target for novel therapies in diseases such as asthma (6, 16). Our group and others have demonstrated the therapeutic potential of the conjugated bile acid (CBA) tauroursodeoxycholic acid (TUDCA) in relieving ER stress and airway inflammatory responses (7, 17, 18). CBAs such as TUDCA are generally well tolerated and have shown positive effects in clinical trials for UPR-related diseases, such as primary biliary cholangitis, amyloidosis, and amyotrophic lateral sclerosis (19–21). Moreover, because different arms of the UPR and their regulated genes have been implicated in various cell types in the inflamed lung, including CHOP in macrophages; IRE1β, PDIA3, and ATF6α in airway epithelial cells; and X-box–binding protein 1 (XBP1) in eosinophils and neutrophils (6, 22–26), it has become clear that therapeutic agents targeting multiple components of the UPR could be more effective at inhibiting allergen-induced airway responses. Whereas CBAs such as TUDCA are established attenuators of the UPR, there are no studies that specifically address the mechanisms of action of CBAs in inhibiting the allergen-induced UPR in the lungs.
The objectives of this study were to evaluate the therapeutic potential of 5 CBAs and 1 unconjugated bile acid and their putative mechanisms of action in inhibiting the allergen-induced UPR and subsequent airway responses. Altogether, the study characterizes additional CBAs as effective attenuators of allergic airway disease via direct interaction with transducers of the UPR and implicates ATF6α in the resolution of airway inflammation. Furthermore, we report that ATF6α-related genes are upregulated in human asthmatic epithelial cells, suggesting a novel target to control allergic airway disease in humans.
Results
TUDCA attenuates inflammation and ER stress in a murine model of allergic asthma.
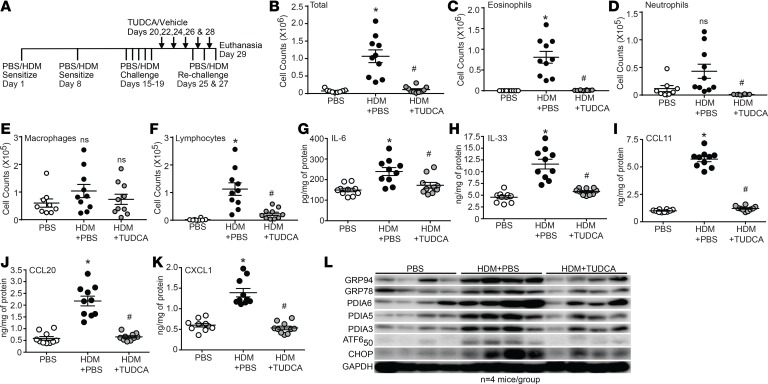
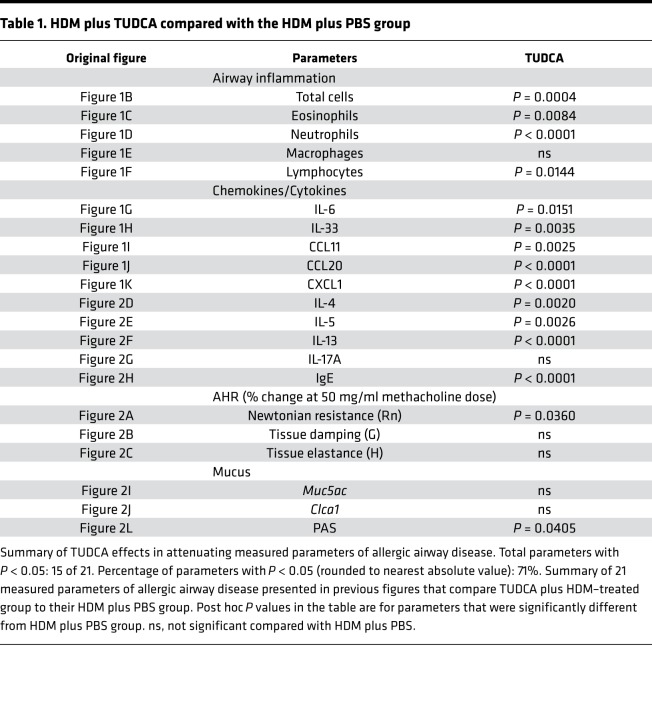
We previously reported that the CBA TUDCA potently attenuates hallmarks of allergic airway disease in C57BL/6J mice (7). The first aim of the current study was to determine whether TUDCA could similarly attenuate pathology in a strain of mouse more strongly skewed toward the Th2 immune response typically associated with allergic airway disease (27). Here, we used our established therapeutic regimen of intranasal HDM and oropharyngeal TUDCA administration in the BALB/cJ strain of mouse (28, 29) (Figure 1A). With the exception of macrophages, HDM-induced increases in total airway inflammatory cell numbers, eosinophils, neutrophils, and lymphocytes were significantly decreased in TUDCA-treated mice (Figure 1, B–F). Increased levels of the inflammatory cytokines IL-6 and IL-33 and the chemokines C-C motif chemokine ligand 11 (CCL11/eotaxin-1), CCL20/macrophage inflammatory protein 3α (MIP3α), and CXCL1/keratinocyte-derived chemokine (KC), measured in lung homogenates of allergic mice, were similarly attenuated by TUDCA (Figure 1, G–K). Western blot analysis and densitometry confirmed the decrease of UPR markers glucose-regulated protein 94 (GRP94), GRP78, subunit 50 of ATF6α (ATF6α50), PDIA5, PDIA3, and CCAAT/enhancer binding protein homologous protein (CHOP) (Figure 1L and Supplemental Figure 1, densitometry; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.98101DS1) in HDM-challenged, TUDCA-treated BALB/cJ mice.
Figure 1. TUDCA attenuates allergic airway inflammation and the UPR.
(A) HDM and TUDCA challenge and treatment regimen. (B–F) Inflammation quantified from the BAL. n = 10 mice/group from 2 experiments (outliers removed in various groups; total cells PBS: n = 1; eosinophils PBS: n = 1; neutrophils PBS: n = 2; TUDCA: n = 2; macrophages PBS: n = 1; lymphocytes PBS: n = 2). (G–K) Quantification of cytokines/chemokines by ELISA from lung homogenates. n = 10 mice/group from 2 experiments. Kruskal-Wallis, 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli. Two comparisons, both with the HDM group. *P < 0.05 vs. PBS group; #P < 0.05 vs. HDM group; ns, not significant. Error bars represent ± SEM. (L) Expression of ER stress markers by Western blot analysis of lung homogenates of HDM-sensitized and -challenged mice treated with TUDCA. n = 4 representative lung lysates for each group for the Western blots. Densitometry in Supplemental Figure 1. ns, not significant. Error bars represent ± SEM.
TUDCA attenuates HDM-induced AHR, several innate/adaptive immune cytokine responses, and mucus-related markers associated with murine allergic asthma.
TUDCA administration to HDM-treated mice attenuated HDM-induced Newtonian resistance (Rn) but not tissue resistance/damping (G) and tissue stiffness/elastance (H) at the highest concentration(s) of methacholine (Figure 2, A–C, and Supplemental Figure 2). Moreover, levels of cytokines IL-4, IL-5, and IL-13, as well as IL-17A, were elevated in the lungs of HDM-treated compared with PBS-treated control mice (Figure 2, D–G). IFN-γ was not detected in any of the samples. With TUDCA, allergic mice produced significantly less IL-4, IL-5, and IL-13, but there was no significant decrease in IL-17A, as compared with untreated allergic mice. Treatment with TUDCA also significantly attenuated levels of HDM-induced IgE in the serum (Figure 2H). Although administration of TUDCA did not decrease HDM-induced expression of the mucin-related genes Muc5ac and Clca1 (Figure 2, I and J), TUDCA treatment significantly reduced mucus metaplasia, as quantified by cells from the lungs stained with periodic acid–Schiff (PAS) (Figure 2, K and L).
Figure 2. TUDCA attenuates AHR, IL-4, IL-5, IL-13, IgE production, and mucus metaplasia.
(A–C) AHR by FlexiVent in response to increasing concentrations of methacholine (PBS n = 11; HDM n = 16; and HDM plus TUDCA n = 7 mice from 2 experiments). One-way ANOVA, Tukey’s post hoc test. *P < 0.05 vs. PBS group; #P < 0.05 vs. HDM group (mice were removed in Rn, G, and H; in HDM plus TUDCA group n = 2 based on noncompliance with coefficient of determination [COD] value). (D–H) Quantification of cytokines by ELISA from lung homogenates and IgE from serum; n = 10 mice/group from 2 experiments (outliers removed in IL-5; PBS: n = 1). (I and J) Muc5ac and Clca1 expression in the lungs assessed by reverse transcription quantitative PCR (RT-qPCR); n = 7 mice from 2 experiments. (K) Representative images of PAS-stained lungs. Scale bar: 25 μm. (L) Quantification of PAS-stained lungs; n = 3–7 representative mice from 2 experiments. (D–H and J) Kruskal-Wallis, 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli. Two comparisons, both with the HDM group. *P < 0.05 vs. PBS group; #P < 0.05 vs. HDM group. Rn, Newtonian resistance; G, tissue damping; H, tissue elastance; ns, not significant vs. HDM plus PBS group. Error bars represent ± SEM.
Effects of other CBAs on allergen-induced inflammation and the UPR in a murine model of allergic asthma.
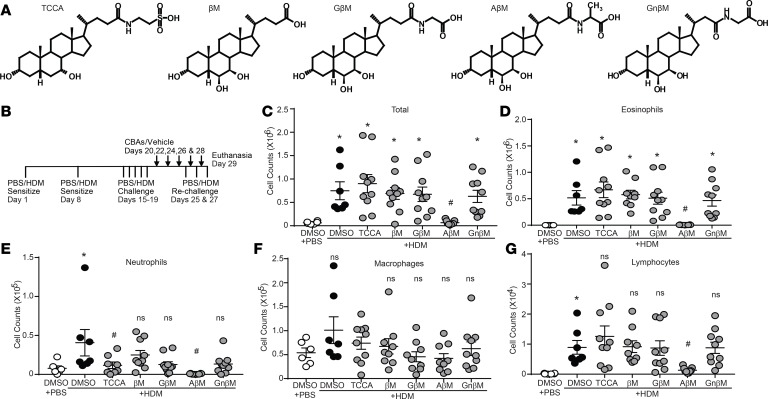
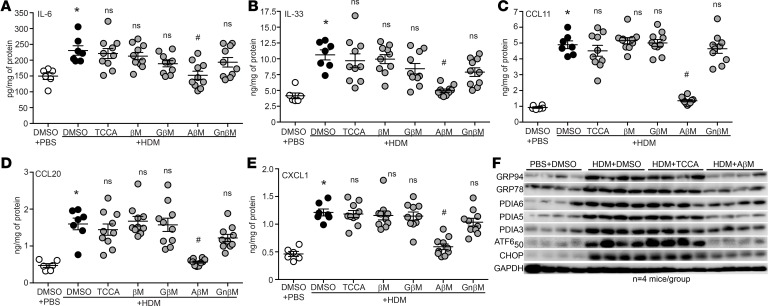
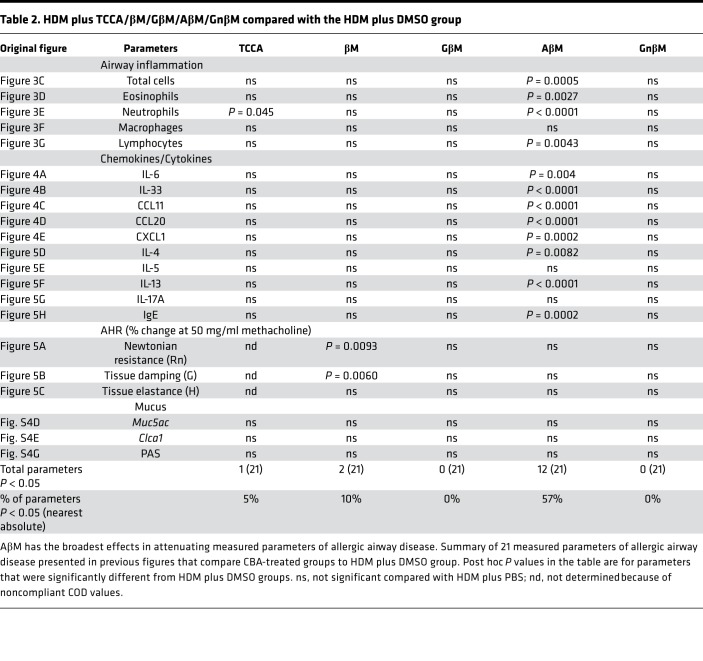
Considering the ability of TUDCA to alleviate allergic airway responses, we decided to evaluate the efficacy of additional CBAs not previously tested in an allergic model of asthma. These include 4 CBAs: taurochenocholic acid (TCCA), glycyl β-muricholic acid (GβM), alanyl β-muricholic acid (AβM), and glycyl-nor-β-muricholic acid (GnβM), along with the unconjugated bile acid (UBA) β-muricholic acid (βM) (Figure 3A). We used our established therapeutic regimen of intranasal HDM (or PBS vehicle) exposure combined with oropharyngeal instillation of CBAs/UBA (or DMSO vehicle) (Figure 3B). Total inflammatory cell numbers, eosinophils, neutrophils, and lymphocytes were all significantly increased in HDM mice treated with vehicle control (HDM plus DMSO) compared with control mice that received PBS plus DMSO (Figure 3, C–G). Only AβM was effective at significantly reducing HDM-induced total inflammatory cell numbers, eosinophils, neutrophils, and lymphocytes. TCCA significantly decreased airway neutrophilia; however, no other CBA/UBA–treated groups showed significant reductions in inflammatory cell numbers (Figure 3, C–G). Analysis of the cytokines IL-6 and IL-33 and chemokines CCL11, CCL20, and CXCL1 showed that they were significantly elevated in the lungs of HDM plus DMSO compared with PBS plus DMSO mice (Figure 4, A–E). However, AβM was the only CBA that significantly reduced levels of IL-6, IL-33, CCL11, CXCL1, and CCL20 (Figure 4, A–E).
Figure 3. CBAs attenuate HDM-induced airway inflammation.
(A) Structures of the CBAs and UBA. (B) HDM and CBA treatment protocol. (C–G) Inflammatory cells quantified from the BAL, PBS, and HDM plus DMSO n = 6 mice/group from 2 experiments; HDM plus TCCA/βM/GM/AβM/GnβM n = 10 mice/group from 2 experiments (outliers removed in various groups; eosinophils PBS: n = 1; βM: n = 1; AM: n = 1. PMNs: TCCA: n = 2; AβM: n = 1. Macrophages: βM: n = 1; GβM: n = 1. Lymphocytes: βM: n = 1). *P < 0.05 vs. PBS plus DMSO group; #P < 0.05 vs. HDM plus DMSO group. ns, not significant. Error bars represent ± SEM.
Figure 4. CBAs attenuate HDM-induced inflammatory cytokines, chemokines, and the UPR.
(A–E) Quantification of cytokines/chemokines by ELISAs from lung homogenates. Kruskal-Wallis, 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli. Six comparisons, all with the HDM plus DMSO group. *P < 0.05 vs. PBS plus DMSO group; #P < 0.05 vs. HDM plus DMSO group; ns, not significant. PBS and HDM plus DMSO n = 6 mice/group from 2 experiments; HDM plus TCCA/βM/GM/AβM/GnβM n = 10 mice/group from 2 experiments. (F) Expression of UPR markers from lung homogenates of HDM-sensitized and -challenged mice treated with AβM, TCCA, or DMSO (vehicle); n = 4 representative lung lysates for each group. Densitometry in Supplemental Figure 3. ns, not significant. Error bars represent ± SEM.
Based on these results, we focused our attention on comparing the expression of several UPR markers by Western blotting in TCCA- and AβM-treated lung samples with those from PBS plus DMSO and HDM plus DMSO controls. Compared with PBS plus DMSO mice, lungs from HDM plus DMSO mice showed significant increases in all measured UPR markers (Figure 4F, Supplemental Figure 3, densitometry). The AβM (HDM plus AβM) group displayed significantly decreased GRP94, GRP78, ATF6α50, PDIA5, and CHOP in comparison with the HDM plus DMSO or HDM plus TCCA groups (Figure 4F and Supplemental Figure 3, densitometry).
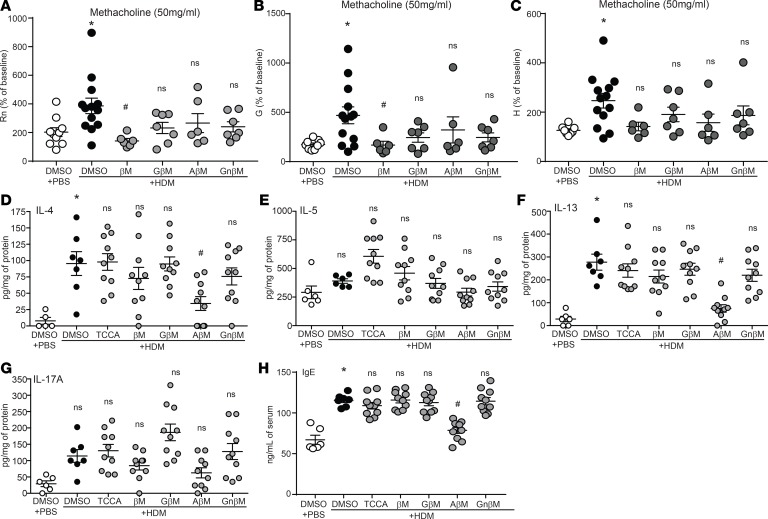
We next examined the effectiveness of CBAs/UBA on AHR, several innate/adaptive immune cytokine responses, and mucus-related markers associated with asthma. We determined that the HDM plus DMSO group significantly increased all 3 measured AHR parameters (i.e., Rn, G, and H). The only compound examined that effectively decreased HDM-induced Rn and G was βM (Figure 5, A and B, and Supplemental Figure 4). In contrast, HDM-induced increases in Rn, G, and H were not significantly reduced by GβM, AβM, or GnβM (Figure 5, A–C, and Supplemental Figure 4) at the highest dose of methacholine (50 mg/ml). The AHR data obtained for TCCA were omitted because of noncompliance for the majority of the mice according to our cutoff COD threshold values (see Methods). HDM-induced production of IL-4 and IL-13 was significantly reduced by AβM, whereas IL-5 and IL-17A were not significantly decreased in any groups (Figure 5, D–G). HDM sensitization and challenge also significantly increased serum IgE levels, and AβM was able to attenuate the HDM-induced IgE as compared with HDM plus DMSO and other CBA/UBA–treated groups (Figure 5H).
Figure 5. CBAs attenuate HDM-induced AHR, IL-4, IL-5, IL-13, and IgE production.
(A–C) AHR measurement by FlexiVent in response to 50 mg/ml methacholine. One-way ANOVA and Tukey’s post hoc test for Rn and Kruskal-Wallis and 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutiel post hoc test for G and H. *P < 0.05 vs. PBS plus DMSO mice; #P < 0.05 vs. HDM plus DMSO mice. PBS plus DMSO n = 10; HDM plus DMSO n = 13; and βM/GM/AβM/GnβM plus HDM n = 7 from 2 experiments (mice were removed based on noncompliance with COD value in Rn/G/H βM: n = 1; AβM: n = 1). (D–H) Quantification of cytokines by ELISA from lung homogenates and IgE from serum. PBS and HDM plus DMSO n = 6 mice/group from 2 experiments; HDM plus TCCA/βM/GβM/AM/GnβM n = 10 mice/group from 2 experiments (outliers removed in various groups; IL-4: PBS plus DMSO n = 1; IL-5: HDM plus DMSO n = 1). Kruskal-Wallis, 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli. Six comparisons, all with the HDM plus DMSO group. *P < 0.05 vs. PBS plus DMSO group; #P < 0.05 vs. HDM plus DMSO group; ns, not significant. Error bars represent ± SEM.
None of the additional 4 CBAs and 1 UBA were particularly effective at reducing mucus-related readouts in BALB/cJ mice. Transcript levels of Muc5ac and Clca1 in the lungs of CBA-treated mice did not differ from their respective HDM controls (Supplemental Figure 4, D and E). Similarly, PAS staining of mucus glycoproteins in the lungs of CBA-treated mice did not differ from staining of their respective controls (Supplemental Figure 4, F and G).
Tables 1 and 2 summarize data on the effects of CBAs/UBA (TCCA, βM, GβM, AβM, GnβM, and TUDCA) on parameters of allergic airway disease that were measured in our HDM model with BALB/cJ mice. AβM and TUDCA had the broadest effects, significantly inhibiting (P < 0.05) 15 and 12 of the 21 measured parameters of allergic airway disease, respectively, as compared with their equivalent HDM plus vehicle groups.
Table 1. HDM plus TUDCA compared with the HDM plus PBS group.
Table 2. HDM plus TCCA/βM/GβM/AβM/GnβM compared with the HDM plus DMSO group.
CBAs do not activate bile acid receptor signaling.
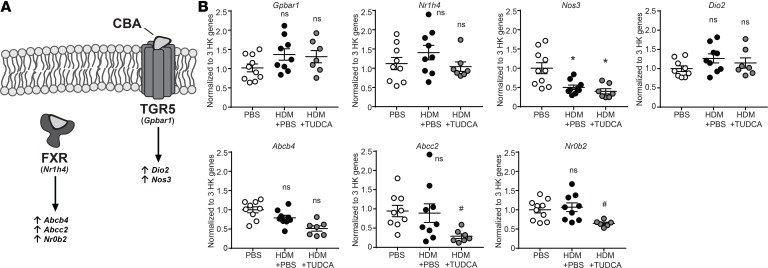
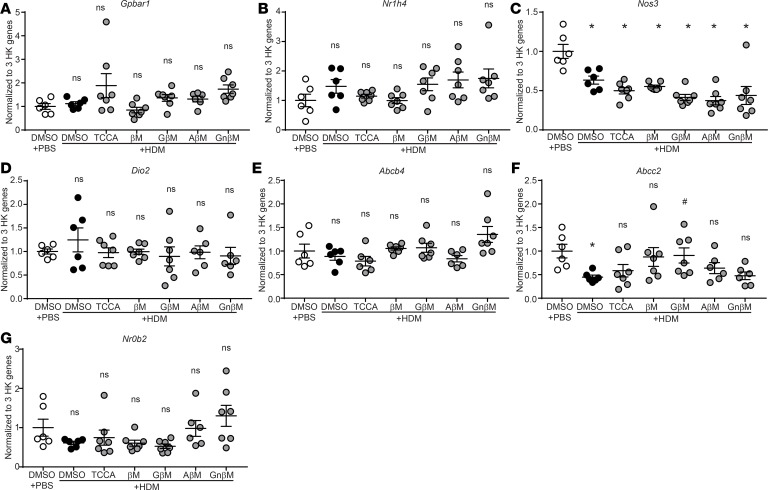
Our next aim was to understand the mechanism whereby CBAs attenuate the UPR and allergic airway responses. We began by investigating the effects of CBAs on canonical bile acid receptors farnesoid X receptor (FXR) and G protein–coupled bile acid receptor 1 (Takeda G protein–coupled receptor 5, TGR5) and their reported downstream mediators (Figure 6A and refs. 30, 31), which are increased by bile acid stimulation in various tissues (e.g., liver and kidney) (32, 33). We found that lung transcript levels of Gpbar1 (TGR5) and downstream mediators Nos3 and Dio2, as well as transcript levels of Nr1h4 (FXR) and downstream mediators Abcb4, Abcc2, and Nr0b2, were not elevated in HDM plus TUDCA–treated mice compared with both the PBS and HDM control groups (Figure 6B). Interestingly, transcripts of Nos3 were significantly decreased in both HDM and HDM plus TUDCA mice compared with PBS control mice, and transcripts for both Abcc2 and Nr0b2 were significantly decreased in HDM plus TUDCA compared with both PBS and HDM groups, suggesting partial inhibition rather than activation of both pathways (Figure 6B). Similarly, lung transcript levels of Gpbar1, Nr1h4, and their downstream mediators did not increase in any of the other CBA/UBA–treated mouse groups compared with the PBS plus DMSO and HDM plus DMSO groups (Figure 7, A–G). Transcript levels of Nos3 were significantly decreased in all HDM-sensitized (CBA-treated and untreated) groups when compared with the PBS plus DMSO group (Figure 7C), indicating a potential allergen-specific inhibitory effect on Nos3 expression in our models. Additionally, transcripts of Abcc2 were also significantly decreased in the HDM plus DMSO group as compared with PBS plus DMSO (Figure 7F). Therefore, the data suggest that the primary mechanism by which the 5 CBAs and 1 UBA inhibit allergic airway responses is unlikely to be via direct activation of the classical bile acid receptors.
Figure 6. TUDCA does not activate the FXR or TGR5 pathways in an allergen-induced model of asthma.
(A) Illustration of mediators regulated downstream of canonical bile acid receptors FXR and TGR5. (B) Gene expression quantified by RT-qPCR from the lungs. One-way ANOVA, Tukey’s post hoc test. *P < 0.05 vs. PBS; #P < 0.05 vs. HDM plus PBS mice. n = 6–10 mice/group from 2 experiments (outliers removed, Nr0b2 in HDM plus TUDCA, n = 1). HK, housekeeping; ns, not significant. Error bars represent ± SEM.
Figure 7. CBAs do not activate the FXR or TGR5 pathways in an allergen-induced model of asthma.
(A–G) Gene expression quantified by RT-qPCR from the lungs. One-way ANOVA, Tukey’s post hoc test. *P < 0.05 vs. DMSO plus PBS mice; #P < 0.05 vs. DMSO plus HDM mice. n = 6–10 mice/group from 2 experiments (outliers removed, Nr0b2 in HDM plus AβM; Abcc2 in HDM plus AβM and HDM plus GnβM; Abcb4 in HDM plus TCCA and HDM plus AβM; Dio2 in HDM plus ABM and HDM plus GnBM; Gpbar1 in HDM plus AβM; n = 1 outlier each). HK, housekeeping; ns, not significant. Error bars represent ± SEM.
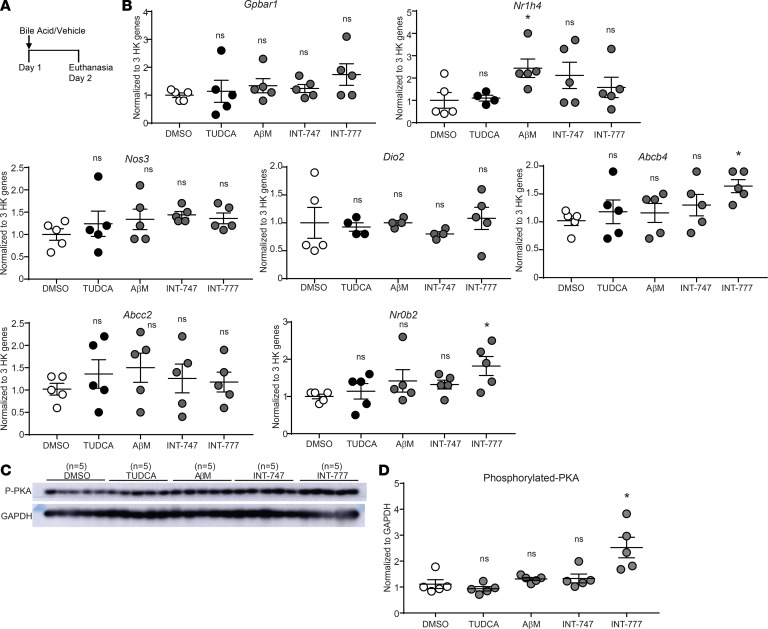
To further explore whether canonical signaling of the bile acid receptors is the primary CBA signaling mechanism in the lungs, we focused on the 2 CBAs with the broadest effects (Tables 1 and 2). TUDCA and AβM, as well as INT-747 and INT-777, which are potent agonists of FXR and TGR5, respectively (34, 35), were all prepared in PBS with DMSO as a vehicle and oropharyngeally instilled without allergen (Figure 8A). Our results showed that direct oropharyngeal administration of CBAs or pharmacological agonists do not activate the bile acid receptor signaling pathways in the lungs (Figure 8B). Bile acid receptor agonists INT-747 and INT-777 failed to increase expression of several downstream mediators of FXR and TGR5 in the lungs, with the exception of INT-777, which elevated Nrob2 and induced phosphorylation of PKA (Figure 8, B–D). These results suggest that even the pharmacological agonists do not robustly upregulate the downstream pathways of FXR and TGR5 in the lungs.
Figure 8. Solitary administration of bile acid-specific agonists and CBAs does not activate the FXR or TGR5 pathways in the lungs.
(A) Bile acid and vehicle treatment regimen. (B) Gene expression quantified by RT-qPCR from the lungs of mice 24 hours after administration of TUDCA, AβM, INT-747, and INT-777 (or DMSO vehicle control). n = 5 mice per group from 1 experiment. (C) Western blot analysis for phospho-PKA (p-PKA). (D) Densitometry for Western blot in C. Data were analyzed by 1-way ANOVA with Tukey’s post hoc test. *P < 0.05 vs. DMSO control group; ns, not significant vs. DMSO control group (Nr1h4 in TUDCA and HDM plus AβM; Dio2 in TUDCA, AβM, and INT-747; n = 1 outlier each). HK, housekeeping; ns, not significant. Error bars represent ± SEM.
CBAs directly bind canonical transducers of the UPR.
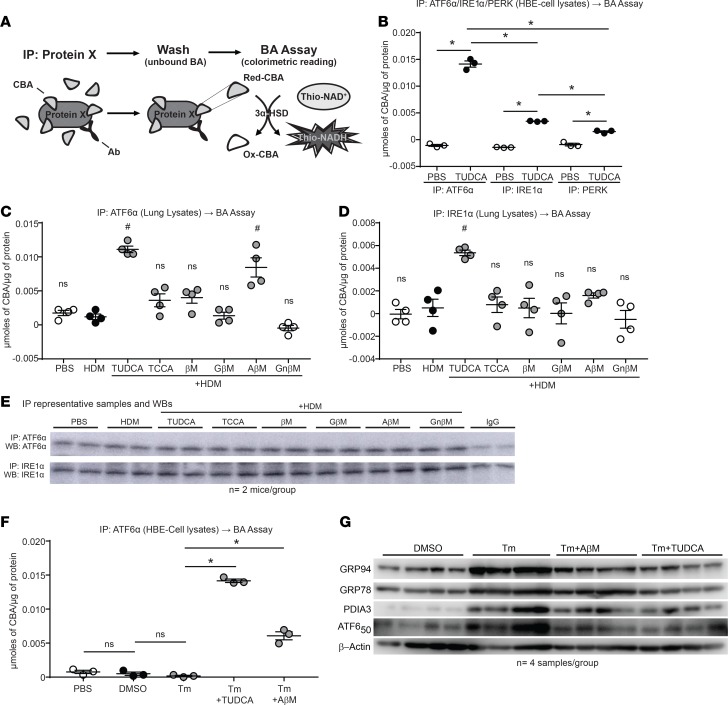
Because CBAs did not activate FXR or TGR5 (Figures 7 and 8), but select CBAs inhibited HDM-induced markers of the UPR (Figure 1L and Figure 4F), we hypothesized that select CBAs attenuate the UPR and allergic airway responses by inhibiting 1 or more of the transducers of UPR signaling, namely ATF6α, PERK, or IRE1α. First, we modified an existing bile acid assay to address whether CBAs bind directly to these UPR transducers (Figure 9A). Briefly, CBA-treated or untreated cell lysates were mixed with antibodies against ATF6α, PERK, or IRE1α; washed of excess unbound CBAs; and pulled down, and then we quantified protein-bound CBAs using a bile acid colorimetric assay.
Figure 9. CBAs inhibit the UPR and airway inflammation via direct binding of the UPR transducer ATF6α.
(A) Outline of the IP and subsequent bile acid assay protocol. (B) Bile acid assay performed on ATF6α, PERK, and IRE1α IP from HBE cells stimulated with or without TUDCA. One-way ANOVA, Tukey’s post hoc test. *P < 0.05; n = 3 samples per group from 1 experiment. (C and D) Bile acid assay performed on ATF6α and IRE1α IP from lung lysates of untreated and CBA/UBA–treated mice challenged with HDM. One-way ANOVA, Tukey’s post hoc test. #P < 0.05 vs. both PBS and HDM control groups; n = 4 mice per group from 2 experiments. (E) Representative Western blots from IPs of ATF6α and IRE1α from mouse lung lysates. (F) IP and subsequent bile acid assay from cell lysates of untreated and TUDCA/AβM–treated cells challenged with Tm. One-way ANOVA, Tukey’s post hoc test. *P < 0.05; n = 3 samples per group from 1 experiment. (G) Western blots for UPR markers from cell lysates of untreated and TUDCA/AβM–treated cells challenged with Tm. IP, immunoprecipitation; UBA, unconjugated bile acid; CBA, conjugated bile acid; Ab, antibody; 3α-HSD, 3α-hydroxysteroid dehydrogenase; Red, reduced; Ox, Oxidized; ns, not significant. Error bars represent ± SEM.
Initially, we applied our modified bile acid assay to cell lysates from human bronchial epithelial (HBE) cells treated with TUDCA or with PBS as a control (Figure 9B). The assay showed TUDCA was bound to all 3 transducers, with increased binding of ATF6α over PERK and IRE1α. We then applied this modified bile acid assay to lung lysates prepared from HDM-sensitized and -challenged mice treated with CBAs or UBA. ATF6α- or IRE1α-immunoprecipitated samples from lungs of CBA/UBA–treated mice had significantly greater ATF6α-bound TUDCA and AβM than untreated PBS and HDM controls (Figure 9C). In addition, there was more TUDCA-bound than AβM-bound IRE1α recovered from the lungs (Figure 9D), despite no variations in immunoprecipitation of ATF6α or IRE1α from the lung lysates (Figure 9E). Inexplicably, we failed to immunoprecipitate PERK from the mouse lung lysates. Furthermore, upon immunoprecipitation of ATF6α in the presence of the UPR-inducing agent, tunicamycin (Tm), the TUDCA- and AβM-treated HBE cells also showed greater recovery of TUDCA and AβM, while significantly more TUDCA was recovered compared with AβM (Figure 9F). We also observed decreases in the Tm-induced UPR in HBE cells treated with TUDCA and AβM (Figure 9G). Collectively, these results suggest that TUDCA and AβM are exerting their anti–airway allergen effects via binding to the UPR transducers in our allergen-induced model of asthma and have direct effects on airway epithelial cells in culture.
UPR markers are increased in asthmatic airway brushings.
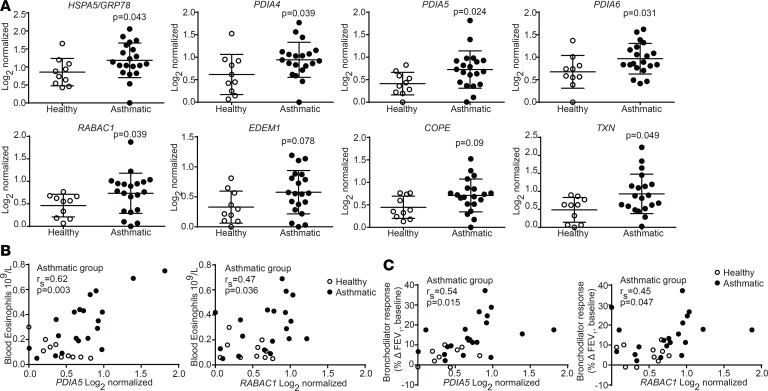
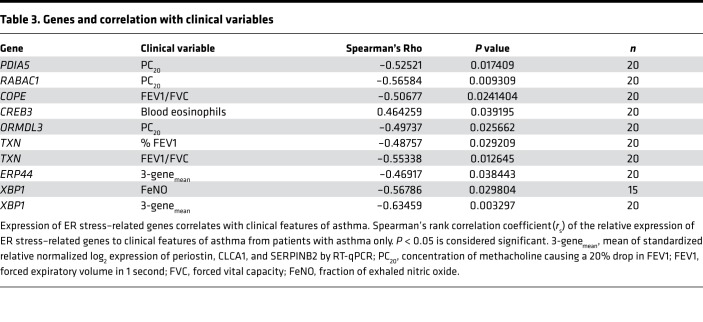
Finally, we analyzed the expression of UPR-related transcripts, with a focus on genes related to ATF6α, in airway epithelial brushings from healthy human and asthmatic subjects (Supplemental Table 1), as previously described (36, 37). Patients with asthma showed significant increases in transcripts for HSPA5, PDIA4, PDIA5, PDIA6, RABAC1, EDEM, COPE, and TXN (38–42) (Figure 10A). Expression of both PDIA5 and RABAC1 in patients with asthma correlated with high blood eosinophils (Figure 10B), impaired reversibility upon bronchodilator treatment (Figure 10C), and hyperresponsiveness to methacholine challenge (PC20, Table 3). The ATF6α-related genes COPE, CREB3, ORMDL3, and TXN similarly correlated with at least 1 pathological feature of asthma, whereas ERP44 and XBP1 were associated with reduced asthma pathology in our analyses (Table 3). Collectively, our results from human subjects and animal models suggest that patients with asthma upregulate the UPR markers associated with the ATF6α pathway. Furthermore, treatment with CBAs, such as TUDCA and AβM, may have the potential to decrease allergen-induced ATF6α activation and subsequent inflammatory and bronchodilator responses.
Figure 10. UPR-related genes are upregulated in the airways of human patients with asthma.
(A) RT-qPCR analysis of ATF6α-related genes from lung tissue. n = 10 healthy and n = 20 asthmatic subjects. Mann-Whitney U test, P values versus healthy subjects. (B and C) Correlation plot of the relative expressions of PDIA5 and RABAC1 to blood eosinophils and bronchodilator response (measured as change in absolute FEV1 relative to baseline). Spearman’s rank correlation coefficient (rs) of the group with asthma (healthy subjects excluded). P < 0.05 is considered significant. Error bars represent ± SD.
Table 3. Genes and correlation with clinical variables.
Discussion
We explored the potential of 5 CBAs and a UBA in alleviating allergen-induced UPR and airway responses in an HDM-induced murine model of allergic asthma. We determined that the CBAs and UBA vary in their ability to inhibit allergic airway disease, with AβM and TUDCA imparting the broadest effects. CBA-regulated inhibition of disease was associated with an attenuated UPR, through a decrease in ATF6α50. Moreover, our experiments demonstrated direct binding of CBAs to ATF6α without activating downstream bile acid receptor signaling. Finally, ATF6α-dependent and -activating transcripts were upregulated in asthmatic airways, and the expression correlated with clinical markers of disease. Taken together, our findings suggest a potentially novel mechanism whereby CBAs inhibit allergic airway responses via suppression of the UPR.
Bile acids are known to regulate cholesterol homeostasis and facilitate digestion and absorption of lipids (43). Early studies reported the therapeutic benefits of bile acids in various diseases through the regulation of distinct signaling pathways, including the UPR (43–45). We previously reported that TUDCA administration in HDM-allergic C57BL/6J mice decreased the UPR, AHR, mucus metaplasia, and airway fibrosis (7). However, the mechanism through which TUDCA reduced allergic airway disease was not directly addressed. Our current study showed that the CBAs AβM and TUDCA are potent inhibitors of the majority of our measured parameters of allergic airway disease, this time using the prototypic Th2-predisposed BALB/cJ mouse strain (Tables 1 And 2). Collectively, these results demonstrate the beneficial effects of CBAs in 2 strains of mice.
The 5 newly tested CBAs and 1 UBA — TCCA, βM, GβM, AβM, and GnβM — were originally dissolved in DMSO and then further diluted in PBS before instillation, unlike TUDCA, which easily dissolved in PBS alone. We therefore kept the TUDCA results separate from the other CBAs. HDM-sensitized and -challenged mice treated with the DMSO-dissolved CBAs showed significant improvement in many parameters of allergic airway disease, including AHR. TUDCA and βM significantly diminished airway resistance (Rn). βM also significantly inhibited peripheral tissue resistance (G) at the highest concentration of methacholine (Figure 5, A and B). TCCA-treated mice did not yield reliable AHR data for reasons currently unknown. Therefore, the TCCA group was excluded from this analysis. It should be noted that AβM decreased many of the measured parameters, without any significant effect on AHR or mucus metaplasia. On the other hand, βM decreased AHR and neutrophilia without affecting any other parameters. This dichotomy in AHR and inflammatory response shows that these compounds may be acting on different (UPR) pathways that are independent of each other. However, additional detailed experiments are required to substantiate this speculation.
Recent publications have identified specific CBAs as alleviating and exacerbating inflammatory and metabolic diseases via activation of the bile acid receptors FXR or TGR5 (46–49). Depending on the disease and the bile acid type, these studies showed that activation of bile acid receptors leads to either activation or inhibition of the NLR family pyrin domain containing 3 inflammasome, which promotes or inhibits inflammation, respectively. However, the mechanism of action of the specific CBAs used in this study and whether the mechanism differs in the setting of the lungs, an organ not normally exposed to high concentrations of bile acids, has not been explored. In our murine model of allergic airway disease, expression of transcripts downstream of FXR and TGR5 were not upregulated in the lungs of mice treated with any of the 5 CBAs or 1 UBA tested, suggesting that the primary mechanism by which CBAs attenuate allergic airway responses may not be through classical bile acid receptor signaling.
We made an effort to elaborate our findings concerning the mechanism of action of CBAs in the lungs by direct administration of the CBAs, TUDCA, and AβM, as well as the FXR-specific agonist INT-747 and TGR5-specific agonist INT-777, into the airways without allergen. Twenty-four hours after administration, expression of downstream mediators of the FXR and TGR5 pathways were not upregulated in the lungs of mice instilled with CBAs or agonists. These results suggest that CBAs and FXR- and TGR5-specific agonists are unable to activate downstream pathways in the lungs at the administered concentrations. There is evidence in the literature of bile acid activity that is FXR and TGR5 independent that occurs in tissue from organs that are nontraditional targets of bile acids, including both cardiomyocytes and airway epithelial cells (50–52). The relative expression of FXR in the lungs is also 2 log scales less than in traditional bile acid target tissues, such as the liver, kidneys, and intestines (53). In addition, there are 2 FXR genes, FXRα and FXRβ, with 4 isoforms of FXRα that are differentially expressed based on tissue type (54). Little is known about the activity of each FXRα isoform and its expression in the lungs. In contrast with FXR, TGR5 is encoded by a single exon and is highly expressed by several organs, including the lungs (55, 56). However, airway epithelial cells grown on an air-liquid interface and stimulated with the TGR5 agonist, INT-777, on the apical and basolateral sides of the cell membrane revealed that TGR5 is preferentially expressed on the basolateral surface (52). Because the CBAs in the present study were administered oropharyngeally, TGR5 would not be accessible unless tight junctions between cells were compromised. Finally, different bile acids can act as agonists or antagonists of FXR and TGR5, and they also have different affinities for each receptor (33). Thus, the inability of TUDCA, AβM, INT-747, and INT-777 to activate downstream FXR and TGR5 signaling could be due to the limited expression of FXR in the lungs, the specific FXRα isoforms expressed, the inaccessibility of the TGR5 receptor to CBAs present in the airway lumen, or their low affinity for both receptors.
We hypothesized that TUDCA and AβM may be inhibiting allergen-induced UPR in allergic airway disease and thus decreasing allergen-induced inflammatory response. To address this hypothesis, we began by immunoprecipitating transducers of the UPR from lysates prepared from the lungs of CBA-treated mice. We conducted a bile acid assay on the samples and recovered more CBAs from samples in which ATF6α was immunoprecipitated compared with IRE1α and PERK, suggesting that CBAs may inhibit the UPR and allergic airway responses via direct interaction with the UPR transducers ATF6α and IRE1α. Interestingly, because latent ATF6α is found as a trimer on the ER membrane (42, 57), we speculate that this ATF6α trimer may be a preferred target for TUDCA and AβM binding as compared with PERK or IRE1α, which appear as monomers in their latent forms (57).
As a regulator of the UPR, ATF6α activation can be both protective and detrimental to disease pathogenesis (38, 58–61). Activated ATF6α acts as a transcription factor, regulating expression of PDIs, chaperones, and antioxidants that aid in the proper folding of proteins, thus reducing ER stress (9, 10). Alternatively, inhibition of the ATF6α arm of the UPR has led to reduced inflammation in an experimental autoimmune encephalomyelitis model of multiple sclerosis (58).
The literature also suggests that the UPR via the IRE1/XBP1 axis or via ATF6α affects the epithelial cell responses to allergens or IL-13 stimulation, respectively (62). Similarly, ATF6α is known to regulate allergen-induced macrophage and airway smooth muscle responses (63–65). These studies indicate that many cell types and facets of allergic asthma are influenced by various pathways of UPR, including ATF6α.
Interestingly, in a study of diabetic mice, the impact of TUDCA on disease pathogenesis was contingent upon the presence of ATF6α and lost in mice with pancreatic β cell–specific deletion of ATF6α (66). Although at this juncture, we have not confirmed the need for ATF6α in CBA-mediated diminished allergic airway responses, we believe that future experiments with lung-specific ATF6α-knockout mice along with CBA treatment should provide insight into the involvement of ATF6α.
Finally, our analysis of ATF6α-dependent and -related genes both upstream and downstream of ATF6α (38–42) showed increased expression and correlated with features of asthma, including poor bronchodilator response and blood eosinophilia.
Collectively, our work indicates that human patients with asthma and a murine model of allergic airway disease exhibit upregulation of ATF6α-dependent genes. CBAs such as AβM and TUDCA potently resolve experimental allergic airway disease via the inhibition of the allergen-induced UPR, by attenuating the UPR transducer ATF6α.
Methods
Mouse models of HDM-induced allergic airway disease.
All mice involved in allergy experiments were anesthetized using isoflurane and exposed to the allergen HDM (item XPB70D3A2.5, lot 279020, normalized to protein content, GREER) suspended in PBS. Mice were administered 25 μg of HDM via the nasopharyngeal route on day 1 and boosted on day 8. Mice were then administered 25 μg of HDM consecutively on days 15 to 19, and days 25 and 27, before being analyzed on day 28 (Figure 1A and Figure 3B). All mice were females, BALB/cJ strain, from The Jackson Laboratory.
Human subjects.
Airway epithelial brushings from patients with physician-diagnosed asthma and from subjects without asthma were obtained from the Department of Medicine of the UCSF. Airway epithelial brushings from 10 nonasthmatic (healthy) subjects and 20 asthmatic patients were from the UCSF Airway Tissue Bank (Supplemental Table 1). All subjects were from a single clinical study (8) and represented a subset with enough remaining RNA quantity for quantitative PCR.
CBAs.
CBAs TCCA, GβM, AβM, and GnβM, along with a UBA, βM, were synthesized and purified in house as previously described (30, 31). TUDCA was purchased from Calbiochem (catalog 580549).
Solubility and treatment of CBAs.
All CBAs/UBA (TCCA, βM, GβM, AβM, and GnβM) with the exception of TUDCA were dissolved in 0.5% DMSO in PBS (pH 7.2), while TUDCA was dissolved in PBS (pH 7.2). Treatment groups received CBAs (0.75 mg/kg body weight) via the oropharyngeal route (intratracheal) on days 20, 22, 24, 26, and 28 (Figure 1A and Figure 3B), and the mice were euthanized 24 hours following the last treatment. The PBS (negative) and HDM (positive) control groups received 0.5% DMSO in PBS (pH 7.2) as the vehicle at all time points (or PBS as the vehicle in experiments involving TUDCA). Administration of CBAs (0.75 mg/kg) and 0.5% DMSO in PBS did not result in any visible distress or mortality.
An additional mouse experiment was conducted to exclude the FXR and TGR5 pathways as the mechanism through which the CBAs attenuate ER stress and allergic airway responses. In this experiment, BALB/cJ mice were administered TUDCA, AβM, INT-747, or INT-777 (or DMSO/PBS as a vehicle control) via oropharyngeal instillation at 0.75 mg/kg. All mice, including TUDCA-treated mice in this experiment, received the 0.5% DMSO in PBS vehicle control. Mice were euthanized and lungs were analyzed 24 hours after administration.
Assessment of AHR.
Mice were anesthetized by the administration of sodium pentobarbital (90 mg/kg, intraperitoneal), followed by an injection of the paralyzing agent pancuronium bromide. Mice were tracheotomized using an 18-gauge cannula and mechanically ventilated at 200 breaths/min using a FlexiVent computer-controlled, small-animal ventilator (SCIREQ). AHR measurements: Newtonian resistance (Rn), tissue damping (G), and tissue elastance (H) were measured from mice exposed to increasing concentrations of aerosolized methacholine. AHR values for each dose are the average of 3 adjacent measurements representing the peak response with a corresponding COD value of greater than or equal to 0.89 obtained at incremental methacholine doses for each animal. AHR values at the saline dose were then set to 100% for each mouse, and all other values for that mouse were normalized to the saline dose. The COD reflects the quality of the fit to the constant phase model. Outliers were removed using a cutoff of Q = 1% in the GraphPad Prism 7 program using the robust regression and outlier removal (ROUT) method.
Bronchoalveolar lavage fluid processing.
Bronchoalveolar lavage fluid (BALF) was collected by washing lungs with 1 ml of sterile PBS. Total and differential cell counts were performed as previously described (6). Briefly, cells were separated by centrifugation, and red blood cells were lysed before counting total cells. Differential cell counts were obtained via cytospins using Hema3 stain reagents (Thermo Fisher Scientific). Differentials were performed on a minimum of 300 cells.
ELISA.
Right lung lobes were flash frozen, pulverized, and lysed in buffer containing 137 mM Tris-HCl (pH 8.0), 130 mM NaCl, and 1% NP-40. Samples were normalized to total lung protein and assayed for IL-13, IL-17A, IL-4 (eBioscience), IL-6, CCL11 (eotaxin-1), CCL20 (MIP-3α) and CXCL1 (KC) (R&D Systems) by ELISA according to manufacturer’s instructions (Supplemental Table 2).
Western blot analysis.
Right lung lobes were flash frozen, pulverized, and lysed in buffer containing 137 mM Tris-HCl (pH 8.0), 130 mM NaCl, and 1% NP-40. Cultured HBE cells were directly lysed on the cell culture plates with the same buffer. Insoluble proteins were pelletized via centrifugation, and the supernatant was used for protein quantification. Furthermore, protein samples resuspended in loading buffer with dithiothreitol were resolved by SDS-PAGE. Proteins were transferred to PVDF and membranes were probed using a standard immunoblotting protocol. The primary antibodies recognizing PDIA3 and GRP94 were purchased from Enzo Life Sciences, CHOP was from Novus Bio, and GRP78, ATF6α50, and PDIA6 were from Abcam. PDIA5 was from Novus Bio, and GAPDH was from BioLegend. Details of the antibodies are provided in Supplemental Table 3. The quantification of protein expression was performed by densitometry using ImageJ software (NIH, https://imagej.nih.gov/ij/). See complete unedited blots in the supplemental material.
Analysis of mRNA expression.
Expression levels of Muc5ac, Clca1, Gpbar1, Nr1h4, Abcb4, Abcc2, Nr0b2, Nos3, and Dio2 were quantified from mouse lungs. Flash-frozen right lung lobes were pulverized, and total RNA was extracted using Qiazol (Qiagen). RNA was purified using the RNeasy kit (Qiagen). We reverse transcribed 1 μg of RNA to cDNA (Promega), for quantification of mRNA expression using SYBR Green Supermix (Bio-Rad). Expression values were normalized to the geometric mean of 3 housekeeping genes (Gapdh, Pp1, and Rp2) using the ΔΔCT method. The primers used in this study are listed in Supplemental Table 4. Transcripts prepared from human airway brushings were analyzed by 2-step, nested-primer RT-qPCR (Supplemental Table 5) as previously described (8, 36, 37). Data were normalized to the mean of GAPDH, PPIA, and RPL37A as previously described (36, 37).
Quantification of mucus metaplasia.
The tissue sections (5 μm) from paraffin blocks of lung were mounted on slides, and mucus metaplasia was assessed by PAS. The images were captured using an Olympus BX50 light microscope, and mucus metaplasia was measured by quantifying the percentage of positively PAS-stained area of airways, using MetaMorph software as described previously (7). Muc5ac and Clca1 mRNA expression were assessed via real-time RT-qPCR analysis.
Cell culture experiments.
The HBE cell line was purchased from ATCC (catalog CRL4051) and cultured as previously described (5). HBE cells used in the bile acid assay were treated with 750 μM of TUDCA or AβM (or PBS as a vehicle control) and harvested after 1 hour using ice-cold PBS, and the cell lysates were centrifuged at 9,400 g for 10 minutes at 4°C. The protein concentration of supernatant samples was quantified.
Immunoprecipitation and the bile acid assay.
Cell lysates from cultured HBE cells treated with or without TUDCA were subjected to immunoprecipitation. ATF6α, IRE1α, and PERK were immunoprecipitated using primary antibodies (ATF6α and IRE1α from Abcam, PERK from Cell Signaling Technology). ATF6α alone was immunoprecipitated from the lung lysates of mice treated with or without TUDCA/AβM. The Protein G Agarose beads (Invitrogen, 15920-010) containing immunoprecipitates were gently washed using lysis buffer (137 mM Tris-HCl, pH 7.0, and 130 mM NaCl) without the detergent (3 times) to remove the unbound CBAs, and the bile acid assay was performed to evaluate the binding capacity of bile acids to ATF6α, PERK, and IRE1α. Total Bile acid assay kit (catalog STA-631) was purchased from Cell Biolabs Inc., and the assay was performed as per the manufacturer’s instructions with slight modifications, including Protein G Agarose beads as controls.
Statistics.
Human data were analyzed as described in the figure and table legends. All mouse data were analyzed as follows: The ROUT method was first used to identify outliers in GraphPad Prism 7 with a cutoff of Q = 1%, and they were removed from analysis. The Shapiro-Wilk normality test was applied, and wherever 1 or more groups did not pass the normality test, the Kruskal-Wallis (nonparametric) test, followed by the 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli, were performed to control for the false discovery rate (FDR). Wherever the data passed the normality test, 1-way analysis of variance (ANOVA) was performed, and Tukey’s post hoc test was applied to adjust for multiple comparisons. Two-tailed Student’s t test was used. The specific statistical analyses used are mentioned in the figure legends. Data were averaged and expressed as the mean ±SEM. The FDR or Tukey’s adjusted P value of less than 0.05 were considered to be statistically significant.
Study approval.
All mice were housed, maintained, and used in the experiments as approved by the Institutional Animal Care and Use Committee of the University of Vermont (protocol no. 15-009). The study (NCT00595153) was approved by the UCSF Committee on Human Research, written informed consent was obtained from all subjects, and all studies were performed in accordance with the principles expressed in the Declaration of Helsinki. The Institutional Review Board of the UCSF approved the provision of data from de-identified patients for presentation in this manuscript.
Author contributions
EMN, BRKM, MEP, CGI, and VA were involved in the experimental design. EMN, NRB, BRKM, AK, SMH, NC, SRB, ND, MA, and VA conducted experiments and acquired data. EMN, NRB, BRKM, AK, DGC, ND, and VA analyzed data. NRB, PGW, SA, CGI, AED, MEP, and DHD provided reagents and critical comments. EMN and VA wrote the manuscript with input from all the authors.
Supplementary Material
Acknowledgments
The authors thank the University of Vermont Larner College of Medicine microscopy facility (P20GM103496), the Vermont Lung Center phenotyping core (P30GM103532), and the UCSF airway tissue core.
Version 1. 05/02/2019
Electronic publication
Version 2. 06/17/2019
The top of Figure 10 was not visible in the PDF.
Version 3. 07/08/2019
This corrected a typographical error in the title.
Footnotes
Acknowledgments
This research was supported by NIH R01 HL122383, HL141364 (to VA), and HL 136917 (to AED); by Department of Pathology and Laboratory Medicine, Parker B. Francis Fellowship, and ATS-Joe Rae Wright (unrestricted) award to VA; by T32 training grant graduate student fellowship (T32HL076122) to NC; by Department of Pathology and Laboratory Medicine graduate student fellowship to SRB; and by a grant from the Pennsylvania Department of Health, Tobacco Commonwealth Universal Research Enhancement Program Funds, to DHD. DGC was supported by a CJ Martin Fellowship from the National Medical Research Council of Australia (NHMRC-ECF 1053790).
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(9):e98101. https://doi.org/10.1172/jci.insight.98101.
Contributor Information
Emily M. Nakada, Email: emily.nakada@mail.mcgill.ca.
Amit Kumar, Email: amit.kumar@uvm.edu.
Nicolas Chamberlain, Email: nicolas.chamberlain@uvm.edu.
David G. Chapman, Email: David.Chapman@med.uvm.edu.
Nirav Daphtary, Email: nirav.daphtary@uvm.edu.
Minara Aliyeva, Email: minara.aliyeva@uvm.edu.
Charles G. Irvin, Email: Charles.Irvin@uvm.edu.
Anne E. Dixon, Email: anne.dixon@uvmhealth.org.
Prescott G. Woodruff, Email: Prescott.Woodruff@ucsf.edu.
Shantu Amin, Email: sga3@psu.edu.
Vikas Anathy, Email: vanathy@uvm.edu.
References
- 1.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18(5):716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- 2.Calderón MA, et al. House dust mite respiratory allergy: an overview of current therapeutic strategies. J Allergy Clin Immunol Pract. 2015;3(6):843–855. doi: 10.1016/j.jaip.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 3.Bousquet PJ, et al. Geographical variation in the prevalence of positive skin tests to environmental aeroallergens in the European Community Respiratory Health Survey I. Allergy. 2007;62(3):301–309. doi: 10.1111/j.1398-9995.2006.01293.x. [DOI] [PubMed] [Google Scholar]
- 4.Gregory LG, Lloyd CM. Orchestrating house dust mite-associated allergy in the lung. Trends Immunol. 2011;32(9):402–411. doi: 10.1016/j.it.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffman SM, et al. Endoplasmic reticulum stress mediates house dust mite-induced airway epithelial apoptosis and fibrosis. Respir Res. 2013;14:141. doi: 10.1186/1465-9921-14-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffman SM, et al. Protein disulfide isomerase-endoplasmic reticulum resident protein 57 regulates allergen-induced airways inflammation, fibrosis, and hyperresponsiveness. J Allergy Clin Immunol. 2016;137(3):822–32.e7. doi: 10.1016/j.jaci.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siddesha JM, et al. Effect of a chemical chaperone, tauroursodeoxycholic acid, on HDM-induced allergic airway disease. Am J Physiol Lung Cell Mol Physiol. 2016;310(11):L1243–L1259. doi: 10.1152/ajplung.00396.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhakta NR, et al. IFN-stimulated gene expression, type 2 inflammation, and endoplasmic reticulum stress in asthma. Am J Respir Crit Care Med. 2018;197(3):313–324. doi: 10.1164/rccm.201706-1070OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13(2):89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 10.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454(7203):455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334(6059):1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schönthal AH. Endoplasmic reticulum stress: its role in disease and novel prospects for therapy. Scientifica (Cairo) 2012;2012:857516. doi: 10.6064/2012/857516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindholm D, Korhonen L, Eriksson O, Kõks S. Recent Insights into the role of unfolded protein response in er stress in health and disease. Front Cell Dev Biol. 2017;5:48. doi: 10.3389/fcell.2017.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13(3):184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hipp MS, Park SH, Hartl FU. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 2014;24(9):506–514. doi: 10.1016/j.tcb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Löser S, et al. Pulmonary ORMDL3 is critical for induction of Alternaria-induced allergic airways disease. J Allergy Clin Immunol. 2017;139(5):1496–1507.e3. doi: 10.1016/j.jaci.2016.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willart MA, et al. Ursodeoxycholic acid suppresses eosinophilic airway inflammation by inhibiting the function of dendritic cells through the nuclear farnesoid X receptor. Allergy. 2012;67(12):1501–1510. doi: 10.1111/all.12019. [DOI] [PubMed] [Google Scholar]
- 18.Shaik FB, Panati K, Narasimha VR, Narala VR. Chenodeoxycholic acid attenuates ovalbumin-induced airway inflammation in murine model of asthma by inhibiting the T(H)2 cytokines. Biochem Biophys Res Commun. 2015;463(4):600–605. doi: 10.1016/j.bbrc.2015.05.104. [DOI] [PubMed] [Google Scholar]
- 19.Nevens F, et al. A placebo-controlled trial of obeticholic acid in primary biliary cholangitis. N Engl J Med. 2016;375(7):631–643. doi: 10.1056/NEJMoa1509840. [DOI] [PubMed] [Google Scholar]
- 20.Elia AE, et al. Tauroursodeoxycholic acid in the treatment of patients with amyotrophic lateral sclerosis. Eur J Neurol. 2016;23(1):45–52. doi: 10.1111/ene.12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Obici L, et al. Doxycycline plus tauroursodeoxycholic acid for transthyretin amyloidosis: a phase II study. Amyloid. 2012;19 Suppl 1:34–36. doi: 10.3109/13506129.2012.678508. [DOI] [PubMed] [Google Scholar]
- 22.Hsu KJ, Turvey SE. Functional analysis of the impact of ORMDL3 expression on inflammation and activation of the unfolded protein response in human airway epithelial cells. Allergy Asthma Clin Immunol. 2013;9(1):4. doi: 10.1186/1710-1492-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino MB, et al. The ER stress transducer IRE1β is required for airway epithelial mucin production. Mucosal Immunol. 2013;6(3):639–654. doi: 10.1038/mi.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, et al. Role of C/EBP homologous protein and endoplasmic reticulum stress in asthma exacerbation by regulating the IL-4/signal transducer and activator of transcription 6/transcription factor EC/IL-4 receptor α positive feedback loop in M2 macrophages. J Allergy Clin Immunol. 2017;140(6):1550–1561.e8. doi: 10.1016/j.jaci.2017.01.024. [DOI] [PubMed] [Google Scholar]
- 25.Bettigole SE, et al. The transcription factor XBP1 is selectively required for eosinophil differentiation. Nat Immunol. 2015;16(8):829–837. doi: 10.1038/ni.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu R, et al. Endoplasmic reticulum stress of neutrophils is required for ischemia/reperfusion-induced acute lung injury. J Immunol. 2015;195(10):4802–4809. doi: 10.4049/jimmunol.1500073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol. 2000;164(12):6166–6173. doi: 10.4049/jimmunol.164.12.6166. [DOI] [PubMed] [Google Scholar]
- 28.Van Hove CL, et al. Comparison of acute inflammatory and chronic structural asthma-like responses between C57BL/6 and BALB/c mice. Int Arch Allergy Immunol. 2009;149(3):195–207. doi: 10.1159/000199715. [DOI] [PubMed] [Google Scholar]
- 29.Gueders MM, et al. Mouse models of asthma: a comparison between C57BL/6 and BALB/c strains regarding bronchial responsiveness, inflammation, and cytokine production. Inflamm Res. 2009;58(12):845–854. doi: 10.1007/s00011-009-0054-2. [DOI] [PubMed] [Google Scholar]
- 30.Jiang C, et al. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J Clin Invest. 2015;125(1):386–402. doi: 10.1172/JCI76738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang C, et al. Intestine-selective farnesoid X receptor inhibition improves obesity-related metabolic dysfunction. Nat Commun. 2015;6:10166. doi: 10.1038/ncomms10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malhi H, Camilleri M. Modulating bile acid pathways and TGR5 receptors for treating liver and GI diseases. Curr Opin Pharmacol. 2017;37:80–86. doi: 10.1016/j.coph.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herman-Edelstein M, Weinstein T, Levi M. Bile acid receptors and the kidney. Curr Opin Nephrol Hypertens. 2018;27(1):56–62. doi: 10.1097/MNH.0000000000000374. [DOI] [PubMed] [Google Scholar]
- 34.Pellicciari R, et al. 6α-Ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J Med Chem. 2002;45(17):3569–3572. doi: 10.1021/jm025529g. [DOI] [PubMed] [Google Scholar]
- 35.Pellicciari R, et al. Discovery of 6α-ethyl-23(S)-methylcholic acid (S-EMCA, INT-777) as a potent and selective agonist for the TGR5 receptor, a novel target for diabesity. J Med Chem. 2009;52(24):7958–7961. doi: 10.1021/jm901390p. [DOI] [PubMed] [Google Scholar]
- 36.Dolganov GM, et al. A novel method of gene transcript profiling in airway biopsy homogenates reveals increased expression of a Na+-K+-Cl– cotransporter (NKCC1) in asthmatic subjects. Genome Res. 2001;11(9):1473–1483. doi: 10.1101/gr.191301. [DOI] [PubMed] [Google Scholar]
- 37.Bhakta NR, et al. A qPCR-based metric of Th2 airway inflammation in asthma. Clin Transl Allergy. 2013;3(1):24. doi: 10.1186/2045-7022-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu J, et al. ATF6α optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13(3):351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Bommiasamy H, et al. ATF6α induces XBP1-independent expansion of the endoplasmic reticulum. J Cell Sci. 2009;122(pt 10):1626–1636. doi: 10.1242/jcs.045625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vekich JA, Belmont PJ, Thuerauf DJ, Glembotski CC. Protein disulfide isomerase-associated 6 is an ATF6-inducible ER stress response protein that protects cardiac myocytes from ischemia/reperfusion-mediated cell death. J Mol Cell Cardiol. 2012;53(2):259–267. doi: 10.1016/j.yjmcc.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belmont PJ, et al. Coordination of growth and endoplasmic reticulum stress signaling by regulator of calcineurin 1 (RCAN1), a novel ATF6-inducible gene. J Biol Chem. 2008;283(20):14012–14021. doi: 10.1074/jbc.M709776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Higa A, et al. Endoplasmic reticulum stress-activated transcription factor ATF6α requires the disulfide isomerase PDIA5 to modulate chemoresistance. Mol Cell Biol. 2014;34(10):1839–1849. doi: 10.1128/MCB.01484-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joyce SA, Gahan CG. Disease-associated changes in bile acid profiles and links to altered gut microbiota. Dig Dis. 2017;35(3):169–177. doi: 10.1159/000450907. [DOI] [PubMed] [Google Scholar]
- 44.Lombardi R, Onali S, Thorburn D, Davidson BR, Gurusamy KS, Tsochatzis E. Pharmacological interventions for non-alcohol related fatty liver disease (NAFLD): an attempted network meta-analysis. Cochrane Database Syst Rev. 2017;3:CD011640. doi: 10.1002/14651858.CD011640.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saffioti F, Gurusamy KS, Eusebi LH, Tsochatzis E, Davidson BR, Thorburn D. Pharmacological interventions for primary biliary cholangitis: an attempted network meta-analysis. Cochrane Database Syst Rev. 2017;3:CD011648. doi: 10.1002/14651858.CD011648.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo C, et al. Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity. 2016;45(4):802–816. doi: 10.1016/j.immuni.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 47.Gong Z, et al. Chenodeoxycholic acid activates NLRP3 inflammasome and contributes to cholestatic liver fibrosis. Oncotarget. 2016;7(51):83951–83963. doi: 10.18632/oncotarget.13796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia-Irigoyen O, Moschetta A. A novel protective role for FXR against inflammasome activation and endotoxemia. Cell Metab. 2017;25(4):763–764. doi: 10.1016/j.cmet.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 49.Bode N, et al. Ursodeoxycholic acid impairs atherogenesis and promotes plaque regression by cholesterol crystal dissolution in mice. Biochem Biophys Res Commun. 2016;478(1):356–362. doi: 10.1016/j.bbrc.2016.07.047. [DOI] [PubMed] [Google Scholar]
- 50.Sheikh Abdul Kadir SH, et al. Bile acid-induced arrhythmia is mediated by muscarinic M2 receptors in neonatal rat cardiomyocytes. PLoS One. 2010;5(3):e9689. doi: 10.1371/journal.pone.0009689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen B, Cai HR, Xue S, You WJ, Liu B, Jiang HD. Bile acids induce activation of alveolar epithelial cells and lung fibroblasts through farnesoid X receptor-dependent and independent pathways. Respirology. 2016;21(6):1075–1080. doi: 10.1111/resp.12815. [DOI] [PubMed] [Google Scholar]
- 52.Hendrick SM, Mroz MS, Greene CM, Keely SJ, Harvey BJ. Bile acids stimulate chloride secretion through CFTR and calcium-activated Cl- channels in Calu-3 airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2014;307(5):L407–L418. doi: 10.1152/ajplung.00352.2013. [DOI] [PubMed] [Google Scholar]
- 53.Comeglio P, et al. Anti-fibrotic effects of chronic treatment with the selective FXR agonist obeticholic acid in the bleomycin-induced rat model of pulmonary fibrosis. J Steroid Biochem Mol Biol. 2017;168:26–37. doi: 10.1016/j.jsbmb.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 54.Lee FY, Lee H, Hubbert ML, Edwards PA, Zhang Y. FXR, a multipurpose nuclear receptor. Trends Biochem Sci. 2006;31(10):572–580. doi: 10.1016/j.tibs.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 55.Kawamata Y, et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278(11):9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 56.Guo C, Chen WD, Wang YD. TGR5, not only a metabolic regulator. Front Physiol. 2016;7:646. doi: 10.3389/fphys.2016.00646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eletto D, Chevet E, Argon Y, Appenzeller-Herzog C. Redox controls UPR to control redox. J Cell Sci. 2014;127(pt 17):3649–3658. doi: 10.1242/jcs.153643. [DOI] [PubMed] [Google Scholar]
- 58.Ta HM, et al. Atf6α deficiency suppresses microglial activation and ameliorates pathology of experimental autoimmune encephalomyelitis. J Neurochem. 2016;139(6):1124–1137. doi: 10.1111/jnc.13714. [DOI] [PubMed] [Google Scholar]
- 59.Yoshikawa A, et al. Deletion of Atf6α impairs astroglial activation and enhances neuronal death following brain ischemia in mice. J Neurochem. 2015;132(3):342–353. doi: 10.1111/jnc.12981. [DOI] [PubMed] [Google Scholar]
- 60.Yamamoto K, et al. Induction of liver steatosis and lipid droplet formation in ATF6α-knockout mice burdened with pharmacological endoplasmic reticulum stress. Mol Biol Cell. 2010;21(17):2975–2986. doi: 10.1091/mbc.e09-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Usui M, et al. Atf6α-null mice are glucose intolerant due to pancreatic β-cell failure on a high-fat diet but partially resistant to diet-induced insulin resistance. Metab Clin Exp. 2012;61(8):1118–1128. doi: 10.1016/j.metabol.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 62.Bhakta NR, et al. IFN-stimulated gene expression, type 2 inflammation, and endoplasmic reticulum stress in asthma. Am J Respir Crit Care Med. 2018;197(3):313–324. doi: 10.1164/rccm.201706-1070OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller M, et al. ORMDL3 transgenic mice have increased airway remodeling and airway responsiveness characteristic of asthma. J Immunol. 2014;192(8):3475–3487. doi: 10.4049/jimmunol.1303047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller M, et al. ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6. Proc Natl Acad Sci U S A. 2012;109(41):16648–16653. doi: 10.1073/pnas.1204151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Unno H, Miller M, Rosenthal P, Beppu A, Das S, Broide DH. Activating transcription factor 6α (ATF6α) regulates airway hyperreactivity, smooth muscle proliferation, and contractility. J Allergy Clin Immunol. 2018;141(1):439–442.e4. doi: 10.1016/j.jaci.2017.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Engin F, et al. Restoration of the unfolded protein response in pancreatic β cells protects mice against type 1 diabetes. Sci Transl Med. 2013;5(211):211ra156. doi: 10.1126/scitranslmed.3006534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.