Abstract
Skeletal muscle weakness in patients suffering from rheumatoid arthritis (RA) adds to their impaired working abilities and reduced quality of life. However, little molecular insight is available on muscle weakness associated with RA. Oxidative stress has been implicated in the disease pathogenesis of RA. Here, we show that oxidative posttranslational modifications of the contractile machinery targeted to actin result in impaired actin polymerization and reduced force production. Using mass spectrometry, we identified the actin residues targeted by oxidative 3-nitrotyrosine (3-NT) or malondialdehyde (MDA) adduct modifications in weakened skeletal muscle from mice with arthritis and patients afflicted by RA. The residues were primarily located in 3 distinct regions positioned at matching surface areas of the skeletal muscle actin molecule from arthritic mice and patients with RA. Moreover, molecular dynamics simulations revealed that these areas, here coined “hotspots,” are important for the stability of the actin molecule and its capacity to generate filaments and interact with myosin. Together, these data demonstrate how oxidative modifications on actin promote muscle weakness in RA patients and may provide novel leads for targeted therapeutic treatment to improve muscle function.
Keywords: Muscle Biology
Keywords: Rheumatology, Skeletal muscle
Oxidative modifications on actin promotes muscle weakness in rheumatoid arthritis patients, suggesting a potential therapeutic target to improve muscle function.
Introduction
Rheumatoid arthritis (RA) is one of the most prevalent chronic inflammatory diseases. Skeletal muscle weakness is a frequent complication in patients with RA; it has debilitating effects on the ability to work and leads to reduced quality of life for the afflicted individuals (1–3). Altered intrinsic muscle function, including a reduction in muscle strength without corresponding loss of muscle mass has been reported in patients with RA (2, 4–6). However, little molecular insight is available on RA-induced muscle weakness.
Oxidative stress arises from a disproportionate amount of reactive oxygen/nitrogen species (ROS/RNS), e.g., peroxynitrite (ONOO–) that has been implicated in the pathogenesis of RA (7–12). ONOO− can induce oxidative posttranslational protein modifications by nitrating tyrosine residues (13) and facilitate the formation of malondialdehyde (MDA) adducts on basic amino acids, such as histidine (13, 14). Increased levels of 3-nitrotyrosine (3-NT) and MDA adducts have been observed in rodent models with arthritis and in patients with RA (7, 12, 15–17). The ROS/RNS sources behind arthritis-induced oxidative stress are not fully established in skeletal muscle (18), but nitric oxide synthase (NOS), NADPH oxidases (NOX), and mitochondria are suggested downstream targets of proinflammatory cytokines in inflammatory processes (18–21). Skeletal muscle is composed of bundles of muscle fibers and each fiber consists of thousands of myofibrils that feature the sarcomeres with the actomyosin contractile machinery (18). Previous work in rodents indicate that arthritis-induced muscle weakness is associated with higher levels of oxidative modifications on actin (15, 16, 18, 22). However, details of how oxidative modifications of actin interfere with the force-producing machinery have remained elusive.
Here, we show that oxidative posttranslational modifications of skeletal muscle actin (α-actin) and myofibrils lead to impaired actin polymerization and decreased force production. Furthermore, using mass spectrometry (MS), we identified the actin 3-NT–modified residues as well as those carrying MDA in weakened skeletal muscle from mice with arthritis and from RA patients. These residues were predominantly located in 3 regions of the actin molecule, and remarkably enough the same regions were recognized in the samples from both the arthritic mice and RA patients. Polymerization studies and molecular dynamics simulations were carried out to analyze functional consequences and provide atomic details of actin after oxidative stress modifications in these regions. This demonstrated the importance of these regions for actin intra- and interdomain bonding and myosin interaction, and provides a molecular explanation for the observations made in arthritic mice and RA patients. Consequently, these regions should be useful starting points for the design of novel specifically targeted therapeutics to treat RA-associated muscle weakness.
Results
Muscle weakness in mice with arthritis.
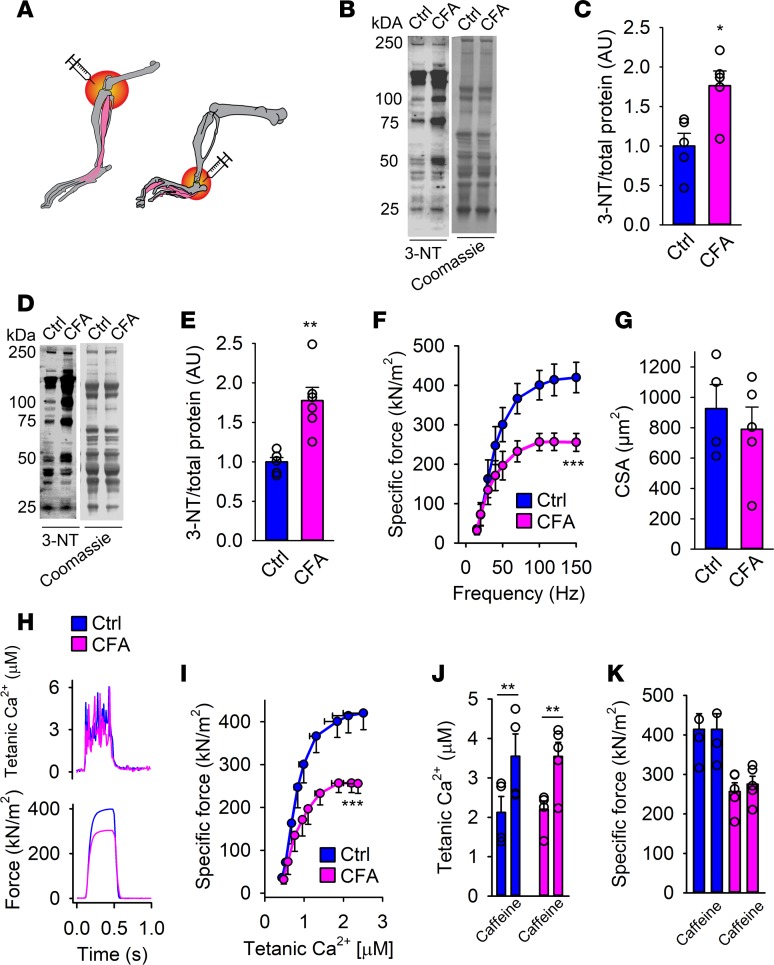
To evoke arthritis, unilateral injections of complete Freund’s adjuvant (CFA) into the knee or ankle of mice were used (Figure 1A). In line with other arthritis models, increased levels of oxidative nitration markers (3-NT) were observed on muscle proteins in adjacent muscle (Figure 1, B and C, interosseous, and Figure 1, D and E, gastrocnemius) at peak inflammation, i.e., 2 weeks after CFA injections with an ankle or knee diameter twice as large as the control leg (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.126347DS1). As compared with the control leg (Ctrl), the arthritic leg developed muscle weakness with both extensor digitorum longus (EDL, Supplemental Figure 1, C and D) and single muscle fibers from flexor digitorum brevis (FDB) producing lower ex vivo muscle-specific force (i.e., force per cross-sectional area [CSA]) (Figure 1F). There was no difference in the CSA between the nonarthritic and arthritic EDL or FDB muscle and the inflammation did not affect total body weight (Figure 1G and Supplemental Figure 1, E and F). Furthermore, the nonarthritic leg showed similar force production as muscle from untreated wild-type mice; hence, the induced arthritis does not interfere with contralateral force production (Supplemental Figure 1C).
Figure 1. Muscle weakness and accompanied oxidative modifications in skeletal muscle from mice with arthritis.
(A) An illustration of the induction site of arthritis by complete Freund’s adjuvant (CFA) (ankle: used for flexor digitorum brevis [FDB] muscle fiber force measurements; knee: used for extensor digitorum longus [EDL] whole-muscle force measurements). Immunoblots and quantification of 3-nitrotyrosine (3-NT) in interosseous (B and C) and gastrocnemius (D and E) muscle from mice with arthritis (CFA) and healthy controls (Ctrl) (n = 6). 3-NT levels were normalized to total protein Coomassie staining. (F) Ex vivo specific force (contractions induced at 15–150 Hz, n = 4–5) of intact individual muscle fibers from arthritis leg (pink) and healthy controls (blue). (G) Calculated cross-sectional area of FDB fibers from CFA and control legs. (H) Typical examples (120-Hz stimulation frequency, 350-ms train duration) of intracellular (tetanic) Ca2+ (upper) and specific force (lower) in control (blue) and CFA (pink) fibers (n = 4–5). (I) Mean (±SEM) force versus intracellular Ca2+ data obtained in 15- to 150-Hz contractions produced in control fibers and CFA fibers. Mean values of intracellular Ca2+ (J) and specific force (K) in the presence or absence of caffeine (5 mM, 2-minute exposure) in CFA and control FDB fibers. Data are mean ± SEM. Statistical analysis in C, E, G, J, and K was performed using 2-tailed Student’s t test and in F and I by 2-way ANOVA. A P value less than 0.05 was considered significant. *P < 0.05; **P < 0.01; ***P < 0.001.
Ca2+ release from the sarcoplasmic reticulum (SR) enables contraction by uncovering active sites on actin for myosin binding (23). Thus, the higher the intracellular free Ca2+ concentration ([Ca2+]i), the greater the force generated until saturation of [Ca2+]i and all actomyosin interactions are activated. However, at the stimulation frequencies used, no difference in tetanic [Ca2+]i or Ca50 ([Ca2+]i at 50% Pmax, 0.8 ± 0.2 vs. 0.7 ± 0.2 μM; not significant) was found between arthritis-induced and Ctrl muscle fibers (Figure 1, H and I). This indicates that SR Ca2+ release and myofibrillar Ca2+ sensitivity are functioning normally in fibers from arthritis muscles.
Caffeine treatment (5 mM) and high stimulation frequency (120 Hz) effectively release all Ca2+ stored in the SR and tetanic [Ca2+]i will reach a level high enough to maximally activate the contractile machinery (24). Indeed, the presence of caffeine at 120-Hz stimulation potentiated SR Ca2+ release (Figure 1J), but had no effect on the force production of either control fibers or the CFA-treated fibers (Figure 1K). Caffeine enhanced SR Ca2+ release in control muscle fibers did not have any notable effect on force production because the tetanic force prior to caffeine application is close to maximal. That the lower force in CFA-treated muscle was not improved by caffeine exposure shows that the weakness is the result of impaired myofibrillar function, rather than impaired SR Ca2+ release or reduced SR Ca2+ storage. Thus, CFA-induced muscle weakness appears to be the result of defects in the contractile machinery.
Declined myofibrillar force and compromised filament formation.
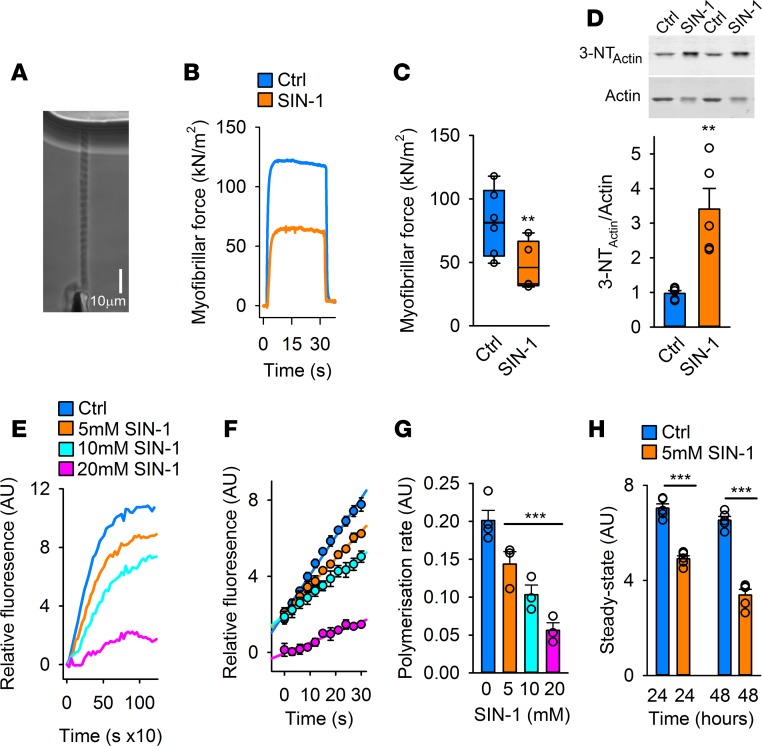
Because muscle weakness is associated with oxidative stress (15, 16, 18, 22, 25–27), we investigated whether inducing oxidative modifications of individual myofibrils (Figure 2A) interfered with force production. Treatment with 5-amino-3-(4-morpholinyl)-1,2,3-oxadiazolium chloride (SIN-1), an NO and O2−• donor that forms ONOO− (28), resulted in markedly weaker myofibrils than untreated myofibrils (Figure 2, B and C), as measured with an atomic force cantilever (16). To avoid formation of nitrosylated cysteine residues, dithiothreitol (DTT) was present during these measurements, and consequently the SIN-1–treated myofibrils showed increased levels of oxidative nitration (3-NT) due to the presence of ONOO− (Figure 2D).
Figure 2. Oxidative modifications introduced by SIN-1 decrease myofibrillar force and impair actin polymerization.
(A) Phase-contrast image of an isolated myofibril set up for force measurement using the atomic force cantilever (AFC). (B) Typical recordings of active force from AFC measurements in myofibrils at pCa2+ of 4.5 with and without SIN-1 (10 mM, 10 minutes) with DTT present (1 mM DTT) (n = 9–10). (C) Mean (±SEM, n = 9–10) of the active isometric force produced by the myofibrils with or without SIN-1 at a sarcomere length of 2.8 μm and pCa2+ 4.5. (D) Immunoblots and quantification of the 3-NT level on myofibrillar actin (n = 9–10). (E) Typical polymerization recordings of the actin polymerization assay using pyrene-labeled actin polymerized with control G-actin (Ctrl) or G-actin preincubated in SIN-1 (5, 10, or 20 mM; 15-minute preincubation time) (n = 3–4). (F) Mean fluorescence intensity (±SEM, n = 3–4) of the polymerization reaction at half maximum level of polymerization of Ctrl and SIN-1 G-actin (5, 10, or 20 mM, 15 minutes). (G) Mean (±SEM, n = 3–4) polymerization rate during linear elongation stage of polymerization of Ctrl and SIN-1 G-actin (5, 10, or 20 mM, 15 minutes). (H) Mean fluorescence (±SEM, n = 3–4) of Ctrl and SIN-1 (5 mM) G-actin at steady state of G-actin polymerization, 24 and 48 hours after induction of the polymerization. Statistical analysis for C, D, and H was performed by applying 2-tailed Student’s t test. For G, 1-way ANOVA with Holm–Sidak post hoc test was used. A P value less than 0.05 was considered significant. **P < 0.01; ***P < 0.001.
A major component of myofibrils is filamentous actin (F-actin) (29), which is formed by polymerization of monomeric actin (G-actin). The polymerization process results in formation of asymmetric helical actin filaments, each characterized by a fast (barbed) and slow (pointed) polymerizing end (30) and by extensive structural cooperativities coupled to ATP hydrolysis and interaction with filament-binding proteins (30–33). Net assembly of ATP-bound G-actin occurs at barbed ends and is followed by nucleotide hydrolysis and subsequent structural transitions of the newly added subunits (33). At steady-state condition, net dissociation of the ADP-carrying monomers occurs from the pointed end. Consequently, the filament is polymorphic, structurally representing an ensemble of different states (34) and with each actin subunit in contact with 4 other subunits, the structure is highly combinatorial and susceptible to protein modification and various molecular interactions.
To investigate whether oxidative modification of G-actin by SIN-1 affects the polymerization process, G-actin was preincubated with SIN-1 and polymerization was then initiated by addition of salt (MgCl2 + KCl) under standardized conditions in the presence of DTT (Supplemental Figure 2, A and B). We found the rate of polymerization of the oxidatively modified actin to be significantly slower than that of unmodified actin (Figure 2, E and F), which is in agreement with previous reports (35, 36). Moreover, the effect of SIN-1 was dose-dependent, as demonstrated by gradual reduction of both filament formation rate and final steady-state polymerization level (Figure 2, G and H). Hence, the polymerization capacity of the modified actin was drastically compromised by the SIN-1 treatment. Prolonged incubation (48 hours) at room temperature after initiation of polymerization led to extensive decline in the steady-state level of F-actin of samples exposed to SIN-1 compared with control samples (31% and 7%, respectively). This suggests that ADP-actin dissociating from the filament pointed end under these conditions exhibits impaired capacity to undergo nucleotide exchange and be recharged with ATP for another round of polymerization, and/or that filaments assembled from modified actin were inherently more unstable than filaments consisting of unmodified subunits.
The oxidative modifications target 3 distinct regions of the actin molecule.
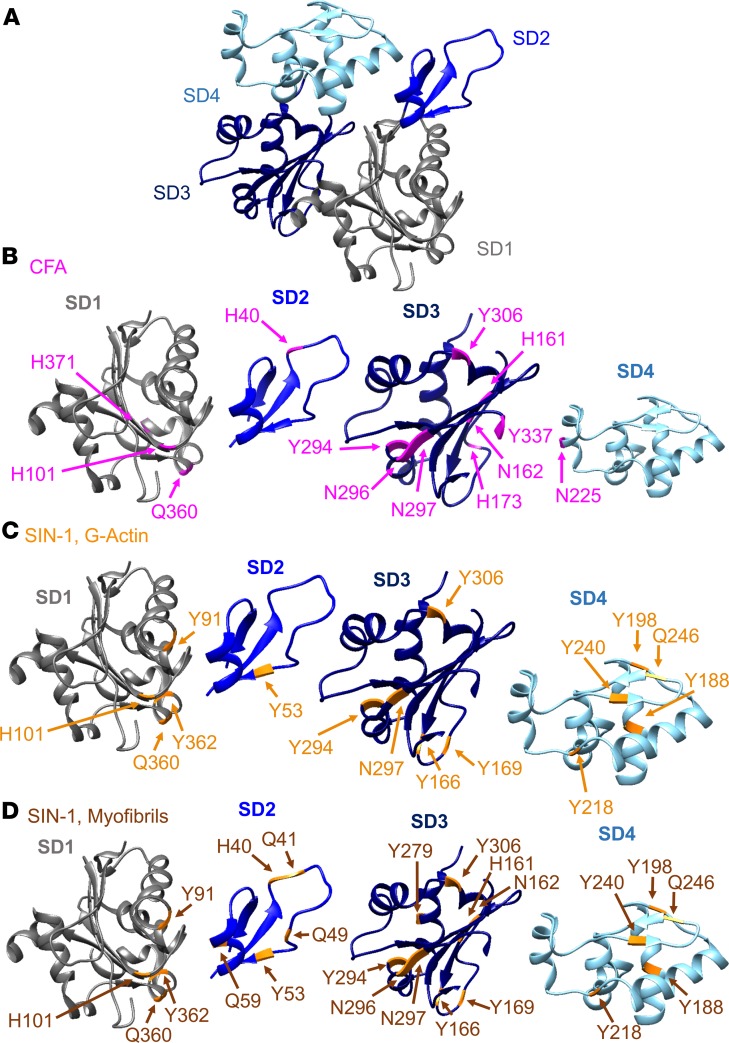
To explore how oxidative modifications of myofibrillar actin and of the isolated protein cause decreased force production and impaired polymerization, MS was used to identify the location and modification of the altered amino acid residues. This approach identified 2 variants of oxidative alterations, 3-NT (NO2; +46 Da) and MDA (C3H3O; +54 Da) consistently occurring in myofibril actin, the purified protein, and in skeletal muscle actin from CFA-treated mice (Supplemental Table 1). Important in this context is that the amino acid sequence of α-actin from mice, rabbits, and humans is identical (Supplemental Table 2). The positions of the 3-NT– and MDA-modified amino acids in the crystal structure of ATP-bound actin (Figure 3A) are visualized in Figure 3, B–D, and all modifications of the 3 different samples are listed in (Supplemental Table 1).
Figure 3. Oxidative 3-NT and MDA modifications on the actin monomer.
(A) Overview of a skeletal muscle α-actin molecule with coloring of subdomain 1 (SD1) in gray, SD2 in light blue, SD3 in dark blue, and SD4 in turquoise, which are kept consistent throughout the figures. (B–D) 3-Nitrotyrosine (3-NT) and malondialdehyde (MDA) oxidative modifications were identified on α-actin with mass spectrometry. SD1–SD4 are shown with the recurrent oxidative modifications highlighted in each subdomain. The actin model is adapted from the 2ZWH crystal structure of the Protein Data Bank Europe (PDBe). Tyrosine residues (Y) were nitrated (3-NT), whereas histidine (H), glutamine (Q), and asparagine (N) were MDA modified. (B) SD1–SD4 of oxidatively modified α-actin residues from mice with arthritis (CFA) (pink, n = 5). (C) Overview of the oxidatively modified amino acids identified in SIN-1–treated (5 mM) G-actin (orange, n = 3) and (D) SIN-1–treated (10 mM) myofibrillar actin (brown, n = 3). The models were generated with UCSF Chimera (82).
Remarkably, a majority of the possible MDA and 3-NT modifications in α-actin from CFA mice and the SIN-1–treated purified protein were either localized to identical positions or in close proximity of each other. The localization similarity suggests that the conserved actin structure exposes distinct regions sensitive to oxidative stress (Figure 4, A–C). The first such “hotspot” was located in subdomain 1 (SD1), where residues H101, Q360, and Y362 underwent MDA, MDA, and 3-NT modifications, respectively (Figure 4A). The second region found to be targeted in all actin samples resides in SD2 (Figure 4B) where H40, Q41, and Y53 went through MDA, MDA, and 3-NT alterations, respectively, which all are likely to affect the flexing of the polypeptide chain proposed to occur upon ATP hydrolysis (37) and actin polymerization (38). Moreover, H40 and Q41 were located in the so-called DNase-binding loop (D-loop), which comprises residues 38–53 and plays a critical role in longitudinal intersubunit contacts in the filament (30, 38) and partakes in the interaction with myosin (39). Consistent with these observations, residue Y53 has previously been shown to be accessible for nitration in vitro by ONOO− (40), and posttranslational modification by phosphorylation of this residue is known to cause slower polymerization and impaired filament stability (41–43).
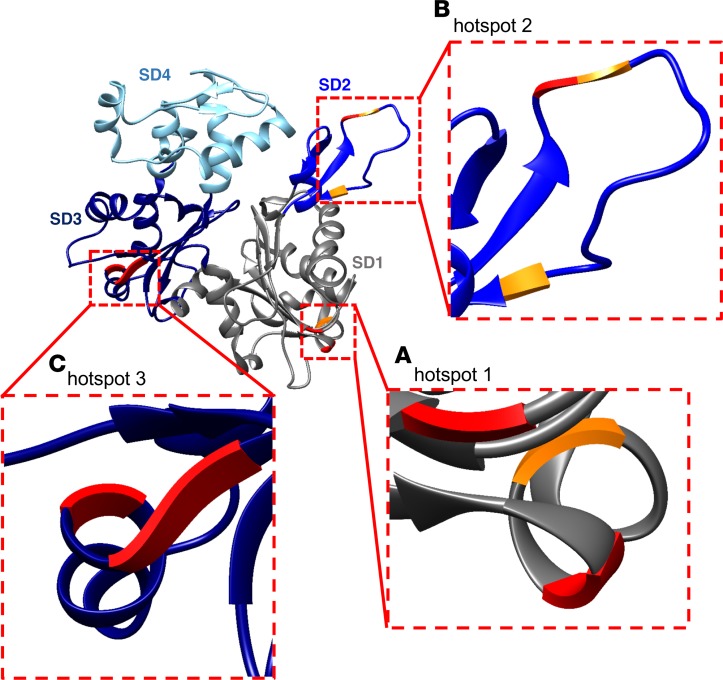
Figure 4. Three oxidative hotspots on the actin monomer.
The actin monomer with the 3 oxidative hotspots identified by mass spectrometry, visualized with zooms. (A) Hotspot 1 in subdomain 1 (SD1) with histidine (H) 101/MDA, glutamine (Q) 360/MDA, and tyrosine (Y) 362/3-NT. (B) Hotspot 2 in SD2 with H40/MDA, Q41/MDA, and Y53/3-NT. (C) Hotspot 3 in SD3 with Y294/3-NT, asparagine (N) 296/MDA, and N297/3-NT. Amino acids in red represent those residues that had the same modification in SIN-1–treated actin and actin from CFA mice (6 out of 9). The models were generated with UCSF Chimera (82).
The third hotspot was localized to Y294, N296, and N297 (modified by 3-NT, MDA, and MDA, respectively) in SD3 (Figure 4C). This was the exact location and kind of oxidative modifications (3-NT and MDA) we found in α-actin from mice with CFA-generated arthritis and SIN-1–treated myofibrillar actin. In addition, the SIN-1–treated G-actin showed modifications in this site (Y294/3-NT and N297/MDA), again pointing to the conserved nature of the actin molecule and its susceptibility to oxidative modification in certain specific regions irrespective of the species origin or preparation of the protein. We conclude that the α-actin molecule has 3 oxidative hotspots formed by a few clustered amino acid residues accessible for MDA and 3-NT modification.
The nonmuscle β/γ-actin isoforms are also expressed in skeletal muscle, but generally are not involved in the contractile machinery (44–46). Prior to MS analysis, actin was digested with trypsin, which created 11 unique peptides when comparing skeletal muscle α-actin (ACTS) and β/γ-actin (ACTB/G). Thus, trypsin digestion enables identification of modifications specific for β/γ-actin in skeletal muscle from CFA mice. Eight modified residues were identified and 6 of them were located in peptides unique for β/γ-actin (Supplemental Table 3). MS analysis of nonmuscle β/γ-actin from the knee joint of CFA mice was also performed. Six modifications (SD1: H87/MDA, Q360/MDA*, and Y362/3-NT*; and in SD3: Y218/3-NT, Y294/3-NT*, and N296/MDA*) were identified. Two of them were exclusive for β/γ-actin and 4 of them (*) matched modified residues found in α-actin (Supplemental Table 3). This shows that 3-NT and MDA modifications are not stringently restricted to α-actin in skeletal muscle from mice with arthritis, but also are present in β/γ-actin in both muscle and nonmuscle tissue.
Human RA patients exhibit muscle weakness and matching oxidative hotspots on actin.
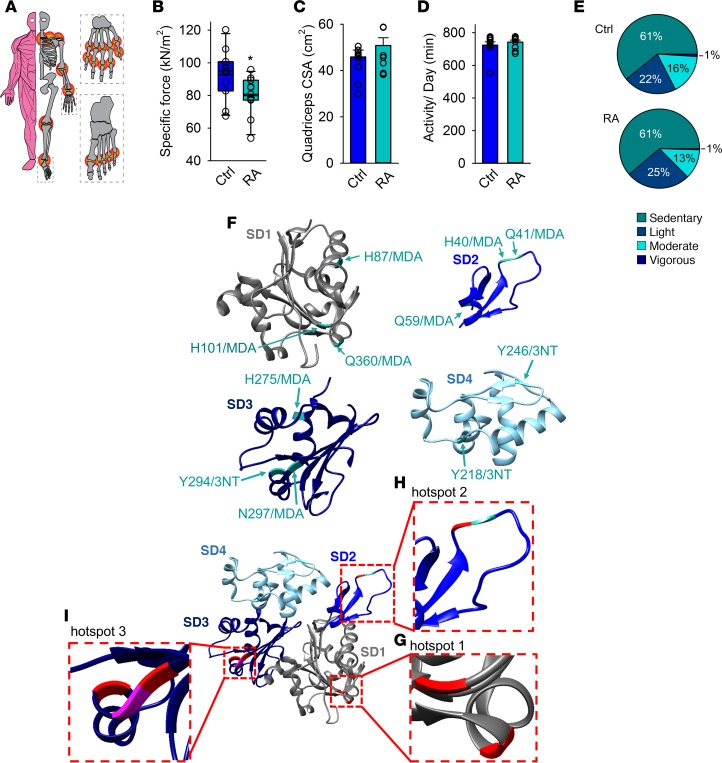
We next investigated if the findings from mice with arthritis were translatable to human patients with RA. Female patients with seropositive RA on stable disease-modifying antirheumatic drug (DMARD) treatment without or in combination with stable low-dose glucocorticoids and displaying a moderate disease activity (average disease activity score [DAS] of 3.3 ± 0.4; n = 11, 53.3 ± 1.5 years old, 8.9 ± 0.4 years disease duration) (Figure 5A and Supplemental Table 4) had significantly weaker leg muscles than age- and weight-matched healthy women (Figure 5B).
Figure 5. Patients with RA exhibit muscle weakness and matching oxidative hotspots on the actin monomer.
(A) An illustration of the composite disease activity score (DAS), with a 44-joint count (red circles) to assess swelling and 53-joint count to assess pain (DAS can range from 0.23 to 9.87). Mean ± SEM DAS was 3.3 ± 0.4 (n = 11, see Supplemental Table 3 for details). (B) Mean ± SEM of isometric specific force of quadriceps femoris from patients with RA and healthy controls (n = 11 per group). (C) Cross-sectional area (CSA) of quadriceps femoris measured by CT scans (mean ± SEM, n = 11). (D) Total daily activity in minutes measured with Actilife accelerometers (mean ± SEM, n = 11) and (E) percentage time spent in each defined activity category. (F) Overview of the oxidative 3-nitrotyrosine (3-NT) and malondialdehyde (MDA) modified amino acids on skeletal muscle actin (SD1–SD4) identified by mass spectrometry in patients with RA (n = 5). The actin model is adapted from the 2ZWH crystal structure of the Protein Data Bank Europe (PDBe). Tyrosine residues (Y) were nitrated (3-NT), whereas histidine (H), glutamine (Q) and asparagine (N) were MDA modified. (G–I) The actin monomer model illustrating that the oxidative hotspots in patients with RA (green) coincide with hotspots in mice with arthritis (pink). Amino acids depicted in red ribbons represent the residues that had the identical modification on actin in RA patients and CFA mice (5 out of 11). Generated with UCSF Chimera (82). Statistical analysis was performed using 2-tailed Student’s t test. A P value less than 0.05 was considered significant. *P < 0.05.
The quadriceps CSA of patients with RA was not significantly different from the quadriceps CSA of healthy individuals (Figure 5C). Further, there were no significant differences in total amount of daily physical activity or the degree of physical intensity (47) performed between patients with RA and healthy control individuals (Figure 5, D and E). Thus, the force decline cannot be explained by a decrease in muscle mass or general muscle inactivity. Instead, the data suggest that the muscle weakness exhibited in RA patients is the result of arthritis-induced intrinsic muscle dysfunction, as observed in mice with arthritis (Figure 1F).
Muscle biopsies from RA patients were analyzed with MS to identify oxidative modifications and revealed 11 consistent oxidative modifications on actin in total (in SD1: H87/MDA, H101/MDA, and Q360/MDA; in SD2: H40/MDA, Q41/MDA, and Q59/MDA; in SD3: H275/MDA, Y294/3-NT, and N297/MDA; and in SD4: Y218/3-NT and Q246/MDA) (Figure 5F). Intriguingly, these modifications were located at matching hotspots, and in some cases even at identical residues in α-actin from mice with arthritis (Figure 5, G–I). Specifically, in hotspot 1: in SD1, H101/MDA and Q360/MDA; in hotspot 2: SD2, H40/MDA; and in hotspot 3: in SD3, Y294/3-NT and N297/MDA were found at precisely the same location in α-actin from mice with arthritis and patients with RA (Figure 5, G–I, red ribbons). The “conserved” location of these α-actin hotspots for oxidative modification reflects the highly conserved nature of all vertebrate actin and illustrates the strength of the mouse model as used above for understanding molecular mechanisms behind human diseases where conserved proteins play a crucial role in the pathogenesis.
Residues important for actin filament formation, stability, and myosin interaction are 3-NT and MDA modified in RA patients.
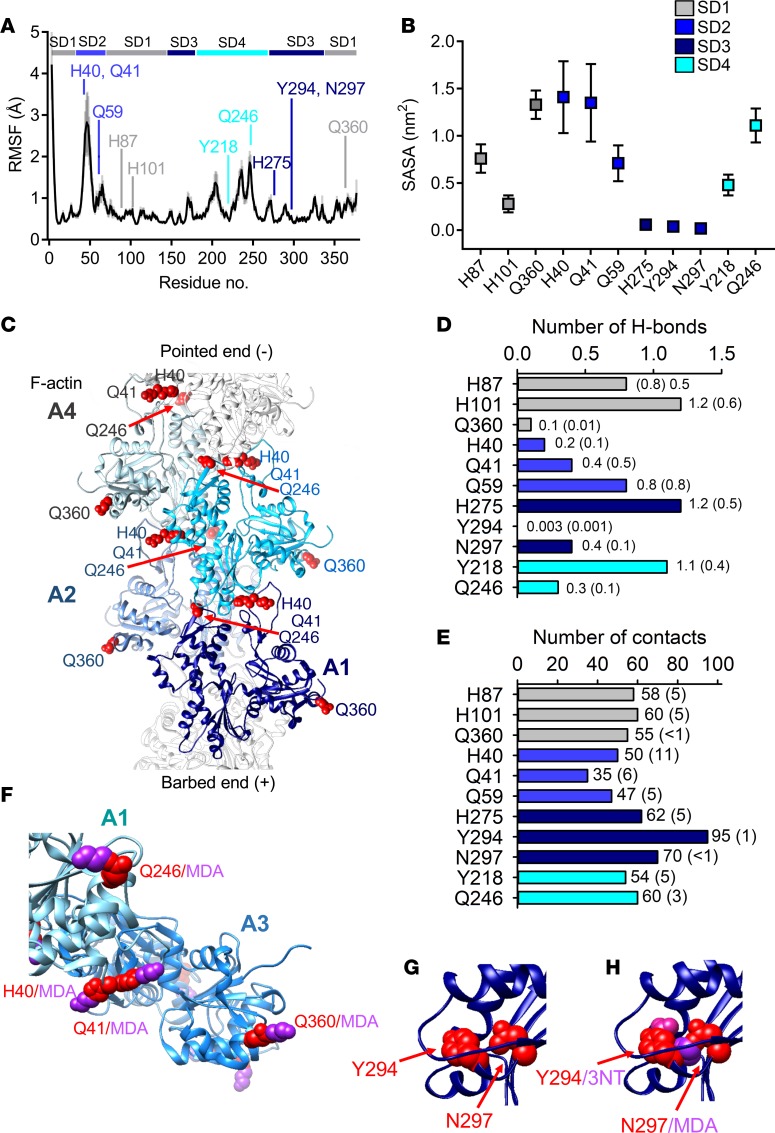
Molecular dynamics simulations (48) of F-actin in the ADP-bound form were carried out to reveal structural details of the polymer at the atomic level, particularly with respect to the modified residues identified from patients with RA. Root mean square fluctuation (RMSF) assesses flexibility of the different domains (Figure 6A); SD1 and SD3 had limited flexibility (except for the residues at the N-terminus); the flexible D-loop region in SD2 showed a large RMSF and holds the second hotspot including the MDA-modified H40 and Q41; SD4 had 2 regions of higher RMSF, including Q246/MDA. The other modified residues were located in less dynamic regions. The simulations of solvent-accessible surface area (SASA) suggest solvent exposure of MDA-modified H40, Q43, Q246, and Q360 (Figure 6B), in accordance with those found to be MDA modified and identified by MS of biopsies from RA patients. Cryo-electron microscopy analyses have highlighted these residues as important for intramolecular bonding (interactions within the actin monomer), filament intersubunit interactions, and for actomyosin interactions during the power stroke and coupled force production (30, 33, 39, 49). Specifically, H40 and Q41 in the D-loop of SD2 are involved in longitudinal bonding with residues in SD1 and SD3 in the adjacent subunit towards the pointed end of the filament, and lateral interactions with SD3 in the neighboring subunit across the filament axes (30, 39). Additionally, residue Q246 in SD4 interacts with residues in the adjacent subunit (SD3) at its pointed end, further contributing to filament stability (Figure 6C).
Figure 6. Altered filament stability, intersubdomain interactions, and myosin interaction with oxidative modifications on actin.
Analysis of molecular dynamics (MD) simulations calculated from trajectories of four 100-ns MD simulations of ATP-bound F-actin. (A) Root mean square fluctuation (RMSF) values of the Cα atoms (black line with gray shadow showing the standard deviation). RMSF values are averages of 2-ns blocks, calculated for the last 60 ns of the simulation time for the four 100-ns simulations. Residues exposed to modifications are pointed out in the sequence by their respective amino acid abbreviation and color coded according to its domain. (B) Solvent-accessible surface area (SASA), with SD shown as error bars. (C) F-actin (PDBe: 5MVA) with 4 G-actin monomers (A1–A4) envisioned in light to dark shade of blue. Histidine (H) 40, glutamine (Q) 41, Q246, and Q360 in red spheres showing their intricate location for inter- and intramolecular bonding. (D) Number of H-bonds and (E) contacts between oxidized hotspot residues and the rest of the actin protein. A contact was defined as when the distance was less than 4 Å between 2 non-hydrogen atoms. Values are averages with error estimates from block averaging in parentheses. (F) A model of a fragment of F-actin (A1–A3) (PDBe: 5H53) with malondialdehyde (MDA) added to the residues. (G and H) Hotspot 2 with and without the presence of modifications on tyrosine (Y) 294 and asparagine (N) 297.
The MDA-modified H275 and N297, and Y294/3-NT are all proximal to a tight electrostatic network with the D-loop in the adjacent subunit formed by H173, D286, and E270 (33). Thus, modification of one or more of these residues could interfere with longitudinal intramolecular bonding. In contrast to the residues discussed above, H275, N297, and Y294 that are all in the less dynamic SD3 were found to be buried throughout the whole simulation (Figure 6B). The biochemistry that makes these residues accessible to chemical modifications despite not being annotated as solvent-accessible remains elusive. However, the local physico-chemical microenvironment in the molecule, including the capacity to propagate tyrosyl radical intermediates and lipid peroxyl radicals, is known to influence the susceptibility of residues to modifications, and hence buried tyrosines can be accessible independently of SASA (13).
Finally, examination of close contacts (hydrogen and non–hydrogen bonds) was performed to further expose what functional effects MDA and 3-NT modifications might impose on the rest of the protein. It was found that H101, H275, and Y218 would lose hydrogen bonds with drastic consequences for their interaction with nearby residues (Figure 6D). Furthermore, each of the 11 residues identified with oxidative modifications in skeletal muscle actin from patients with RA exhibits approximately 35–95 close contacts between non-hydrogen atoms separated by less than 4 Å (Figure 6E), and altering the local chemical environment by the bulky covalent MDA (C3H3O) or 3-NT (NO2) is likely to dramatically impede these contacts by creating local steric restrictions and triggering of conformational changes (Figure 6, F–H). Together, the above results demonstrate the presence of oxidative modifications of residues in skeletal muscle actin from patients with RA, which are critical to actin filament stability and myosin interaction, and therefore represent strong contributing causes to the muscle weakness from which these patients suffer.
Discussion
Impaired muscle capacity has a negative influence on the quality of life of RA patients and is a contributing factor to long-term sick leaves associated with RA and therefore important also from a socioeconomic perspective (1–6, 50, 51). Here, we show that oxidative stress–induced intrinsic muscle dysfunction is a major contributor to muscle weakness associated with RA and is specifically due to modifications of actin with impeding consequences for actin filament formation, stability, and actomyosin force generation. The modified residues are found in discrete regions of the actin molecule, referred to as oxidative hotspots, that provide molecular insights into how oxidative stress promotes skeletal muscle weakness in RA without loss of muscle mass or reduced overall physical activity in the patients.
With 3-NT and MDA oxidative modifications being nonenzymatic mechanisms, the selectivity for the targeted residues on actin in vivo is not evident (13, 52, 53). There is currently no consensus regarding the selectivity of oxidative modifications. However, the local microenvironment and the structural features of the protein are believed to affect the selectivity, i.e., residues located near charged amino acids or on loop structures appear more prone to become modified (13, 52, 54, 55), which harmonizes with our results. Supplemental Figure 3 shows that our hotspots are primarily located in loop regions and that the modifications were all closely located to charged amino acid residues, i.e., to arginine (R), lysine (K), aspartic acid (D), glutamic acid (E), and histidine (H). Due to the short biological half-life of ROS/RNS (e.g., ~5–20 ms for ONOO−, ~1 μs for O2−•), the protein also needs to be in close vicinity of the subcellular sites of ROS/RNS generation for the modification to occur (52, 56). Indeed, NOX2 and the MICAL flavoprotein monooxygenase has been shown to interact with actin and directly affect actin dynamics and cause instability of F-actin, respectively (57–59). These enzymes diffusely release ROS/RNS and the actin isoforms represent only one set of proteins that are modified by these ROS/RNS generators. Thus, the specificity and selectivity of these ROS/RNS sources for particular actin residues remain poorly understood.
In addition to actin, other proteins directly involved in skeletal muscle sarcomere function, such as myosin, tropomyosin, and troponin might be targeted by oxidative stress that may interfere with muscle force production. Nevertheless, in line with previous reports (15, 16, 22, 60), our results emphasize actin as a prime target for intracellular oxidative stress modifications. This appears valid also for nonmuscle actin with 3-NT and MDA modifications found in joints from arthritic mice (Supplemental Table 3). Moreover, specific actin oxidation has been reported in brain regions from patients with Alzheimer’s disease, in skin biopsies from Friedrich’s ataxia patients (61, 62), and in spinal cords from a mouse model of inflammatory pain (63). Moreover, the oxidative modifications identified here were consistently present on α-actin from RA patients on DMARD treatment in combination with or without low doses of glucocorticoids (see Supplemental Table 4). The patients in our study used conventional synthetic DMARDs (mostly methotrexate) with a broad immunosuppressive activity, and biological DMARDs that target a specific protein structure with proinflammatory activity in the attempt to lower the rheumatic disease activity, e.g., rituximab (Mabthera), etanercept (Embrel), abatacept (Orencia), and tocilizumab (RoActerma). This implies that muscle weakness is a comorbidity that cannot be counteracted by DMARDs and glucocorticoids alone and suggests that supplemental pharmacological treatment targeted to improve intrinsic muscle function is needed.
Currently, there is a serious deficit in available clinical therapeutic approaches to counteract muscle weakness. However, the data presented here provide new leads for the development of such targeted treatments. It is well known that ROS/RNS have both beneficial (23, 64–66) and detrimental (25, 67–69) effects on skeletal muscle function. Antioxidant treatment of illnesses related to oxidative stress ought therefore to be directed towards the negatively affected protein functions, instead of having a broad-scoped effect on ROS/RNS signaling, as is common with antioxidant treatment today. Hence, the use of general antioxidants, e.g., vitamin E, vitamin C, and N-acetylcysteine (NAC), which will localize to the intracellular space and interact with surrounding molecules in a nonspecific and uncontrolled manner, is a likely reason for the failure of numerous clinical trials (70–72), including a recent attempt to reduce RA disease activity with NAC (73). Therefore, the aim should be to specifically target proteins identified as being negatively affected by the oxidative stress. In this regard, a SOD2/catalase mimetic (EUK-134) that supposedly targets mitochondria has shown promising effects in the attempt to prevent muscle weakness in rats with arthritis (15).
Here, we have identified a limited set of oxidative hotspots in actin that seriously interfere with its function when it has undergone 3-NT and MDA modifications, 2 common markers of oxidative stress. Thus, to develop actin-targeted compounds with the capacity to reduce the local chemical environment, analogous to targeted synthetic DMARDs, which are designed to attack a particular molecular structure, may be a way to counteract RA-induced muscle weakness. Notably, oxidative stress–induced loss of force production without a reduction in muscle mass has been reported in rodent models of heart failure and cancer, suggesting that intrinsic muscle weakness might be a present comorbidity across several chronic diseases. Moreover, because actin is a highly conserved protein essential to all cells and targeted by oxidative stress in both muscle and nonmuscle tissue, the development of an actin-directed drug has the potential to be of wider use than to improve muscle function in patients suffering from muscle weakness.
Methods
Study approval.
All animal experiments complied with the Swedish Animal Welfare Act, the Swedish Welfare ordinance, and applicable regulations and recommendations from the Swedish authorities. The study was approved by the Stockholm North Ethical Committee on Animal Experiments (N108/13 and N273/15). All human experiments were approved by the regional ethical review board in Stockholm (2014/516-31/2) and complied with the Declaration of Helsinki. Oral and written informed consent were obtained from all patients and control subjects prior to participation in the study.
Animals.
In all studies, mice were handled and euthanized in accordance with approved institutional, national, and international guidelines. All efforts were made to follow the Replacement, Refinement, and Reduction guidelines. Arthritis was induced in female C57BL/6JRj (Janvier) mice by intra-articular injection of CFA (10 mg/ml, Chondrex) in the ankle (5 μl) or in the knee joint (10 μl). Injections were performed on animals anesthetized with isoflurane. Mice were housed at the local animal facility with a 12-hour light/dark cycle, and were provided with standard rodent chow and water ad libitum. Mice were monitored daily to ensure that a sufficient health and nutritional status was maintained. Knee and ankle joint diameters were measured with a caliper every other day. Mice were sacrificed 14 days after injection by cervical dislocation.
Patients with RA and healthy controls.
Eleven women with seropositive RA diagnosis according to the criteria of the American College of Rheumatology (ACR, 1987) participated in the study. All patients were on stable antirheumatic treatment. RA-disease activity was determined by calculating DAS. Detailed information about all patients included in this study can be found in Supplemental Table 4. The control group consisted of 11 age-matched (age range 45–62 years) women without inflammatory disease. All participants were subjected to Biodex force measurements and a CT scan of the legs, and an accelerometer analysis of their daily activity. Biopsies of the quadriceps vastus lateralis muscle were obtained from a subset of the RA patients by the Bergström percutaneous needle biopsy technique (74).
Ex vivo force measurements.
EDL muscles from both legs (arthritic and nonarthritic) were rapidly excised under a microscope after sacrifice, as described previously (75). Muscles were kept in Tyrode solution (temperature was set at 31°C) containing (in mM): 121 NaCl, 5 KCl, 1.8 CaCl2, 0.4 NaH2PO4, 0.5 MgCl2, 24 NaHCO3, 0.1 EDTA, and 5.5 glucose, gassed with 95% O2/5% CO2 to achieve a bath pH of 7.4. The muscles were mounted between a force transducer and an adjustable holder (World Precision Instruments). Muscles were stimulated with supramaximal current pulses (0.5-ms duration; 150% of current required for maximal force response) via plate electrodes lying parallel to the muscles. Muscles were set to the length at which tetanic force was maximum (optimal length L0) and the force-frequency relationship was determined using the following stimulus frequencies: 1, 20, 30, 40, 50, 70, 100, 120, and 150 Hz (300-msec tetanic duration). At least 1 minute of recovery separated electrical stimulations. Electrically stimulated force production was expressed as absolute force (mN) and as specific force (kN/m2). Muscle CSA was assessed by dividing muscle mass by the product of muscle length and muscle density (1.06 g/cm3).
Single intact FDB fibers were obtained by dissection and thereafter mounted in a chamber between a force transducer and an adjustable holder, as described previously (16). Fibers were superfused with Tyrode bubbled with 95% O2/5% CO2 to achieve a bath pH of 7.4. The fiber length was adjusted to obtain maximal tetanic force. The diameter of the fiber at this length was used to calculate the CSA. The fiber was stimulated with supramaximal electrical pulses (0.5-ms in duration) delivered via platinum electrodes placed along the long axis of the fiber. [Ca2+]i was measured with the fluorescent Ca2+ indicator Indo-1 (Thermo Fisher Scientific). The mean fluorescence of Indo-1 at rest and during tetanic contractions was measured and converted to [Ca2+]i using an intracellularly established calibration curve (76).
Immunoblots.
Muscles (gastrocnemius, tibialis anterior, and interosseous) were homogenized in buffer containing (in mM): 10 Tris-maleate, 100 KCl, 2 MgCl2, 2 EGTA, 2 Na4P2O7, 1 NaVO4, 25 KF, and protease inhibitor (Roche, 1 tablet/50 ml). The lysates were cleared by 10-minute centrifugation at 1000 g (4°C). Equal amounts of protein were separated by electrophoresis and transferred onto membranes (Immobilon-FL, Millipore). Membranes were incubated with primary antibody (anti–3-NT, ab52309, Abcam; anti-MDA, ab2764-2, Abcam) and then washed and incubated with infrared-labeled secondary antibody (IRDye 680 and IRDye 800 LI-COR Biosciences). Immunoreactive bands were analyzed with the LI-COR Odyssey Infrared Imaging System and normalized against total protein or actin.
Myofibrillar force measured with atomic force cantilevers.
Myofibrils were isolated from rabbit psoas muscle as described previously (16, 77) and suspended in rigor buffer (10 mM imidazole, 100 mM KCl, 2 mM MgCl2, 1 mM EGTA, pH 7.2) with DTT (1 mM). Myofibrils were incubated with SIN-1 (10 mM, 10 minutes) and transferred to the experimental chamber placed on the top of an inverted microscope equipped with a system of atomic force cantilevers. The chamber was washed with rigor buffer followed by relaxing buffer (pH 7, pCa2+ 9; 20 mM imidazole, 7.2 mM EGTA, 13.74 μM CaCl2, 5.4 mM MgCl2, 68.7 mM KCl, 5.6 mM ATP, 14.4 mM creatine phosphate). An atomic force cantilever with a known stiffness was attached to one end of a myofibril and a glass needle to the other end of the myofibril acted as a force transducer. The length of the myofibril sarcomere was adjusted to 2.8 μm and contraction was induced by addition of activating buffer with pCa2+ 4.5 (20 mM imidazole, 7.2 mM EGTA, 7 mM CaCl2, 5.4 mM MgCl2, 52.3 mM KCl, 5.7 mM ATP, 14.4 mM creatine phosphate, pH 7). The deflection of the cantilever due to myofibrillar contraction was measured by shining a laser light on the cantilever, which then was translated to force in kN/μm2, as described previously (16, 77).
Actin polymerization assay.
Actin polymerization was measured using the pyrenyl assay (78). G-actin (Cytoskeleton, AKL99) in 5 mM Tris-HCl (pH 7.6 at room temperature) containing 0.1 mM CaCl2, 0.5 mM ATP, and 0.5 mM DTT (G-buffer) was incubated with SIN-1 at indicated concentrations for 15 minutes at room temperature. Actin concentration was determined by absorbance measurement at 290 nm using E = 0.63 ml/mg × cm as described by Houk and Ue (79). Ten minutes prior to initiation of polymerization, EGTA and MgCl2 were added to final concentrations of 0.2 mM and 50 μM, respectively, to replace actin-bound Ca2+ with Mg2+ (80). Polymerization was induced by addition of MgCl2 and KCl to 1 and 100 mM, respectively (final concentrations). The assay was performed in 96-well plates with 8 μM actin in the presence of 4% pyrene-actin and in a final volume of 130 μl (accomplished by pre-addition of G-buffer). The process was followed by excitation at 365 nm and emission detection at 410 nm (Fluoroskan II, Labsystems). Steady-state fluorescence was measured 24 and 48 hours after polymerization initiation.
MS analysis.
MS was performed on purified rabbit skeletal muscle actin (Cytoskeleton, AKL99), isolated myofibrillar actin from rabbit psoas muscle (myofibrillar actin), actin from CFA mice, and actin from patients with RA (human RA actin). Samples were digested by trypsin (overnight), cleaned on a C-18 HyperSep plate, and dried in a speedvac centrifugal evaporator. The peptides were then separated by chromatography in a C18 column (Silica Tip 360 μm OD, 75 μm ID, New Objective) connected to the UltiMate 3000 RSLCnano chromatography system (Thermo Fisher Scientific). Peptides ionized by electrospray were injected into an Orbitrap QExactive Plus mass spectrometer (Thermo Fisher Scientific). The survey MS spectrum was acquired at the resolution of 60.000 in the range of m/z 200–2,000.
MS/MS data were extracted with RawtoMGF software and analyzed using Mascot (Matrix Science, version 2.5.1) and X!Tandem (thegpm.org; version CYCLONE 2010.12.01.1) software. Fragment ion mass tolerance and parent ion tolerance were set to 0.020 Da and 9.0 PPM, respectively. Variable modifications of amino acids were specified as Tyr-nitration, Cys-nitrosylation, and MDA modifications, with fixed modifications formed by the sample preparation specified as Cys-carbamidomethylations. Carbonylation (DNP) could not be detected as it requires a targeted approach with labeled reaction sites. Validation of the MS/MS-based peptide and protein identifications was performed in Scaffold (version Scaffold_4.4.5, Proteome Software Inc.) and protein probabilities were assigned by the protein prophet algorithm (81). All posttranslational modifications were validate and localized by the Scaffold PTM software. The UCSF Chimera program (82) version 1.1rc was used to visualize the posttranslational modifications identified with MS. The monomeric G-actin model in its ATP-bound state was obtained from the 2ZWH (RSCS Protein Databank) protein crystal structure. All modifications are presented with their 1-letter code and specifier. The homology between the skeletal muscle actin sequences was confirmed with ClustalW (83) sequence alignment (EMBL-EBI).
Human accelerometer and force measurements.
The daily activity of patients with RA and control subjects was recorded with accelerometers (GT, ActiGraph). Accelerometer data were collected from the subjects over 7 consecutive days and analyzed with Actilife software. Total daily activity was calculated by dividing the total minutes of activity by the validated wearing period (e.g., >90 minutes of consecutive inactivity were classified as non-worn periods and were filtered out from data analysis by using a previously defined algorithm) (81). The degree of activity was categorized as sedentary, light, moderate, or vigorous (47). A Biodex System 4 Pro (Biodex Medical Systems Inc.) was used to measure isometric force from quadriceps femoris of RA patients and healthy control persons. The subjects were carefully positioned in the Biodex chair and performed 3 repetitions of maximal isometric contractions (at knee angle 120°). The specific force (kN/m2) was calculated by converting the peak torque (Nm) into kN by using the defined length of the cantilever in which the leg was fixed, which was then divided by the CSA of the quadriceps muscle as determined by a CT scan of the leg. ImageJ (NIH) was used to measure CSA of the quadriceps at the intermediate of the femur bone.
Molecular dynamics simulations.
The 3.3-Å resolution (PDBe: 2ZWH) structure of rabbit skeletal muscle F-actin with ADP and Ca2+ was used as starting structure for the molecular dynamics simulations. This structure includes all amino acid residues and has previously been used in molecular dynamics–simulation studies (84–86). Simulations and analysis were performed by the Gromacs simulation package (87) version 5.1.2. The actin structure was solvated in a truncated dodecahedron box so that the minimal distance between any protein atom and the edge of the box was 1.0 nm and the TIP3P (88) water model was applied. Fifty-two K+ and 39 Cl− ions were added to neutralize the protein and mimic an ionic strength of 0.15 M. The CHARMM36 force field (89) was applied for all atoms except Ca2+, K+, and Cl− for which force-field parameters were obtained from previous studies (90–92). Prior to the simulations, internal constraints were relaxed by energy minimization until the maximal force on individual atoms was smaller than 100 kJ/mol/nm. The system was equilibrated for 5 ns before data collection for analysis. During the molecular dynamics runs, the LINCS (93) algorithm was used to constrain the lengths of bonds. The temperature was kept constant at 300K by use of the velocity-rescaling thermostat (94) (τT = 0.1 ps). The pressure was coupled to an external bath with the Parrinello-Rahman algorithm (95) (Pref = 1 bar, τP = 1 ps). Van der Waals forces were truncated at 1.0 nm with a plain cutoff and long-range dispersion corrections were applied for the energy and pressure. Long-range electrostatic forces were treated using the particle mesh Ewald method (96, 97). Four replicates were run for 100 ns each. Analysis of the simulations was carried out by automatic tools available in Gromacs. Error estimate is a block averaging method and the error estimate is the variance between averages of all blocks. RMSF of the Cα atoms were calculated as averages of 2-ns blocks for the last 60 ns of the simulation time for the four 100-ns simulations.
Statistics.
The data are presented as mean ± SEM. For all statistical analysis the normal distribution was tested with a Shapiro-Wilk test and equal variance was tested with Brown-Forsythe test. In case of no equal variance a Mann-Whitney rank-sum test was applied. Differences among experimental groups were analyzed by 2-tailed Student’s t test or analysis of variance (ANOVA) with appropriate post hoc and multiple-comparisons tests. P values less than 0.05 were considered significant (*P < 0.05; **P < 0.01; ***P < 0.001). All statistical analysis was performed with help of Sigma plot 13.0 (Systat Software) or Prism 7.0 (GraphPad).
Author contributions
CIS, NB, RK, TY, BCGK, RF, PT, TG, and JTL designed and conceptualized the research. MMS, MP, BA, KO, AJC, EA, ML, TRL, ER, KÄM, KS, SA, BCGK, and EK performed research. MMS, MP, AJC, EA, TRL, ER, PT, DER, RK, RF, TG, and JTL analyzed data. MMS, KÄM, KS, and CIS provided the animals. MMS, MP, AJC, EA, TRL, ER, PT, DER, RK, RF, TG, and JTL performed statistical analyses and power calculations. MMS, RK, and JTL wrote the original draft of the manuscript. All authors reviewed and edited the manuscript.
Supplementary Material
Acknowledgments
The authors thank Akos Vegvari, Alexey Chernobrovkin, and coworkers at the Proteomics Biomedicum at Karolinska Institutet for assistance with the mass spectrometry analyses. The study has been supported by The Swedish Research Council (to JTL), Reumatikerfonden (to JTL), KI grant for rheumatology research (to EK), Knut and Alice Wallenberg Foundation (to CIS), and the Carl Trygger Foundation (to RK) for studies of actin polymerization. The computations were performed on resources provided by SNIC through LUNARC, the center for scientific and technical computing at Lund University under Project SNIC 2018/3-47.
Version 1. 03/28/2019
In-Press Preview
Version 2. 05/02/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(9):e126347. https://doi.org/10.1172/jci.insight.126347.
Contributor Information
Maarten M. Steinz, Email: maarten.steinz@ki.se.
Malin Persson, Email: malincharlottpersson@gmail.com.
Bejan Aresh, Email: bejan.aresh@su.se.
Karl Olsson, Email: karl.olsson@ki.se.
Arthur J. Cheng, Email: ajcheng@yorku.ca.
Emma Ahlstrand, Email: emma.ahlstrand@hotmail.se.
Mats Lilja, Email: mats.lilja.1@ki.se.
Tommy R. Lundberg, Email: tommy.lundberg@ki.se.
Eric Rullman, Email: eric.rullman@ki.se.
Kristina Ängeby Möller, Email: kristina.angeby.moller@ki.se.
Katalin Sandor, Email: katalin.sandor@ki.se.
Sofia Ajeganova, Email: sofia.ajeganova@ki.se.
Takashi Yamada, Email: takashi.yamada1976@sapmed.ac.jp.
Nicole Beard, Email: Nicole.Beard@canberra.edu.au.
Björn C.G. Karlsson, Email: bjorn.karlsson@lnu.se.
Pasi Tavi, Email: pasi.tavi@uef.fi.
Ellinor Kenne, Email: ellinor.kenne@ki.se.
Camilla I. Svensson, Email: camilla.svensson@ki.se.
Dilson E. Rassier, Email: dilson.rassier@mcgill.ca.
Roger Karlsson, Email: roger.karlsson@su.se.
Ran Friedman, Email: ran.friedman@lnu.se.
Thomas Gustafsson, Email: Thomas.Gustafsson@ki.se.
Johanna T. Lanner, Email: johanna.lanner@ki.se.
References
- 1.van Vilsteren M, et al. Productivity at work and quality of life in patients with rheumatoid arthritis. BMC Musculoskelet Disord. 2015;16:107. doi: 10.1186/s12891-015-0562-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fraser A, Vallow J, Preston A, Cooper RG. Predicting ‘normal’ grip strength for rheumatoid arthritis patients. Rheumatology (Oxford) 1999;38(6):521–528. doi: 10.1093/rheumatology/38.6.521. [DOI] [PubMed] [Google Scholar]
- 3.Sokka T, et al. Physical inactivity in patients with rheumatoid arthritis: data from twenty-one countries in a cross-sectional, international study. Arthritis Rheum. 2008;59(1):42–50. doi: 10.1002/art.23255. [DOI] [PubMed] [Google Scholar]
- 4.Helliwell PS, Jackson S. Relationship between weakness and muscle wasting in rheumatoid arthritis. Ann Rheum Dis. 1994;53(11):726–728. doi: 10.1136/ard.53.11.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Palma L, Chillemi C, Albanelli S, Rapali S, Bertoni-Freddari C. Muscle involvement in rheumatoid arthritis: an ultrastructural study. Ultrastruct Pathol. 2000;24(3):151–156. doi: 10.1080/01913120050132886. [DOI] [PubMed] [Google Scholar]
- 6.Lemmey AB, et al. Tight control of disease activity fails to improve body composition or physical function in rheumatoid arthritis patients. Rheumatology (Oxford) 2016;55(10):1736–1745. doi: 10.1093/rheumatology/kew243. [DOI] [PubMed] [Google Scholar]
- 7.Grönwall C, et al. Autoreactivity to malondialdehyde-modifications in rheumatoid arthritis is linked to disease activity and synovial pathogenesis. J Autoimmun. 2017;84:29–45. doi: 10.1016/j.jaut.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Hassan SZ, Gheita TA, Kenawy SA, Fahim AT, El-Sorougy IM, Abdou MS. Oxidative stress in systemic lupus erythematosus and rheumatoid arthritis patients: relationship to disease manifestations and activity. Int J Rheum Dis. 2011;14(4):325–331. doi: 10.1111/j.1756-185X.2011.01630.x. [DOI] [PubMed] [Google Scholar]
- 9.Wang G, Pierangeli SS, Papalardo E, Ansari GA, Khan MF. Markers of oxidative and nitrosative stress in systemic lupus erythematosus: correlation with disease activity. Arthritis Rheum. 2010;62(7):2064–2072. doi: 10.1002/art.27442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mishra R, et al. A comparative analysis of serological parameters and oxidative stress in osteoarthritis and rheumatoid arthritis. Rheumatol Int. 2012;32(8):2377–2382. doi: 10.1007/s00296-011-1964-1. [DOI] [PubMed] [Google Scholar]
- 11.Nakajima A, et al. Identification of clinical parameters associated with serum oxidative stress in patients with rheumatoid arthritis. Mod Rheumatol. 2014;24(6):926–930. doi: 10.3109/14397595.2014.891495. [DOI] [PubMed] [Google Scholar]
- 12.Kaur H, Halliwell B. Evidence for nitric oxide-mediated oxidative damage in chronic inflammation. Nitrotyrosine in serum and synovial fluid from rheumatoid patients. FEBS Lett. 1994;350(1):9–12. doi: 10.1016/0014-5793(94)00722-5. [DOI] [PubMed] [Google Scholar]
- 13.Radi R. Protein tyrosine nitration: biochemical mechanisms and structural basis of functional effects. Acc Chem Res. 2013;46(2):550–559. doi: 10.1021/ar300234c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ayala A, Muñoz MF, Argüelles S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid Med Cell Longev. 2014;2014:360438. doi: 10.1155/2014/360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamada T, et al. Muscle dysfunction associated with adjuvant-induced arthritis is prevented by antioxidant treatment. Skelet Muscle. 2015;5:20. doi: 10.1186/s13395-015-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamada T, et al. Nitrosative modifications of the Ca2+ release complex and actin underlie arthritis-induced muscle weakness. Ann Rheum Dis. 2015;74(10):1907–1914. doi: 10.1136/annrheumdis-2013-205007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wadley AJ, et al. Three months of moderate-intensity exercise reduced plasma 3-nitrotyrosine in rheumatoid arthritis patients. Eur J Appl Physiol. 2014;114(7):1483–1492. doi: 10.1007/s00421-014-2877-y. [DOI] [PubMed] [Google Scholar]
- 18.Yamada T, Steinz MM, Kenne E, Lanner JT. Muscle weakness in rheumatoid arthritis: the role of Ca. EBioMedicine. 2017;23:12–19. doi: 10.1016/j.ebiom.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blaser H, Dostert C, Mak TW, Brenner D. TNF and ROS crosstalk in inflammation. Trends Cell Biol. 2016;26(4):249–261. doi: 10.1016/j.tcb.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Cangemi R, et al. Role of platelets in NOX2 activation mediated by TNFα in heart failure. Intern Emerg Med. 2014;9(2):179–185. doi: 10.1007/s11739-012-0837-2. [DOI] [PubMed] [Google Scholar]
- 21.Fischer R, Maier O. Interrelation of oxidative stress and inflammation in neurodegenerative disease: role of TNF. Oxid Med Cell Longev. 2015;2015:610813. doi: 10.1155/2015/610813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowen TS, et al. Diaphragm muscle weakness in mice is early-onset post-myocardial infarction and associated with elevated protein oxidation. J Appl Physiol. 2015;118(1):11–19. doi: 10.1152/japplphysiol.00756.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng AJ, Yamada T, Rassier DE, Andersson DC, Westerblad H, Lanner JT. Reactive oxygen/nitrogen species and contractile function in skeletal muscle during fatigue and recovery. J Physiol (Lond) 2016;594(18):5149–5160. doi: 10.1113/JP270650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allen DG, Westerblad H. The effects of caffeine on intracellular calcium, force and the rate of relaxation of mouse skeletal muscle. J Physiol (Lond) 1995;487(Pt 2):331–342. doi: 10.1113/jphysiol.1995.sp020883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qaisar R, et al. Oxidative stress-induced dysregulation of excitation-contraction coupling contributes to muscle weakness. J Cachexia Sarcopenia Muscle. 2018;9(5):1003–1017. doi: 10.1002/jcsm.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waning DL, et al. Excess TGF-β mediates muscle weakness associated with bone metastases in mice. Nat Med. 2015;21(11):1262–1271. doi: 10.1038/nm.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamada T, et al. Electrical stimulation prevents preferential skeletal muscle myosin loss in steroid-denervation rats. Front Physiol. 2018;9:1111. doi: 10.3389/fphys.2018.01111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin-Romero FJ, Gutiérrez-Martin Y, Henao F, Gutiérrez-Merino C. Fluorescence measurements of steady state peroxynitrite production upon SIN-1 decomposition: NADH versus dihydrodichlorofluorescein and dihydrorhodamine 123. J Fluoresc. 2004;14(1):17–23. doi: 10.1023/B:JOFL.0000014655.89256.bd. [DOI] [PubMed] [Google Scholar]
- 29.Frontera WR, Ochala J. Skeletal muscle: a brief review of structure and function. Calcif Tissue Int. 2015;96(3):183–195. doi: 10.1007/s00223-014-9915-y. [DOI] [PubMed] [Google Scholar]
- 30.Dominguez R, Holmes KC. Actin structure and function. Annu Rev Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehrer SS, Geeves MA. The muscle thin filament as a classical cooperative/allosteric regulatory system. J Mol Biol. 1998;277(5):1081–1089. doi: 10.1006/jmbi.1998.1654. [DOI] [PubMed] [Google Scholar]
- 32.Schüler H. ATPase activity and conformational changes in the regulation of actin. Biochim Biophys Acta. 2001;1549(2):137–147. doi: 10.1016/S0167-4838(01)00255-2. [DOI] [PubMed] [Google Scholar]
- 33.Merino F, et al. Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM. Nat Struct Mol Biol. 2018;25(6):528–537. doi: 10.1038/s41594-018-0074-0. [DOI] [PubMed] [Google Scholar]
- 34.Galkin VE, Orlova A, Schröder GF, Egelman EH. Structural polymorphism in F-actin. Nat Struct Mol Biol. 2010;17(11):1318–1323. doi: 10.1038/nsmb.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clements MK, et al. Inhibition of actin polymerization by peroxynitrite modulates neutrophil functional responses. J Leukoc Biol. 2003;73(3):344–355. doi: 10.1189/jlb.0802401. [DOI] [PubMed] [Google Scholar]
- 36.DalleDonne I, Milzani A, Colombo R. H2O2-treated actin: assembly and polymer interactions with cross-linking proteins. Biophys J. 1995;69(6):2710–2719. doi: 10.1016/S0006-3495(95)80142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graceffa P, Dominguez R. Crystal structure of monomeric actin in the ATP state. Structural basis of nucleotide-dependent actin dynamics. J Biol Chem. 2003;278(36):34172–34180. doi: 10.1074/jbc.M303689200. [DOI] [PubMed] [Google Scholar]
- 38.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467(7316):724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 39.Fujii T, Namba K. Structure of actomyosin rigour complex at 5.2 Å resolution and insights into the ATPase cycle mechanism. Nat Commun. 2017;8:13969. doi: 10.1038/ncomms13969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aslan M, et al. Nitric oxide-dependent generation of reactive species in sickle cell disease. Actin tyrosine induces defective cytoskeletal polymerization. J Biol Chem. 2003;278(6):4194–4204. doi: 10.1074/jbc.M208916200. [DOI] [PubMed] [Google Scholar]
- 41.Baek K, Liu X, Ferron F, Shu S, Korn ED, Dominguez R. Modulation of actin structure and function by phosphorylation of Tyr-53 and profilin binding. Proc Natl Acad Sci USA. 2008;105(33):11748–11753. doi: 10.1073/pnas.0805852105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X, Shu S, Hong MS, Yu B, Korn ED. Mutation of actin Tyr-53 alters the conformations of the DNase I-binding loop and the nucleotide-binding cleft. J Biol Chem. 2010;285(13):9729–9739. doi: 10.1074/jbc.M109.073452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X, Shu S, Hong MS, Levine RL, Korn ED. Phosphorylation of actin Tyr-53 inhibits filament nucleation and elongation and destabilizes filaments. Proc Natl Acad Sci USA. 2006;103(37):13694–13699. doi: 10.1073/pnas.0606321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hájková L, Nyman T, Lindberg U, Karlsson R. Effects of cross-linked profilin:beta/gamma-actin on the dynamics of the microfilament system in cultured cells. Exp Cell Res. 2000;256(1):112–121. doi: 10.1006/excr.1999.4786. [DOI] [PubMed] [Google Scholar]
- 45.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112(4):453–465. doi: 10.1016/S0092-8674(03)00120-X. [DOI] [PubMed] [Google Scholar]
- 46.Gunning PW, Ghoshdastider U, Whitaker S, Popp D, Robinson RC. The evolution of compositionally and functionally distinct actin filaments. J Cell Sci. 2015;128(11):2009–2019. doi: 10.1242/jcs.165563. [DOI] [PubMed] [Google Scholar]
- 47.Freedson PS, Melanson E, Sirard J. Calibration of the Computer Science and Applications, Inc. accelerometer. Med Sci Sports Exerc. 1998;30(5):777–781. doi: 10.1097/00005768-199805000-00021. [DOI] [PubMed] [Google Scholar]
- 48.Friedman R, Boye K, Flatmark K. Molecular modelling and simulations in cancer research. Biochim Biophys Acta. 2013;1836(1):1–14. doi: 10.1016/j.bbcan.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 49.Behrmann E, Müller M, Penczek PA, Mannherz HG, Manstein DJ, Raunser S. Structure of the rigor actin-tropomyosin-myosin complex. Cell. 2012;150(2):327–338. doi: 10.1016/j.cell.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olofsson T, et al. Predictors of work disability during the first 3 years after diagnosis in a national rheumatoid arthritis inception cohort. Ann Rheum Dis. 2014;73(5):845–853. doi: 10.1136/annrheumdis-2012-202911. [DOI] [PubMed] [Google Scholar]
- 51.Burton W, Morrison A, Maclean R, Ruderman E. Systematic review of studies of productivity loss due to rheumatoid arthritis. Occup Med (Lond) 2006;56(1):18–27. doi: 10.1093/occmed/kqi171. [DOI] [PubMed] [Google Scholar]
- 52.Bayden AS, Yakovlev VA, Graves PR, Mikkelsen RB, Kellogg GE. Factors influencing protein tyrosine nitration--structure-based predictive models. Free Radic Biol Med. 2011;50(6):749–762. doi: 10.1016/j.freeradbiomed.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Terman JR, Kashina A. Post-translational modification and regulation of actin. Curr Opin Cell Biol. 2013;25(1):30–38. doi: 10.1016/j.ceb.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Souza JM, Daikhin E, Yudkoff M, Raman CS, Ischiropoulos H. Factors determining the selectivity of protein tyrosine nitration. Arch Biochem Biophys. 1999;371(2):169–178. doi: 10.1006/abbi.1999.1480. [DOI] [PubMed] [Google Scholar]
- 55.Ishii T, et al. Site-specific modification of positively-charged surfaces on human serum albumin by malondialdehyde. Biochem Biophys Res Commun. 2008;371(1):28–32. doi: 10.1016/j.bbrc.2008.03.140. [DOI] [PubMed] [Google Scholar]
- 56.Heijnen HF, et al. Subcellular localization of tyrosine-nitrated proteins is dictated by reactive oxygen species generating enzymes and by proximity to nitric oxide synthase. Free Radic Biol Med. 2006;40(11):1903–1913. doi: 10.1016/j.freeradbiomed.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 57.Tamura M, Itoh K, Akita H, Takano K, Oku S. Identification of an actin-binding site in p47phox an organizer protein of NADPH oxidase. FEBS Lett. 2006;580(1):261–267. doi: 10.1016/j.febslet.2005.11.080. [DOI] [PubMed] [Google Scholar]
- 58.Munnamalai V, et al. Bidirectional interactions between NOX2-type NADPH oxidase and the F-actin cytoskeleton in neuronal growth cones. J Neurochem. 2014;130(4):526–540. doi: 10.1111/jnc.12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hung RJ, Pak CW, Terman JR. Direct redox regulation of F-actin assembly and disassembly by Mical. Science. 2011;334(6063):1710–1713. doi: 10.1126/science.1211956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Canton M, Menazza S, Sheeran FL, Polverino de Laureto P, Di Lisa F, Pepe S. Oxidation of myofibrillar proteins in human heart failure. J Am Coll Cardiol. 2011;57(3):300–309. doi: 10.1016/j.jacc.2010.06.058. [DOI] [PubMed] [Google Scholar]
- 61.Pastore A, et al. Actin glutathionylation increases in fibroblasts of patients with Friedreich’s ataxia: a potential role in the pathogenesis of the disease. J Biol Chem. 2003;278(43):42588–42595. doi: 10.1074/jbc.M301872200. [DOI] [PubMed] [Google Scholar]
- 62.Aksenov MY, Aksenova MV, Butterfield DA, Geddes JW, Markesbery WR. Protein oxidation in the brain in Alzheimer’s disease. Neuroscience. 2001;103(2):373–383. doi: 10.1016/S0306-4522(00)00580-7. [DOI] [PubMed] [Google Scholar]
- 63.Lu J, Katano T, Uta D, Furue H, Ito S. Rapid S-nitrosylation of actin by NO-generating donors and in inflammatory pain model mice. Mol Pain. 2011;7:101. doi: 10.1186/1744-8069-7-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ristow M, et al. Antioxidants prevent health-promoting effects of physical exercise in humans. Proc Natl Acad Sci USA. 2009;106(21):8665–8670. doi: 10.1073/pnas.0903485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paulsen G, et al. Vitamin C and E supplementation hampers cellular adaptation to endurance training in humans: a double-blind, randomised, controlled trial. J Physiol (Lond) 2014;592(8):1887–1901. doi: 10.1113/jphysiol.2013.267419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gomez-Cabrera MC, et al. Oral administration of vitamin C decreases muscle mitochondrial biogenesis and hampers training-induced adaptations in endurance performance. Am J Clin Nutr. 2008;87(1):142–149. doi: 10.1093/ajcn/87.1.142. [DOI] [PubMed] [Google Scholar]
- 67.Lanner JT, et al. AICAR prevents heat-induced sudden death in RyR1 mutant mice independent of AMPK activation. Nat Med. 2012;18(2):244–251. doi: 10.1038/nm.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khairallah RJ, et al. Microtubules underlie dysfunction in Duchenne muscular dystrophy. Sci Signal. 2012;5(236):ra56. doi: 10.1126/scisignal.2002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sakellariou GK, et al. Mitochondrial ROS regulate oxidative damage and mitophagy but not age-related muscle fiber atrophy. Sci Rep. 2016;6:33944. doi: 10.1038/srep33944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Steinhubl SR. Why have antioxidants failed in clinical trials? Am J Cardiol. 2008;101(10A):14D–19D. doi: 10.1016/j.amjcard.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 71.Bjelakovic G, Nikolova D, Gluud LL, Simonetti RG, Gluud C. Mortality in randomized trials of antioxidant supplements for primary and secondary prevention: systematic review and meta-analysis. JAMA. 2007;297(8):842–857. doi: 10.1001/jama.297.8.842. [DOI] [PubMed] [Google Scholar]
- 72.Bjelakovic G, Nikolova D, Gluud LL, Simonetti RG, Gluud C. Antioxidant supplements for prevention of mortality in healthy participants and patients with various diseases. Cochrane Database Syst Rev. 2012;(3):CD007176. doi: 10.1002/14651858.CD007176.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Batooei M, et al. Evaluating the effect of oral N-acetylcysteine as an adjuvant treatment on clinical outcomes of patients with rheumatoid arthritis: a randomized, double blind clinical trial. Rev Recent Clin Trials. 2018;13(2):132–138. doi: 10.2174/1574887113666180307151937. [DOI] [PubMed] [Google Scholar]
- 74.Bergström J, Hultman E. A study of the glycogen metabolism during exercise in man. Scand J Clin Lab Invest. 1967;19(3):218–228. doi: 10.3109/00365516709090629. [DOI] [PubMed] [Google Scholar]
- 75.Chaillou T, McPeek A, Lanner JT. Docetaxel does not impair skeletal muscle force production in a murine model of cancer chemotherapy. Physiol Rep. 2017;5(11):null. doi: 10.14814/phy2.13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng AJ, Westerblad H. Mechanical isolation, and measurement of force and myoplasmic free [Ca2+] in fully intact single skeletal muscle fibers. Nat Protoc. 2017;12(9):1763–1776. doi: 10.1038/nprot.2017.056. [DOI] [PubMed] [Google Scholar]
- 77.Rassier DE. Pre-power stroke cross bridges contribute to force during stretch of skeletal muscle myofibrils. Proc Biol Sci. 2008;275(1651):2577–2586. doi: 10.1098/rspb.2008.0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kouyama T, Mihashi K. Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur J Biochem. 1981;114(1):33–38. [PubMed] [Google Scholar]
- 79.Houk TW, Ue K. The measurement of actin concentration in solution: a comparison of methods. Anal Biochem. 1974;62(1):66–74. doi: 10.1016/0003-2697(74)90367-4. [DOI] [PubMed] [Google Scholar]
- 80.Schüler H, Nyåkern M, Schutt CE, Lindberg U, Karlsson R. Mutational analysis of arginine 177 in the nucleotide binding site of beta-actin. Eur J Biochem. 2000;267(13):4054–4062. doi: 10.1046/j.1432-1033.2000.01466.x. [DOI] [PubMed] [Google Scholar]
- 81.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Med Anal Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 82.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 83.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 84.Lorenz M, Holmes KC. The actin-myosin interface. Proc Natl Acad Sci U S A. 2010;107(28):12529–12534. doi: 10.1073/pnas.1003604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saunders MG, Voth GA. Water molecules in the nucleotide binding cleft of actin: effects on subunit conformation and implications for ATP hydrolysis. J Mol Biol. 2011;413(1):279–291. doi: 10.1016/j.jmb.2011.07.068. [DOI] [PubMed] [Google Scholar]
- 86.Baker JL, Voth GA. Effects of ATP and actin-filament binding on the dynamics of the myosin II S1 domain. Biophys J. 2013;105(7):1624–1634. doi: 10.1016/j.bpj.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 88.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Physics. 1983;79(2):926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 89.Best RB, et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J Chem Theory Comput. 2012;8(9):3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Church AT, Hughes ZE, Walsh TR. Improving the description of interactions between Ca2+ and protein carboxylate groups, including γ-carboxyglutamic acid: revised CHARMM22* parameters. RSC Advances. 2015;5(83):67820–67828. doi: 10.1039/C5RA11268K. [DOI] [Google Scholar]
- 91.Hess B, van der Vegt NF. Cation specific binding with protein surface charges. Proc Natl Acad Sci USA. 2009;106(32):13296–13300. doi: 10.1073/pnas.0902904106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Weerasinghe S, Smith PE. A Kirkwood–Buff derived force field for sodium chloride in water. J Chem Physics. 2003;119(21):11342–11349. doi: 10.1063/1.1622372. [DOI] [Google Scholar]
- 93.Hess B. P-LINCS: A parallel linear constraint solver for molecular simulation. J Chem Theory Comput. 2008;4(1):116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 94.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126(1):014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 95.Parrinello M, Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Physics. 1981;52(12):7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 96.Ewald PP. Die Berechnung optischer und elektrostatischer Gitterpotentiale. Annalen der Physik. 1921;369(3):253–287. doi: 10.1002/andp.19213690304. [DOI] [Google Scholar]
- 97.Darden T, York D, Pedersen L. Particle mesh Ewald: An N log(N) method for Ewald sums in large systems. J Chem Physics. 1993;98(12):10089–10092.92. doi: 10.1063/1.464397. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.