Abstract
Autoimmune disease is 4 times more common in women than men. This bias is largely unexplained. Female skin is “autoimmunity prone,” showing upregulation of many proinflammatory genes, even in healthy women. We previously identified VGLL3 as a putative transcription cofactor enriched in female skin. Here, we demonstrate that skin-directed overexpression of murine VGLL3 causes a severe lupus-like rash and systemic autoimmune disease that involves B cell expansion, autoantibody production, immune complex deposition, and end-organ damage. Excess epidermal VGLL3 drives a proinflammatory gene expression program that overlaps with both female skin and cutaneous lupus. This includes increased B cell–activating factor (BAFF), the only current biologic target in systemic lupus erythematosus (SLE); IFN-κ, a key inflammatory mediator in cutaneous lupus; and CXCL13, a biomarker of early-onset SLE and renal involvement. Our results demonstrate that skin-targeted overexpression of the female-biased factor VGLL3 is sufficient to drive cutaneous and systemic autoimmune disease that is strikingly similar to SLE. This work strongly implicates VGLL3 as a pivotal orchestrator of sex-biased autoimmunity.
Keywords: Autoimmunity, Dermatology
Keywords: Autoimmune diseases, Lupus, Mouse models
VGLL3, a factor enriched in female skin, may prime women to develop cutaneous and systemic autoimmune disease, such as systemic lupus erythematosus (SLE).
Introduction
Autoimmune disease is common, can be deadly, and affects women disproportionately. The prevalence of systemic lupus erythematosus (SLE) is 9 times higher among women than men, and many other autoimmune diseases show similar profound female skewing (1). However, the cause of this female bias remains elusive. Previous work has focused primarily on the influence of sex hormones, yet female bias in autoimmunity is observed even prior to puberty and following menopause (2), indicating alternative mechanisms at play. As most autoimmune diseases remain incurable, investigating the causes of autoimmunity is critical, and the drivers of female-biased autoimmunity are logical targets.
We previously identified the conserved putative transcription cofactor vestigial like family member 3 (VGLL3) as a candidate female-biased immune regulator (3). VGLL3 is more abundant in the epidermis of women than men and shows female-specific nuclear localization, suggesting a role in sex-biased transcriptional regulation. In cell culture, VGLL3 knockdown decreased expression of select female-biased immune transcripts, including B cell–activating factor (BAFF, also known as TNFSF13B), the target of the only currently approved biologic therapy for SLE. Intriguingly, men who have SLE showed loss of sex-specific regulation of VGLL3, demonstrating upregulation and nuclear localization of VGLL3 in inflamed skin (3). This suggested that VGLL3 may govern a key upstream regulatory program promoting autoimmunity, but this hypothesis had yet to be explored.
Results
Human and murine VGLL3 share 87% sequence homology, including an identical putative transcription cofactor domain (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.127291DS1). Similar to human women, female mice showed 2.8-fold higher Vgll3 expression than male mice in the skin (P = 0.053) (Supplemental Figure 1B), suggesting conserved sex-biased dynamics.
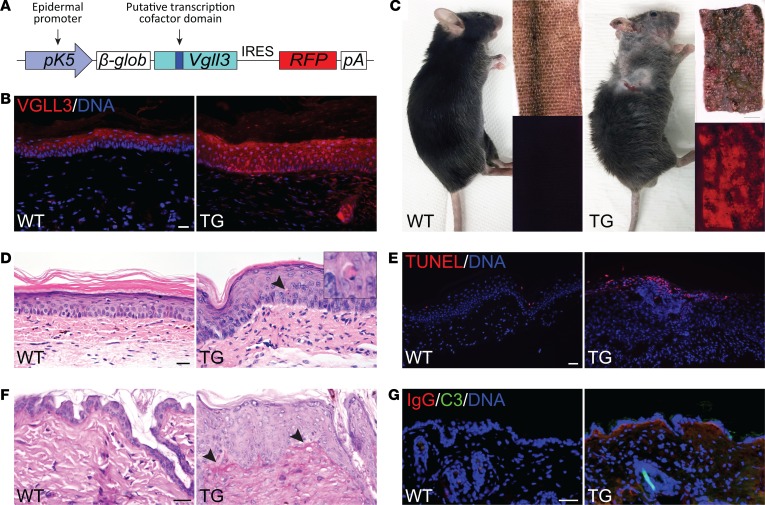
To test for a causative role for cutaneous VGLL3 in promoting autoimmune disease, we generated transgenic mice overexpressing Vgll3 under the control of the bovine keratin 5 (K5) promoter (Figure 1A), which drives gene expression primarily in the epidermis but also in some other stratified squamous epithelia (4). Levels of Vgll3, as shown by quantitative reverse transcription PCR (qRT-PCR), in transgenic skin varied both across and within founder lines, with transgenic mice generally showing 5- to 50-fold expression relative to WT mice, and no consistent variation by sex (data not shown). Compared with WT mice, these K5-Vgll3–transgenic mice showed increased epidermal VGLL3 staining, with conspicuous nuclear localization (Figure 1B) analogous to human VGLL3 distribution in affected skin of patients with SLE (3). Transgenic pups were indistinguishable from WT pups at birth. Within 6–12 weeks of life, they began developing progressive skin thickening and scaling prominently involving the face and ears, common sites for human discoid lupus erythematosus (DLE) lesions (Figure 1C and Supplemental Figure 1C). No consistent skin phenotypic variation by sex was noted among transgenic littermates (data not shown).
Figure 1. Overexpression of VGLL3 in the epidermis produces a skin phenotype with gross and histologic features of cutaneous lupus.
(A) Transgenic (TG) cassette. The bovine keratin 5 (K5) promoter drives polycistronic expression of the full-length mouse VGLL3 and mCherry red fluorescent protein linked by an internal ribosome entry site (IRES). β-glob, rabbit β-globin intronic sequence; pA, polyadenylation signal. (B) Detection of VGLL3 protein (red) by immunofluorescence (IF) in skin of female WT and TG mice. Scale bar: 20 μm. Images are representative of sections from 3 WT and 3 TG animals examined. (C) Left: WT mouse compared with age-matched TG mouse with lupus-like skin rash. Right: Bright-field and fluorescence images of WT and lesional TG tail skin. Scale bar: 2 mm. (D) H&E staining of WT and TG volar skin sections, demonstrating epidermal hyperplasia, basal cell vacuolization, apoptotic keratinocytes (arrowhead, magnified on inset), and dermal inflammatory infiltrate. Scale bar: 20 μm. (E) TUNEL (red) staining of WT and TG tail skin sections. Scale bar: 50 μm. (F) Periodic acid–Schiff staining of WT and TG dorsal skin sections. Arrowheads indicate subtle basement membrane thickening. Scale bar: 20 μm. (G) Detection of IgG and complement factor C3 by IF in WT and TG nonlesional neck skin. Scale bar: 50 μm. In E–G, images are representative of sections from 3 WT and 3 TG animals examined.
Histologically, early skin lesions showed epidermal thickening and focal interface dermatitis, an inflammatory reaction pattern seen in cutaneous lupus (Figure 1D and Supplemental Figure 1, D and E). TUNEL staining confirmed increased keratinocyte apoptosis (Figure 1E), subtle basement membrane thickening was evident on periodic acid–Schiff (PAS) staining (Figure 1F), and direct immunofluorescence (IF) revealed IgG and C3 deposition at the dermal-epidermal junction (Figure 1G): all are characteristic features of cutaneous lupus in humans.
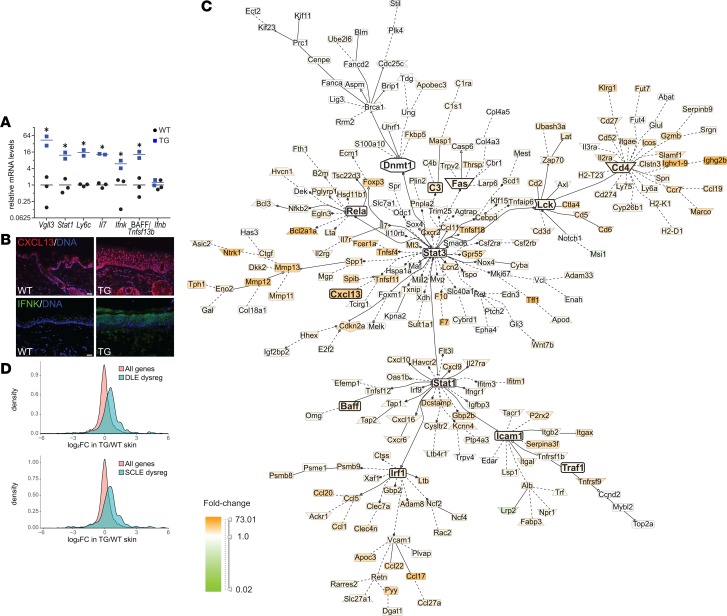
To examine the effects of VGLL3 that were driving this lupus-like cutaneous phenotype, we evaluated WT mice and robustly expressing transgenic mice by qRT-PCR for transcript levels of a panel of proinflammatory and lupus-related factors. Many of these transcripts showed significant elevation in transgenic mice (Figure 2A and Supplemental Figure 2A), including Tnfsf13b (encoding BAFF); IFN-κ (Ifnk), the predominant type I IFN in cutaneous lupus (5); and Cxcl13, a biomarker of early-onset SLE, heightened disease activity, and renal involvement (6). IF studies of key VGLL3-regulated factors (Figure 2B) corroborated our qRT-PCR findings. Thus, epidermal VGLL3 overexpression drives immunological gene dysregulation, similar to what we previously observed in the skin of healthy women (3).
Figure 2. Overexpression of VGLL3 in the epidermis recapitulates the autoimmunity-prone genetic signature of human female skin.
(A) Detection of Vgll3 and enriched immune transcripts versus example nonenriched transcript Ifnb (IFN-β) by quantitative reverse transcription PCR (qRT-PCR) in skin of WT (n = 3) and TG mice (n = 2) with high Vgll3 expression (more than tenfold WT average). Horizontal bars represent the mean. *P < 0.05 by 2-tailed Student’s t test. (B) Detection of VGLL3 targets CXCL13 (top, red) and IFN-κ (bottom, green) by IF in WT and TG skin. Blue, DNA. Scale bar: 20 μm. Images are representative of sections from 3 WT and 3 TG animals examined. (C) Literature-based network analysis of genes differentially expressed in nonlesional, normal-appearing TG skin relative to WT skin by RNA-seq. (D) Expression in nonlesional TG versus WT skin of genes dysregulated (dysreg) in discoid lupus erythematosus (DLE; P = 4.0 × 10–10) or subacute cutaneous lupus erythematosus (SCLE; P = 2.3 × 10–8) versus all genes. x axis, log2 fold change (FC) in TG versus WT. See Methods for additional statistical details.
For a broader examination of VGLL3 effects, we performed RNA-seq of normal-appearing dorsal skin from WT and transgenic mice to identify differentially expressed genes (transgenic DEGs) (Supplemental Table 1). Results largely affirmed our qRT-PCR data (Supplemental Figure 2B) and revealed that the panel of transcripts examined in Supplemental Figure 2B represent only a fraction of the VGLL3-regulated transcripts identified in transgenic mice. Of the 120 gene ontology terms significantly enriched (FDR ≤ 10%) among transgenic DEGs, nearly half were related to immunological processes (Supplemental Figure 2C). Importantly, these included multiple key pathways involved in SLE pathogenesis, such as IFN responses. Literature-based network analysis of transgenic DEGs revealed additional nodes of autoimmune pathogenesis (Figure 2C). To further explore our hypothesis that female-biased VGLL3 expression in human skin drives gene changes that may predispose women to autoimmunity, we compared transgenic DEGs with the set of genes upregulated in healthy human female skin relative to male skin (3) and found a significant overlap (P = 0.032).
To evaluate for a direct effect of VGLL3 overexpression in keratinocytes of our mouse model, we cultured primary keratinocytes from WT and transgenic mouse tails and performed RNA-seq. Genes differentially expressed in transgenic keratinocytes also demonstrated enrichment for immunological gene ontology terms, such as immune response (P = 6.8 × 10–9) and cytokine activity (P = 1.2 × 10–8), and showed even more significant overlap with female-biased genes (P = 4.0 × 10–7). Thus, epidermal overexpression of VGLL3 is a prominent driver of immunological dysregulation and sex-biased gene expression in keratinocytes.
We then compared our mouse skin RNA-seq results to transcriptomic data from skin of cutaneous lupus patients (7). Genes dysregulated in lesional skin of patients with DLE or subacute cutaneous lupus erythematosus (SCLE) were overrepresented among transgenic DEGs (DLE, P = 1.1 × 10–13; SCLE, P = 5.0 × 10–9) and showed widespread upregulation in transgenic mice (Figure 2D), revealing a shared pattern of gene dysregulation in skin of K5-Vgll3–transgenic mice and lupus patients. Together, these in vivo data demonstrate that VGLL3 overexpression in the epidermis is sufficient to drive a female-biased gene expression signature with dysregulation of pathways implicated in human autoimmune disease.
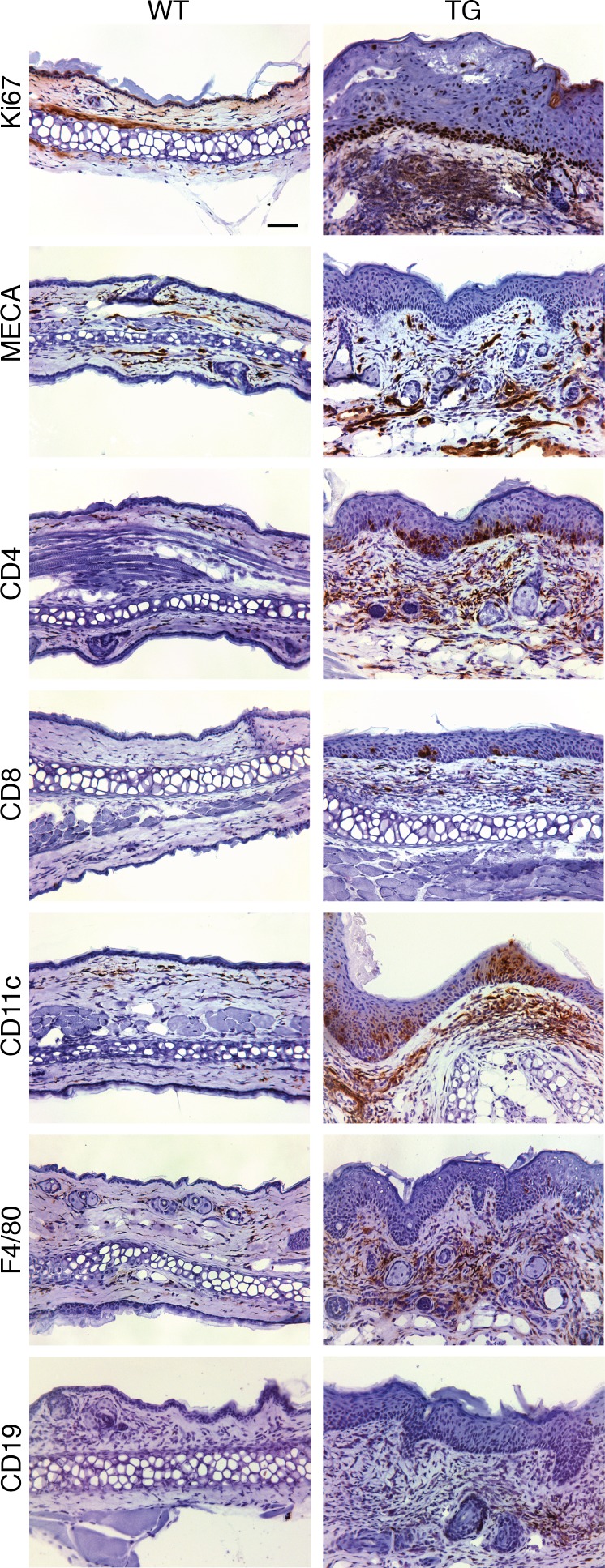
Immunohistochemistry of K5-Vgll3–transgenic skin revealed a mixed inflammatory infiltrate (Figure 3). T and B cells were increased, as were DCs, which promote lymphocyte expansion, autoantibody production by B cells, and tissue damage in SLE (8). Flow cytometry of cells isolated from skin corroborated these findings, demonstrating increased plasmacytoid DCs, as well as IgM+ and IgG+ cells, consistent with B cell expansion (Supplemental Figure 3). Neutrophils were also elevated, consistent with the neutrophilic inflammation noted on some lesional sections (Supplemental Figure 1F) and observed in a significant subset of cutaneous lupus manifestations (9). These results illustrate that epidermal overexpression of VGLL3 results in a cutaneous phenotype with gross, histologic, and inflammatory features of cutaneous lupus.
Figure 3. Overexpression of VGLL3 in the epidermis causes skin inflammation with features of cutaneous lupus.
Detection of the indicated cell markers by immunohistochemistry in WT and TG ear sections. Ki67, cell proliferation marker; MECA, mouse endothelial cell antigen. Scale bar: 50 μm. Images are representative of sections from 4 WT and 4 TG animals examined.
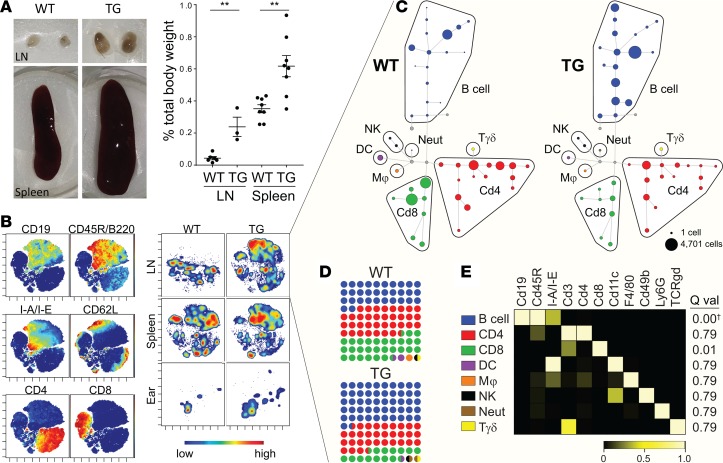
To further interrogate the inflammatory response in the K5-Vgll3–transgenic mouse, we analyzed skin-draining lymph nodes, spleen, and ear tissue from WT and transgenic mice with mass cytometry (CyTOF) using a 37-marker panel (Supplemental Figure 4 and Supplemental Table 2). Transgenic mice showed significant lymphadenopathy and splenomegaly (Figure 4A), features common in SLE and some lupus mouse models (10, 11). CyTOF data visualized using the dimensional reduction tool viSNE (12) showed expansion of B cell populations that were most prominent in skin-draining lymph nodes (Figure 4B and Supplemental Figure 4). Key populations emerging from CyTOF are highlighted in a SPADE tree (13) (Figure 4, C–E, and Supplemental Figure 5). B cells were significantly overrepresented in transgenic lymph nodes (q = 6.2 × 10–4; blue circles in Figure 4D) and spleen (q = 0.024). Together, these findings suggest that skin-directed VGLL3 overexpression drives a systemic inflammatory response with B cell expansion.
Figure 4. Skin-directed VGLL3 overexpression drives a systemic inflammatory response with B cell expansion.
(A) Left: Representative images of WT and TG skin-draining lymph nodes (LN) and spleens. Right: LN and spleen weights represented as a percentage of total body weight. Error bars represent mean ± SEM. **P < 0.01 by 2‑tailed Student’s t test (LN, n = 6 WT and 3 TG; spleen, n = 8 WT and 8 TG). (B) CyTOF data derived from a single experiment consisting of 3 age- and sex-matched WT and TG mice visualized by viSNE. Left: viSNE maps depicting expression of select markers. Each dot is a cell. Color reflects the level of expression of marker from low (blue) to high (red). Right: Contour plot of viSNE maps colored by density of cells isolated from the specified tissues in WT and TG mice. Here, color reflects cell density from low (blue) to high (red). Data shown correspond to 1 WT and 1 TG mouse that are representative of the experiment. LN and spleen data represent approximately 20,000 CD45+ live singlets per sample. Ear skin samples represent all recorded CD45+ live singlets (WT, 262 and TG, 264) for each sample. The complete viSNE analysis for all markers and samples for the experiment can be found in Supplemental Figure 4. Data are representative of 2 independent CyTOF experiments. (C) SPADE tree of LN samples depicted in B. Node size represents the number of cells in the population derived from the experiment represented in B and Supplemental Figure 4B. Mϕ, macrophage; Neut, neutrophil; Tγδ, γ δ T cell. (D) Percentage dot plot showing the proportion of each cell type in aggregated WT and TG lymph node samples for the CyTOF experiment. Colors correspond to the populations indicated in C and E. (E) Heatmap showing marker expression of the SPADE populations specified in C across aggregated LN, spleen, and skin samples included in the CyTOF experiment. Q val, q values for differential detection in WT versus TG LN. †q = 6.2 × 10–4, significantly upregulated in TG LN.
Peripheral blood mononuclear cells (PBMCs) of patients with SLE show altered gene expression, with prominent dysregulation of genes in IFN and cytokine signaling pathways that likely contributes to systemic inflammation (14). Evaluating gene expression in blood of WT versus K5-Vgll3–transgenic mice, we observed a significantly higher effect size for genes whose human orthologs are dysregulated in blood of SLE patients (P = 1.6 × 10–22) (Figure 5A and Supplemental Table 3), indicating a shared pattern of gene dysregulation in circulating immune cells of K5-Vgll3–transgenic mice and patients with SLE.
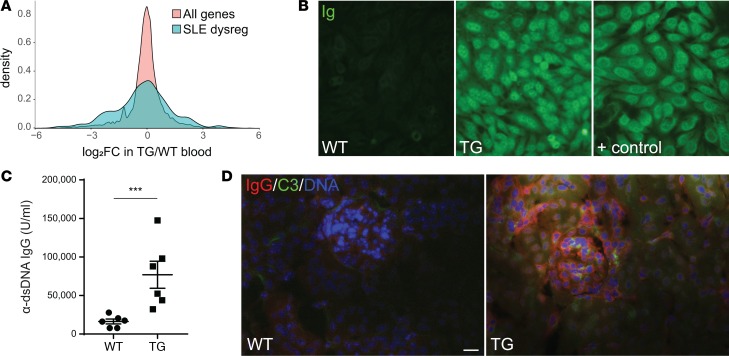
Figure 5. Mice with skin-directed VGLL3 overexpression develop manifestations of systemic autoimmune disease.
(A) Differential expression in WT (n = 4) and TG (n = 4) mouse blood of genes dysregulated in SLE peripheral blood mononuclear cells (aqua) versus all genes (peach). (B) Detection at 1:50 dilution of circulating antinuclear antibodies (Ig) by indirect IF in HEp-2 cells in WT, TG, and positive control (+ control, aged NZM2328) serum. In total, 7 TG and 3 WT animals were evaluated; all TG mouse sera showed positive autoantibody staining at a dilution of 1:25 or higher, whereas all WT mouse sera were negative. (C) Detection of circulating anti–double-stranded DNA antibodies (α-dsDNA) by ELISA. Error bars represent mean ± SEM. ***P = 0.0005 by 2-tailed Student’s t test. n = 6 WT and 6 TG. (D) Detection of IgG (red) and complement factor C3 (green) by IF in WT and TG glomeruli. Blue, DNA. Scale bar: 20 μm. Images are representative of glomeruli visualized on sections from 6 WT and 6 TG animals examined. 0 of 6 WT and 6 of 6 transgenic mice scored positive for renal immune complex deposition.
Finally, we investigated whether the B cell expansion of the K5-Vgll3 mouse was associated with autoantibody production. Autoantibodies are integral to pathogenesis of SLE and associated with increased risk of progression from cutaneous to systemic lupus (15). Commonly used SLE classification systems include their detection as a diagnostic criterion. By indirect IF of HEp-2 cells, antinuclear antibodies were detected in sera of transgenic mice with developed phenotypes (Figure 5B). A homogeneous pattern of nuclear staining was observed, which is commonly found in SLE patients. Anti–double-stranded DNA (anti-dsDNA) antibodies, which are a specific marker for SLE and fluctuate with disease activity (16), were abundant in sera of transgenic mice (Figure 5C). As anti-dsDNA antibodies are pathogenic to the kidney (17), we examined kidneys of transgenic mice and found IgG and C3 deposition in glomeruli (Figure 5D). Most transgenic mice were harvested by 4–5 months of age due to the severity of the skin phenotype, limiting time for evolution of fulminant nephritis; nonetheless, examination of the transgenic kidneys revealed a subtle trend toward increased inflammation relative to WT kidneys (P = 0.20; Supplemental Figure 6, A and B). As expected based on the mild histology, significant proteinuria was not detected (P = 0.19; Supplemental Figure 6C). Thus, skin-directed VGLL3 overexpression stimulates development of lupus-specific anti-dsDNA autoantibodies that results in immune complex deposition in the kidney, the purported initiating event in development of lupus nephritis (17).
Discussion
Female sex alone carries a greater risk for developing SLE than any genetic or environmental factor identified to date (18). Previously, we determined that women express an autoimmunity-prone genetic signature in their skin. We hypothesized that this program was driven by the putative transcription cofactor VGLL3, which is upregulated and nuclear localized in the skin of healthy women and SLE patients of both sexes (3). Here, we establish that skin-directed overexpression of murine VGLL3 is sufficient to drive an autoimmunity-prone transcriptional signature similar to that observed in female skin, causing inflammation and activation of type I IFN signaling that mimics cutaneous lupus. Intriguingly, although VGLL3 overexpression in our model was restricted by the bovine K5 promoter to keratinocytes and some other squamous epithelia (4), K5-Vgll3–transgenic mice developed systemic inflammation, with B cell activation and autoantibody production that culminated in renal immune complex deposition analogous to that seen in lupus nephritis. Thus, overexpression of a single female-biased gene is sufficient to trigger cutaneous autoimmune disease accompanied by a systemic autoimmune response with striking similarities to SLE. This includes development of anti-dsDNA antibodies, whose presence constitutes an independent risk factor for lupus nephritis (19), and detection of immune complexes in the skin and kidneys.
A growing body of evidence suggests that, in autoimmune disease pathogenesis, the initial break in self-tolerance — the inciting “first hit” — occurs at epithelial surfaces. In rheumatoid arthritis, localized mucosal tissue damage may lead to posttranslational modification of peptides, resulting in formation of anti-citrullinated peptide antibodies (20), autoantibodies that often precede the onset of disease and in susceptible individuals may incite it (21). In antinuclear antibody–associated diseases, such as SLE and other connective tissue diseases, the first hit may occur in the skin. In patients with SLE, clearance of apoptotic cells is impaired (22). Insults, such as ultraviolet light, which induces immunological activation and apoptosis of keratinocytes, cause release of endogenous nuclear antigens. Presence of these autoantigens results in elaboration of cytokines and immune cell recruitment, and cytotoxic inflammation perpetuates their release, with resulting accumulation and possible entry into circulation as proinflammatory microparticles. Inflammation in the skin may therefore prime or exacerbate antinuclear antigen-focused autoimmunity. There are sparse data suggesting that treatment of cutaneous lupus may prevent progression to SLE (23). The results herein demonstrate that cutaneous disease may be sufficient to trigger a break in self-tolerance with evolution of systemic autoimmune disease. These findings are complemented by reports in mice describing evolution of SLE-like disease with development of anti-dsDNA antibodies and immune complex nephritis in response to epidermal IFN-γ overexpression (24, 25). Further study of the IFN-γ epidermal overexpression and K5-Vgll3 lupus mouse models may reveal that treatment of cutaneous lupus is sufficient to ameliorate or even prevent progression to systemic disease. If so, this could prompt a paradigm shift in our understanding of the pathogenesis of SLE.
Our results do not distinguish a principal pathway promoting autoimmunity in the K5-Vgll3–transgenic mouse. As the VGLL3-regulated factor IFN-κ is the predominant type I IFN in cutaneous lupus (5), this presents a parsimonious explanation; however, VGLL3 alters the expression of genes that act in multiple independent inflammatory pathways, and the lupus-like phenotype of the K5-Vgll3–transgenic mouse may represent the cumulative manifestation of widespread immunological VGLL3-induced dysregulation. This is reflected in the diverse inflammatory pathways activated in the skin of transgenic mice (Figure 2C), many of which have key roles in autoimmunity: MHC class I, antigen binding, cytokine activation, humoral immune responses, cellular response to IFN, and regulation of T cell–mediated cytotoxicity (26). The significant overlap of VGLL3 targets with genes dysregulated in female skin suggests that the transgenic mouse phenotype represents a fully developed autoimmune disease of which female-biased autoimmunity is the forme fruste. Additional studies of the component inflammatory pathways in the K5-Vgll3 lupus model are required to dissect the pathogenesis further.
Finally, one of the principal challenges of treating lupus is disease heterogeneity. Broad immunosuppressants address disease in a majority of patients but carry serious risks of infection and malignancy that contribute substantially to morbidity and mortality. In contrast, narrower agents, such as belimumab, the biologic therapy that targets BAFF, are effective only in a subset of patients where presumably the disease is driven more heavily by the targeted autoimmune pathway. As VGLL3 appears to be not only constitutively active in women but also turned on in men with SLE (3), targeting VGLL3 may prove beneficial in patients of both sexes. Additionally, the low levels of VGLL3 in healthy men suggest that it may be amenable to depletion without adverse effects. Given stimulation of diverse inflammatory pathways — including those involving type I IFN and BAFF — observed in the K5-Vgll3–transgenic mouse, VGLL3 depletion may successfully treat more subgroups within this heterogeneous patient population.
In conclusion, our data support the assertion that enrichment of VGLL3 in female skin primes women for autoimmunity. This positions VGLL3 as a master orchestrator of sex bias in autoimmune disease, providing potentially novel avenues for future research and therapeutic development.
Methods
Mice, primary keratinocyte isolation, and culture
The K5-Vgll3-IRES-tdTomato–transgenic (K5-Vgll3–transgenic) cassette was generated as follows. The Vgll3-IRES-mCherry insert was synthesized de novo by GenScript and subcloned into the pBK5 vector, in which the bovine K5 promoter drives expression primarily in epidermis but also in some other stratified squamous epithelia (4). Of note, expression of this specific promoter has been reported to be absent in thymus and spleen (4, 27), indicating no significant expression in the immune cells therein. The Vgll3 coding sequence was taken from NCBI reference sequence NM_028572.1. The mCherry coding sequence was taken from GenBank (accession AY678264, nt 1–711) (28). Following sequence verification, the transgenic cassette was isolated by restriction enzyme digest, purified, and injected into C57BL/6 mouse oocytes at the University of Michigan Transgenic Core. Founders were identified by PCR using the following PCR primers: forward, 5′-ATCGTGCCAAGTGTGGGCTTCGATACA-3′ (located in the Vgll3 coding sequence), reverse, 5′-CACATTGCCAAAAGACGGCAATATGG-3′ (located in the IRES) and were crossed with C57BL/6J breeders (The Jackson Laboratory) to establish transgenic lines.
Transgene-positive offspring were screened for cutaneous phenotypes. Nine independent founder lines were generated that demonstrated spontaneous development of cutaneous phenotypes; progeny of 6 additional independent founders either failed to develop phenotypes or were harvested for experimental purposes prior to development of phenotypes. Phenotypes observed were overall fairly uniform and typically involved alopecia and ulceration that progressed with age, often necessitating euthanasia; transgenic animals from 6 independent founder lines are pictured at time of euthanasia in Supplemental Figure 1C. Transgenic mice were observed scratching and excessively barbering, and some transgenic animals were noted to be excessively barbering nontransgenic littermates. K5-Vgll3–transgenic mouse skin harvested from multiple sites was evaluated by fluorescent microscopy of transgenic mCherry to verify epidermal transgene expression (tail, Figure 1C). For RNA-seq analysis of nonlesional (posterior dorsal) skin and blood, 4 transgenic animals representing 3 independent founder lines were included (see below for associated methods). For RNA-seq analysis of transgenic and WT primary keratinocytes, all animals were from a single litter representing 1 founder line to enable simultaneous isolation, culture, and harvest. RNA‑seq analysis of transgenic nonlesional skin and primary keratinocytes confirmed significant Vgll3 mRNA overexpression relative to WT skin and primary keratinocytes, respectively. qRT-PCR also confirmed significant Vgll3 mRNA overexpression, although the variation and upper limit of Vgll3 overexpression was greater (generally 5- to 50-fold), possibly due to increase in epidermal thickness. Consistent with prior reports of absent expression in lymphoreticular organs (4, 27), RNA-seq analysis of K5-Vgll3–transgenic blood showed no Vgll3 upregulation that would be suggestive of leakage of transgene expression in immune cells (data not shown). Given the overall similarity in phenotype and gene expression changes noted by RNA-seq analysis of nonlesional skin of 3 different founder lines, multiple founder lines were included throughout the manuscript, including in qRT-PCR analyses.
Primary keratinocytes were isolated as previously described from tails of 3 WT and 3 transgenic 23-day-old weanlings from a single litter (29) with the following change: skin was incubated for 1.5 hours at 37°C rather than overnight at 4°C. Cells were expanded in culture for 3 days and harvested for RNA using Buffer RLT (Qiagen) per the manufacturer’s protocol.
Tissue collection
For harvest of tissues for generating RNA and frozen or paraffin sectioning, transgenic mice and WT controls were euthanized at 8 weeks of age or older. Whole blood was obtained by cardiac stick and RNA was isolated using the Mouse RiboPure-Blood RNA Isolation Kit (Thermo Fisher Scientific) or TriPure Isolation Reagent (MilliporeSigma) per the manufacturer’s protocols. Nonlesional posterior dorsal skin was removed by punch biopsy. Ears were removed in entirety. Skin from the indicated sites was otherwise harvested and processed as previously described (30).
For harvest of tissues for mass cytometry (CyTOF) and flow cytometry, sex-matched transgenic and WT littermates were sacrificed at 8 weeks of age or older. Ears, spleen, and skin-draining (cervical and inguinal) lymph nodes were removed in entirety. The ear and dorsal skin samples were minced with a sterile razor and transferred to 18 ml RPMI (Gibco) containing 10% fetal calf serum (Atlanta Biologicals) and 1% Penicillin-Streptomycin (Gibco) (hereafter, RPMI complete media). To each sample, 2 ml of a skin digestion solution consisting of Hanks’ Balanced Salt Solution (Gibco) with 1 mg/ml DNAse I (MilliporeSigma), 1 mg/ml Hyaluronidase type V (MilliporeSigma), and 5 mg/ml Collagenase type IV (MilliporeSigma) was added. Samples were rotated for 2 hours at 37°C to digest. Following digestion, samples were filtered through a 40-μm cell strainer, washed with fresh RPMI complete media, and incubated on ice. Spleens and lymph nodes (4 lymph nodes combined for each animal) were weighed before being ground through a 70-μm cell strainer and suspended in RPMI complete media. For spleen samples, media were removed, and cells were incubated in RBC lysis buffer (Invitrogen) for 2 minutes. Lysis buffer was removed, and spleen samples were resuspended in fresh RPMI complete media.
qRT-PCR
RNA was converted to cDNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qRT-PCR was performed in technical duplicates for the biological replicate numbers indicated in the figure legends using TaqMan Universal PCR Master Mix (Applied Biosystems). TaqMan primer sets and probes were purchased from Applied Biosystems by Life Technologies and are listed in Supplemental Table 4. All values were normalized to the housekeeping gene 18S. All qRT-PCR data are presented with y axis log2-scaled and autolog numbered.
RNA-seq and gene expression analyses
Stranded mRNA libraries were prepared from nonlesional (posterior dorsal) skin, cultured keratinocytes, and whole blood from age- and sex-matched WT and transgenic mice (2 male and 2 female each) using the TruSeq RNA library prep kit (Illumina) and sequenced on the Illumina HiSeq 4000 sequencer at the University of Michigan DNA Sequencing Core. Data were quality controlled and analyzed using the pipeline described previously for RNA-seq analysis, including adapter trimming (31), read mapping, and quantification of gene expression (32). We used the GENCODE release m18 (GRCm38) from mouse. We filtered for average abundance of ≥1 read per gene across all samples. DESeq was used for expression normalization. Generalized linear regression was used for differential expression analysis using negative binomial distribution to model the count data (33). FDR was used to control the multiple testing. DEGs were defined as having FDR ≤ 10% and with |log2fold change| ≥ 1 (Supplemental Tables 1 and 3). For Supplemental Figure 2B, data are presented with y axis after log2 scaling and autologing of numbers for ease of comparison to qPCR data. Software‑extracted literature‑based network analysis was performed using Genomatix Pathway System Software.
For identification of genes dysregulated in DLE and SCLE, microarray data from 26 DLE and 23 SCLE lesional skin biopsies and 7 normal skin biopsies (GEO GSE81071) were utilized to identify genes that are differentially expressed in DLE and SCLE skin (termed dysregulated in text to avoid confusion with transgenic DEGs) at the FDR ≤ 10% and with |log2fold change| ≥ 1 threshold. For identification of genes dysregulated in blood of SLE patients, RNA-seq data from whole blood of 99 SLE and 18 healthy control patients were analyzed as above and used to identify genes that were differentially expressed in blood of SLE patients (termed dysregulated in text to avoid confusion with transgenic DEGs). We performed analysis only for genes expressed in at least 20% of the normal samples. For investigation of target overlap with DLE and SCLE skin and SLE blood, only genes with identical names in mouse and human were included.
Histology and immunostaining
Hematoxylin (Surgipath, 3801540) and eosin (Surgipath, 3801600) staining was performed per standard protocols. Epidermal thickness was quantified on the H&E-stained sections using Adobe Photoshop. For each animal, approximately 100 measurements were taken from 8 different fields of view for each section analyzed. Epidermal thickness was measured from the stratum basale to stratum granulosum, excluding the stratum corneum and hair follicles for each animal.
IF to detect VGLL3 and target proteins was performed as follows. For CXCL13 detection, formalin-fixed, paraffin-embedded tissue sections were deparaffinized, rehydrated, and heated at 95°C for 20 minutes in pH 6 antigen retrieval buffer. Slides were blocked and incubated with goat anti-mouse CXCL13 antibody (R&D System, AF470-SP) overnight at 4°C. For VGLL3 and IFN-κ detection, cryosections were subjected to acetone fixation. Slides were blocked and incubated with rabbit anti-mouse VGLL3 antibody (St. John’s Laboratory, STJ115228) or sheep anti-mouse IFN-κ antibody (R&D System, AF5206), respectively. All slides were then incubated with biotinylated secondary antibodies (Vector Laboratories) as appropriate, incubated with fluorochrome-conjugated streptavidin (Streptavidin, Alexa Fluor 488 conjugate [Thermo Fisher Scientific, S32354] or Streptavidin, Alexa Fluor 594 conjugate [Life Technologies, S32356]) as appropriate and mounted. Images were acquired using Zeiss Axioskop 2 microscope and analyzed by SPOT software V.5.1.
PAS staining of paraffin sections was performed using the Periodic acid–Schiff (PAS) staining system (MilliporeSigma) per the manufacturer’s instructions (see procedure 395).
TUNEL staining of paraffin sections was performed using the In Situ Cell Death Detection Kit, TMR red (Roche). Slides were mounted with ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific).
IF of immune complex components was performed as follows. Slides with tissue cryosections were dried for 1 hour at 37°C, incubated in 4% paraformaldehyde for 20 minutes, and blocked. C3-FITC (ICL, GC3-90F-Z) and IgG-Texas Red-X (Thermo Fisher Scientific, T-862) goat anti-mouse antibodies were then added, and slides were incubated at 4°C for 1 hour. Hoechst stain (BD Biosciences, 561906) was added. Slides were dried and mounted using ProLong Gold Antifade Mountant (Thermo Fisher Scientific). Images were captured as above. For renal immune complex deposition, 10 glomeruli were assessed for each mouse, and mice were considered positive if more than one glomerulus demonstrated deposition of C3 and IgG.
Immunohistochemistry of frozen sections was performed as previously described (30) using antibodies specific for CD4 (BD Biosciences, 550280), CD8a (BD Biosciences, 550281), CD11c (BD Biosciences, 550283), CD19 (BD Biosciences, 550284), F4/80 (eBioscience, 14-4801-82), Ki-67 (DAKO, M7249), and MECA-32 (Developmental Studies Hybridoma Bank, MECA-32-s). Antibodies were detected using either rabbit anti-rat IgG biotinylated (Vector Laboratories), goat anti-rabbit IgG biotinylated (Vector Laboratories), or rabbit anti-hamster IgG biotinylated (Southern Biotech) secondary antibodies, amplified with Avidin/Biotinylated Enzyme Complex (Vector Laboratories), and visualized using the enzyme substrate diaminobenzidine (Vector Laboratories). Slides were counterstained with hematoxylin. Images were captured using a Leica DM L82 microscope with an attached QImaging MicroPublisher 3.3 Mega Pixel camera and Q-capture Pro software.
For detection of antinuclear antibodies, serum was taken undiluted, diluted 1:25, or diluted 1:50 and used for indirect IF on Kallestad HEp-2 Slides (Bio-Rad) with FITC goat anti-mouse Ig antibody (BD Biosciences) as the detection reagent. Imaging was performed as for IF above.
Mass cytometry (CyTOF)
Stimulation.
Tissue was harvested and cell suspensions were prepared as above. All samples were then stimulated for 3 hours at 37°C in RPMI complete media with 5 ng/ml phorbol 12-myristate 13-acetate (MilliporeSigma), 1 μg/ml ionomycin (MilliporeSigma), 3 μg/ml Golgistop with Brefeldin A (BioLegend), and 0.67 μl/ml BD GolgiStop Protein Transport Inhibitor with Monensin (Thermo Fisher Scientific), and then incubated on ice until staining.
Antibody conjugation with lanthanide metal.
The lanthanide metals (Fluidigm) were conjugated to the antibodies using the Maxpar Antibody Labeling Kit (Fluidigm) per the manufacturer’s instructions. Briefly, 2.5 mM Maxpar polymer was preloaded with lanthanide metal at 37°C for 1 hour and buffer exchanged to conjugation buffer using a 3 K Nanosep Centrifugal Device (Pall Life Sciences). The antibody was partially reduced using TCEP at 37°C for 30 minutes and buffer exchanged to conjugation buffer using an Amicon Ultra 0.5-ml 50-kDa Centrifugal Filter Concentrator (MilliporeSigma). The lanthanide-loaded polymer was conjugated to partially reduced antibody following overnight incubation at room temperature. The unbound metal was removed from the metal-tagged antibody by washing with wash buffer in a 50-kDa concentrator. The final concentration of metal-tagged antibody was determined by measuring the absorbance at 280 nm against the wash buffer. See Supplemental Table 2 for the antibodies.
Staining of cells with metal-tagged antibodies.
Cell-ID Cisplatin-195Pt and Cell-ID Intercalator Iridium-191/193 (Fluidigm) were used to identify live cells. The cells were washed once with prewarmed serum-free media by pelleting at 300 g for 5 minutes at room temperature and stained with 1.25 μM live/dead stain (Cell-ID Cisplatin-195Pt diluted in serum-free media from 500 mM stock) at room temperature for 5 minutes. Free cisplatin was quenched by washing the cells with serum-containing media. CyPBS (1× PBS without heavy metal contaminants, prepared from 10 × PBS stock [Invitrogen] in deionized distilled Milli-Q water [MilliporeSigma]) was used to prepare CyFACS buffer (CyPBS containing 0.1% BSA, 2 mM EDTA and 0.05% sodium azide). The cells were then washed with CyFACS buffer and incubated with TruStain FcX (anti-mouse CD16/32, Biolegends) for 10 minutes at room temperature to block the Fc receptors. For cells surface marker staining, the metal-tagged antibody cocktail was made in CyFACS buffer and added to the cells in the presence of TruStain FcX (BioLegend) and incubated on ice for 60 minutes. Following cell surface marker staining, the cells were washed twice with CyFACS buffer and fixed with 1.6% paraformaldehyde in CyPBS for 20 minutes at room temperature. The cells were then washed and stored overnight at 4°C in CyFACS buffer. The following day, the cells were pelleted at 800 g for 5 minutes and permeabilized with eBioscience permeabilization buffer (Thermo Fisher Scientific) for 30 minutes at room temperature. The cells were then stained with intracellular staining metal-tagged antibody cocktail (made in eBioscience permeabilization buffer) at room temperature for 60 minutes. Following intracellular staining, the cells were washed once with permeabilization buffer and twice with CyFACS buffer and stained with 62.5 nM Cell-ID Intercalator Iridium-191/193 (diluted in 1.6% paraformaldehyde in PBS from 500 μM stock) at room temperature for 40 minutes or left at 4°C until ready for acquisition on CyTOF.
CyTOF analysis of samples stained with metal-tagged antibodies.
The samples were acquired using CyTOF Helios system (Fluidigm). The system was maintained and tuned according to the manufacturer’s instructions. In addition, internal vendor-set calibration was performed before acquiring samples. The fixed cells were washed twice with CyPBS and deionized distilled Milli-Q water and filtered through a 40-μM cell strainer. EQ Four Element Calibration Beads (Fluidigm) were added at the recommended concentration to the samples before acquisition on CyTOF. The samples were acquired on CyTOF at approximately 50–300 events/s. After acquisition, the instrument software applied a signal correction algorithm based on the calibration bead signal to correct for any temporal variation in detector sensitivity.
CyTOF data analysis
Total events were gated to remove noncellular events (negative for DNA intercalator), dead cells (uptake of cisplatin), and doublets (event length greater than 25). A viSNE was performed using combined lymph node (~20,000 events/sample), spleen (~20,000 events/sample), and ear (between 262 and 1234 events/sample) samples using Cytobank (34). All antibody channels were included in the viSNE analysis except for IL-17f, as it was suspected this marker was staining nonspecifically (37 included markers). The viSNE run was performed with 2000 iterations, a perplexity of 30, and a θ of 0.5. A spanning-tree progression analysis of density-normalized events (SPADE) clustering algorithm was performed on the same events used in the viSNE analysis using the Cytobank platform (13). As with the viSNE analysis, all antibody channels were included except for IL-17f, as we suspected this marker was staining nonspecifically (37 included markers). After some experimentation, it was found that 50 nodes with 100% event downsampling gave an adequate resolution of cellular subsets. SPADE nodes were manually bubbled based on defined phenotypic markers of major cellular populations (summarized in Figure 4E). Samples were compared based on the percentage of total cells from that sample contained within a bubble or individual node.
Flow cytometry
WT and transgenic ear tissue was harvested and cell suspensions were prepared as above. Cells were resuspended in blocking reagent of 1% BSA (Fisher Bioreagents) and 1% Horse Serum (Corning) in PBS. Antibody master mixes were prepared using antibodies specific for IgG-PerCP (BioLegend, 405334), IgM-APC (BioLegend, 406509), PDCA-1-APC (BioLegend, 127016), Cd11c-Pacific Blue (BioLegend, 117322), F4/80-Pacific Blue (BioLegend, 123124), and Ly-6G-PE (BioLegend, 127606) and added to each sample. Samples were incubated at room temperature in a dark environment for 45 minutes, washed with PBS, and resuspended in 4% paraformaldehyde in PBS (Affymetrix). Flow cytometry was performed with a BD LSR II (BD Biosciences). After manual gating to exclude dead cells and debris, data corresponding to all remaining cells were analyzed and visualized using FlowJo software (Tree Star).
Detection of autoantibodies
Anti–dsDNA antibodies were detected in serum using the Mouse anti-dsDNA IgG2a ELISA Kit (Alpha Diagnostic International) according to manufacturer’s instructions.
Renal pathology scoring
Kidneys from 4 WT and 4 transgenic animals were analyzed. A semiquantitative scoring system (0, no involvement; 0.5, minimal involvement of <10% per section; 1, mild involvement of 10%–30% of section; 2, moderate involvement of 31%–60% of section; and 3, severe involvement >60% of section) was used to assess 13 different parameters of activity and chronicity: mesangial hypercellularity, mesangial deposits, mesangial sclerosis, endocapillary cellular infiltrate, subepithelial deposits, subendothelial deposits, capillary thrombi, capillary sclerosis, cellular crescents, organized crescents, synechiae, tubular atrophy, and interstitial fibrosis. An activity and chronicity index was generated by compiling scores from groups of related parameters. For activity, mesangial hypercellularity, mesangial deposits, endocapillary cellular infiltrate, and cellular crescents were considered; for chronicity, interstitial fibrosis, tubular atrophy, organized crescents, and capillary sclerosis were considered.
Analysis of urine albumin/creatinine ratio
Urine from 8 WT and 5 transgenic animals was analyzed. Urine was collected from animals within 1 week of euthanasia. Technical duplicates were performed for every sample. To calculate albumin-to-creatinine ratios, urinary albumin was measured using the Albuwell M Kit (Exocell) and urinary creatinine using the QuantiChrom Creatinine Assay Kit (BioAssay Systems), both according to the manufacturer’s protocol.
Data availability
RNA-seq data have been deposited in GEO (GSE128453). CyTOF data have been made publically available in FlowReposity under repository ID FR‑FCM‑Z2Y6 (35).
Statistics
qRT-PCR data were tested for statistical significance using 2-tailed Student’s t test assuming homoscedasticity. Significance was defined as P ≤ 0.05. RNA-seq data were tested for normality, and statistical significance was calculated using a 2-tailed Student’s t test, Mann-Whitney U test, or Friedman’s test, as appropriate. For CyTOF data, statistical analysis on the bubbled SPADE populations was performed using Prism 8.0 (GraphPad). P values were computed using unpaired Student’s t tests assuming homoscedasticity. P values were considered discoveries if they fell below an FDR of 10% using the 2-stage step-up method of Benjamini, Krieger, and Yekutieli (36).
Study approval
Procedures involving the care and the use of mice in this study were reviewed and approved by the University of Michigan Institutional Animal Care and Use Committee (protocol PRO00006657).
Author contributions
ACB, MGK, MEV, AAD, JMK, and JEG conceived the study and designed experiments; LCT, BDH, and CCB developed computational methods and analyzed the data; MGK, JF, DG, JL, XX, SE, SJW, SMR, JBH, YL, PWH, and MEV conducted the experiments; and ACB, JMK, and JEG wrote the paper with input from all authors, including MAB, MKS, RU, SS, CZ, JJV, FW, and NLW.
Supplementary Material
Acknowledgments
This work was supported by the A. Alfred Taubman Medical Research Institute’s Taubman Institute Innovation Projects program (to JEG, JMK, LCT, and FW), the Parfet Emerging Scholar Award (to JMK), and the Frances and Kenneth Eisenberg Emerging Scholar Award (to JEG); the University of Michigan Babcock Endowment Fund (to LCT and JEG); the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the NIH (5T32AR007197-40 to ACB; K01AR072129 to LCT; R01-AR071384 to JMK; R01-AR069071 to JEG; and P50-AR070590, R01-AR062546, R01-AR063437, and R01-AR073196 to NLW); the Office of the Director of the NIH (S10-OD020053 to FW); a Rogel Cancer Center Support Grant (NIH P30-CA046592 to BDH, SMR, and FW); the National Science Foundation (1511720, 1645229, and 1653611 to FW); a Rheumatology Research Foundation Innovative Research Award (to JMK); the Dermatology Foundation (to LCT); the Arthritis National Research Foundation (to LCT); and the National Psoriasis Foundation (to LCT and NLW).
Version 1. 04/18/2019
Electronic publication
Funding Statement
JEG, JMK, LCT, and FW
JMK
JEG
LCT, JEG
ACB
LCT
JMK
JEG
NLW
FW
BDH, SMR, and FW
FW
JMK
LCT
LCT
LCT and NLW
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(8):e127291. https://doi.org/10.1172/jci.insight.127291.
Contributor Information
Allison C. Billi, Email: acbilli@med.umich.edu.
Mehrnaz Gharaee-Kermani, Email: nazy@med.umich.edu.
Joseph Fullmer, Email: jmfull@umich.edu.
Brett D. Hill, Email: labrett@umich.edu.
Dennis Gruszka, Email: drg59@case.edu.
Jessica Ludwig, Email: jel151@case.edu.
Xianying Xing, Email: xingx@med.umich.edu.
Sonya J. Wolf, Email: sonwolf@med.umich.edu.
Syed Monem Rizvi, Email: smrizvi@umich.edu.
Maria A. Beamer, Email: mbeamer@med.umich.edu.
Mrinal K. Sarkar, Email: mksarkar@umich.edu.
Yun Liang, Email: liang95@wisc.edu.
Ranjitha Uppala, Email: uranjith@med.umich.edu.
Shuai Shao, Email: 857746654@qq.com.
Chang Zeng, Email: changz@med.umich.edu.
Paul W. Harms, Email: paulharm@med.umich.edu.
Fei Wen, Email: feiwenum@umich.edu.
Andrzej A. Dlugosz, Email: dlugosza@umich.edu.
References
- 1.Fish EN. The X-files in immunity: sex-based differences predispose immune responses. Nat Rev Immunol. 2008;8(9):737–744. doi: 10.1038/nri2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tedeschi SK, Bermas B, Costenbader KH. Sexual disparities in the incidence and course of SLE and RA. Clin Immunol. 2013;149(2):211–218. doi: 10.1016/j.clim.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Liang Y, et al. A gene network regulated by the transcription factor VGLL3 as a promoter of sex-biased autoimmune diseases. Nat Immunol. 2017;18(2):152–160. doi: 10.1038/ni.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramírez A, Bravo A, Jorcano JL, Vidal M. Sequences 5’ of the bovine keratin 5 gene direct tissue- and cell-type-specific expression of a lacZ gene in the adult and during development. Differentiation. 1994;58(1):53–64. doi: 10.1046/j.1432-0436.1994.5810053.x. [DOI] [PubMed] [Google Scholar]
- 5.Sarkar MK, et al. Photosensitivity and type I IFN responses in cutaneous lupus are driven by epidermal-derived interferon kappa. Ann Rheum Dis. 2018;77(11):1653–1664. doi: 10.1136/annrheumdis-2018-213197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ezzat M, El-Gammasy T, Shaheen K, Shokr E. Elevated production of serum B-cell-attracting chemokine-1 (BCA-1/CXCL13) is correlated with childhood-onset lupus disease activity, severity, and renal involvement. Lupus. 2011;20(8):845–854. doi: 10.1177/0961203311398513. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Berthier CC, Kahlenberg JM. Enhanced Inflammasome Activity in Systemic Lupus Erythematosus Is Mediated via Type I Interferon-Induced Up-Regulation of Interferon Regulatory Factor 1. Arthritis Rheumatol. 2017;69(9):1840–1849. doi: 10.1002/art.40166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teichmann LL, Ols ML, Kashgarian M, Reizis B, Kaplan DH, Shlomchik MJ. Dendritic cells in lupus are not required for activation of T and B cells but promote their expansion, resulting in tissue damage. Immunity. 2010;33(6):967–978. doi: 10.1016/j.immuni.2010.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ribero S, Sciascia S, Borradori L, Lipsker D. The Cutaneous Spectrum of Lupus Erythematosus. Clin Rev Allergy Immunol. 2017;53(3):291–305. doi: 10.1007/s12016-017-8627-2. [DOI] [PubMed] [Google Scholar]
- 10.Cohen PL, Eisenberg RA. Lpr and gld: single gene models of systemic autoimmunity and lymphoproliferative disease. Annu Rev Immunol. 1991;9:243–269. doi: 10.1146/annurev.iy.09.040191.001331. [DOI] [PubMed] [Google Scholar]
- 11.Andrews BS, et al. Spontaneous murine lupus-like syndromes. Clinical and immunopathological manifestations in several strains. J Exp Med. 1978;148(5):1198–1215. doi: 10.1084/jem.148.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amir el-AD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31(6):545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu P, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29(10):886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rai R, Chauhan SK, Singh VV, Rai M, Rai G. RNA-seq Analysis Reveals Unique Transcriptome Signatures in Systemic Lupus Erythematosus Patients with Distinct Autoantibody Specificities. PLoS One. 2016;11(11):e0166312. doi: 10.1371/journal.pone.0166312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wieczorek IT, Propert KJ, Okawa J, Werth VP. Systemic symptoms in the progression of cutaneous to systemic lupus erythematosus. JAMA Dermatol. 2014;150(3):291–296. doi: 10.1001/jamadermatol.2013.9026. [DOI] [PubMed] [Google Scholar]
- 16.ter Borg EJ, Horst G, Hummel EJ, Limburg PC, Kallenberg CG. Measurement of increases in anti-double-stranded DNA antibody levels as a predictor of disease exacerbation in systemic lupus erythematosus. A long-term, prospective study. Arthritis Rheum. 1990;33(5):634–643. doi: 10.1002/art.1780330505. [DOI] [PubMed] [Google Scholar]
- 17.Raz E, Brezis M, Rosenmann E, Eilat D. Anti-DNA antibodies bind directly to renal antigens and induce kidney dysfunction in the isolated perfused rat kidney. J Immunol. 1989;142(9):3076–3082. [PubMed] [Google Scholar]
- 18.Voskuhl R. Sex differences in autoimmune diseases. Biol Sex Differ. 2011;2(1):1. doi: 10.1186/2042-6410-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moroni G, et al. The value of a panel of autoantibodies for predicting the activity of lupus nephritis at time of renal biopsy. J Immunol Res. 2015;2015:106904. doi: 10.1155/2015/106904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Catrina AI, Deane KD, Scher JU. Gene, environment, microbiome and mucosal immune tolerance in rheumatoid arthritis. Rheumatology (Oxford) 2016;55(3):391–402. doi: 10.1093/rheumatology/keu469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.England BR, Thiele GM, Mikuls TR. Anticitrullinated protein antibodies: origin and role in the pathogenesis of rheumatoid arthritis. Curr Opin Rheumatol. 2017;29(1):57–64. doi: 10.1097/BOR.0000000000000356. [DOI] [PubMed] [Google Scholar]
- 22.Kuhn A, et al. Accumulation of apoptotic cells in the epidermis of patients with cutaneous lupus erythematosus after ultraviolet irradiation. Arthritis Rheum. 2006;54(3):939–950. doi: 10.1002/art.21658. [DOI] [PubMed] [Google Scholar]
- 23.James JA, et al. Hydroxychloroquine sulfate treatment is associated with later onset of systemic lupus erythematosus. Lupus. 2007;16(6):401–409. doi: 10.1177/0961203307078579. [DOI] [PubMed] [Google Scholar]
- 24.Carroll JM, Crompton T, Seery JP, Watt FM. Transgenic mice expressing IFN-gamma in the epidermis have eczema, hair hypopigmentation, and hair loss. J Invest Dermatol. 1997;108(4):412–422. doi: 10.1111/1523-1747.ep12289702. [DOI] [PubMed] [Google Scholar]
- 25.Seery JP, Carroll JM, Cattell V, Watt FM. Antinuclear autoantibodies and lupus nephritis in transgenic mice expressing interferon gamma in the epidermis. J Exp Med. 1997;186(9):1451–1459. doi: 10.1084/jem.186.9.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsokos GC, Lo MS, Costa Reis P, Sullivan KE. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat Rev Rheumatol. 2016;12(12):716–730. doi: 10.1038/nrrheum.2016.186. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, et al. Chronic skin-specific inflammation promotes vascular inflammation and thrombosis. J Invest Dermatol. 2012;132(8):2067–2075. doi: 10.1038/jid.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22(12):1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 29.Lichti U, Anders J, Yuspa SH. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat Protoc. 2008;3(5):799–810. doi: 10.1038/nprot.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnston A, et al. Keratinocyte overexpression of IL-17C promotes psoriasiform skin inflammation. J Immunol. 2013;190(5):2252–2262. doi: 10.4049/jimmunol.1201505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotecha N, Krutzik PO, Irish JM. Web-based analysis and publication of flow cytometry experiments. Curr Protoc Cytom. 2010;Chapter 10:Unit10.17. doi: 10.1002/0471142956.cy1017s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spidlen J, Breuer K, Rosenberg C, Kotecha N, Brinkman RR. FlowRepository ‑ A Resource of Annotated Flow Cytometry Datasets Associated with Peer‑reviewed Publications. Cytometry A. 2012;81(9):727–731. doi: 10.1002/cyto.a.22106. [DOI] [PubMed] [Google Scholar]
- 36.Benjamini Y, Krieger AM, Yekutieli D. Adaptive linear step-up procedures that control the false discovery rate. Biometrika. 2006;93(3):491–507. doi: 10.1093/biomet/93.3.491. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in GEO (GSE128453). CyTOF data have been made publically available in FlowReposity under repository ID FR‑FCM‑Z2Y6 (35).