Abstract
Background
Composition and diversity of intestinal microbial communities (microbiota) are generally accepted as a risk factor for poor outcomes; however, we cannot yet use this information to prevent adverse outcomes.
Methods
Stool was collected from eight long-term acute care hospital (LTACH) patients experiencing diarrhea and two fecal microbiota transplant donors; 16S rDNA V1-V2 hypervariable regions were sequenced. Composition and diversity of each sample were described. Stool was also tested for Clostridium difficile, vancomycin-resistant enterococci (VRE), and carbapenem-resistant Enterobacteriaceae. Associations between microbiota diversity and demographic and clinical characteristics, including antibiotic use, were analyzed.
Results
Antibiotic exposure and Charlson Comorbidity Index were inversely correlated with diversity (Spearman = −0.7). Two patients were positive for VRE; both had microbiomes dominated by Enterococcus faecium, accounting for 67–84% of their microbiome.
Discussion
Antibiotic exposure correlated with diversity; however, other environmental and host factors not easily obtainable in a clinical setting are also known to impact the microbiota. Therefore, direct measurement of microbiome disruption by sequencing, rather than reliance on surrogate markers, might be most predictive of adverse outcomes.
Conclusions
If and when microbiome characterization becomes a standard diagnostic test, improving our understanding of microbiome dynamics will allow for interpretation of results to improve patient outcomes.
Introduction
In recent years, research on the collective genome of microbial communities, known as the microbiome, living in or on humans has accelerated.1,2 A healthy intestinal microbiota assists in digestion and metabolism, and protects against pathogen invasion and overgrowth of pathobionts, which are commensal bacteria that can intermittently reside as minor members of the microbiota and also can act as pathogens when that microbiota becomes disrupted.3 Loss of microbial diversity or protective species, and overgrowth or dominance by a single organism are characteristic of microbiome disruption.
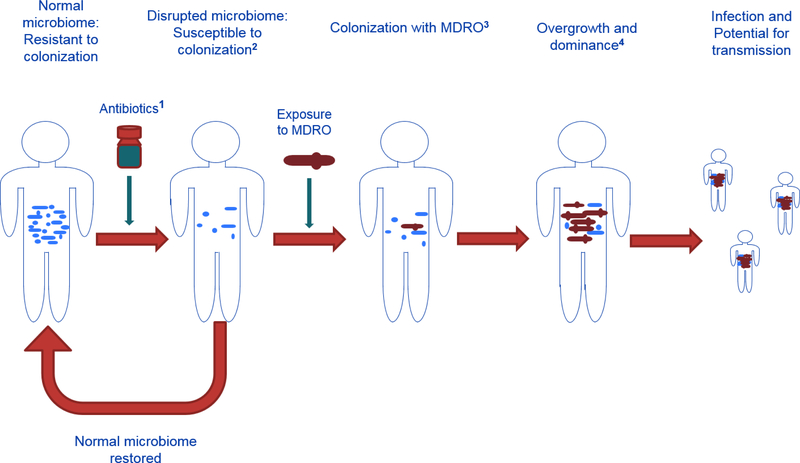
From birth, environment and host factors impact a person’s microbiota. However, capturing a lifetime of exposures is not feasible. Microbiome disruption is generally accepted as a risk factor for poor outcomes, such as infection and, as recently suggested, sepsis.4 However, we cannot yet use microbiome status to predict or prevent poor outcomes. One way to translate increasing understanding of the microbiome to the field of infection control is via the development and use of ‘Microbiome Disruption Indices’ (MDIs) (Figure 1). Such indices could become standardized criteria for not only characterizing the status of a patient’s microbiome but also evaluating and communicating the disruptive potential of various drugs, including antibiotics. Applications for MDIs range from improving antibiotic stewardship, infection control, and clinical management of patients, to assigning a risk index to antibiotics and other microbiota disruptive drugs during the drug approval process.
Figure 1.
Causal Pathway from health to disease: Microbiome Disruption Indices (MDI)
1. Antibiotic MDI indicates the potential an antibiotic has for disrupting the intestinal microbiome
2. Disrupted microbiome status MDI characterizes the degree and type of disruption in the intestinal microbiome, as well as the susceptibility to colonization by a MDRO
3. MDRO colonization MDI indicates susceptibility to overgrowth and dominance by a MDRO
4. MDI characterizing overgrowth and dominance by a MDRO indicates susceptibility for infection with a MDRO, and the potential for transmission to others through skin/environment contamination.
Among the host and environmental factors that lead to microbiota shifts,5–8 antibiotic exposures cause dramatic disruptions, lasting six months or more.9 Not only do antibiotic-induced disruptions lead to a loss of colonization resistance to MDROs, but once colonization does occur, further disruptions can lead to dominance (defined as a single MDRO constituting ≥30% of the microbiota), which is associated with the occurrence of invasive infection and increased transmission risk through skin and environmental contamination.10,11 MDROs, such as Clostridium difficile, vancomycin-resistant enterococci (VRE), and carbapenem-resistant Enterobacteriaceae (CRE), are major public health concerns in healthcare settings, where they are transmitted between patients and can colonize the lower intestine in more individuals than they infect.12,13
Long-term acute care hospital (LTACH) patients are a population with high antibiotic consumption,14 likely leading to severe intestinal microbiota disruption. In an effort to make a case for the potential impact of MDIs in improving infection control, we describe and compare the microbiomes from LTACH patients with prior antibiotic exposure, when individuals are most susceptible to MDRO colonization, to those of fecal microbiota transplant donors from a small pilot study, described below. We also examine associations between intestinal microbiome diversity, and clinical and demographic characteristics.
METHODS
Study design and participants
The study was a cross-sectional pilot evaluation of the clinical characteristics and intestinal microbiome from a convenience sample of eight LTACH patients with new onset diarrhea and two healthy fecal microbiota transplant donors.15,16 The donors had no history of antibiotics in at least the previous 90 days and were not taking any other medications. Neither the LTACH patients nor the donors had histories of Crohn’s disease, ulcerative colitis, or other inflammatory bowel disease.
Patients were enrolled sequentially at first diarrheal episode during December 2013 through February 2014 when stool was being collected for C. difficile diagnostic PCR testing (GenExpert, Cepheid). Providers (S.L., J.M.) consented patients (or a family member for patients unable to consent) to have stool collected for microbiome analysis.
Data collected on each patient during retrospective chart review (A.C.) included demographics, proton pump inhibitor use, previous C. difficile infection, comorbidities, and antibiotic use in both the LTACH and acute care settings. Each antibiotic was classified by when it was administered in reference to the date of stool collection: day of or day before, during the seven days before, and during the 30 days before stool collection. Data were used to calculate cumulative antibiotic days17 and the number of days exposed to any (i.e., ≥ 1) antibiotic. Antibiotics were categorized into classes: carbapenems, cephalosporins (first-generation), cephalosporins (third- and fourth-generation), beta-lactam/beta-lactamase inhibitor (BL/BLI) combinations, fluoroquinolones, glycopeptides (vancomycin), metronidazole, or other antibiotics. For vancomycin, route of administration was documented.
The Emory University Institutional Review Board (IRB) approved this study protocol. No incentives were provided for participation.
MDRO Colonization Status
In addition to C. difficile diagnostic PCR testing, patient stool was cultured for VRE using Spectra VRE chromogenic agar (Remel, Lenexa, KS) and screened for CRE using a selective medium containing ertapenem (http://www.cdc.gov/hai/pdfs/labSettings/Klebsiella_or_Ecoli.pdf). At the time of collection, stool specimens were de-identified; linkage to the clinical specimen was known only to C.K.
Sample processing and next generation sequencing analyses
Because the majority of the microbiota cannot be cultured using current laboratory methods,18 next generation sequencing analysis to describe the bacteria present was performed on either residual stool (i.e., stool remaining after C. difficile diagnostic PCR testing) or stool collected within 7–10 days of C. difficile diagnostic PCR testing if insufficient residual stool was available. Stool was stored at 4°C and processed within 24 hours. DNA was extracted; the target 16S ribosomal gene hypervariable V1 and V2 regions were PCR amplified and PCR products were sequenced using an Illumina MiSeq instrument (Illumina, San Diego, CA). Within each sample, any DNA sequences sharing ≥97% similarity were clustered into operational taxonomic units (OTUs) (equivalent to a bacterial species, genus, or family).19 To assess within-sample microbial composition, or alpha diversity, we calculated values for Shannon Diversity Index9 and Observed Species, represented by the number of unique OTUs within a single sample. The Shannon Index accounts for both abundance and evenness to measure non-redundancy within a sample. To measure between-sample composition, we completed beta-diversity community analyses, using weighted UniFrac distance,20 which accounts for OTU abundances and phylogenetic distances. For additional details on sample processing, next generation sequencing, and sequence analyses, see Supplemental Methods.
Data analysis
For univariable analyses, relationships between demographic and clinical variables and alpha diversity were evaluated using nonparametric methods: Spearman’s correlation for continuous variables and Wilcoxon Rank Sum exact for categorical variables. All analyses were performed using SAS 9.3 (Cary, NC) or R statistical software (version 3.0.2).
RESULTS
Study design and participants
Summary demographic and clinical characteristics are in Table 1. Both donors were male, ages 29 and 30 years. Four of eight patients were male, and median age at stool collection was 66.5 (range: 50–75) years. At the time of stool collection, patients had been in the facility for a median of 3 (range: 0–52) days and were hospitalized in a short-stay acute care hospital a median 41.5 (range: 18–85) days before admission to the LTACH. All patients received antibiotics in the 30 days before stool collection. Patients received a median 34 (range: 4–76) cumulative antibiotic days and spent a median of 17.5 (range: 3–30) days on any antibiotic in the 30 days before stool sample collection.
Table 1.
Characteristics of population and health status
| Characteristic | No. (%) of subjects | No. (%) LTACH patients | No. (%) Stool donors |
|---|---|---|---|
| Total | 10 | 8 (80) | 2 (20) |
| Median age at collection (range) | 61.5 (29–75) | 66.5 (50–75) | 29.5 (29–30) |
| Gender | |||
| Male | 6 (60) | 4 (50) | 2 (100) |
| Female | 4 (40) | 4 (50) | |
| Proton pump inhibitor use | 7 (70) | 7 (87.5) | |
| Acid reducer (H2 blocker) use | 1 (10) | 1 (12.5) | |
| Steroid use | 3 (30) | 3 (37.5) | |
| Feeding tube | 8 (80) | 8 (100) | |
| Charlson Comorbidity Index Score | |||
| 0 | 2 (20) | 0 (0) | 2 (100) |
| 1–3 | 1 (10) | 1 (12.5) | |
| 4–6 | 4 (40) | 4 (50) | |
| 7–9 | 3 (30) | 3 (37.5) | |
| Number of observed species (OTUs), Median (range) | 339.2 (119.5–901.6) | 242.55 (119.5–827) | 829.75 (757.9–901.6) |
| Shannon Diversity Index, Median (range) | 3.08 (0.51–6.05) | 2.3 (0.51–5.11) | 5.99 (5.95–6.05) |
MDRO colonization status
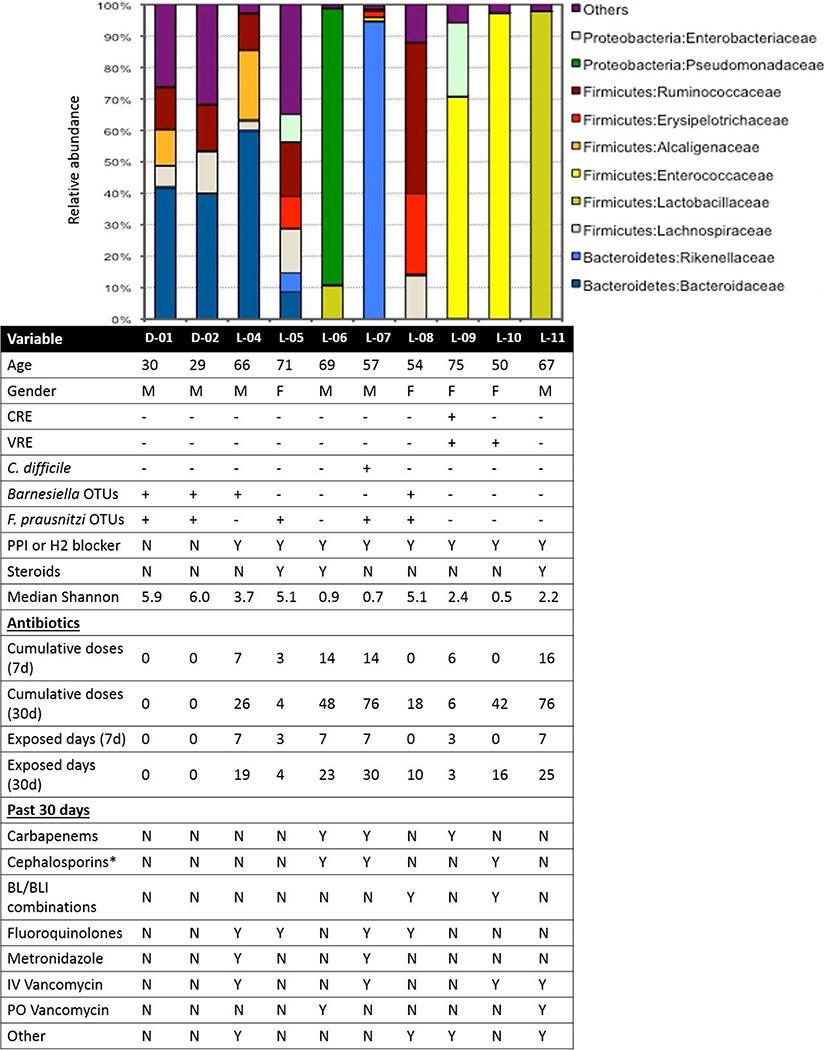
Patient-07 had a positive C. difficile PCR diagnostic test. Two of eight patient stools were positive for VRE by culture (Patient-09, 10), and one was positive for CRE (Patient-09) (Figure 2).
Figure 2.
OTUs identified in each intestinal microbiota were aggregated at the Family taxonomic level and those that constituted ≥1% of sequences in the dataset are plotted. Each color represents a different Family. Sequences present at <1% were grouped into “Others”. The bottom portion of the figure aligns the demographic and clinical data of each patient with their intestinal microbiota.
*Third and fourth generation cephalosporins only.
Next generation sequencing
Family-level microbial community composition for each sample is depicted in Figure 2. Seven of the patient microbiomes examined were dominated (≥30%) by a single OTU. Patient-04 was dominated by Bacteroides fragilis (32%), two patients were dominated by Enterococcus faecium (Patient-09, 67%; Patient-10, 84%), and five were dominated by other OTUs: Patient-06, 88% Pseudomonas aeruginosa; Patient-07, 98% Alistipes finegoldii; Patient-08, 48% Faecalibacterium prausnitzi; Patient-11, 98% Lactobacillus OTUs (57% Lactobacillus salivarius, 41% Lactobacillus rhamnosus). No single OTU dominated the microbiome of Patient-05.
To assess internal validity of our findings, we compared next generation sequencing results and clinical microbiology results. C. difficile sequences (<1%) were identified in Patient-07 stool, who had a positive C. difficile diagnostic PCR. Patient-08 and Patient-09 stools contained OTU sequences matching C. difficile (2%, 9%, respectively), although C. difficile diagnostic PCR performed approximately one week before stool was collected for next generation sequencing was negative. No subsequent C. difficile infections were documented in these two patients. E. faecium and E. faecalis sequences were identified in both patient stools that were culture positive for VRE (Patient-09, 10). Because the genus or species was not identified during CRE screening, we could only confirm whether the clinical microbiology results matched the next generation sequencing results at the family-level. Sequences matching several members of the Enterobacteriaceae family were identified in the CRE-positive patient (Patient-09), including Escherichia coli, Serratia marcescens, and Salmonella enterica.
Microbiome Composition: Diversity
Median Shannon Index was 3.1 (range: 0.5–6.1) overall, 2.3 (range: 0.5–5.1) among patient samples, and 6.0 (range: 6.0–6.1) among donor samples (Table 1, Figure 2). Median Observed Species (OTU count) overall was 339.2 (range: 119.5–901.6), 242.55 (range: 119.5–827.0) among patient samples, and 829.8 (range: 757.9–901.6) among donor samples.
When all individuals were included, the number of Observed Species was inversely correlated with cumulative antibiotic days in the previous 30 days (Spearman correlation coefficient = −0.8, P = 0.009), days exposed to antibiotics in the previous 30 days (Spearman = −0.7, P = 0.02), cumulative antibiotic days in the previous seven days (Spearman = −0.6, P = 0.05), days exposed to antibiotics in the previous seven days (Spearman = −0.6, P = 0.05), and Charlson Comorbidity Index Score (Spearman = −0.8, P = 0.008) (Table 2). Shannon Index correlated inversely with cumulative antibiotic days in the previous 30 days (Spearman = −0.8, P = 0.002), days exposed to antibiotics in the previous 30 days (Spearman = −0.8, P = 0.01), and Charlson Comorbidity Index Score (Spearman = −0.9, P = 0.002).
Table 2.
Correlation between demographic and clinical data, and diversity measures
| Population | Spearman Correlation (P-value) | ||||||
|---|---|---|---|---|---|---|---|
| All individuals | Cumulative antibiotics (30 days) | Days exposed (30 days) | Cumulative Antibiotics (7 days) | Days exposed (7 days) | Age (years) | Charlson Comorbidity Index Score | Days hospitalized before LTACH admission |
| Observed Species (OTUs) | −0.77 (0.009) | −0.72 (0.02) | −0.64 (0.05) | −0.62 (0.05) | −0.44 (0.2) | −0.78 (0.008) | −0.66 (0.04) |
| Shannon Diversity Index | −0.84 (0.002) | −0.77 (0.01) | −0.54 (0.1) | −0.51 (0.1) | −0.35 (0.3) | −0.85 (0.002) | −0.58 (0.08) |
| LTACH patients only | |||||||
| Observed Species (OTUs) | −0.65 (0.08) | −0.55 (0.2) | −0.46 (0.3) | −0.41 (0.3) | 0.05 (0.9) | −0.65 (0.08) | −0.38 (0.4) |
| Shannon Diversity Index | −0.69 (0.06) | −0.55 (0.2) | −0.31 (0.4) | −0.23 (0.6) | 0.29 (0.5) | −0.7 (0.05) | −0.19 (0.7) |
Among patients only, alpha diversity measures only correlated with cumulative antibiotic days in the previous 30 days although not significant (Shannon Index: Spearman = −0.7, P = 0.06; Observed Species: Spearman = −0.6, P = 0.08), and Charlson Comorbidity Index Score (Shannon Index: Spearman = −0.7, P = 0.05; Observed Species: Spearman = −0.7, P = 0.08). Alpha diversity was significantly lower among patients who had received third or fourth-generation cephalosporins during the 30 days before stool collection (Shannon Index: 0.68 versus 3.75; Wilcoxon, P = 0.04; Observed Species: 122.0 versus 404.8; Wilcoxon, P = 0.04). The number of Observed Species was also lower among patients who had received carbapenems in the past 30 days (122.0 versus 404.8, P = 0.07) (Table 2).
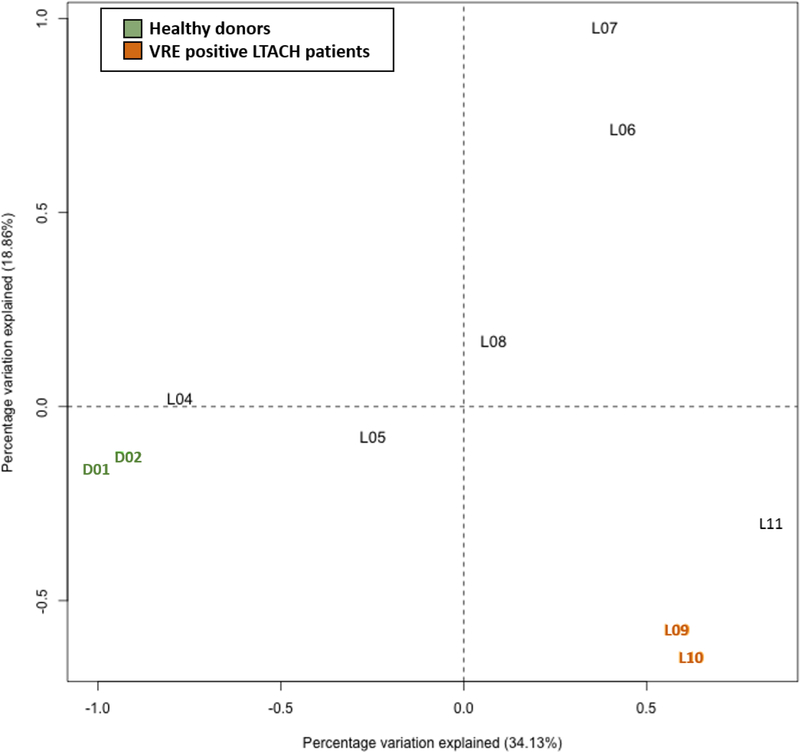
Weighted UniFrac beta diversity analysis results are plotted in Figure 3. The VRE positive patients (LTACH-09, 10) were closest (weighted UniFrac distance=0.26), followed by LTACH-10, 11 (0.53), and the two healthy donors (0.65). Small distances indicates similarity between microbial communities from different individuals.
Figure 3.
Weighted UniFrac Beta diversity results. ‘D’ indicates a donor microbiome. ‘L’ indicates an LTACH patient microbiome.
Discussion
We describe intestinal microbial communities among LTACH patients with diarrhea, in an effort to highlight the potential impact of MDIs on infection control and prevention. In addition to Charlson Comorbidity Index Score, we found that antibiotic exposure in the previous month was inversely correlated with diversity measures, further strengthening the conclusion that cumulative antibiotic exposure is a major driver of microbiome disruption. Disruption could result in poor future outcomes, including infections and transmission (Figure 1). Additionally, although we did not assess outcomes, the significant inverse correlation with the Charlson Comorbidity Index further supports our hypothesis that microbiota characterization might also predict poor future outcomes. Before microbiome characterization or MDIs can become a standard diagnostic test that drives improved patient outcomes, our understanding of the dynamics of microbiome composition needs to be advanced.
The spectrum of microbiome composition among patients ranged from resembling healthy donors, such as in Patient-04 which had a weighted UniFrac distance of 0.81 from the healthy donors), to partial loss of Bacteroidetes (Patient-05) in a patient receiving only inhaled tobramycin, to dominance by E. faecium (Patient-09) and virtual mono-dominance by E. faecium (Patient-10), Lactobacillus (Patient-11), or Rikenellaceae (Patient-07) (Figures 2, 3). Although dominance by Enterococcus in LTACH patients has not been previously described, dominance by Enterococcus has been reported in patients undergoing allogeneic hematopoietic stem cell transplantation and is associated with increased risk for bacteremia.10
The impact of mono-dominance by other bacteria on morbidity and mortality is not well-understood and might vary by patient population. For example, Lactobacillus species are typically thought of as a non-dominant part of the commensal microbiome, including along the digestive tract; however, we found L. rhamnosus and L. salivarius dominated the microbiome of Patient-11. Lactobacillus are frequently resistant to vancomycin (10%−27% susceptible),21 and Patient-11 received oral and intravenous vancomycin before stool collection, which might have enabled overgrowth of Lactobacillus. Intravenous vancomycin has been identified as an independent risk factor for C. difficile infection,22 demonstrating its disruptive potential. Although Lactobacillus can cause infection in sterile sites among highly debilitated patients, they cause infection much less frequently than well-recognized pathobiont MDROs, such as VRE. Consequently, it will be important to gain increased understanding of how dominance by low virulence commensals, such as Lactobacillus, impact outcomes. The genus or species dominating the intestine, and the metabolic functions performed by these dominating bacteria (e.g., Enterococcus versus Bacteroides) may be impacting overall health of the host and could be translated into a MDI. Determining how often dominance by different bacteria leads to infection in specific patient populations will help us better elucidate the impact of mono-dominance by bacteria not generally considered pathogens.
Recent exposures to third- or fourth-generation cephalosporins and carbapenems were common in microbiomes that tended towards lower diversity. This is biologically plausible for carbapenems, which have broad spectrum and distribution in tissues and feces,23 as well as for intravenous ceftriaxone, which is excreted in stool at high concentrations.24 Receipt of third- and fourth-generation cephalosporins has also been identified as an important and widespread risk factor for C. difficile-associated diarrhea.25 The inverse correlation between antibiotic exposures and microbiome disruption reiterates the value and importance of implementing and maintaining an antimicrobial stewardship program to improve prescribing practices, in particular optimizing duration.
This study had several limitations, including that the design was a pilot study in a convenience sample, which prevents us from drawing major, generalizable conclusions, establishing whether C. difficile or MDRO colonization or infection preceded disruption, or performing multivariable analyses controlling for factors previously identified as impacting microbial diversity. However, the intent of the study was to allow us to present the concept of the importance of development of microbiome disruption indices and their potential implications on infection control and prevention efforts. In addition, by targeting patients with diarrhea, we selected for individuals likely to have more disrupted intestinal microbial communities than asymptomatic patients. Donors were younger than patients. Compared to younger adults, intestinal microbial communities vary widely in older populations26,27; however, this may be due to increasing exposure to antibiotics over time..
Previous research showed a relationship between presence of specific obligate anaerobes, Barnesiella species and C. scindens, and absence of VRE and C. difficile, respectively.28,29 In our sample, only the healthy donors, as well as Patient-04 and Patient-08 had Barnesiella species (Figure 2). As predicted, these individuals simultaneously lacked any Enterococcus sequences in their intestinal microbiomes and may be protected from colonization if transmission were to occur. C. scindens was not identified in any of the microbiomes analyzed. In addition, Faecalibacterium prausnitzi, identified as an anti-inflammatory intestinal community member,30 was also found in both healthy donors and three LTACH patients.
Our cross-sectional pilot highlighted several potential MDIs beyond diversity metrics: relative absence of healthy components of the microbial flora, loss of keystone components that fill an ecologic niche and prevent colonization by a MDRO, and presence or dominance by microbial community components that are indicators of microbiome disruption. One example of the need for multiple MDIs, interpreted in context, is Donor-02, whose microbiome had a high proportion of Bacteroides vulgatus (33%), which previous research has suggested can be pro-inflammatory when present at levels higher than observed in the healthy intestinal microbiomes.31 However, this individual’s microbiome also had an alpha diversity score higher than any of the other dominated microbiomes (Figure 2) and contained sequences matching bacteria previously identified as protective and anti-inflammatory (Barnesiella species and F. prausnitzi, Figure 2), indicating that a single measure of the microbial composition is likely insufficient for determining risk for poor outcomes.
There are many host and environmental drivers of microbial composition, including some not easily captured or described, such as antibiotics received early in life, whether breastfed or not during infancy, mode of delivery at birth, diet, co-morbidity history, a history of appendectomy or other gastrointestinal surgery, receipt of certain drugs (other than antibiotics), and the intestinal microbial composition of close contacts.32,33 Because the selection of MDRO genetic determinants within a bacterial species is a relatively rare event following genetic mutation, rearrangement, or translocation, the more common widespread effect of antibiotics on MDRO transmission is mediated via microbial community disruption.34 Even when compared to a reliable, comprehensive antibiotic exposure history dating back several years, MDIs are the more proximate and therefore comprehensive indicators of risk for not only infection, but also transmission. MDIs determined for patients at critical points throughout their course of care hold promise for guiding infection control and clinical management (Figure 1).
We anticipate that MDIs will someday be used to predict the degree and type of disruption that typically result from administration of a specific antibiotic (Figure 1: Footnote 1), as well as describe the risk status of a patient at a given point in time for subsequent colonization (Figure 1: Footnote 2), dominance (Figure 1: Footnote 3), transmission and infection (Figure 1: Footnote 4) by a MDRO. As microbiome analyses become fiscally feasible, interpretation of patients’ MDIs at admission or during care could help identify modifiable patient-, unit- or facility-level factors, including informing infection control measures, such as appropriate use of cohorting, antibiotic selection, and treatments to help patients with poor MDIs return to a more healthy state (e.g., fecal microbiota transplant).
Supplementary Material
Acknowledgements
We would like to thank Duncan MacCannell for early input on bioinformatics tool selection, Jessica Ingersoll and Deborah Abdul-Ali for assistance in the laboratory.
Financial Support: This work was supported in part by the Center for AIDS Research (P30 AI050409 awarded to C.K.)
Footnotes
Potential conflicts of interest: All authors report no conflicts of interest relevant to this article.
Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Data Availability: Raw sequences were placed in the NCBI Sequence Read Archive (SRA) under BioProject ID, PRJNA271791.
References
- 1.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(12):6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foster JA, Bunge J, Gilbert JA, Moore JH. Measuring the microbiome: perspectives on advances in DNA-based techniques for exploring microbial life. Briefings in bioinformatics. 2012;13(4):420–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow J, Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell host & microbe. 2010;7(4):265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prescott HC, Dickson RP, Rogers MA, Langa KM, Iwashyna TJ. Hospitalization Type Predicts Risk of Subsequent Severe Sepsis. American journal of respiratory and critical care medicine. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freedberg DE, Salmasian H, Friedman C, Abrams JA. Proton Pump Inhibitors and Risk for Recurrent Clostridium difficile Infection Among Inpatients. Am J Gastroenterol. 2013;108(11):1794–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez A, Stombaugh J, Lozupone C, Turnbaugh PJ, Gordon JI, Knight R. The mind-body-microbial continuum. Dialogues in clinical neuroscience. 2011;13(1):55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS biology. 2008;6(11):e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taur Y, Xavier JB, Lipuma L, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2012;55(7):905–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donskey CJ, Sunkesula VCK, Jencson AL, et al. Utility of a Commercial Polymerase Chain Reaction Assay and a Clinical Prediction Rule for Detection of Asymptomatic Carriers of Toxigenic Clostridium difficile. Journal of Clinical Microbiology. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siegel JD, Rhinehart E, Jackson M, Chiarello L. Management of multidrug-resistant organisms in health care settings, 2006. American Journal of Infection Control.35(10):S165–S193. [DOI] [PubMed] [Google Scholar]
- 13.Weber DJ, Rutala WA, Miller MB, Huslage K, Sickbert-Bennett E. Role of hospital surfaces in the transmission of emerging health care-associated pathogens: Norovirus, Clostridium difficile, and Acinetobacter species. American Journal of Infection Control. 2010;38(5, Supplement):S25–S33. [DOI] [PubMed] [Google Scholar]
- 14.Gould CV, Rothenberg R, Steinberg JP. Antibiotic resistance in long-term acute care hospitals: the perfect storm. Infection control and hospital epidemiology. 2006;27(9):920–925. [DOI] [PubMed] [Google Scholar]
- 15.Wang XJ, Kraft CS, Dhere T. Use of standard donors in fecal microbiotal transplants. Southern medical journal. 2015;108(1):68–69. [DOI] [PubMed] [Google Scholar]
- 16.Friedman-Moraco RJ, Mehta AK, Lyon GM, Kraft CS. Fecal microbiota transplantation for refractory Clostridium difficile colitis in solid organ transplant recipients. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2014;14(2):477–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevens V, Dumyati G, Fine LS, Fisher SG, van Wijngaarden E. Cumulative antibiotic exposures over time and the risk of Clostridium difficile infection. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2011;53(1):42–48. [DOI] [PubMed] [Google Scholar]
- 18.Dave M, Higgins PD, Middha S, Rioux KP. The human gut microbiome: current knowledge, challenges, and future directions. Translational research : the journal of laboratory and clinical medicine. 2012;160(4):246–257. [DOI] [PubMed] [Google Scholar]
- 19.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. [DOI] [PubMed] [Google Scholar]
- 20.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Applied and environmental microbiology. 2005;71(12):8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cannon JP, Lee TA, Bolanos JT, Danziger LH. Pathogenic relevance of Lactobacillus: a retrospective review of over 200 cases. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2005;24(1):31–40. [DOI] [PubMed] [Google Scholar]
- 22.Dubberke ER, Reske KA, Yan Y, Olsen MA, McDonald LC, Fraser VJ. Clostridium difficile--associated disease in a setting of endemicity: identification of novel risk factors. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2007;45(12):1543–1549. [DOI] [PubMed] [Google Scholar]
- 23.Kager L, Brismar B, Malmborg AS, Nord CE. Imipenem concentrations in colorectal surgery and impact on the colonic microflora. Antimicrobial agents and chemotherapy. 1989;33(2):204–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison CJ, Bratcher D. Cephalosporins: a review. Pediatrics in review / American Academy of Pediatrics. 2008;29(8):264–267; quiz 273. [DOI] [PubMed] [Google Scholar]
- 25.Arango JI, Restrepo A, Schneider DL, et al. Incidence of Clostridium difficile-associated diarrhea before and after autologous peripheral blood stem cell transplantation for lymphoma and multiple myeloma. Bone marrow transplantation. 2006;37(5):517–521. [DOI] [PubMed] [Google Scholar]
- 26.Claesson MJ, Cusack S, O’Sullivan O, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proceedings of the National Academy of Sciences. 2011;108(Supplement 1):4586–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biagi E, Nylund L, Candela M, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PloS one. 2010;5(5):e10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buffie CG, Bucci V, Stein RR, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517(7533):205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ubeda C, Bucci V, Caballero S, et al. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infection and immunity. 2013;81(3):965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(43):16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dicksved J, Halfvarson J, Rosenquist M, et al. Molecular analysis of the gut microbiota of identical twins with Crohn’s disease. The ISME journal. 2008;2(7):716–727. [DOI] [PubMed] [Google Scholar]
- 32.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Claesson MJ, Jeffery IB, Conde S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488(7410):178–184. [DOI] [PubMed] [Google Scholar]
- 34.Tosh PK, McDonald LC. Infection control in the multidrug-resistant era: tending the human microbiome. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2012;54(5):707–713. [DOI] [PubMed] [Google Scholar]
- 35.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for illumina sequences. BMC bioinformatics. 2012;13:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME journal. 2012;6(8):1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and environmental microbiology. 2006;72(7):5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome biology. 2014;15(3):R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a Web browser. BMC bioinformatics. 2011;12:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26(2):266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Molecular biology and evolution. 2009;26(7):1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.