Abstract
Stem cells (SCs) have been proven to possess regenerative and immunomodulatory properties and can be utilized to treat diseases that involve loss of cells due to tissue damage or inflammation. For this approach to succeed, SCs or their derivatives should be able to engraft in the target tissue at least for at a short period of time. Unfortunately, once injected, therapeutic SCs will encounter a hostile environment, including hypoxia, lack of nutrients and stromal support, and cells may also be targeted and rejected by the immune system. Therefore, SC’s stress-response mechanisms likely play a significant role in survival of injected cells and possibly contribute to their therapeutic efficacy. Autophagy, a stress-response pathway, is involved in many different cellular processes such as survival during hypoxia and nutrient deprivation, cellular differentiation and de-differentiation, and it can also contribute to their immunovisibility by regulating antigen presentation and cytokine secretion. Autophagy machinery interacts with many proteins and signaling pathways that regulate SC properties including PI3K/Akt, mTOR, Wnt, Hedgehog, Notch and it is also involved in regulating intracellular ROS levels. In this review, we contend that autophagy is an important therapeutic target that can be utilized in order to improve the outcome of SC-based tissue repair and regeneration. Further research should reveal whether inhibition or stimulation of autophagy increases the therapeutic utility of SCs and it should also identify appropriate therapeutic regiments that can be applied in the clinic.
Introduction
Many organisms possess high regenerative capacity and can replace lost body parts or damaged tissues. Some invertebrates such as planaria or hydra are capable of regenerating whole bodies, while numerous vertebrates maintain mechanisms for efficient regeneration of specialized tissues, including muscle, bone, nerves, and blood vessels [1]. Higher vertebrates and mammals, however, do not have a corresponding capacity for tissue replacement and the limited existing regenerative properties diminish with aging [2]. In humans, primarily liver and skin demonstrate a high regenerative capacity while the remainder of human organs do not demonstrate this capacity. More significant injuries and degenerative disease in organs such as the heart are therefore frequently associated with a high mortality [3, 4]. Consequently, novel approaches such as treatments with therapeutic SCs are needed in order to improve structure and function of damaged tissues and organs due to injuries and/or diseases. Therapeutic potential of SCs and thus overall tissue regeneration capacity can be improved by pre-treatment with small molecules, biologics or combinations with biomaterials that target specific aspects of SC biology. This will modulate their interactions with other cells and tissue components to avoid the immune response, facilitate cell engraftment and/or cell expansion, accelerate cell differentiation, or de-differentiation. Current research efforts are directed toward understanding what aspect(s) of SC biology should be targeted to improve their therapeutic properties.
Therapeutic application of SCs
Therapeutic SCs are a key component of regenerative medicine; they have the ability to differentiate into many different cell types and tissues and thus, can be utilized for numerous clinical applications, including tissue regeneration [5]. Hematopoietic SCs (HSC) transplantation, in which autologous or allogeneic HSCs are collected from bone marrow and injected into a patient in order to restore healthy immune system, is a well-established procedure [6]. Numerous clinical trials are now testing other therapeutic cells for injuries and diseases such as neurological disorders, cancer, heart disease, diabetes and other conditions [7]. The primary strategy of cell therapies is to increase the number of functional cells in the damaged or diseased tissue [8]. The US National Library of Medicine reports 7,031 clinical trials (January 2019), either ongoing or completed, that utilize “stem cells” (http://www.clinicaltrials.gov/).
There are three main types of SCs: embryonic, adult, and induced pluripotent SCs [9]. Human embryonic SCs (hESCs) are derived from developing blastocyst-stage embryos and can be cultured in an undifferentiated state [10]. Compared to other SC types, hESCs are more suitable for cell-based tissue regeneration therapies due to their differentiation potential. However, formation of benign tumor tissues (teratomas) has been observed in some pre-clinical studies raising the question of potential side effects of such therapies [11]. Nevertheless, hESCs can potentially be used in a wide range of conditions either in their undifferentiated or differentiated states. For example, formation of myocardial grafts was observed in a rat model of cardiac infraction upon injection of hESC-derived cardiomyocytes [12] which was also observed in mice transplanted with human cardiomyocytes [13]. Similarly, murine model of liver damage was treated with hepatocyte-like cells derived from hESCs and both cell replacement and delivery of trophic factors that contributed to liver regeneration were observed [14].
Adult SCs (ASCs) can be obtained from bone marrow, dental pulp, umbilical cord blood, amniotic fluid, and several other tissues and can be differentiated into a variety of cell types [15]. However, the range of differentiation properties of ACSs is limited; hence, typically such cells can only form cell types from the same lineage as the tissue of origin [16]. ASCs have been extensively studied as therapeutics. Several preclinical trials demonstrated that transplantation of autologous bone marrow cells or precursor cells improved cardiac function after myocardial infarction and in chronic coronary heart disease. In animal models of spinal cord injuries, ASCs provided sensorimotor benefits [16]. Mesenchymal stromal cells (MSCs) are a sub-type of adult SCs found throughout adult tissues that have attracted attention in clinics due to their abundance and pleiotropic effects [17]. These MSC properties allow for their use in multiple therapeutic applications, in addition to tissue regeneration. MSCs possess high immunomodulatory capacity and could aid in the treatment of injury associated with or caused by inflammatory activit y[18]. Such immune modulatory properties of MSCs were successfully utilized in a seminal study by Le Blanc et al. in which haploidentical MSCs were used to treat a patient with severe treatment-resistant grade IV acute graft-versus-host disease (GVHD) of the gut and liver [19]. Immunomodulatory benefits of MSCs were also observed in a wide-range of pre-clinical models of inflammatory diseases such as graft-versus-host disease (GVHD), multiple sclerosis (MS), rheumatoid arthritis (RA), and sepsis [20–28]. A number of other studies also evaluated MSC-mediated immunomodulation and established that administration of MSCs, either systemically or locally, induce T cell anergy, reduce the numbers of pro-inflammatory T helper 17 cells and also increase the numbers of immunosuppressive regulatory T cells, capable of suppressing T-effector responses [29].
Irrespective of their tissue of origin or downstream application, therapeutic cells are typically stressed during the isolation and expansion procedures as well as during and after injection, resulting in increased number of senescent and apoptotic cells followed by disappointingly low tissue retention after delivery [30–32]. Current research efforts are directed toward an increased understanding of the effects of hypoxia, nutrient deprivation, immune responses, and other stress-response mechanism(s) on survival of therapeutic cells [33–36]. These efforts will likely improve our understanding of the cell fate dynamics following systemic administration and explain the effects of variables in the production of such cells, which may be critical in maintaining of their therapeutic properties [37]. Moreover, stress-response mechanisms likely have an important function in survival of therapeutic cells as they contribute to cell differentiation, attachment to surrounding tissues and their immunovisibility. Autophagy plays pleiotropic roles in cellular homeostasis, the most important of which is allowing cell survival during nutrient deprivation; this pathway can also prolong survival in other adverse situations such as toxic insult(s) or detachment from extracellular matrix. In this review we address potential roles of this pro-survival stress-response pathway in determining the outcomes of SC-based therapies.
Mechanistic aspects of autophagy
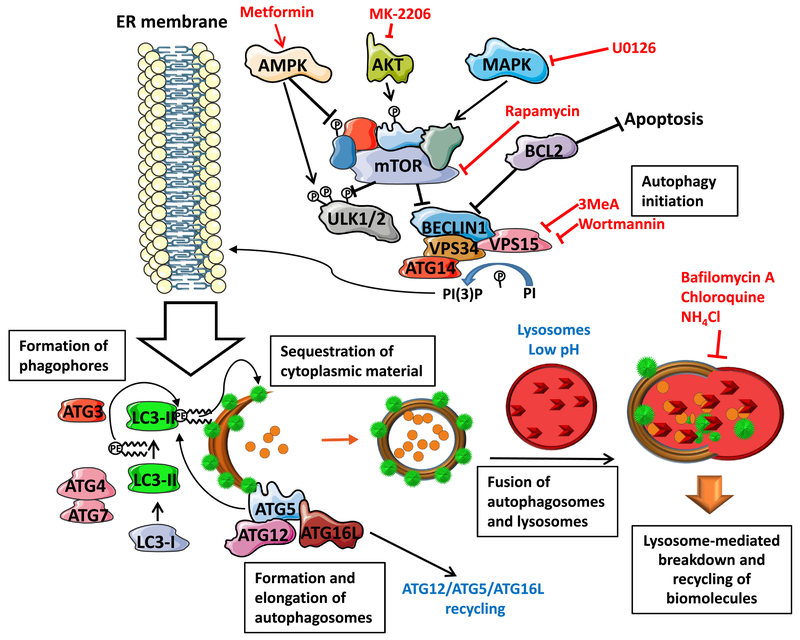
Macroautophagy (referred to as autophagy herein) is an evolutionary conserved stress-response homeostatic mechanism responsible for selective or non-selective breakdown of cytoplasmic constituents (reviewed in [38]). While autophagy was originally described and studied as the pathway upregulated during starvation, it is now recognized that autophagy plays pleiotropic roles in regulating metabolism, cellular remodeling, quiescence, response to pathogen invasion, antigen presentation and in many other processes [39–43]. Mechanistically, autophagy is remarkably similar between yeast and mammals and genetic experiments in Saccharomyces cerevisiae allowed for reconstitution of the pathway which exists in both unicellular and multicellular eukaryotes [44]. Autophagy delivers diverse substrates such as cytoplasmatic components, damaged organelles, protein aggregates, or infectious particles engulfed in double-membrane vesicles (autophagosomes) to lysosomes for degradation (Figure 1). Engulfed substrates can be either non-specific cytoplasmic constituents or organelles such as damaged mitochondria or peroxisomes [45]. Recycling of intracellular components is especially important in SCs as it facilitates cytoplasmic remodeling during differentiation and de-differentiation (see below).
Figure 1:
Key players in autophagosome formation. mTOR protein complex responds to the metabolic state of the cell via multiple signaling axes. Such signaling molecules sense changes in cellular homeostasis, and, by default, mTOR blocks induction of autophagy. However, increased cellular stress blocks mTOR signaling initiating the pathway, which, in addition to ATG1 kinase requires several other kinases such as PI3K III and VPS34. BECLIN1 is a scaffold protein inhibited by the oncogene BCL2 that inhibits both autophagy and apoptosis initiation. Lipid molecules incorporated into phagophore membrane are not synthesized de-novo but originate in the endoplasmic reticulum (ER). The elongation of autophagosomes is facilitated by formation of ATG12-ATG5-ATG16L protein complex and by LC3-II protein (indicated by green circles) which is lipidated and associated with the autophagosomal membranes. Proper autophagosomes processing also requires ubiquitin-like conjugating enzymes ATG3, ATG7 and ATG10 and protease ATG4 which catalyzes LC3-I to LC3-II conversion. After the membrane enclosure, only ATG12-ATG5-ATG16L1 protein complex dissociates from the autophagosomes - LC3-II remains associated with autophagosomal membrane and is degraded in lysosomes together with other cytoplasmic molecules targeted for degradation. Various small molecules targeting distinct molecules are listed in red.
Autophagy proteins function in complexes and the degradation of substrates is achieved by dynamic regulation of autophagy protein synthesis, their degradation as well as post-translational modifications, leading to temporal and spatial control of the pathway (Figure 1). The most upstream kinase in the pathway, uncoordinated 51-like kinase 1 (ULK1) is responsible for the recruitment of phosphatidylinositol (PI) 3-kinase (PI3K) catalytic subunit 3 (VPS34) to the pre-autophagosomal membrane structure known as phagophore [46]. Beclin-1 is a scaffold protein which forms a complex with the anti-apoptotic oncogene BCL-2 leading to suppression of autophagy. The c-Jun N-terminal kinase 1 phosphorylates BCL-2 during starvation resulting in its dissociation from Beclin-1 and autophagy induction [47]. Moreover, autophagy-related gene 14L (ATG14L) promotes ULK1-mediated phosphorylation of Beclin-1 to facilitate autophagy [48]. ATG7 and ATG10 are E1- and E2-like enzymes, respectively, that activate ATG12 C-terminal to form a conjugate with ATG5, and subsequently form a protein complex with ATG16L1 [49, 50]. This complex acts as an E3-like enzyme for the conjugation of ATG8 family of proteins to the phosphatidylethanolamine (PE), which results in its tight association with autophagosomes [51]. The cysteine protease ATG4 cleaves C-terminal arginine residues from ATG8 leading to activation of the C-terminal glycine residue of ATG8 which conjugates to PE in the presence of ATG5/ATG12/ATG16 complex [52]. This complex is recycled upon autophagosome closure, but ATG8 (represented as a green dot on Figure 1) remains associated with the autophagosome and is degraded in an autolysosome after fusion of autophagosomes with lysosomes [53].
Because autophagy is upregulated during starvation and deprivation of pro-survival signaling, it is not surprising that pro-proliferation/survival signaling pathways such as PI3K/AKT or MAPK pathways suppress autophagy pathway (Figure 1) [54]. AKT is a positive regulator of the mammalian target of rapamycin (mTOR) complex; this complex, when activated, blocks transcriptional response to starvation and also inactivates the autophagy-initiating kinase ULK1 [55]. AKT also phosphorylates and inactivates the FOXO3a transcription factor which primarily controls expression of genes involved in cell growth and differentiation [56]. This AKT-mediated phosphorylation also prevents FOXO3a from promoting autophagy by suppressing expression of numerous core ATG genes involved in autophagic vesicle formation [57, 58]. On the other hand, 5’ adenosine monophosphate-activated protein kinase (AMPK), an intracellular energy sensor which detects changes in the AMP:ATP ratio, inhibits mTOR complex and activates autophagy when ATP levels are low by phosphorylating ULK1 at Ser317 and Ser777 [59].
In addition to low energy sensors, many other stressors are also known to regulate autophagy, either alone or parallel with starvation. Likewise, many transcription factors that regulate gene expression upon stress induction are also associated with autophagy regulation. For example, hypoxia-inducible factor 1 which regulates gene expression under hypoxic conditions, also regulates mitophagy (see below) under low oxygen [60]. Nuclear factor kappa B (NFκB) family of transcription factors regulate responses to inflammation, immune responses, development, and metabolism, also regulate autophagy indirectly by interacting with transcription factors directly involved in controlling genes with roles in autophagy regulation. Interestingly, it was also shown that autophagy can impact on NFκB signaling, which could provide a signaling feedback loop. For example, in autophagy-competent mouse embryonic fibroblasts (MEFs), autophagy triggering upon starvation or treatment with Tumor Necrosis Factor a led to nuclear translocation of NFκB, but in MEFs lacking autophagy genes ATG5 or ATG7 no such translocation was observed [61].
Due to its involvement in diverse signaling pathways important for SC homeostasis, it is possible to use modulation of autophagy as a tool to change the properties of SCs, from changing of how they respond to inflammation, or their ability to differentiate or de-differentiate, or to re-program their transcription profile to make them more prepared for survival upon injection into diseased/damaged tissues. Next, we summarize some key relationships between autophagy and cellular processes/pathwavs which are thought to be important for the therapeutic efficacy of SCs.
Self-eating facilitates self-regeneration and survival of SCs under stress conditions
The classical role for autophagy is to provide energy during nutrient depravation (starvation) by non-selective recycling of cytoplasmic material which results in increased ATP/ADP levels (as described above). Moreover, autophagy also contributes to normal homeostasis by removal of aggregated or toxic macromolecules and/or damaged organelles, thus acting as a “quality control” mechanism[62]. For example, one of the key specialized functions of autophagy, mitophagy, removes damaged and leaky mitochondria, which are targeted for transport into autophagosomes and degraded in lysosomes resulting in control of intracellular reactive oxygen species levels (ROS) [63]. Mitophagy plays a protective role in cells by removing leaky and de-polarized mitochondria following mitochondrial permeability transition pore opening. This ability to remove organelles and larger pieces of cytoplasm is also useful for cells which need to undergo “remodeling and refreshing” such as endogenous SCs that differentiate at the site of tissue injury. For example, in a mouse model of partial hepatectomy with a liver-specific knockout of a key autophagy gene Atg5, liver regeneration was severely impaired by 70%. Furthermore, increases in size of hepatocytes were observed in mice together with accumulation of damaged mitochondria, p62 and ubiquitinated proteins and a decrease in ATP levels [64]. Similarly, pharmacologic inhibition of autophagy by 3-methyladenine or stimulation by β-Guanidinopropionic acid [65] in a mouse model of muscle injury had a profound effect on mitochondrial remodeling and the speed of the overall muscle regeneration in vivo [66].
This unique dual role of autophagy in both providing energy during starvation and removal of cytoplasmic “waste” has been reported for both human and mouse muscle SCs (“satellite cells”), which are responsible for regeneration of skeletal muscle [67]. At the geriatric age, satellite cells enter senescence resulting in a loss of regenerative capacity and muscle mass. Basal autophagy flux in nondividing cells such as satellite cell has been identified as a key factor that contributes to maintenance of stemness by attenuating both proteotoxicity and ROS levels [68]. Consistently, genetic or pharmacological stimulation of autophagy in geriatric satellite cells reversed senescence and conversely, autophagy inhibition in young cells caused their entry into senescence. Therefore, autophagy in SCs is directly responsible for their regenerative capacity and increased autophagy translates into a higher capacity for regeneration [69, 70].
Ischemia and hypoxia/serum deprivation (HSD) can be unregulated in exogenously administered therapeutic SCs leading to cell death. This can be prevented by pharmacological stimulation of autophagy which has been demonstrated in cells treated with the AMPK stimulator atorvastatin [71]. While HSD induced autophagosome formation in MSCs, treatment with atorvastatin further increased autophagic activity and decreased apoptosis, and the autophagy inhibitor 3-methyladenine consequently blocked this effect. Altogether, these results support the hypothesis that manipulation of autophagy might enhance survival of MSC during HSD [71]. Similarly, HSCs reside in tissues where their homeostasis is adapted to low nutrient supply. Therefore, in order to expand HSCs ex-vivo, autophagy stimulation with mTOR inhibitors is used to improve long-term maintenance of HSCs [72].
Autophagy contributes to “stemness” and facilitates SC differentiation
SCs have a capacity to differentiate into specialized cells but they can also remain undifferentiated for longer periods of time. This propensity to remain in an immature state without exhaustion is frequently referred to as “stemness” or “self-renewal capacity.” Self-renewal is one of the key properties that defines SCs and is mediated to a large extent by evolutionary conserved signaling pathways such as Hedgehog. Wnt and Notch. These pathways are increasingly recognized for their key roles in SC maintenance, development and cell fate. Autophagy-developmental pathway cross-talk has been established previously indicating that modulation of autophagy can impact on CSs’ ability to both self-renew and differentiate and repopulate damaged tissue(s).
The existence of cross-regulation between developmental pathway signaling and autophagy was demonstrated in both mammalian cells and in Drosophila where the Hedgehog signaling pathway regulates autophagy [73]. This regulation depends on transcription factors Gli2 and Ci and was mediated by the two transmembrane receptors Ptchl and Ptch2, and also includes the ER stress response protein PERK and its downstream effector, eukaryotic translation initiation factor 2A (eIF2a). These results implicate Hedgehog signaling in controlling protein homeostasis via regulation of autophagy.
Wnt signaling is important for the maintenance of cell stemness. Wnt binds to its receptor Frizzled, which leads to activation of proteins Dishevelled and β-catenin, resulting in activation of transcription factor TCF4 and increased transcription of its target genes [74]. It was shown that the loss of TCF4 leads to depletion of SCs in the crypt of the colon [75, 76]. Moreover, the evidence also implicates Wnt pathway in self-renewal of the hematopoietic system as well as several other organ systems [77]. It was shown that autophagy regulates Wnt signaling by facilitating Dishevelled degradation via its ubiquitylation by Von Hippel–Lindau protein, leading to binding of p62, its aggregation, and LC3-mediated recruitment to autophagosomes under starvation, which ultimately results in its degradation via the autophagy-mediated lysosomal degradation pathway [78].
The Notch pathway plays crucial roles both in disease and development, from embryogenesis to adulthood [79]. Additionally, it has an important function in neural SC maintenance and neuronal development where activation of Notch signaling induces the expression of transcriptional repressor genes leading to maintenance of neural progenitor cells [80]. Upon activation by proteolytic cleavage, Notch1 protein is internalized via endocytosis and targeted to the nucleus. In addition to endocytosis, activated Notch1 co-localizes with the autophagy protein ATG16L and is sequestered into autophagosome-precursor vesicles and subsequently degraded. Moreover, a mouse model with an Atg16L hypomorphic mutation shows delays in differentiation and development in several tissues, indicating that even modest changes in autophagy can impact tissue development [81].
Not surprisingly, these developmental signaling pathways have been investigated (mostly) for cancer therapy and a number of inhibitors have been tested in pre-clinical or clinical trials. For example. Vismodegib was the first Hedgehog signaling pathway targeting molecule to gain FDA approval in 2012 for the treatment of basal-cell carcinoma. Bone-derived Wnt inhibitor Sclerostin can be targeted with an antibody to treat osteoporosis and other inhibitors are in pre-clinical trials now. However, whether autophagy plays a role in the activity of such therapeutics or how they can be utilized to “boost” therapeutic properties of SCs remains unknown at this time.
While throughout their existence SCs need to maintain “stemness”, they also need to be able to quickly undergo differentiation into specialized cells when needed. Dynamic and rapidly inducible, autophagy can quickly facilitate changes that are needed during cellular differentiation when intracellular remodeling is needed. Upon injury, or as a consequence of a disease, bone marrow or tissue resident multipotent SCs are recruited to sites of injury to support tissue regeneration by differentiation and re-populating damaged tissues or by secretion of factors that support tissue repair [82]. This activity puts a certain pressure on migrating cells that may increase the requirements on cellular homeostatic mechanisms to provide additional energy and activation of cell survival mechanisms [83]. Prior to ex-vivo differentiation into osteocytes and adipocytes, bone marrow MSCs have been shown to accumulate autophagosomes without a simultaneous increase in autophagic flux, whereas induction of differentiation has been demonstrated to decrease the number of autophagosomes. Additional stimulation of the autophagic flux with Rapamycin decreased the number of autophagosomes and delayed adipogenic differentiation indicating that well-regulated autophagy facilitates efficient cell differentiation [84].
SCs possess a high capacity for differentiation and recent data indicate an essential role for autophagy in these processes. Pharmacological blocking of autophagy by 3-methyladenine for 24 h or shRNA-mediated knockdown of an essential autophagy gene ATG5 in epidermal, dermal, or hematopoietic SCs, blocked both their ability to self-renew and completely blocked cell differentiation, confirming essential roles for autophagy in both maintenance of cellular stemness and differentiation [85]. Epithelial-mesenchymal transition (EMT) is a process opposite to cellular differentiation and represents a critical response during cancer cell metastasis. Pharmacologic or genetic disruption of mTORCl signaling significantly enhanced EMT in A549 non-small lung cancer cells, indicating a crucial role of autophagy in both differentiation and de-differentiation, i.e. cellular “re-modeling” [86].
Autophagy and SCs’ response to inflammation
Because successful application of SCs in clinics depends, in part, on their ability to modulate inflammation by decreasing immune cell activation, it is quite surprising that very few studies addressed the role of autophagy in this context. Autophagy has prominent regulatory roles in both innate and adaptive immunity in multiple cell types and current data indicate that it can participate in both anti- and pro-inflammatory immune responses (reviewed in [40]). A balance between these activating and inhibitory signals determines the net outcomes in terms of autophagy-mediated induction or inhibition of immune responses. For example, molecular pattern recognition receptors such as NOD-like, RIG-I-like or Toll-like receptors all play a role in invading pathogen recognition and elimination and they were also shown to interact with the autophagy machinery [87]. Moreover, cytoplasmic debris of endogenous or exogenous origin can be cleared by autophagy and thereby prevent activation of the inflammasome. There are two instances where autophagy plays a main role in immune responses that are highly relevant to therapeutic cells: antigen presentation and cytokine secretion. Autophagy supports processing of intracellular antigens for MHC class II presentation: in starved cells, MHC class II presentation of cytosolic and nuclear antigens was increased by 50% after starvation [88]. Furthermore, when proteins of various origin were fused to LC3B, their MFIC class II presentation to T cells was increased 4-20 fold [89]. Presence of cytokines such as IFN-γ, TNF-α, TGF-β, IL-1, IL-2, IL-6 (pro-inflammatory), as well as IL-4, IL-10 and IL-13 (immunomodulatory) can affect formation of autophagosomes [90, 91]. Moreover, production and secretion of cytokines such as Type I IFN, TNF-α, IL-1, and IL-18 can be regulated by autophagy [92]. Autophagy can also contribute to cell survival in the presence of inflammation stimulation. For example, MSCs treated with a combination of tumor necrosis factor-α and cyclohexamide elevated both autophagic flux and apoptosis. Furthermore, pharmacologic or genetic inhibition of autophagy during this treatment accelerated apoptosis; however, induction of autophagy with rapamycin treatment decreased apoptosis [93]. Such data indicate protective role of autophagy in inflammation-mediated apoptosis supporting the notion that survival of MSCs may be enhanced by autophagy manipulation. This is both relevant to endogenous and exogenous (injected) SCs that respond and home to the site of inflammation to both engage in cell-cell contact with immune cells and to secrete factors with a role in inflammation dampening. Furthermore, because autophagy can contribute to both antigen presentation and secretion of cvtokines/extracellular vesicles in stromal cells, it may also influence immunovisibilitv of SCs.
Cell-cell and cell-extracellular matrix-mediated autophagy regulation
Any change and re-structuring of SCs in stratified tissues requires extensive reprogramming of both signaling networks and gene expression - cells must be able to adapt, differentiate, and survive. This is especially important for therapeutic cells which are typically injected without the matrix support into tissues, and, in addition to the high rate of apoptosis during cell isolation and engraftment, apoptosis can also occur in SCs due to the lack of proper attachment to surrounding tissues. Autophagy is induced upon loss of integrin-mediated cell attachments to the surrounding extracellular matrix to protect cells from detachment-induced apoptotic mechanism termed “anoikis.” Fleterodimeric transmembrane proteins termed integrins mediate cell adhesion by holding basal cells to the basement membrane, facilitating the interaction with signaling molecules and maintain the connection between the extracellular matrix and the cytoskeleton[94]. Disruption of integrin-interactions and loss of cell adhesion activates anoikis[95]. In differentiating epithelium, anoikis can be delayed by induction of autophagy; for example, suspended human mammary epithelial MCF10A and canine kidney epithelial cells survived anoikis by upregulation of autophagy and, consequently, genetic inhibition of autophagy in suspended cells increased caspase-3 cleavage[96]. Remarkably, cellular attachment may also promote autophagy, although this effect is the most prominent during starvation. Blocking integrin subunits with antibodies inhibited both autophagy and survival in primary basal prostate epithelial cells in a mechanism that does not depend on PI3K pathway and thus likely proceeds via non-canonical autophagy[97].
MSCs are usually cultured as adherent cells in plastic flasks. Before injection these cells are detached by trypsination, where surface receptors and integrins are removed. This preparation procedure could affect the anchoring capacity of the infused MSCs and thereby the induction of apoptosis. One approach to increasing MSCs survival after infusion might be manipulating autophagy and/or combining MSCs with matrix support. Autophagy is also activated during the process of entosis when a cell-cell adhesion in the absence of integrin adhesion leads to entry of one cell into another, a process that is reminiscent of pathogen engulfment [98]. Therefore, autophagy may be a key mechanism that can prolong survival of injected cells without extracellular matrix support and provides an opportunity for therapeutic intervention.
While autophagy is mostly regulated and executed at the cellular level, it can also be regulated via factors secreted by adjacent cells by secreted factor interactions with membrane receptors[99]. For example, autophagy can contribute to cell-cell interactions and thus modulate adaptive immune responses by contributing to both dendritic cell (DC)–T cell interactions and DC-epithelial cell interactions [100]. DC–T cell interactions upregulate autophagy in DCs and the molecules mediating this interaction are degraded by the autophagosome. Suppression of autophagy renders this interaction hyperstable resulting in increased T cell activation [101].
Several studies have also shown that MSCs can trigger autophagy in damaged cells to enhance regeneration and/or to promote cell survival. A model of acute rat liver injury by intraperitoneal injection of carbon tetrachloride (CCl4) was used to investigate apoptosis and autophagy in injured hepatic cells following transplantation with chorionic plate-derived MSCs (CP-MSCs) isolated from human placenta [102]. Transplantation of CP-MSCs to CCl4-injured liver tissue decreased caspase 3/7 activities but increased expression of several autophagy proteins and cell survival, up to three weeks after cell injection. Similarly, in co-cultures of CCl4-injured primary rat hepatocytes and CP-MSCs, a decrease in the number of necrotic cells and increase in autophagic markers was observed, indicating that administration of CP-MSCs promotes repair in part via autophagy-related mechanism(s).
While the exact mechanistic basis for cell-cell interaction-mediated autophagy regulation is unclear, it is known that integrins can activate PI3K/AKT signaling [103]; moreover, protein serine/threonine kinase 11 activates AMPK signaling upon co-localization with integrin-interacting protein E-cadherin [104]. These signaling mechanisms could explain the connection between autophagy and cell-cell contact maintenance and detachment.
Targeting autophagy for improvement of SC-based therapies
The roles of autophagy in human health, disease, and responses to therapies are now hot topics that are being studied extensively and with good reason - this pathway mitigates a wide range of stress conditions by removing various cytoplasmic components, including toxic metabolites and its targeting could have important application in the clinics. The most efficient approach to targeting autophagy in vivo is still being evaluated but the first deliberate attempts to modulate autophagy therapeutically (based on pre-clinical data) are currently being examined in clinical trials involving hydroxychloroquine (HCQ) in cancer patients [105, 106]. Based on the data from these trials, autophagy can be inhibited in vivo with chloroquine derivatives leading to therapy-associated increases in autophagic vesicles and LC3 cleavage in both peripheral blood mononuclear cells and tumor cells in a concentration-dependent manner. No evidence of metabolic toxicity, liver injury or neurologic side-effects were observed in these trials and less than 10% of patients developed severe side-effects. While these data are collected from clinical trials involving development of combination treatments for cancer, pharmaceutical targeting of autophagy for treatments of other conditions can be further complicated by the roles that autophagy plays in different cell types which, to a large extent, depends on cellular metabolic requirements. Because of such diverse (and frequently opposing) roles autophagy plays in these processes, the overall sum of the effects is difficult to predict without experimental assessment. Therefore, extensive set of experiments are required in pre-clinical disease models to generate enough data that can be used for optimizing autophagy-targeting treatments in clinical trials.
Therapeutic SCs have an immense potential in the clinics as SCs can carry out many functions which cannot be performed by small molecules and thus have a potential to address critical, unmet needs in the development of regenerative medicine approaches. The success of SC-mediated tissue regeneration relies on the capacity of donor cells to integrate with the damaged tissue, promote tissue healing via secretion of paracrine factors, and decrease inflammation-associated cell injury. Upon injection, SCs sense their surroundings and adapt responses to appropriate physiologic conditions, which includes differentiation and immunomodulation among other functions. Such adaptation may require cytoplasmic re-modeling and cellular pre-conditioning to increase re-modeling capacity may also lead to improvement of responses upon SC injection. Because therapeutic SCs encounter hostile environment upon injection, stress-response mechanisms are responsible for prolonging cellular survival and likely contribute to therapeutic properties of SCs. Therefore, cellular pre-conditioning or administration of therapeutic SCs together with stress-response modulators should be considered as means to increase therapeutic benefits of SC therapies. Thus far, SC therapies have mostly been administered as a single therapy. Future pre- and clinical studies should consider combining SCs with therapies which can improve cell survival by up-regulating stress response pathways such as autophagy. Complexity of this approach also arises from the potential to target autophagy only in SCs before injection, only in the host, or simultaneously in both injected cells and the host. The effects of such approaches can be dramatically different. For example, stimulation of autophagy in SCs prior to injection may prolong their survival after administration but it can also make them more immunovisible by stimulating antigen presentation in SCs. Furthermore, because autophagy is reversible, the effect of autophagy modulation in SCs could be only temporary and may not have any measurable impact on their therapeutic efficacy. Similarly, pre-treatment of the host with autophagy modulators may suppress immune responses toward injected cells but it may not shield them from apoptosis. For example, both autophagy inducer rapamycin and autophagy inhibitor chloroquine can suppress immune responses and could be used to prolong SC engraftment. However, this will not impact on other adverse factors (outlined above) which can limit the efficacy of SCs by promoting their apoptosis. The third approach in which small molecules are used to modulate autophagy immediately upon SC administration could be the most efficient one: this would allow for simultaneous and controlled changes in metabolism in all cells involved and it could be facilitated by sets of ex vivo experiments that will assess efficient combinations as the interplay between these factors is rather complex but can be quickly evaluated experimentally. Along those lines, autophagy alone has been shown to bear some regenerative benefits - in Table 1 we list autophagy modulating drugs that are shown to impact tissue regeneration. However, it is still unclear how much autophagy actually contributes to the observed effects on tissue regeneration.
Table 1:
Autophagy modulating drugs and their effect on tissue regeneration; The mechanistic aspects of the compounds and their effects on autophagy are explained in detail in [117]; S – stimulator, I – inhibitor
| Drug | I/S | Effect | Reference |
|---|---|---|---|
| Rapamycin | S | Prevented loss of adult SCs and reversed age-related loss of stem cells in mouse trachea | [107] |
| Chloroquine | I | Inhibits autophagy-associated inflammation and ER stress in rats during their recovery from acute spinal cord injury | [108] |
| Resveratrol | S | Enhances the functionality and improves the regeneration of periodontal ligament mesenchymal stem cell aggregates | [109] |
| Bafilomycin A | I | Impairs the regeneration of amputated caudal fins in the zebrafish | [110] |
| Metformin | S | Treatment stimulated bone lesion regeneration in control and diabetic rats | [111] |
| Retinoic acid | S | Induces the complete regeneration of alveoli that have been destroyed by various noxious treatments | [112] |
| Melatonin | S | Treatment significantly reduces scar formation in nerve stump and collagen synthesis in the granulation tissue | [113] |
| Compound C | I | Blocks MSC-mediated repair of tight junction by inhibiting AMP-activated kinase | [114] |
| Mdivi-1 | I | Treatment ameliorated ischemia-reperfusion injury-induced functional deterioration with improvement in angiogenesis | [115] |
| Spermidine | S | Enhances and rejuvenates cardiac cells and improves mechano-elastical properties of cardiomyocytes | [116] |
Unanswered questions and future directions
Despite tremendous progress made towards mechanistic elucidation of the autophagy, its value in translational research has not been properly assessed. This review highlights potential importance of autophagy in interactions of therapeutic SCs and the host which are summarized in Figure 2. The complexity of cell-cell interactions and the possible multiple ways autophagy modulation can be applied led to new questions which will need to be addressed. Some of these are: What are the crucial interactions between tissues and SCs that we need to focus on in order to maximize clinical relevance/impact? What is the relationship between mechanisms mediating regenerative properties of SCs and stress response pathways such as autophagy? To what extent does the biological heterogeneity of cellular responses to autophagy modulation affects outcomes? Does autophagy in SCs alone contribute to tissue repair? To what extent does metabolic modification of SCs can improve their regenerative properties? Are there specific SC stress-response pathways that can be targeted more efficiently than others to improve SC’s therapeutic efficacy? Is it better to apply genetically modified CSs or combine them with small molecules? Answering some of these questions will pave the way for applying these therapeutic modalities in clinic.
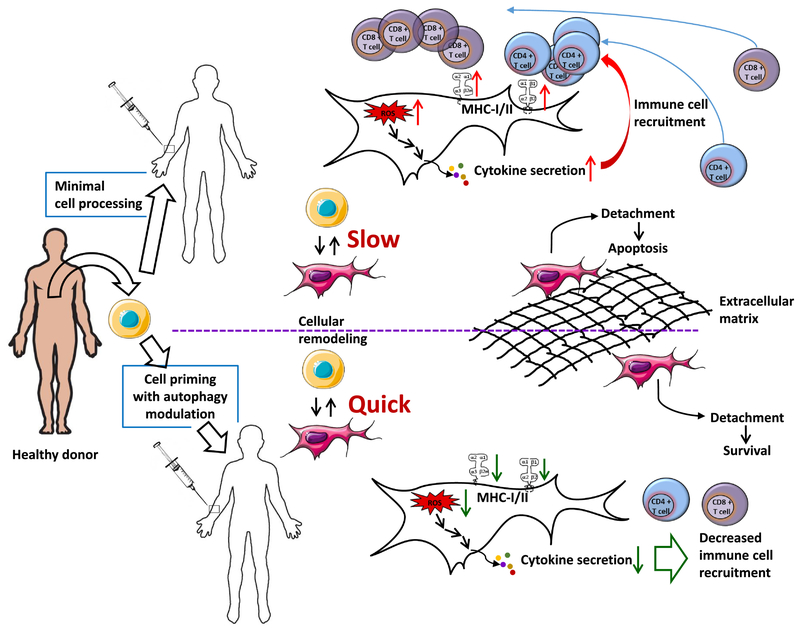
Figure 2:
Key aspects of MSC-based therapies that could be improved with autophagy modulation. Upon injection, the cells will likely encounter lack of both nutrients and stromal support, adverse inflammation response, and increased levels of damaged proteins due to increased levels of Reactive Oxygen Species (ROS). Therapeutic modulation of autophagy prior to or during cell administration may offset some or all these adverse effects by preparing MSCs for adverse conditions they will encounter and by facilitating cellular remodeling that may be needed for increased therapeutic effects.
Regardless of whether SCs are exposed to instructive cues before administration (priming) or not, cells will respond to the environment present in the tissue into which they are injected. This represents two opportunities: first, to both prepare SCs to stressful environment they will encounter and second, to make them more therapeutically active by, for example, stimulating their secretome in vivo or ex vivo. There are also opportunities for “dual priming” where two (or more) factors/processes are stimulated in parallel. The complex role autophagy plays in cellular homeostasis and the inadequate understanding of factors that determine efficacy of therapeutic SCs, calls for a concerted effort towards identifying how manipulation of autophagy can be applied for improvement of SC-based therapies. Availability of autophagy-modulating drugs in clinics will facilitate translation of pre-clinical data and facilitate development of combination therapies for tissue regeneration.
ACKNOWLEDGEMENTS
This research was supported, in part, by funding from Nova Southeastern University provided to The Cell Therapy Institute, by a President’s Faculty Research and Development Grant and by NIH 1R15GM128189-01 to V.B., and by The Swedish Research Council grant to K-H.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Stoick-Cooper CL, Moon RT, Weidinger G, Advances in signaling in vertebrate regeneration as a prelude to regenerative medicine, Genes Dev 21(11) (2007) 1292–315. [DOI] [PubMed] [Google Scholar]
- [2].Sousa-Victor P, Garcia-Prat L, Serrano AL, Perdiguero E, Munoz-Canoves P, Muscle stem cell aging: regulation and rejuvenation, Trends Endocrinol Metab 26(6) (2015) 287–96. [DOI] [PubMed] [Google Scholar]
- [3].Michalopoulos GK, Liver regeneration, J Cell Physiol 213(2) (2007) 286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Metcalfe AD, Ferguson MW, Tissue engineering of replacement skin: the crossroads of biomaterials, wound healing, embryonic development, stem cells and regeneration, J R Soc Interface 4(14) (2007) 413–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sen CK, Expanding horizons of cellular plasticity in regenerative medicine, Am J Pathol 185(10) (2015) 2592–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Passweg JR, Baldomero H, Bader P, Bonini C, Cesaro S, Dreger P, Duarte RF, Dufour C, Kuball J, Farge-Bancel D, Gennery A, Kroger N, Lanza F, Nagler A, Sureda A, Mohty M, Hematopoietic stem cell transplantation in Europe 2014: more than 40 000 transplants annually, Bone marrow transplantation (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Trounson A, McDonald C, Stem Cell Therapies in Clinical Trials: Progress and Challenges, Cell Stem Cell 17(1) (2015) 11–22. [DOI] [PubMed] [Google Scholar]
- [8].Wankhade UD, Shen M, Kolhe R, Fulzele S, Advances in Adipose-Derived Stem Cells Isolation, Characterization, and Application in Regenerative Tissue Engineering, Stem cells international 2016 (2016) 3206807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Alvarez CV, Garcia-Lavandeira M, Garcia-Rendueles ME, Diaz-Rodriguez E, Garcia-Rendueles AR, Perez-Romero S, Vila TV, Rodrigues JS, Lear PV, Bravo SB, Defining stem cell types: understanding the therapeutic potential of ESCs, ASCs, and iPS cells, J Mol Endocrinol 49(2) (2012) R89–111. [DOI] [PubMed] [Google Scholar]
- [10].Romito A, Cobellis G, Pluripotent Stem Cells: Current Understanding and Future Directions, Stem Cells Int 2016 (2016) 9451492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Herberts CA, Kwa MS, Hermsen HP, Risk factors in the development of stem cell therapy, J Transl Med 9 (2011) 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O’Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE, Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts, Nat Biotechnol 25(9) (2007) 1015–24. [DOI] [PubMed] [Google Scholar]
- [13].Forrester JS, Shah PK, Makkar RR, Myocardial regeneration by stem cells: seeing the unseeable, J Am Coll Cardiol 48(8) (2006) 1722–4. [DOI] [PubMed] [Google Scholar]
- [14].Tsolaki E, Yannaki E, Stem cell-based regenerative opportunities for the liver: State of the art and beyond, World J Gastroenterol 21(43) (2015) 12334–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Thirumala S, Goebel WS, Woods EJ, Clinical grade adult stem cell banking, Organogenesis 5(3) (2009) 143–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Buzhor E, Leshansky L, Blumenthal J, Barash H, Warshawsky D, Mazor Y, Shtrichman R, Cell-based therapy approaches: the hope for incurable diseases, Regen Med 9(5) (2014) 649–72. [DOI] [PubMed] [Google Scholar]
- [17].Mabuchi Y, Matsuzaki Y, Prospective isolation of resident adult human mesenchymal stem cell population from multiple organs, Int J Hematol 103(2) (2016) 138–44. [DOI] [PubMed] [Google Scholar]
- [18].Ma S, Xie N, Li W, Yuan B, Shi Y, Wang Y, Immunobiology of mesenchymal stem cells, Cell Death Differ 21(2) (2014) 216–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Le Blanc K, Rasmusson I, Sundberg B, Gotherstrom C, Hassan M, Uzunel M, Ringden O, Treatment of severe acute graft-versus-host disease with third party haploidentical mesenchymal stem cells, Lancet 363(9419) (2004) 1439–41. [DOI] [PubMed] [Google Scholar]
- [20].Girdlestone J, Pido-Lopez J, Srivastava S, Chai J, Leaver N, Galleu A, Lombardi G, Navarrete CV, Enhancement of the immunoregulatory potency of mesenchymal stromal cells by treatment with immunosuppressive drugs, Cytotherapy 17(9) (2015) 1188–99. [DOI] [PubMed] [Google Scholar]
- [21].Castro-Manrreza ME, Montesinos JJ, Immunoregulation by mesenchymal stem cells: biological aspects and clinical applications, J Immunol Res 2015 (2015) 394917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Auletta JJ, Eid SK, Wuttisarnwattana P, Silva I, Metheny L, Keller MD, Guardia-Wolff R, Liu C, Wang F, Bowen T, Lee Z, Solchaga LA, Ganguly S, Tyler M, Wilson DL, Cooke KR, Human mesenchymal stromal cells attenuate graft-versus-host disease and maintain graft-versus-leukemia activity following experimental allogeneic bone marrow transplantation, Stem Cells 33(2) (2015) 601–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Llufriu S, Sepulveda M, Blanco Y, Marin P, Moreno B, Berenguer J, Gabilondo I, Martinez-Heras E, Sola-Valls N, Arnaiz JA, Andreu EJ, Fernandez B, Bullich S, Sanchez-Dalmau B, Graus F, Villoslada P, Saiz A, Randomized placebo-controlled phase II trial of autologous mesenchymal stem cells in multiple sclerosis, PLoS One 9(12) (2014) e113936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Glenn JD, Smith MD, Calabresi PA, Whartenby KA, Mesenchymal stem cells differentially modulate effector CD8+ T cell subsets and exacerbate experimental autoimmune encephalomyelitis, Stem Cells 32(10) (2014) 2744–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Alvaro-Gracia JM, Jover JA, Garcia-Vicuna R, Carreno L, Alonso A, Marsal S, Blanco F, Martinez-Taboada VM, Taylor P, Martin-Martin C, DelaRosa O, Tagarro I, Diaz-Gonzalez F, Intravenous administration of expanded allogeneic adipose-derived mesenchymal stem cells in refractory rheumatoid arthritis (Cx611): results of a multicentre, dose escalation, randomised, single-blind, placebo-controlled phase Ib/IIa clinical trial, Ann Rheum Dis (2016). [DOI] [PubMed] [Google Scholar]
- [26].Lopez-Santalla M, Mancheno-Corvo P, Menta R, Lopez-Belmonte J, DelaRosa O, Bueren JA, Dalemans W, Lombardo E, Garin MI, Human Adipose-Derived Mesenchymal Stem Cells Modulate Experimental Autoimmune Arthritis by Modifying Early Adaptive T Cell Responses, Stem Cells 33(12) (2015) 3493–503. [DOI] [PubMed] [Google Scholar]
- [27].Rocheteau P, Chatre L, Briand D, Mebarki M, Jouvion G, Bardon J, Crochemore C, Serrani P, Lecci PP, Latil M, Matot B, Carlier PG, Latronico N, Huchet C, Lafoux A, Sharshar T, Ricchetti M, Chretien F, Sepsis induces long-term metabolic and mitochondrial muscle stem cell dysfunction amenable by mesenchymal stem cell therapy, Nat Commun 6 (2015) 10145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chao YH, Wu HP, Wu KH, Tsai YG, Peng CT, Lin KC, Chao WR, Lee MS, Fu YC, An increase in CD3+CD4+CD25+ regulatory T cells after administration of umbilical cord-derived mesenchymal stem cells during sepsis, PLoS One 9(10) (2014) e110338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Qi H, Chen G, Huang Y, Si Z, Li J, Foxp3-modified bone marrow mesenchymal stem cells promotes liver allograft tolerance through the generation of regulatory T cells in rats, J Transl Med 13 (2015) 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rennert RC, Sorkin M, Garg RK, Gurtner GC, Stem cell recruitment after injury: lessons for regenerative medicine, Regen Med 7(6) (2012) 833–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Malliaras K, Marban E, Cardiac cell therapy: where we’ve been, where we are, and where we should be headed, Br Med Bull 98 (2011) 161–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Heneidi S, Simerman AA, Keller E, Singh P, Li X, Dumesic DA, Chazenbalk G, Awakened by cellular stress: isolation and characterization of a novel population of pluripotent stem cells derived from human adipose tissue, PLoS One 8(6) (2013) e64752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liu J, Hao H, Huang H, Tong C, Ti D, Dong L, Chen D, Zhao Y, Liu H, Han W, Fu X, Hypoxia regulates the therapeutic potential of mesenchymal stem cells through enhanced autophagy, Int J Low Extrem Wounds 14(1) (2015) 63–72. [DOI] [PubMed] [Google Scholar]
- [34].Wang XM, Yang YJ, Wu YJ, Zhang Q, Qian HY, Attenuating Hypoxia-Induced Apoptosis and Autophagy of Mesenchymal Stem Cells: the Potential of Sitagliptin in Stem Cell-Based Therapy, Cell Physiol Biochem 37(5) (2015) 1914–26. [DOI] [PubMed] [Google Scholar]
- [35].Brodarac A, Saric T, Oberwallner B, Mahmoodzadeh S, Neef K, Albrecht J, Burkert K, Oliverio M, Nguemo F, Choi YH, Neiss WF, Morano I, Hescheler J, Stamm C, Susceptibility of murine induced pluripotent stem cell-derived cardiomyocytes to hypoxia and nutrient deprivation, Stem Cell Res Ther 6 (2015) 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Coulson-Thomas VJ, Coulson-Thomas YM, Gesteira TF, Kao WW, Extrinsic and Intrinsic Mechanisms by Which Mesenchymal Stem Cells Suppress the Immune System, Ocul Surf 14(2) (2016) 121–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Deskins DL, Bastakoty D, Saraswati S, Shinar A, Holt GE, Young PP, Human mesenchymal stromal cells: identifying assays to predict potency for therapeutic selection, Stem Cells Transl Med 2(2) (2013) 151–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Noda NN, Inagaki F, Mechanisms of Autophagy, Annu Rev Biophys 44 (2015) 101–22. [DOI] [PubMed] [Google Scholar]
- [39].Ponpuak M, Mandell MA, Kimura T, Chauhan S, Cleyrat C, Deretic V, Secretory autophagy, Curr Opin Cell Biol 35 (2015) 106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Deretic V, Saitoh T, Akira S, Autophagy in infection, inflammation and immunity, Nat Rev Immunol 13(10) (2013) 722–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Deretic V, Autophagy: an emerging immunological paradigm, J Immunol 189(1) (2012) 15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Maiuri MC, Kroemer G, Autophagy in stress and disease, Cell Death Differ 22(3) (2015) 365–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Galluzzi L, Pietrocola F, Levine B, Kroemer G, Metabolic control of autophagy, Cell 159(6) (2014) 1263–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Noda NN, Inagaki F, Mechanisms of Autophagy, Annu Rev Biophys 44 (2015) 101–22. [DOI] [PubMed] [Google Scholar]
- [45].Schreiber A, Peter M, Substrate recognition in selective autophagy and the ubiquitin-proteasome system, Biochim Biophys Acta 1843(1) (2014) 163–81. [DOI] [PubMed] [Google Scholar]
- [46].Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL, ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase, Nat Cell Biol 15(7) (2013) 741–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wei Y, Pattingre S, Sinha S, Bassik M, Levine B, JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy, Mol Cell 30(6) (2008) 678–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Park JM, Jung CH, Seo M, Otto NM, Grunwald D, Kim KH, Moriarity B, Kim YM, Starker C, Nho RS, Voytas D, Kim DH, The ULK1 complex mediates MTORC1 signaling to the autophagy initiation machinery via binding and phosphorylating ATG14, Autophagy 12(3) (2016) 547–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Noda NN, Fujioka Y, Hanada T, Ohsumi Y, Inagaki F, Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation, EMBO Rep 14(2) (2013) 206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y, The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy, J Biol Chem 282(52) (2007) 37298–302. [DOI] [PubMed] [Google Scholar]
- [51].Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T, LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing, EMBO J 19(21) (2000) 5720–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Xie Y, Kang R, Sun X, Zhong M, Huang J, Klionsky DJ, Tang D, Posttranslational modification of autophagy-related proteins in macroautophagy, Autophagy 11(1) (2015) 28–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Carlsson SR, Simonsen A, Membrane dynamics in autophagosome biogenesis, J Cell Sci 128(2) (2015) 193–205. [DOI] [PubMed] [Google Scholar]
- [54].Qin L, Wang Z, Tao L, Wang Y, ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy, Autophagy 6(2) (2010) 239–47. [DOI] [PubMed] [Google Scholar]
- [55].Lazarus MB, Novotny CJ, Shokat KM, Structure of the human autophagy initiating kinase ULK1 in complex with potent inhibitors, ACS Chem Biol 10(1) (2015) 257–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zhang X, Tang N, Hadden TJ, Rishi AK, Akt, FoxO and regulation of apoptosis, Biochim Biophys Acta 1813(11) (2011) 1978–86. [DOI] [PubMed] [Google Scholar]
- [57].Mammucari C, Schiaffino S, Sandri M, Downstream of Akt: FoxO3 and mTOR in the regulation of autophagy in skeletal muscle, Autophagy 4(4) (2008) 524–6. [DOI] [PubMed] [Google Scholar]
- [58].Zhu WL, Tong H, Teh JT, Wang M, Forkhead box protein O3 transcription factor negatively regulates autophagy in human cancer cells by inhibiting forkhead box protein O1 expression and cytosolic accumulation, PLoS One 9(12) (2014) e115087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kim J, Kundu M, Viollet B, Guan KL, AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1, Nat Cell Biol 13(2) (2011) 132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL, Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia, J Biol Chem 283(16) (2008) 10892–903. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [61].Criollo A, Chereau F, Malik SA, Niso-Santano M, Marino G, Galluzzi L, Maiuri MC, Baud V, Kroemer G, Autophagy is required for the activation of NFkappaB, Cell Cycle 11(1) (2012) 194–9. [DOI] [PubMed] [Google Scholar]
- [62].Nivon M, Fort L, Muller P, Richet E, Simon S, Guey B, Fournier M, Arrigo AP, Hetz C, Atkin JD, Kretz-Remy C, NFkappaB is a central regulator of protein quality control in response to protein aggregation stresses via autophagy modulation, Mol Biol Cell 27(11) (2016) 1712–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Yoshii SR, Mizushima N, Autophagy machinery in the context of mammalian mitophagy, Biochim Biophys Acta 1853(10 Pt B) (2015) 2797–801. [DOI] [PubMed] [Google Scholar]
- [64].Toshima T, Shirabe K, Fukuhara T, Ikegami T, Yoshizumi T, Soejima Y, Ikeda T, Okano S, Maehara Y, Suppression of autophagy during liver regeneration impairs energy charge and hepatocyte senescence in mice, Hepatology 60(1) (2014) 290–300. [DOI] [PubMed] [Google Scholar]
- [65].Yang S, Long LH, Li D, Zhang JK, Jin S, Wang F, Chen JG, beta-Guanidinopropionic acid extends the lifespan of Drosophila melanogaster via an AMP-activated protein kinase-dependent increase in autophagy, Aging Cell 14(6) (2015) 1024–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Nichenko AS, Southern WM, Atuan M, Luan J, Peissig KB, Foltz SJ, Beedle AM, Warren GL, Call JA, Mitochondrial maintenance via autophagy contributes to functional skeletal muscle regeneration and remodeling, Am J Physiol Cell Physiol 311(2) (2016) C190–200. [DOI] [PubMed] [Google Scholar]
- [67].Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, Partridge T, Bucingham M, Direct isolation of satellite cells for skeletal muscle regeneration, Science 309(5743) (2005) 2064–7. [DOI] [PubMed] [Google Scholar]
- [68].Garcia-Prat L, Martinez-Vicente M, Perdiguero E, Ortet L, Rodriguez-Ubreva J, Rebollo E, Ruiz-Bonilla V, Gutarra S, Ballestar E, Serrano AL, Sandri M, Munoz-Canoves P, Autophagy maintains stemness by preventing senescence, Nature 529(7584) (2016) 37–42. [DOI] [PubMed] [Google Scholar]
- [69].Saera-Vila A, Kish PE, Louie KW, Grzegorski SJ, Klionsky DJ, Kahana A, Autophagy Regulates Cytoplasmic Remodeling During Cell Reprogramming in a Zebrafish Model of Muscle Regeneration, Autophagy (2016) 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Gonzalez-Estevez C, Felix DA, Aboobaker AA, Salo E, Gtdap-1 promotes autophagy and is required for planarian remodeling during regeneration and starvation, Proc Natl Acad Sci U S A 104(33) (2007) 13373–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zhang Q, Yang YJ, Wang H, Dong QT, Wang TJ, Qian HY, Xu H, Autophagy activation: a novel mechanism of atorvastatin to protect mesenchymal stem cells from hypoxia and serum deprivation via AMP-activated protein kinase/mammalian target of rapamycin pathway, Stem Cells Dev 21(8) (2012) 1321–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Huang J, Nguyen-McCarty M, Hexner EO, Danet-Desnoyers G, Klein PS, Maintenance of hematopoietic stem cells through regulation of Wnt and mTOR pathways, Nat Med 18(12) (2012) 1778–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Jimenez-Sanchez M, Menzies FM, Chang YY, Simecek N, Neufeld TP, Rubinsztein DC, The Hedgehog signalling pathway regulates autophagy, Nat Commun 3 (2012) 1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].MacDonald BT, Tamai K, He X, Wnt/beta-catenin signaling: components, mechanisms, and diseases, Dev Cell 17(1) (2009) 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].van Es JH, Jay P, Gregorieff A, van Gijn ME, Jonkheer S, Hatzis P, Thiele A, van den Born M, Begthel H, Brabletz T, Taketo MM, Clevers H, Wnt signalling induces maturation of Paneth cells in intestinal crypts, Nat Cell Biol 7(4) (2005) 381–6. [DOI] [PubMed] [Google Scholar]
- [76].Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H, Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4, Nat Genet 19(4) (1998) 379–83. [DOI] [PubMed] [Google Scholar]
- [77].Ring A, Kim YM, Kahn M, Wnt/catenin signaling in adult stem cell physiology and disease, Stem Cell Rev 10(4) (2014) 512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T, Fu W, Zhang J, Wu W, Zhang X, Chen YG, Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation, Nat Cell Biol 12(8) (2010) 781–90. [DOI] [PubMed] [Google Scholar]
- [79].Guruharsha KG, Kankel MW, Artavanis-Tsakonas S, The Notch signalling system: recent insights into the complexity of a conserved pathway, Nat Rev Genet 13(9) (2012) 654–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R, Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains, J Neurosci 30(9) (2010) 3489–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wu X, Fleming A, Ricketts T, Pavel M, Virgin H, Menzies FM, Rubinsztein DC, Autophagy regulates Notch degradation and modulates stem cell development and neurogenesis, Nat Commun 7 (2016) 10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Eggenhofer E, Luk F, Dahlke MH, Hoogduijn MJ, The life and fate of mesenchymal stem cells, Front Immunol 5 (2014) 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Altomare DA, Khaled AR, Homeostasis and the importance for a balance between AKT/mTOR activity and intracellular signaling, Curr Med Chem 19(22) (2012) 3748–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Nuschke A, Rodrigues M, Stolz DB, Chu CT, Griffith L, Wells A, Human mesenchymal stem cells/multipotent stromal cells consume accumulated autophagosomes early in differentiation, Stem Cell Res Ther 5(6) (2014) 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Salemi S, Yousefi S, Constantinescu MA, Fey MF, Simon HU, Autophagy is required for self-renewal and differentiation of adult human stem cells, Cell Res 22(2) (2012) 432–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Kim YH, Baek SH, Kim EK, Ha JM, Jin SY, Lee HS, Ha HK, Song SH, Kim SJ, Shin HK, Yong J, Kim DH, Kim CD, Bae SS, Uncoordinated 51-like kinase 2 signaling pathway regulates epithelial-mesenchymal transition in A549 lung cancer cells, FEBS Lett 590(9) (2016) 1365–74. [DOI] [PubMed] [Google Scholar]
- [87].Delgado M, Singh S, De Haro S, Master S, Ponpuak M, Dinkins C, Ornatowski W, Vergne I, Deretic V, Autophagy and pattern recognition receptors in innate immunity, Immunol Rev 227(1) (2009) 189–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, Muller M, Kreymborg K, Altenberend F, Brandenburg J, Kalbacher H, Brock R, Driessen C, Rammensee HG, Stevanovic S, Autophagy promotes MHC class II presentation of peptides from intracellular source proteins, Proc Natl Acad Sci U S A 102(22) (2005) 7922–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Munz C, Antigen Processing for MHC Class II Presentation via Autophagy, Front Immunol 3 (2012) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Wu TT, Li WM, Yao YM, Interactions between Autophagy and Inhibitory Cytokines, Int J Biol Sci 12(7) (2016) 884–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Lapaquette P, Guzzo J, Bretillon L, Bringer MA, Cellular and Molecular Connections between Autophagy and Inflammation, Mediators Inflamm 2015 (2015) 398483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Jiang S, Dupont N, Castillo EF, Deretic V, Secretory versus degradative autophagy: unconventional secretion of inflammatory mediators, J Innate Immun 5(5) (2013) 471–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Yang R, Ouyang Y, Li W, Wang P, Deng H, Song B, Hou J, Chen Z, Xie Z, Liu Z, Li J, Cen S, Wu Y, Shen H, Autophagy Plays a Protective Role in Tumor Necrosis Factor-alpha-Induced Apoptosis of Bone Marrow-Derived Mesenchymal Stem Cells, Stem Cells Dev 25(10) (2016) 788–97. [DOI] [PubMed] [Google Scholar]
- [94].Delon I, Brown NH, Integrins and the actin cytoskeleton, Curr Opin Cell Biol 19(1) (2007) 43–50. [DOI] [PubMed] [Google Scholar]
- [95].Paoli P, Giannoni E, Chiarugi P, Anoikis molecular pathways and its role in cancer progression, Biochim Biophys Acta 1833(12) (2013) 3481–98. [DOI] [PubMed] [Google Scholar]
- [96].Fung C, Lock R, Gao S, Salas E, Debnath J, Induction of autophagy during extracellular matrix detachment promotes cell survival, Mol Biol Cell 19(3) (2008) 797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Edick MJ, Tesfay L, Lamb LE, Knudsen BS, Miranti CK, Inhibition of integrin-mediated crosstalk with epidermal growth factor receptor/Erk or Src signaling pathways in autophagic prostate epithelial cells induces caspase-independent death, Mol Biol Cell 18(7) (2007) 2481–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M, Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes, Nat Cell Biol 13(11) (2011) 1335–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Wauson EM, Dbouk HA, Ghosh AB, Cobb MH, G protein-coupled receptors and the regulation of autophagy, Trends Endocrinol Metab 25(5) (2014) 274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Strisciuglio C, Duijvestein M, Verhaar AP, Vos AC, van den Brink GR, Hommes DW, Wildenberg ME, Impaired autophagy leads to abnormal dendritic cell-epithelial cell interactions, J Crohns Colitis 7(7) (2013) 534–41. [DOI] [PubMed] [Google Scholar]
- [101].Wildenberg ME, Vos AC, Wolfkamp SC, Duijvestein M, Verhaar AP, Te Velde AA, van den Brink GR, Hommes DW, Autophagy attenuates the adaptive immune response by destabilizing the immunologic synapse, Gastroenterology 142(7) (2012) 1493–503 e6. [DOI] [PubMed] [Google Scholar]
- [102].Jung J, Choi JH, Lee Y, Park JW, Oh IH, Hwang SG, Kim KS, Kim GJ, Human placenta-derived mesenchymal stem cells promote hepatic regeneration in CCl4 -injured rat liver model via increased autophagic mechanism, Stem Cells 31(8) (2013) 1584–96. [DOI] [PubMed] [Google Scholar]
- [103].Pece S, Chiariello M, Murga C, Gutkind JS, Activation of the protein kinase Akt/PKB by the formation of E-cadherin-mediated cell-cell junctions. Evidence for the association of phosphatidylinositol 3-kinase with the E-cadherin adhesion complex, J Biol Chem 274(27) (1999) 19347–51. [DOI] [PubMed] [Google Scholar]
- [104].Han D, Li SJ, Zhu YT, Liu L, Li MX, LKB1/AMPK/mTOR signaling pathway in non-small-cell lung cancer, Asian Pac J Cancer Prev 14(7) (2013) 4033–9. [DOI] [PubMed] [Google Scholar]
- [105].Manic G, Obrist F, Kroemer G, Vitale I, Galluzzi L, Chloroquine and hydroxychloroquine for cancer therapy, Mol Cell Oncol 1(1) (2014) e29911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Rebecca VW, Amaravadi RK, Emerging strategies to effectively target autophagy in cancer, Oncogene 35(1) (2016) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Haller S, Kapuria S, Riley RR, O’Leary MN, Schreiber KH, Andersen JK, Melov S, Que J, Rando TA, Rock J, Kennedy BK, Rodgers JT, Jasper H, mTORC1 Activation during Repeated Regeneration Impairs Somatic Stem Cell Maintenance, Cell Stem Cell 21(6) (2017) 806–818 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Wu F, Wei X, Wu Y, Kong X, Hu A, Tong S, Liu Y, Gong F, Xie L, Zhang J, Xiao J, Zhang H, Chloroquine Promotes the Recovery of Acute Spinal Cord Injury by Inhibiting Autophagy-Associated Inflammation and Endoplasmic Reticulum Stress, J Neurotrauma 35(12) (2018) 1329–1344. [DOI] [PubMed] [Google Scholar]
- [109].Wang YJ, Zhao P, Sui BD, Liu N, Hu CH, Chen J, Zheng CX, Liu AQ, Xuan K, Pan YP, Jin Y, Resveratrol enhances the functionality and improves the regeneration of mesenchymal stem cell aggregates, Exp Mol Med 50(6) (2018) 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Varga M, Sass M, Papp D, Takacs-Vellai K, Kobolak J, Dinnyes A, Klionsky DJ, Vellai T, Autophagy is required for zebrafish caudal fin regeneration, Cell Death Differ 21(4) (2014) 547–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Molinuevo MS, Schurman L, McCarthy AD, Cortizo AM, Tolosa MJ, Gangoiti MV, Arnol V, Sedlinsky C, Effect of metformin on bone marrow progenitor cell differentiation: in vivo and in vitro studies, J Bone Miner Res 25(2) (2010) 211–21. [DOI] [PubMed] [Google Scholar]
- [112].Maden M, Hind M, Retinoic acid, a regeneration-inducing molecule, Dev Dyn 226(2) (2003) 237–44. [DOI] [PubMed] [Google Scholar]
- [113].Majidinia M, Reiter RJ, Shakouri SK, Mohebbi I, Rastegar M, Kaviani M, Darband SG, Jahanban-Esfahlan R, Nabavi SM, Yousefi B, The multiple functions of melatonin in regenerative medicine, Ageing Res Rev 45 (2018) 33–52. [DOI] [PubMed] [Google Scholar]
- [114].Rowart P, Erpicum P, Krzesinski JM, Sebbagh M, Jouret F, Mesenchymal Stromal Cells Accelerate Epithelial Tight Junction Assembly via the AMP-Activated Protein Kinase Pathway, Independently of Liver Kinase B1, Stem Cells Int 2017 (2017) 9717353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Veeranki S, Tyagi SC, Mdivi-1 induced acute changes in the angiogenic profile after ischemia-reperfusion injury in female mice, Physiol Rep 5(11) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Eisenberg T, Abdellatif M, Schroeder S, Primessnig U, Stekovic S, Pendl T, Harger A, Schipke J, Zimmermann A, Schmidt A, Tong M, Ruckenstuhl C, Dammbrueck C, Gross AS, Herbst V, Magnes C, Trausinger G, Narath S, Meinitzer A, Hu Z, Kirsch A, Eller K, Carmona-Gutierrez D, Buttner S, Pietrocola F, Knittelfelder O, Schrepfer E, Rockenfeller P, Simonini C, Rahn A, Horsch M, Moreth K, Beckers J, Fuchs H, Gailus-Durner V, Neff F, Janik D, Rathkolb B, Rozman J, de Angelis MH, Moustafa T, Haemmerle G, Mayr M, Willeit P, von Frieling-Salewsky M, Pieske B, Scorrano L, Pieber T, Pechlaner R, Willeit J, Sigrist SJ, Linke WA, Muhlfeld C, Sadoshima J, Dengjel J, Kiechl S, Kroemer G, Sedej S, Madeo F, Cardioprotection and lifespan extension by the natural polyamine spermidine, Nat Med 22(12) (2016) 1428–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Galluzzi L, Bravo-San Pedro JM, Levine B, Green DR, Kroemer G, Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles, Nat Rev Drug Discov 16(7) (2017) 487–511. [DOI] [PMC free article] [PubMed] [Google Scholar]