Abstract
Background and Purpose:
The genetic relationships between stroke risk, stroke severity, and early neurological changes are complex and not completely understood. Genetic studies have identified 32 all stroke risk loci. Polygenic Risk Scores (PRS) can be used to compare the genetic architecture of related traits. In this study, we compare the genetic architecture of stroke risk, stroke severity, and early neurological changes with that of two stroke risk factors: Type 2 Diabetes Mellitus (T2DM) and Hypertension (HTN).
Methods:
We assessed the degree of overlap in the genetic architecture of stroke risk, T2DM, HTN, and two acute stroke phenotypes based on the NIH Stroke Scale (NIHSS), which ranges from 0 for no stroke symptoms to 21–42 for a severe stroke: baseline (within 6h after onset) and change in NIHSS (ΔNIHSS=NIHSS at baseline minus NIHSS at 24h). This was done by: 1) SNP by SNP comparison, 2) weighted PRS with sentinel variants and 3) whole genome PRS using multiple p-values thresholds.
Results:
We found evidence of genetic architecture overlap between stroke risk and T2DM (p=2.53×10−169), HTN (p=3.93×10−04) and baseline NIHSS (p=0.03). However, there was no evidence of overlap between ΔNIHSS and stroke risk, T2DM or HTN.
Conclusions:
The genetic architecture of stroke risk is correlated with that of T2DM, HTN and initial stroke severity (NIHSS within 6h of stroke onset). However, the genetic architecture of early neurological change after stroke (ΔNIHSS) is not correlated with that of ischemic stroke risk, T2DM or HTN. Thus, stroke risk and early neurological change after stroke have distinct genetic architectures.
Term list: Stroke, Delta NIHSS, Genetic Overlap, Polygenic Risk Score, Ischemic Stroke, Genetics
INTRODUCTION
Hypertension (HTN) is the most common risk factor for stroke along with any form of diabetes.1 Type 2 Diabetes Mellitus (T2DM), is also related to the development of HTN2 and unfavorable functional outcomes after stroke.3 Controlling both HTN and T2DM reduces the risk of stroke.4, 5 Stroke risk factors have been described using epidemiological studies, which are not designed to infer causality. Therefore, it is not clear if T2DM or HTN are part of the causal pathway for stroke or merely comorbid diseases.
Inherited susceptibility through genetic variants has long been postulated to underlie some of the classic risk factors for stroke as well as to potentially explain the missing risk.6 Studies have demonstrated that there is a genetic component to both HTN and T2DM.7 In addition, genetic studies have described a number of loci for stroke risk, some with overlap between stroke and its risk factors.8, 9
The hours after stroke onset are crucial for long term outcomes. There are currently no published GWAs studies for early neurological outcomes after ischemic stroke, or studies comparing the genetic architecture of stroke risk with that of early neurological outcomes. To our knowledge, the largest genetic study for early neurological outcomes is the Genetics of Early Neurological InStability after Ischemic Stroke (GENISIS). This study uses the difference between the baseline NIHSS (collected within six hours after stroke onset) and NIHSS at 24h after onset (ΔNIHSS), as an endophenotype to capture a variety of mechanisms related to ischemic brain injury and early recovery. To date, the study includes more than 2,300 stroke subjects from four countries. The epidemiological analyses showed that T2DM was associated with baseline and ΔNIHSS. HTN was not consistently collected across all sites, but systolic and diastolic blood pressure at the time of initial presentation were found associated with baseline NIHSS. Only systolic blood pressure was found associated with ΔNIHSS (Laura Heitsch, personal communication, 12/12/2018).
Polygenic Risk Scores (PRS) have been successfully used to collapse the effects of common variants in order to calculate the overall risk of an individual or to identify individuals at risk.10 Even though, the predictive power and accuracy of PRS are still insufficient to be applied in a clinical setting, they are becoming more informative.10–14 PRS can also be employed as a measure to identify the extent of overlap between the genetic architecture of co-morbid complex traits.15, 16 In this manuscript we explore the relationships between the genetic architecture of stroke risk, stroke severity, and early neurological changes (ΔNIHSS) with that of two stroke risk factors: T2DM and HTN.
MATERIALS AND METHODS
Summary statistics of the GENISIS dataset used for this analyses are available upon request. Individual data for the full GENISIS dataset will be uploaded to dbGAP in the near future under the name: “Genetics of Early Neurological Instability After Ischemic Stroke (GENISIS)”.
Study Design
The aim of this study is to identify the potential overlap in the genetic architecture of stroke-related phenotypes. First, we wanted to examine if early neurological outcomes after ischemic stroke share common genetic risk factors with stroke risk. Two early stroke phenotypes (ΔNIHSS and baseline NIHSS) were available from the GENISIS cohort to perform the comparison with the MEGASTROKE (stroke risk) cohort. Secondly, we wanted to investigate phenotypes relevant to both stroke and ΔNIHSS. During the clinical characterization of the GENISIS population, T2DM and HTN but not the other known stroke risk factors such as lipid levels or smoking were found to be associated with ΔNIHSS. For this reason, we decided to investigate the potential overlap between the genetic architecture of early neurological changes and T2DM and HTN.
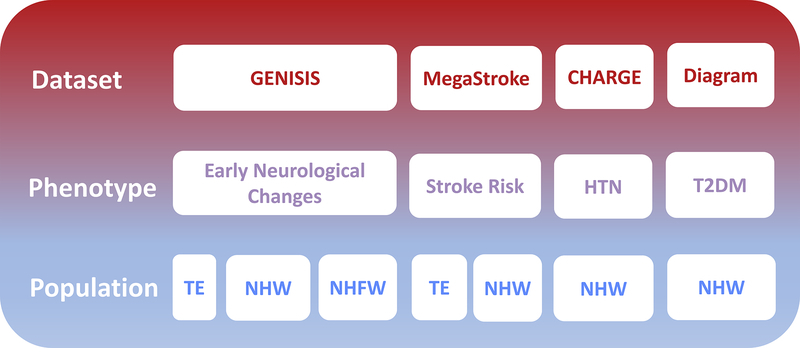
After bibliographic revision, no summary statistics for trans-ethnic (TE) meta-analyses were available for T2DM and HTN. In consequence, we focused on the Non-Hispanic Whites (NHW). We used the summary statistics from the Diagram consortia for T2DM and the ones from CHARGE consortia for HTN. All the comparisons were performed by matching the studies ethnically (Figure 1).
Figure 1. Study design scheme.
The study design scheme shows the different datasets with the phenotypes extracted from each of them (HTN – Hypertension, T2DM – Type 2 Diabetes Mellitus). The last row shows all the available populations depending on the study (TE – Trans-Ethnic, NHW – Non-Hispanic Whites and NHFW – Non-Hispanic, Non-Finnish Withes).
Datasets
Early Neurological Changes: GENESIS
The GENISIS population was composed of five sub-cohorts (Table 1). For a more detailed description of the populations and the distribution of Delta NIHSS and Baseline NIHSS see supplementary materials.
Table 1.
Demographics Characteristics of the GENISIS cohort
| Barcelona (N=1,198) |
Helsinki (N=391) |
Krakow (N=111) |
St Louis EuA (N=430) |
St. Louis AfA (N=187) |
|
|---|---|---|---|---|---|
| Age(years)* | 78.0(75.0–76.5) | 67.0(64.5–67.5) | 69.0(68.0–73.0) | 70.0(68.0–70.5) | 62.0(61.0–65.5) |
| Females(%) | 46.77 | 38.94 | 47.75 | 43.91 | 57.67 |
| Baseline NIHSS* | 9.00(9.5–10.5) | 5.00(5.5–6.5) | 6.00(6.5–9.0) | 7.0(8.0–9.0) | 7.0(7.0–9.0) |
| tPA Treatment(%) | 64.83 | 48.85 | 51.35 | 79.35 | 79.23 |
| Delta NIHSS | 2.66±5.67 | 2.33±5.95 | 2.23±4.42 | 1.93±6.11 | 2.43±6.04 |
| TOAST Classification**(%): | |||||
| Cardioembolic | 39.51 | 43.96 | 35.92 | 41.53 | 37.16 |
| Large Artery | 12.00 | 15.42 | 11.65 | 13.69 | 9.29 |
| Small Vessel Disease | 10.91 | 7.71 | 2.91 | 8.82 | 3.83 |
| Other | 4.03 | 8.23 | 1.94 | 1.62 | 15.30 |
| Undetermined | 33.56 | 24.68 | 47.57 | 34.34 | 34.43 |
Values are expressed as mean±Standard Deviation. EuA: European American Ancestry, AfA: African American Ancestry,
Values expressed as median (95% confidence interval)
TOAST classification criteria17
Stroke Risk: MEGASTROKE
The MEGASTROKE study9 is a meta-analysis of numerous GWAs studies on stroke risk. The GENISIS study is not part of the MEGASTROKE meta-analyses. However, there is an overlap of 200 individuals from the NINDS-Stroke Genetics Network (SiGN) that are part of both studies. In the MEGASTROKE study 32 loci were associated with stroke risk. In this study we have focused only in the 20 loci that were associated with ischemic stroke. We restricted our analyses to the ischemic stroke loci because the GENESIS cohort only includes ischemic strokes. We did not test the stroke subtypes separately due to the limited power of the GENISIS population.
All participants from the different studies included in the MEGASTROKE meta-analysis provided written informed consent. All individual studies were reviewed and approved by the corresponding ethics committees.
Stroke Risk Factors: T2DM (Diagram) and HTN (CHARGE)
We explored the possible genetic overlap between the GENISIS phenotypes and GWAs for T2DM and HTN. We used the GWAs catalog (https://www.ebi.ac.uk/gwas/ - Accessed April 3rd 2018) to select GWAs studies related to HTN and T2DM. We reviewed all the original manuscripts from the GWAs catalog to confirm the effect size, and reference alleles. We selected one GWAs study for each trait based on publicly available summary statistics and ethnicity. We gave preference the studies that had the greatest sample size. We utilized data from the Diagram consortia to map the genetic architecture of T2DM7, 18, and the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortia to map the HTN genetic architecture (accession number:pha004258). T2DM cases were defined as having fasting glucose ≥ 126 mg/dl or random blood sugar ≥ 200 mg/dl. HTN cases were defined as any individual with one or more of the following characteristic: systolic blood pressure ≥ 140 mmHg; diastolic blood pressure ≥ 90 mmHg and/or currently taking antihypertensive or blood pressure-lowering medication.
Statistical Analyses
Analysis of variance
To test if the early neurological changes phenotype (ΔNIHSS) from the GENESIS study had a genetic component, we used the software genome-wide complex traits analysis (GCTA).19 Briefly, GCTA estimates the amount of phenotypic variance explained by all the SNPs in the genome for a complex trait fitting the effects of all SNPs as random effects in a linear mixed model. We were unable to evaluate baseline NIHSS using this approach as it did not follow a normal distribution.
Single Variant Analyses
The associations between the sentinel SNPs from the studies used to model the PRS and the two phenotypes from the GENISIS cohort were tested using an additive linear model with PLINK 1.9.20 The model included age, sex and the two first principle components (PCs) as covariates. In the case of ΔNIHSS, baseline NIHSS is used to calculate ΔNIHSS, but does not predict it, thus it was also included in the model to adjust for initial stroke severity.21 We performed the analyses of the Non-Hispanic Whites (NHW), Non-Hispanic Non-Finnish Whites (NHFW) and African Ancestry populations separately. The results were meta-analyzed using METAL22 and MANTRA.23
Polygenic Risk Score using sentinel SNPs
We modeled a PRS using the sentinel SNPs from the original studies (MEGASTROKE, Diagram and CHARGE), using the method described by the Psychiatric Consortium10 and tested it on the phenotypes from the GENISIS population (ΔNIHSS and baseline NIHSS). Our group has successfully used this method previously.16, 24 Briefly, only the genetic variants corresponding to each GWAs hit (sentinel SNP) reported in the different studies that were available in the GENISIS dataset with an overall call rate >85% were included in the PRS calculation (Supplementary Table I). If available, proxies with R2>0.90 were used for the variants that were not present in the GENISIS dataset. The weighted PRS was computed using the binary logarithm transformation of the reported ORs. The PRS values were computed using PLINK 1.9.20
The effect and statistical significance of the PRS was tested using general linear models (R) adjusting by age, sex and population structure (first and second PCs) and baseline NIHSS when ΔNIHSS was being used as phenotype.21 To adjust for multiple testing, we applied a Bonferroni correction. We also corrected each PRS by the three tested phenotypes. Only p values < 0.017 were considered significant.
Genome-wide Polygenic Risk Score (PRSice)
We used the PRSice software to calculate the genome-wide PRS using multiple p-value thresholds.25 We tested the possible association between the three PRSs (Stroke Risk, Type 2 Diabetes Mellitus and Hypertension) with the two phenotypes of the GENISIS cohort (ΔNIHSS and baseline NIHSS) using a linear regression adjusting by age, sex and population structure (first two PCs). In the case of ΔNIHSS, baseline NIHSS was also included in the model to correct for initial severity.21 Briefly, SNPs present in only one dataset, ambiguous SNPs (A/T or C/G) and all SNPs in linkage disequilibrium (LD) were removed prior to PRS calculation. The PRS is calculated as the sum of risk alleles weighted by the effect size estimates.
Using PRSice, we also tested the possible association between the PRS of stroke risk with T2DM and HTN. We used the sumsum option implemented in PRSice that uses summary statistics from both datasets to evaluate if there is any evidence of shared genetic architecture between the target and base phenotypes.
Additionally to PRSice, we performed the same comparisons using the genetic covariance analyzer, GNOVA.26 Instead of PRS, GNOVA is an annotation-stratified analysis that provides more power to detect moderate genetic correlations.
RESULTS
Analysis of variance (GCTA)
We used GCTA to compute the amount of phenotypic variance explained by common SNPs in the NHW and NHFW populations in the GENISIS study. Since GCTA exploits linkage disequilibrium (LD) patterns to calculate the explained variance, it is not advisable to mix ethnicities. Therefore, we did not test the overall trans-ethnic population, but restricted our analysis to NHW and NHFW. The common genetic variants explained between 7–9% of ΔNIHSS (Supplementary Table II), suggesting that there is a genetic component for early stroke outcome phenotypes.
Single Variants
Prior to the constructing any PRS, we determined if any of the sentinel SNPs from stroke risk, T2DM and HTN GWAs were associated with any early neurological change phenotypes (Supplementary Table III). See supplemental results for details.
Polygenic Risk Score
After checking single variants, we aimed to test the possible overlap between the genetic architecture of T2DM and HTN, acute stroke phenotypes and stroke risk. We constructed PRS based on the genome-wide loci for each MEGASTROKE, Diagram and CHARGE and tested against the GENISIS population (Supplementary Table IV). We found no evidence of association between the T2DM or HTN PRS with any of the acute stroke phenotypes. The stroke risk PRS was not associated with baseline NIHSS, or ΔNIHSS in the TE (trans-ethnic) or NHW populations.
The lack of association of the sentinel-PRS with the different acute stroke phenotypes could be because the sentinel SNPs may lack the power to detect a modest overlap between the genetic architecture of these complex traits. To address this issue, we constructed a PRS using the whole genome summary statistics from each of the three meta-analyses using PRSice. In summary, several PRSs were modeled, each including a different number of independent locus depending on different p-value thresholds. Then, all the PRSs were tested to identify the best fit (Table 2 and Supplementary Figure III). Only the best fit is presented in Table 2. Full results can be found in Supplementary Table V.
Table 2.
PRSice best fit for Stroke risk, HTN and T2DM for the two GENISIS phenotypes (ΔNIHSS and baseline NIHSS) in each subpopulation (Trans-ethnic, Non-Hispanic White and Non-Hispanic Non-Finnish White)
| Population | Phenotype | P-Value | Variance | SNPs |
|---|---|---|---|---|
| Stroke Risk (MEGASTROKE) | ||||
| Trans-Ethnic | Delta | 0.154 | <0.001 | 7,907 |
| Baseline | 0.238 | <0.001 | 27,378 | |
| Non-Hispanic White | Delta | 0.055 | <0.001 | 1,003 |
| Baseline | 0.006 | 0.003 | 136,023 | |
| Non-Hispanic Non-Finnish White | Delta | 0.014 | 0.003 | 1,003 |
| Baseline | 0.030 | 0.003 | 136,023 | |
| Type 2 Diabetes Mellitus (Diagram) | ||||
| Non-Hispanic White | Delta | 0.007 | 0.003 | 4,618 |
| Baseline | 0.447 | <0.001 | 268 | |
| Non-Hispanic Non-Finnish White | Delta | 0.024 | 0.003 | 4,618 |
| Baseline | 0.530 | <0.001 | 268 | |
| Hypertension (CHARGE) | ||||
| Non-Hispanic White | Delta | 0.066 | 0.001 | 1,257 |
| Baseline | 0.027 | 0.002 | 6 | |
| Non-Hispanic Non-Finnish White | Delta | 0.057 | 0.002 | 205 |
| Baseline | 0.103 | 0.001 | 205 | |
Nominally significant p-values are in bold
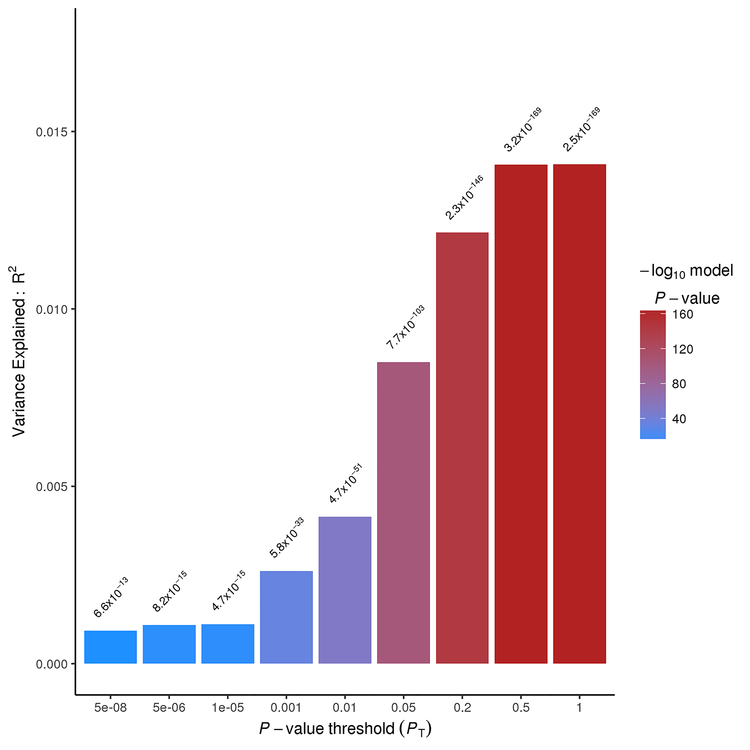
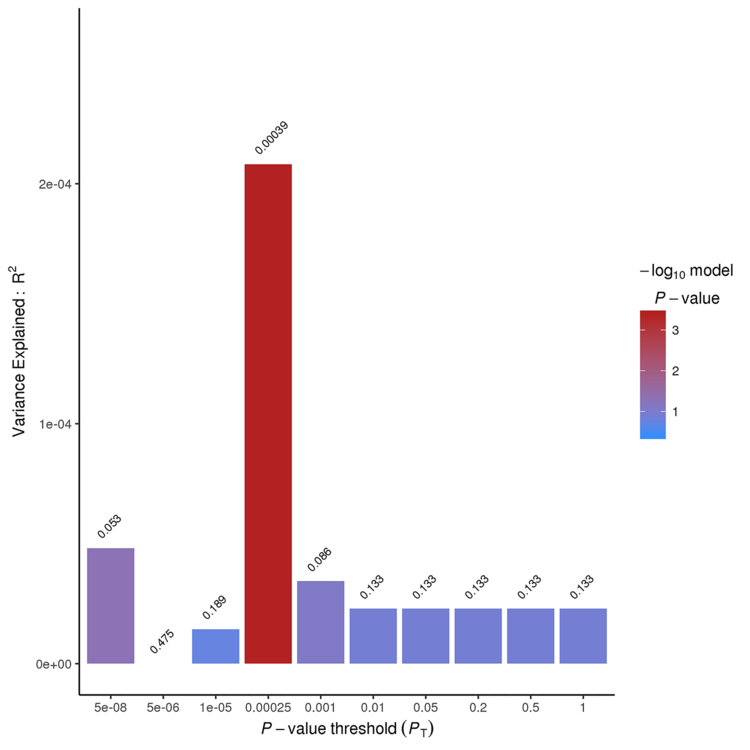
Both T2DM and HTN PRSs were highly associated with ischemic stroke risk in the NHW population. The T2DM PRS explained 1.4% of the variance stroke risk (p=2.53×10−169; SNPs=102,455) (Figure 2–Panel A), which is in line with other studies comparing complex traits.13 We also found that the genetic architecture of HTN and stroke risk showed a significant overlap (p=3.93×10−04) (Figure 2–Panel B). It is important to highlight that the genetic architecture of T2DM overlaps that of HTN (p=2.2×10−16) when using the same approach.
Figure 2. PRSice sumsum Bar Plots for the Type 2 Diabetes Mellitus (T2DM) and Hypertension (HTN) and risk of ischemic stroke, Nagelkerke’s fit.
A - Nagelkerke’s fit for the model: T2DM PRS ~ Ischemic Stroke Risk using the option sumsum from PRSice.
B - - Nagelkerke’s fit for the model: HTN PRS ~Ischemic Stroke Risk using the option sumsum from PRSice.
T2DM PRS was nominally associated with ΔNIHSS in both NHW (pDelta=0.007; r2=0.3%) (Supplementary Figure IV-Panel G) and NHFW populations (pDelta=0.024; r2=0.3%) (Supplementary Figure IV-Panel I) (Table 2). However, when using the trans-ethnic GWAs summary statistics18 the results were not significant (data not shown), probably due to differences in the ethnicities included in the different analyses. Finally, the HTN PRS was found to be nominally associated with baseline NIHSS in the NHW population (p=0.027; r2=0.2%) (Supplementary Figure IV-Panel L and Table 2). None of those associations passed the multiple test correction.
The risk for ischemic stroke PRS modeled with the MEGASTROKE results from the NHW were nominally associated with baseline NIHSS in the NHW and NHFW GENISIS populations (pNHW=0.006; r2=0.3% and pNHFW=0.030; r2=0.3%) (Supplementary Figure IV Panels D and F and Table 2) when including 136,023 SNPs. This PRS was also nominally associated with ΔNIHSS (p=0.014; r2=0.3%) in the NHFW population (Supplementary Figure IV-Panel E and Table 2).
Similar results were found when using GNOVA (Supplementary Table VI), supporting the findings from PRSice in the European populations. We did not test the TE populations since GNOVA is based on linkage disequilibrium structure, and the TE population was confounded by the effects of multiple ethnicities in the trans-ethnic population.
DISCUSSION
Long-term outcome after stroke is influenced by initial stroke severity as well as early neurological changes within the first 24h — a period of great instability.27–30 Moreover, it is known that the underlying stroke etiology can influence initial stroke severity.31, 32 However, little is known about the relationship between ischemic stroke risk, acute stroke severity, and early neurological changes. To our knowledge, this is the first study investigating the possible overlap between the genetic architecture of stroke risk and acute stroke-related phenotypes.
The involvement of T2DM and HTN genetics in stroke risk genetics has been largely suspected, even though both of them are considered modifiable risk factors.4, 5 The MEGASTROKE study was the first, to our knowledge to give evidence of the genetic correlation between these two stroke risk factors and stroke risk. Statistically, they performed weighted genetic risk scores and LD-score regression analyses using the cohorts Diagram and CHARGE (the same used in the present study). When using weighted genetic risk scores, they reported that both T2DM and HTN were associated to the risk of suffering ischemic stroke (p=1×10−10 for both). When using LD-scores, only T2DM was reported to be associated with stroke risk (p=1×10−5).9 Another study used a third statistical approach. They used Mendelian Randomization to test the implication of T2DM in cerebral small vessel disease (p=7×10−3).33 In this study, by using PRS, we also found that the genetic architecture of T2DM and HTN overlap with the genetic architecture of the ischemic stroke risk (p=2.53×10−169 and p=3.93×10−4; respectively). Taken together, this suggests that the PRS approach is the most powerful. All the results implicate the genetic architecture of T2DM and HTN in stroke risk. Furthermore, our results suggest that T2DM is also linked genetically to HTN, suggesting that there is a deleterious feedback loop between stroke risk factors and actual stroke risk. T2DM and HTN may be controllable stroke risk factors, but they may also be implicated genetically in the causality, thus being partly-modifiable factors.
In this study, we also determined the overlap of early neurological changes after ischemic stroke (ΔNIHSS) with T2DM and HTN, but failed to find any strong associations. At face value, these results suggest that early neurological changes after ischemic stroke (ΔNIHSS) are not influenced by stroke risk factors.
We found that the genetic architecture of stroke risk overlaps with that of stoke severity as measured by baseline NIHSS in both NHW (p=0.006) and NHFW cohorts (p=0.030). This association might suggest that the genetic load of stroke risk alleles could be additive, as has been suggested for other diseases34, 35 and may influence the initial stroke severity. However, additional analyses are needed to demonstrate this. The amount of explained variance is small, but it is similar to that of other studies.12, 25 These results, also suggest that the GENESIS study provides enough power to identify the genetic overlap between stroke outcomes, T2DM and HTN. Since all traits are different and not all the genetic studies have the same power, this might not be true for other complex traits.
Additionally, the lack of association of ΔNIHSS with T2DM and HTN might also be explained by other limitations. First, the sample size for the ΔNIHSS (GENISIS) is not as large as those for T2DM and HTN, and therefore has less power to capture the entire genetic architecture of this complex phenotype. Second, even though the GWAs studies for stroke risk, stroke recovery, T2DM and HTN, include very large samples sizes, they may not capture all the genetic variants associated with those phenotypes due to the complexity of the traits. Third, the lack of association can also be caused by the stringent p values threshold set in the GWAs studies (p=5×10−08); thus we may not be capturing the full complexity of the genetic architecture of stroke risk. In addition, the risk of stroke T2DM and HTN is highest in the African American population, which were not represented in sufficient numbers in these studies. Future studies with greater numbers of African American participants may increase our understanding of the genetic architecture of stroke risk. Finally, we also have to take into account that GWAs studies capture a sentinel SNP as a proxy of the causal variant. Each population has its own unique LD structure, and will therefore have different sentinel SNPs. A PRS constructed with the causal variants would have a clearer biological basis.
We attempted to solve the above limitations by creating PRSs using a whole genome approach. However, the results indicate that early neurological change after stroke (ΔNIHSS) does not seem to share a genetic architecture with stroke risk. A trend towards association was observed in the NHFW (p=0.014) but not in the NHW population. It should be noted that the NHW population from MEGASTROKE includes only 1% of subjects from Finland, whereas in the GENISIS population, 18.4% of the NHW population are from Finland. When the Finnish cohort is removed from GENISIS, the populations become more homogeneous, and the power to detect the genetic overlap seems to increase. This association should be further tested in larger populations.
False positive errors due to a smaller sample size and statistical power reduction is also possible. Here, we are able to detect overlap between baseline NIHSS and stroke risk suggesting that the power of this study is sufficient to detect overlap of at least 0.3%, which may be insufficient for ΔNIHSS and stroke risk. There may be other known or unknown variables that may be affecting ΔNIHSS or baseline NIHSS, but are not captured in this study.
ΔNIHSS is a complex phenotype that captures early neurological deterioration or improvement after acute ischemic stroke. It is a quantitative phenotype where larger values (either positive or negative) may be more clinically meaningful than smaller values. While small changes in scores across different domains (e.g. language vs. motor) may result from different mechanisms, large changes may reflect common mechanisms regardless of domains affected. Indeed, a recent analysis of a GENISIS sub-cohort reveals that extreme improvement (positive 2–3*SD) is highly associated with recanalization, while extreme deterioration (negative 2–3*SD) is associated with hemorrhagic transformation. In addition, ΔNIHSS must be considered in the context of its baseline NIHSS (e.g. a 4-point improvement in a patient with a baseline score of 4, may be very different from a 4-point improvement in a patient with a baseline score of 24). Thus, we have included baseline NIHSS in our model to account for this difference in context.
SUMMARY
Our data suggests that the genetic architecture of T2DM and HTN overlaps with that of ischemic stroke risk. Moreover, the genetic architecture of ischemic stroke risk overlaps that of initial stroke severity. This suggests that the presence of certain risk alleles for stroke risk may have additive effects on stroke severity. We found no genetic overlap between stroke risk and early neurological changes after stroke, suggesting that the mechanisms contributing to stroke risk are distinct from those that influence early neurological changes after stroke. Therefore, GWAs of early neurological changes will likely reveal a distinct set of variants from that of stroke risk.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank the individuals and their families for making possible all the genetic studies included in this manuscript. We also want to thank the MEGASTROKE consortium for access to the data (see full list of MEGASTROKE authors in supplementary data). We would also like to thank Kathie. A. Mihindukulasuriya, PhD for reviewing the spelling and grammar of this manuscript.
SOURCES OF FUNDING
Emergency Medicine Foundation Career Development Grant; AHA Mentored Clinical & Population Research Award (14CRP18860027); NIH/NINDS-R01-NS085419; NIH/NINDS-K23-NS099487–01; Barnes-Jewish Hospital Foundation; Helsinki University Central Hospital; Finnish Medical Foundation; Finland government subsidiary funds; Spanish Ministry of Science and Innovation; Instituto de Salud Carlos III (grants “Registro BASICMAR” Funding for Research in Health (PI051737), “GWALA project” from Fondos de Investigación Sanitaria ISC III (PI10/02064, PI12/01238 and PI15/00451)); Fondos FEDER/EDRF Red de Investigación Cardiovascular (RD12/0042/0020); Fundació la Marató TV3; Genestroke Consortium (76/C/2011); Recercaixa’13 (JJ086116). Israel Fernandez is supported by Miguel Servet II Program, Generacion project (PI15/01978), Pre-test project (PMP15/00022), Invictus plus Network (RD16/0019) from Instituto de Salud Carlos III and Fondos Feder; Agaur; and Epigenesis project from Marató TV3 Foundation. The MEGASTROKE project received funding from sources specified at http://www.megastroke.org/acknowledgments.html.
DISCLOSURES
CC receives research support from: Biogen, EISAI, Alector and Parabon. The funders of the study had no role in the collection, analysis, or interpretation of data; in the writing of the report; or in the decision to submit the paper for publication. CC is a member of the advisory board of ADx Healthcare and Vivid Genomics
REFERENCES
- 1.Alloubani A, Saleh A, Abdelhafiz I. Hypertension and diabetes mellitus as a predictive risk factors for stroke. Diabetes & metabolic syndrome. 2018 [DOI] [PubMed] [Google Scholar]
- 2.Ago T, Matsuo R, Hata J, Wakisaka Y, Kuroda J, Kitazono T, et al. Insulin resistance and clinical outcomes after acute ischemic stroke. Neurology. 2018 [DOI] [PubMed] [Google Scholar]
- 3.Kamouchi M, Matsuki T, Hata J, Kuwashiro T, Ago T, Sambongi Y, et al. Prestroke glycemic control is associated with the functional outcome in acute ischemic stroke: The fukuoka stroke registry. Stroke. 2011;42:2788–2794 [DOI] [PubMed] [Google Scholar]
- 4.Ravenni R, Jabre JF, Casiglia E, Mazza A. Primary stroke prevention and hypertension treatment: Which is the first-line strategy? Neurology international. 2011;3:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (ukpds 33). Uk prospective diabetes study (ukpds) group. Lancet. 1998;352:837–853 [PubMed] [Google Scholar]
- 6.Hankey GJ. Potential new risk factors for ischemic stroke: What is their potential? Stroke. 2006;37:2181–2188 [DOI] [PubMed] [Google Scholar]
- 7.Scott RA, Scott LJ, Magi R, Marullo L, Gaulton KJ, Kaakinen M, et al. An expanded genome-wide association study of type 2 diabetes in europeans. Diabetes. 2017;66:2888–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chauhan G, Debette S. Genetic risk factors for ischemic and hemorrhagic stroke. Curr Cardiol Rep. 2016;18:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nature genetics. 2018;50:524–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vassos E, Di Forti M, Coleman J, Iyegbe C, Prata D, Euesden J, et al. An examination of polygenic score risk prediction in individuals with first-episode psychosis. Biological psychiatry. 2016 [DOI] [PubMed] [Google Scholar]
- 11.Power Dudbridge F. and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9:e1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.International Schizophrenia C, Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatterjee N, Wheeler B, Sampson J, Hartge P, Chanock SJ, Park JH. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat Genet. 2013;45:400–405, 405e401–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke TK, Hall LS, Fernandez-Pujals AM, MacIntyre DJ, Thomson P, Hayward C, et al. Major depressive disorder and current psychological distress moderate the effect of polygenic risk for obesity on body mass index. Transl Psychiatry. 2015;5:e592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cruchaga C, Del-Aguila JL, Saef B, Black K, Fernandez MV, Budde J, et al. Polygenic risk score of sporadic late-onset alzheimer’s disease reveals a shared architecture with the familial and early-onset forms. Alzheimer’s & dementia : the journal of the Alzheimer’s Association. 2017; 14:205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams HP, Bendixen BH Jr, Kappelle LJ, Biller, Love BB, Gordon DL, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. Toast. Trial of org 10172 in acute stroke treatment. Stroke. 1993;24:35–41 [DOI] [PubMed] [Google Scholar]
- 18.Replication DIG, Meta-analysis C, Asian Genetic Epidemiology Network Type 2 Diabetes C, South Asian Type 2 Diabetes C, Mexican American Type 2 Diabetes C, Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples C, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nature genetics. 2014;46:234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, Lee SH, Goddard ME, Visscher PM. Gcta: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation plink: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glymour MM, Weuve J, Berkman LF, Kawachi I, Robins JM. When is baseline adjustment useful in analyses of change? An example with education and cognitive change. Am J Epidemiol. 2005;162:267–278 [DOI] [PubMed] [Google Scholar]
- 22.Willer CJ, Li Y, Abecasis GR. Metal: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris AP. Transethnic meta-analysis of genomewide association studies. Genetic epidemiology. 2011;35:809–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Del-Aguila JL, Fernandez MV, Schindler S, Ibanez L, Deming Y, Ma S, et al. Assessment of the genetic architecture of alzheimer’s disease risk in rate of memory decline. Journal of Alzheimer’s disease : JAD. 2018;62:745–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Euesden J, Lewis CM, O’Reilly PF. Prsice: Polygenic risk score software. Bioinformatics. 2015;31:1466–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Q, Li B, Ou D, Erlendsdottir M, Powles RL, Jiang T, et al. A powerful approach to estimating annotation-stratified genetic covariance via gwas summary statistics. American journal of human genetics. 2017;101:939–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saver JL, Altman H. Relationship between neurologic deficit severity and final functional outcome shifts and strengthens during first hours after onset. Stroke. 2012;43:1537–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams HP, Davis PH Jr, Leira EC, Chang KC, Bendixen BH, Clarke WR, et al. Baseline nih stroke scale score strongly predicts outcome after stroke: A report of the trial of org 10172 in acute stroke treatment (toast). Neurology. 1999;53:126–131 [DOI] [PubMed] [Google Scholar]
- 29.Yeo LL, Paliwal P, Teoh HL, Seet RC, Chan BP, Wakerley B, et al. Early and continuous neurologic improvements after intravenous thrombolysis are strong predictors of favorable long-term outcomes in acute ischemic stroke. J Stroke Cerebrovasc Dis. 2013;22:e590–596 [DOI] [PubMed] [Google Scholar]
- 30.Sajobi TT, Menon BK, Wang M, Lawal O, Shuaib A, Williams D, et al. Early trajectory of stroke severity predicts long-term functional outcomes in ischemic stroke subjects: Results from the escape trial (endovascular treatment for small core and anterior circulation proximal occlusion with emphasis on minimizing ct to recanalization times). Stroke. 2017;48:105–110 [DOI] [PubMed] [Google Scholar]
- 31.Pan YT, Lee JD, Lin YH, Huang YC, Weng HH, Lee M, et al. Comparisons of outcomes in stroke subtypes after intravenous thrombolysis. Springerplus. 2016;5:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mustanoja S, Meretoja A, Putaala J, Viitanen V, Curtze S, Atula S, et al. Outcome by stroke etiology in patients receiving thrombolytic treatment: Descriptive subtype analysis. Stroke. 2011;42:102–106 [DOI] [PubMed] [Google Scholar]
- 33.Liu J, Rutten-Jacobs L, Liu M, Markus HS, Traylor M. Causal impact of type 2 diabetes mellitus on cerebral small vessel disease: A mendelian randomization analysis. Stroke. 2018;49:1325–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nordmark G, Kristjansdottir G, Theander E, Eriksson P, Brun JG, Wang C, et al. Additive effects of the major risk alleles of irf5 and stat4 in primary sjogren’s syndrome. Genes Immun. 2009;10:68–76 [DOI] [PubMed] [Google Scholar]
- 35.Yiannakouris N, Katsoulis M, Trichopoulou A, Ordovas JM, Trichopoulos D. Additive influence of genetic predisposition and conventional risk factors in the incidence of coronary heart disease: A population-based study in greece. BMJ open. 2014;4:e004387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.