Abstract
Inhibition of type 1 IGF receptor (IGF-1R) sensitizes to DNA-damaging cancer treatments, and delays repair of DNA double strand breaks (DSBs) by non-homologous end-joining and homologous recombination (HR). In a recent screen for mediators of resistance to IGF-1R inhibitor AZ12253801, we identified RAD51, required for the strand invasion step of HR. These findings prompted us to test the hypothesis that IGF-1R-inhibited cells accumulate DSBs formed at endogenous DNA lesions, and depend on residual HR for their repair. Indeed, initial experiments showed time-dependent accumulation of γH2AX foci in IGF-1R -inhibited or -depleted prostate cancer cells. We then tested effects of suppressing HR, and found that RAD51 depletion enhanced AZ12253801 sensitivity in PTEN wild-type prostate cancer cells but not in cells lacking functional PTEN. Similar sensitization was induced in prostate cancer cells by depletion of BRCA2, required for RAD51 loading onto DNA, and in BRCA2-/- colorectal cancer cells, compared with isogenic BRCA2+/- cells. We also assessed chemical HR inhibitors, finding that RAD51 inhibitor BO2 blocked RAD51 focus formation and sensitized to AZ12253801. Finally, we tested CDK1 inhibitor RO-3306, which impairs HR by inhibiting CDK1-mediated BRCA1 phosphorylation. R0-3306 suppressed RAD51 focus formation consistent with HR attenuation, and sensitized prostate cancer cells to IGF-1R inhibition, with 2.4-fold reduction in AZ12253801 GI50 and 13-fold reduction in GI80. These data suggest that responses to IGF-1R inhibition are enhanced by genetic and chemical approaches to suppress HR, defining a population of cancers (PTEN wild-type, BRCA mutant) that may be intrinsically sensitive to IGF-1R inhibitory drugs.
Introduction
Type 1 insulin-like growth factor receptor (IGF-1R) signals via multiple effectors including phosphatidylinositol 3 kinase (PI3K)-AKT to promote cell survival, and IGF-1R overexpression is associated with clinical radioresistance1–3. IGF-1R targeting enhances chemo- and radio-sensitivity, attributed to apoptosis induction4–7. Data from our group and others indicate that IGF-1R targeting influences the DNA damage response (DDR), with evidence for delayed repair of DNA double strand breaks (DSBs) by both non-homologous end-joining (NHEJ) and homologous recombination (HR)8–11.
Despite activity in preclinical models and early phase clinical trials, IGF-1R inhibitors have shown limited benefit in Phase 2/3 trials of unselected patients, and there are no biomarkers to predict response3. We recently screened for regulators of response to IGF-1R inhibition; of the hits, the only authentic repair protein was RAD51, the recombinase that catalyzes the strand invasion step of HR12. Here, we aimed to validate RAD51 as a screen hit, and understand how IGF-1R inhibition sensitizes to RAD51 depletion.
Materials and Methods
Prostate cancer cell lines DU145 and PC3 were from Cancer Research UK Laboratories (Clare Hall, Hertfordshire, UK), and 22Rv1 and LNCaP from Professor Sir Walter Bodmer (Dept. of Oncology, University of Oxford, UK). Cell line identity was validated by STR genotyping. DLD-1 colorectal cancer cells expressing (BRCA2+/-) or lacking (BRCA2-/-) BRCA2 were from Dr. Scott Kern (Laboratory of Cellular and Molecular Biology, NIH, Baltimore, MA, USA). We used IGF-1R inhibitor AZ12253801 (AstraZeneca, Alderley Park, UK), described in11, human R3 IGF-1 (Sigma-Aldrich, USA), CDK1 inhibitor RO-3306 and RAD51 inhibitors RL-1 and BO2 (Calbiochem, Merck Millipore, Watford, UK). Cells underwent Cesium-137 irradiation in an IBL 637 irradiator (CIS Bio International, Bagnols/Ceze, France). Gene silencing and western blotting were performed as11 using siRNAs and antibodies listed in Supplementary information. Cell viability was quantified by CellTiter-Glo (CTG) Luminescent assay (Promega, USA), and clonogenic survival, cell cycle distribution and immunofluorescent detection of γH2AX and RAD51 foci as in11 and Supplementary information.
Results and Discussion
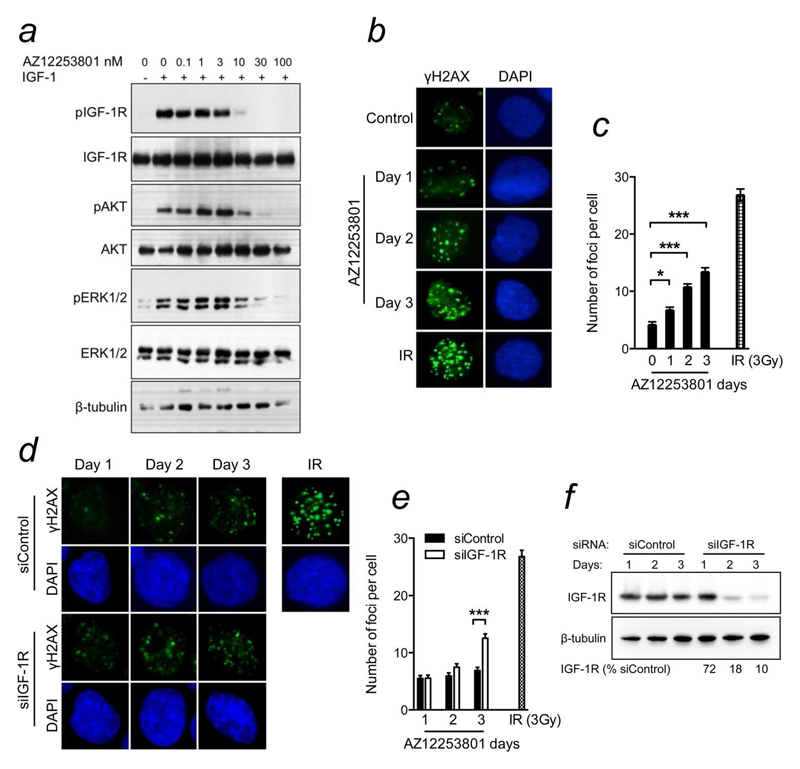
Data from our group and others indicate that IGF-1R targeting sensitizes tumor cells to ionizing radiation and cytotoxic drugs, and delays DSB repair by NHEJ and HR4, 7, 10, 11. DSBs also arise from endogenous damage, typically following collapse of stalled replication forks, and depend on HR for repair due to their one-ended structure13. Given these findings and our identification of RAD51 as a candidate mediator of resistance to IGF-1R inhibition in a screen conducted in the absence of exogenous DNA damage12, we speculated that IGF-1R inhibition leads to accumulation of DSBs that form at endogenous DNA lesions. To investigate this hypothesis, we used AZ12253801, an IGF-1R inhibitor that shows ~10 fold selectivity over the insulin receptor11. In DU145 cells, AZ12253801 inhibited phosphorylation of IGF-1R and its downstream effectors (Figure 1a), confirming previous results11. Using γH2AX as a DSB marker, we assessed whether IGF-1R inhibition influences accumulation of endogenous damage. Compared with controls, AZ12253801-treated cells showed progressive accumulation of γH2AX foci over a 3-day time-course (Figure 1b,c). To assess the specificity of this effect, we repeated the experiment using siRNA to deplete IGF-1R. There was no difference 1-2 days after siRNA transfection, but by 3 days IGF-1R depleted cells contained more γH2AX foci than controls (figure 1d,e). The relative delay compared with effects of AZ12253801 may be because IGF-1R depletion was achieved only after 2-3 days (Figure 1f), consistent with the relatively long half-life (~16-20hr) of IGF-1R protein14.
Figure 1. IGF-1R influences repair of endogenous DNA damage.
a) Serum-starved DU145 cells were treated with AZ12253801 for 1hr and in the final 15min with 50nM IGF-1. b) DU145 cells were treated with solvent (control) or 100nM AZ12253801, fixed 0-3 days later and stained for γH2AX (green) and DAPI (blue). Irradiated cells (3Gy, 6hr) served as positive controls for γH2AX foci. c) Graph: mean ± SEM foci per cell from 2 independent experiments. Foci increased with time in IGF-1R inhibited cells (*p<0.05, ***p<0.001). d) DU145 were transfected with siControl or siIGF1R, and fixed and stained 1-3 days later as b), with irradiated cells as positive controls. e) Cells transfected as d) were analysed as c). Graph: mean ± SEM foci per cell. Three days post-siRNA transfection, IGF-1R depleted cells contained more γH2AX foci than controls (***p<0.001). f) DU145 cells were siRNA-transfected and lysed on days 1-3 for western blotting. Figures below blot: IGF-1R levels corrected for tubulin loading, expressed as % of siControl-transfectants on the equivalent day.
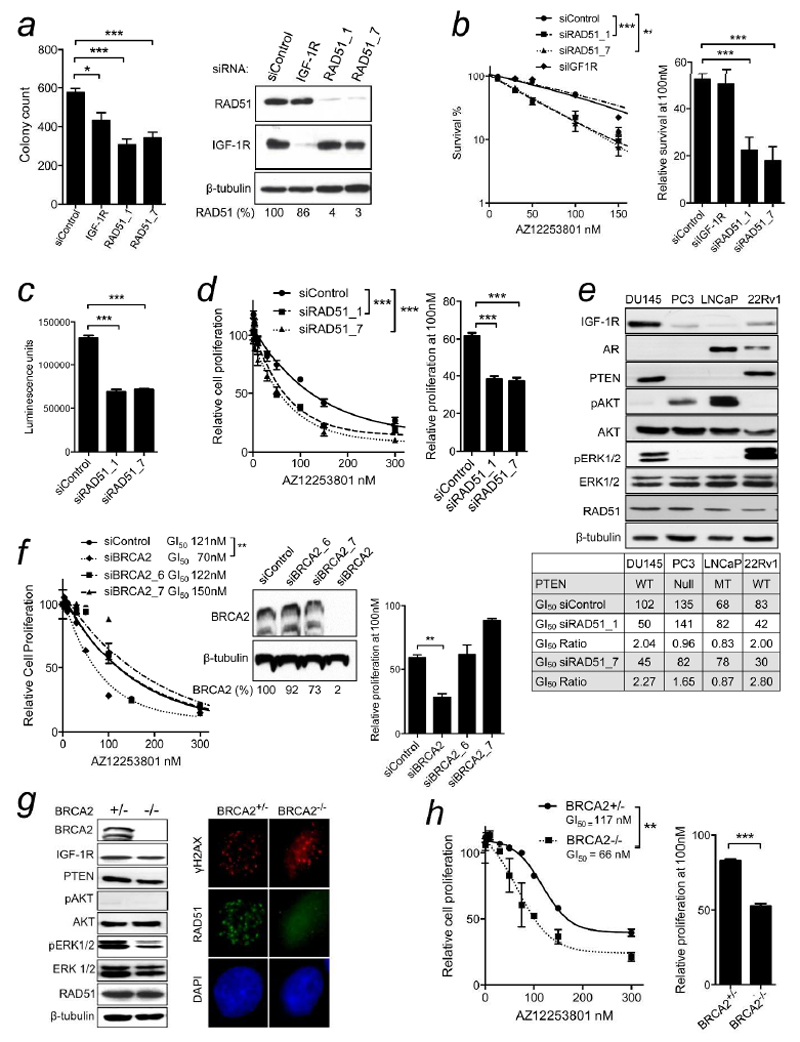
While γH2AX foci can form at single-stranded DNA, their detection correlates closely with numbers of DSBs15. Therefore, these results suggest that IGF-1R inhibition or depletion enhances accumulation of DSBs induced by endogenous DNA damage. These data are consistent with the initial report from Reiss and colleagues, and later work from our group, showing that IGF signaling influences HR, and consequently HR is impaired in cells in which IGF-1R is depleted or inhibited8, 10, 11. This effect may contribute to reduced cell survival in IGF-1R depleted cells (Figure 2a). In light of the identification of RAD51 in a screen for proteins whose depletion sensitizes to IGF-1R inhibition12, we speculated that IGF-1Rinhibited cells are capable of residual HR, albeit with reduced efficiency, and are susceptible to manoeuvres that further comprise repair. Therefore, we tested effects of depleting RAD51 using two RAD51 siRNAs from the screen. Both mediated effective gene silencing, and RAD51-depleted DU145 cells showed reduced cell survival (Figure 2a), consistent with the known toxicity of RAD51 loss in mammalian cells16. As in other siRNA screens17, our screen included a cut-off for hits that were toxic in the absence of IGF-1R inhibition. RAD51 had not been excluded on this basis, suggesting that RAD51 silencing had not crossed a threshold that impairs fitness, and that IGF-1R inhibited cells are more sensitive to RAD51 depletion than controls12. Indeed, both RAD51 siRNAs sensitized DU145 cells to AZ12253801 (Figure 2b). Depletion of IGF-1R itself did not affect AZ12253801 sensitivity (Figure 2b), consistent with findings that IGF-1R expression has little predictive value for response to IGF-1R inhibition3, 12. RAD51 depletion also inhibited cell viability, and induced ~2-fold reduction in AZ12253801 GI50 (Figure 2c,d), comparable to the siRNA screen results12. We then tested three further prostate cancer cell lines, confirming RAD51 depletion by western blotting (Supporting Information Figure S1a). The 22Rv1 cells were sensitized to IGF-1R inhibition by both RAD51 siRNAs, with ~2-fold reduction in AZ12253801 GI50, as in DU145. There was no change in AZ12253801 sensitivity in RAD51-depleted LNCaP cells, and a minor effect in PC3, unlikely to be biologically significant (Figures 2e, S1b-d). PC3 and LNCaP lack functional PTEN, while DU145 and 22Rv1 express wild-type (WT) PTEN (Figure 2e), as do ≥40% of prostate cancers18. Given that PTEN encodes a phosphatase that inactivates PI3K18, we tested whether RAD51 depletion affects IGF signalling, as we found for other screen hits12, but did not detect altered activation of IGF-1R or its effectors in RAD51-depleted DU145 cells (Supplementary Figure S2e). It is possible that a link with PTEN status is provided by the reported ability of AKT and PTEN to influence DSB repair, although the nature of the effect apparently varies with cellular context17, 19.
Figure 2. PTEN wild-type tumor cells are sensitized to IGF-1R inhibition by depletion or loss of RAD51 or BRCA2.
a) Survival of DU145 cells transfected with siControl, IGF-1R or RAD51 siRNAs. Graph: mean ± SEM colonies showing reduced survival on depletion of IGF-1R (*p<0.05) or RAD51 (***p<0.001). To right: parallel cultures blotted for IGF-1R and RAD51. Figures below blot: RAD51 levels corrected for loading, expressed as % siControl. b) DU145 cells were siRNA-transfected as a) and treated with solvent or AZ12253801. Graph: survival (% untreated) from two independent experiments (6 data points; ***p<0.001 by two-way ANOVA). Data were curve-fitted to interpolate AZ12253801 SF50: 102nM for controls, and 108, 50 and 45 nM for IGF-1R, RAD51_1, RAD51_7 siRNA transfectants respectively. Graph to right: relative survival at 100nM AZ12253801, showing sensitization by RAD51-depletion (***p<0.001). c) DU145 cells were reverse-transfected with siControl, siRAD51_1, or siRAD51_7 and viability assayed 5 days later. Graph: mean ± SEM luminescence(***p<0.001). d) DU145 cells were siRNA-transfected as c), 48hr later treated with AZ12253801, and after 5 days viability assayed. Graph: mean ± SEM viability as % siControl, pooled data from 3 independent assays (***p<0.001), generating AZ12253801 GI50 values in controls 122nM, RAD51_1 transfectants 65nM, RAD51_7 transfectants 55nM. Graph to right: relative viability at 100nM AZ12253801 (***p<0.001). e) Cell lysates analyzed for components of IGF axis, androgen receptor (AR), RAD51. Table below: PTEN status, AZ12253801 GI50 values (3 experiments in each cell line as d), fold sensitization as GI50 Ratio in siControl/siRAD51 transfectants. f) DU145 cells were transfected with siControl or BRCA2 siRNAs (siBRCA2_6, siBRCA2_7, siBRCA2), and AZ12253801-treated and viability assayed as d). Graph: mean ± SEM of 2 independent experiments. Only siBRCA2 sensitized to AZ12253801 (** p<0.01). Inset: parallel cultures analysed by western blotting. Figures below blot: BRCA2 levels corrected for loading, expressed as % siControl. Graph to right: relative viability at 100nM AZ12253801 (**p<0.01). g) BRCA2+/- and BRCA2-/- DLD-1 cells analysed by: left, western blotting; right, immunofluorescent staining for ɣH2AX (red), RAD51 (green), DAPI (blue), 6hr after 5Gy irradiation. h) DLD-1 cells were treated with AZ12253801 and viability assayed as d). Graph: mean ± SEM viability, pooled data from 3 experiments. BRCA2-/- cells were more sensitive to AZ12253801 than BRCA2+/- cells (** p<0.01). Graph to right: relative viability at 100 nM AZ12253801 (***p<0.001).
A key step in HR is the loading of RAD51 nucleofilaments onto DNA, a process that requires BRCA220. BRCA2 was present in the DNA repair set used in our siRNA screen, but was not identified as a hit12. We used two models to investigate whether BRCA2 influences sensitivity to IGF-1R inhibition. Aiming to deplete DU145 cells of BRCA2, we tested screen siRNAs BRCA2_6 and _7, and found that both were ineffective, while a third (siBRCA2) did mediate effective target depletion (Figure 2f), as previously reported21. In viability assays, the response of DU145 cells to AZ12253801 was enhanced by siBRCA2 but not by the ineffective screen siRNAs (Figure 3f). This suggests that depletion of BRCA2, like RAD51, is capable of sensitizing to IGF-1R inhibition, and also that BRCA2 was a false negative in the screen12. As a second approach, we used isogenic DLD1 colorectal cancer cells that express WT PTEN and are BRCA2 WT (BRCA2+/-) or null (BRCA2-/-)22. The cell lines had similar expression of IGF-1R and downstream effectors, undetectable AKT phosphorylation consistent with WT PTEN status, and detectable BRCA2 only in BRCA2+/- cells (Figure 2g, left). The effect of BRCA2 loss was tested by measuring the ability to generate irradiation-induced repair foci. Both cell lines formed γH2AX foci, indicative of damage induction, and BRCA2+/- cells contained RAD51 foci that co-localized with γH2AX, while RAD51 foci were clearly reduced in BRCA2-/- cells, indicating defective HR (Figure 2g, right). Consistent with effects of BRCA2 knockdown in DU145 cells, BRCA2-/- DLD1 cells were more sensitive than isogenic BRCA2+/- cells to AZ12253801 (Figure 2h), raising the possibility that patients with BRCA2 mutant cancers may be sensitive to IGF-1R inhibition.
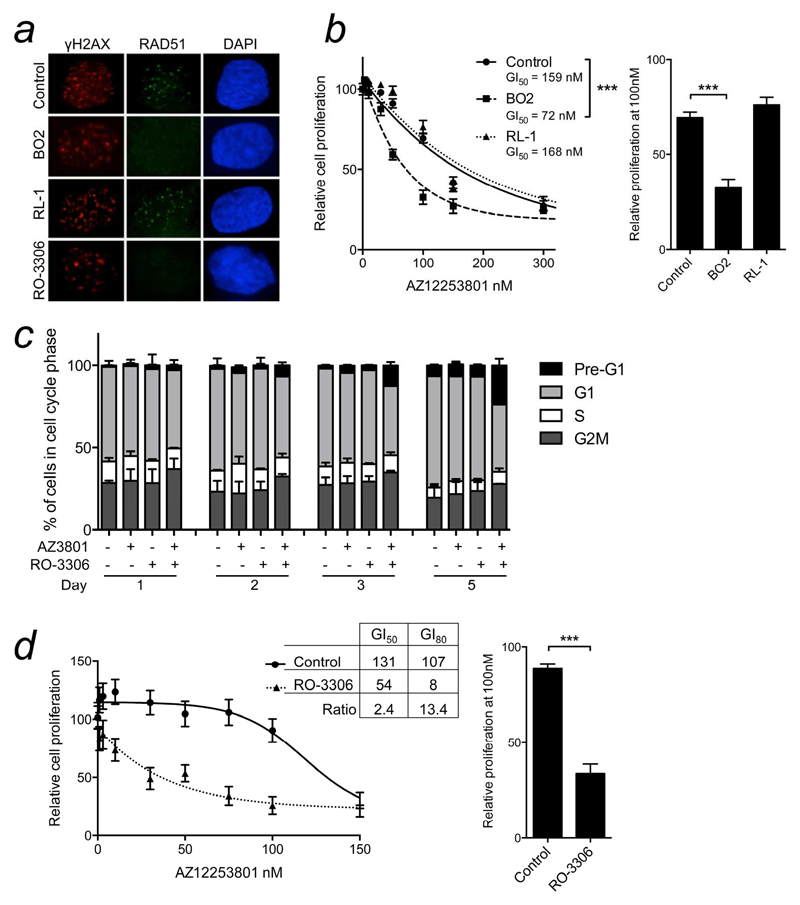
Figure 3. Prostate cancer cells are sensitized to IGF-1R inhibition by inhibitors of RAD51 or CDK1.
a) DU145 cells were treated with solvent (Control), BO2 (10μM), RL-1 (10μM), or RO-3306 (1μM), after 1hr irradiated (5Gy), and after 6hr stained for γH2AX (red), RAD51 (green), DAPI (blue). b) DU145 cells were treated with AZ12253801 with solvent or 10μM BO2 or RL-1, and viability assessed 5 days later. Graph: mean ± SEM of 3 independent experiments. Sensitivity to AZ12253801 was enhanced by BO2 (*** p<0.001 by 2-way ANOVA) but not RL-1. Graph to right: relative viability at 100nM AZ12253801 (*** p<0.001). c) DU145 cells were treated for 1-5 days with 100nM AZ12253801 (AZ3801) and/or 1μM RO- 3306 and analyzed by flow cytometry. d) DU145 cells were treated with AZ12253801 alone or with 1μM RO-3306 and viability assayed after 5 days. Graph: mean ± SEM viability from 3 independent experiments. Sensitivity to AZ12253801 was enhanced by RO-3306 (p<0.001 by two-way ANOVA). Legend shows GI50, GI80 and fold sensitization (ratio of GI50 or GI80 values). Graph to right: relative viability at 100nM AZ12253801 (***p<0.001).
We next explored chemical approaches to inhibit HR. First, we used small molecule drugs BO2 that inhibits the DNA strand exchange activity of human RAD51, and RL-1, which binds covalently to RAD51 protein and is thought to destabilize the formation of RAD51 nucleofilaments23, 24. Viability assays were used to determine GI50 values for use in subsequent experiments (Supplementary Figure S2a-d). In DU145 cells, radiation-induced RAD51 foci were inhibited by BO2 but not by RL-1 (Figure 3a). BO2 sensitized to IGF-1R inhibition, with 2.2 fold reduction in AZ12253801 GI50 (Figure 3b), similar to effects of RAD51 depletion (Figure 2d), while RL-1 did not affect the response to AZ12253801 (Figure 3b). These data suggest that the sensitization effect tracks with the ability to suppress RAD51 foci. BO2 also suppressed RAD51 focus formation in PC3, LNCaP and 22Rv1 cells (Supplementary Figure S2e), and induced AZ12253801 sensitization in 22Rv1 and PC3 cells, and no change in LNCaP (Figure S3a-c).
Finally, we exploited the knowledge that cyclin-dependent kinase (CDK) activity is required for serine phosphorylation of BRCA1 and 2 prior to RAD51 nucleofilament formation20. CDK1 inhibitor RO-3306 was reported to impair BRCA1 localization to DSBs25, and consistent with this, suppressed RAD51 focus formation in DU145 cells when applied at the GI50 (1μM; Figures 3a, S2c-e). Analysis of cell cycle distribution showed that cells co-treated with 100nM AZ12253801 and 1μM RO-3306 manifested a marked increase in pre-G1 DNA, indicating apoptosis induction (Figure 3c). In viability assays, RO-3306 induced a significant shift to the left of the AZ12253801 dose-response curve, with 2.4-fold reduction in GI50, comparable to RAD51 depletion or inhibition. At sub-50nM AZ12253801 concentrations there was more striking sensitization, with 13-fold reduction in GI80 (Figure 3d). Paralleling effects of RAD51 knockdown, RO-3306 did not affect AZ12253801 sensitivity of PC3 or LNCaP cells (Supplementary Figure S3 d, e), but did sensitize 22Rv1 cells (Supplementary Figure S3f); as in DU145, CDK1 inhibition was more effective than RAD51 depletion at sensitizing to low AZ12253801 concentrations, suggesting that CDK1 may influence response to IGF-1R inhibition via processes in addition to HR modulation.
These results indicate that the response to IGF-1R inhibition is enhanced in PTEN WT cells by manipulations that suppress BRCA2 and RAD51 function. To explore the clinical relevance of these data, it will be important to test these findings in primary human tumor cells and in vivo models. In terms of mechanistic insights, we recently reported that IGF-1R inhibited cells show delayed repair of radiation-induced damage, with inhibition of repair by both NHEJ and HR in reporter assays11. IGF-1R inhibition was phenotypically silent in cells with an NHEJ defect induced by inhibition or loss of DNA-PK11, implying that with respect to its role in DSB repair, IGF-1R acts primarily through NHEJ. This is consistent with the results of the current study, which support the concept that IGF-1R inhibited cells depend on residual HR for repair of endogenous damage, and are sensitized to its inhibition. These findings suggest that HR-deficient tumors may be intrinsically sensitive to IGF-1R inhibitory drugs.
Brief description of novelty and impact.
In a screen for biomarkers that predict response to IGF-1R inhibition, we identified RAD51, required for homologous recombination (HR) repair of DNA damage. Here, we show that sensitivity to IGF-1R inhibition is enhanced by multiple approaches to suppress HR, including RAD51 depletion, depletion/loss of BRCA2, or inhibition of RAD51 or CDK1. These data suggest that IGF-1R-inhibited cells depend on HR to repair endogenous damage, and that HR-deficient tumors may be sensitive to IGF-1R inhibitory drugs.
Acknowledgements
We thank Elaine Kilgour, AstraZeneca, for AZ12253801, Walter Bodmer for LNCaP and 22Rv1 cells, Scott Kern for DLD-1 cells, and Tim Humphrey and Chris Lord for comments on the manuscript. This work was supported by NIHR Oxford Biomedical Research Centre, Prostate Cancer UK (KL and TA), NIHR Research Capability Funding (KL), Breast Cancer Campaign (SG), MRC and HEFCE Clinical Senior Lectureship (VMM), and Wellcome Trust Senior Research Fellowship (FE).
Footnotes
Conflict of interest: None declared.
Author contributions: The study was designed by KAL, SG, TA, FE, VMM, experiments were performed by KAL, data analysis was by KAL and VMM, the manuscript was written by KAL and VMM and edited by all authors.
References
- 1.Turner BC, Haffty BG, Narayanan L, Yuan J, Havre PA, Gumbs AA, Kaplan L, Burgaud JL, Carter D, Baserga R, Glazer PM. Insulin-like growth factor-I receptor overexpression mediates cellular radioresistance and local breast cancer recurrence after lumpectomy and radiation. Cancer Res. 1997;57:3079–83. [PubMed] [Google Scholar]
- 2.Moreno-Acosta P, Gamboa O, Sanchez de Gomez M, Cendales R, Diaz GD, Romero A, Balart Serra J, Conrado Z, Levy A, Chargari C, Magne N. IGF1R gene expression as a predictive marker of response to ionizing radiation for patients with locally advanced HPV16-positive cervical cancer. Anticancer Res. 2012;32:4319–25. [PubMed] [Google Scholar]
- 3.King H, Aleksic T, Haluska P, Macaulay VM. Can we unlock the potential of IGF-1R inhibition in cancer therapy? Cancer Treat Rev. 2014;40:1096–1105. doi: 10.1016/j.ctrv.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rochester MA, Riedemann J, Hellawell GO, Brewster SF, Macaulay VM. Silencing of the IGF1R gene enhances sensitivity to DNA-damaging agents in both PTEN wild-type and mutant human prostate cancer. Cancer Gene Ther. 2005;12:90–100. doi: 10.1038/sj.cgt.7700775. [DOI] [PubMed] [Google Scholar]
- 5.Riesterer O, Yang Q, Raju U, Torres M, Molkentine D, Patel N, Valdecanas D, Milas L, Ang KK. Combination of anti-IGF-1R antibody A12 and ionizing radiation in upper respiratory tract cancers. Int J Radiat Oncol Biol Phys. 2011;79:1179–87. doi: 10.1016/j.ijrobp.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Awasthi N, Zhang C, Ruan W, Schwarz MA, Schwarz RE. BMS-754807, a small-molecule inhibitor of insulin-like growth factor-1 receptor/insulin receptor, enhances gemcitabine response in pancreatic cancer. Mol Cancer Ther. 2012;11:2644–53. doi: 10.1158/1535-7163.MCT-12-0447. [DOI] [PubMed] [Google Scholar]
- 7.Ferte C, Loriot Y, Clemenson C, Commo F, Gombos A, Bibault JE, Fumagalli I, Hamama S, Auger N, Lahon B, Chargari C, et al. IGF-1R targeting increases the antitumor effects of DNA damaging agents in SCLC model: an opportunity to increase the efficacy of standard therapy. Mol Cancer Ther. 2013;12:1213–22. doi: 10.1158/1535-7163.MCT-12-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trojanek J, Ho T, Del Valle L, Nowicki M, Wang JY, Lassak A, Peruzzi F, Khalili K, Skorski T, Reiss K. Role of the insulin-like growth factor I/insulin receptor substrate 1 axis in Rad51 trafficking and DNA repair by homologous recombination. Mol Cell Biol. 2003;23:7510–24. doi: 10.1128/MCB.23.21.7510-7524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cosaceanu D, Budiu RA, Carapancea M, Castro J, Lewensohn R, Dricu A. Ionizing radiation activates IGF-1R triggering a cytoprotective signaling by interfering with Ku-DNA binding and by modulating Ku86 expression via a p38 kinase-dependent mechanism. Oncogene. 2007;26:2423–34. doi: 10.1038/sj.onc.1210037. [DOI] [PubMed] [Google Scholar]
- 10.Turney BW, Kerr M, Chitnis MM, Lodhia K, Wang Y, Riedemann J, Rochester M, Protheroe AS, Brewster SF, Macaulay VM. Depletion of the type 1 IGF receptor delays repair of radiation-induced DNA double strand breaks. Radiother Oncol. 2012;103:402–9. doi: 10.1016/j.radonc.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Chitnis MM, Lodhia KA, Aleksic T, Gao S, Protheroe AS, Macaulay VM. IGF-1R inhibition enhances radiosensitivity and delays double-strand break repair by both non-homologous end-joining and homologous recombination. Oncogene. 2013 doi: 10.1038/onc.2013.460. epub 4 Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao S, Bajrami I, Verrill C, Kigozi A, Ouaret D, Aleksic T, Asher R, Han C, Allen P, Bailey D, Feller S, et al. Dsh homolog DVL3 mediates resistance to IGF-1R inhibition by regulating IGF signaling to RAS. Cancer Res. 2014 doi: 10.1158/0008-5472.CAN-14-0806. epub Aug 28. [DOI] [PubMed] [Google Scholar]
- 13.Helleday T. Pathways for mitotic homologous recombination in mammalian cells. Mutation Research. 2003;532:103–15. doi: 10.1016/j.mrfmmm.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 14.Yuen JS, Cockman ME, Sullivan M, Protheroe A, Turner GD, Roberts IS, Pugh CW, Werner H, Macaulay VM. The VHL tumor suppressor inhibits expression of the IGF1R and its loss induces IGF1R upregulation in human clear cell renal carcinoma. Oncogene. 2007;26:6499–508. doi: 10.1038/sj.onc.1210474. [DOI] [PubMed] [Google Scholar]
- 15.Lobrich M, Shibata A, Beucher A, Fisher A, Ensminger M, Goodarzi AA, Barton O, Jeggo PA. gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle. 2010;9:662–9. doi: 10.4161/cc.9.4.10764. [DOI] [PubMed] [Google Scholar]
- 16.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci U S A. 1996;93:6236–40. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendes-Pereira AM, Martin SA, Brough R, McCarthy A, Taylor JR, Kim JS, Waldman T, Lord CJ, Ashworth A. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med. 2009;1:315–22. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phin S, Moore MW, Cotter PD. Genomic Rearrangements of PTEN in Prostate Cancer. Front Oncol. 2013;3:240. doi: 10.3389/fonc.2013.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu N, Lao Y, Zhang Y, Gillespie DA. Akt: a double-edged sword in cell proliferation and genome stability. J Oncol. 2012;2012 doi: 10.1155/2012/951724. 951724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 21.Yata K, Lloyd J, Maslen S, Bleuyard JY, Skehel M, Smerdon SJ, Esashi F. Plk1 and CK2 act in concert to regulate Rad51 during DNA double strand break repair. Mol Cell. 2012;45:371–83. doi: 10.1016/j.molcel.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hucl T, Rago C, Gallmeier E, Brody JR, Gorospe M, Kern SE. A syngeneic variance library for functional annotation of human variation: application to BRCA2. Cancer Res. 2008;68:5023–30. doi: 10.1158/0008-5472.CAN-07-6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Budke B, Logan HL, Kalin JH, Zelivianskaia AS, Cameron McGuire W, Miller LL, Stark JM, Kozikowski AP, Bishop DK, Connell PP. RI-1: a chemical inhibitor of RAD51 that disrupts homologous recombination in human cells. Nucleic Acids Res. 2012;40:7347–57. doi: 10.1093/nar/gks353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang F, Mazina OM, Zentner IJ, Cocklin S, Mazin AV. Inhibition of homologous recombination in human cells by targeting RAD51 recombinase. J Med Chem. 2012;55:3011–20. doi: 10.1021/jm201173g. [DOI] [PubMed] [Google Scholar]
- 25.Johnson N, Li YC, Walton ZE, Cheng KA, Li D, Rodig SJ, Moreau LA, Unitt C, Bronson RT, Thomas HD, Newell DR, et al. Compromised CDK1 activity sensitizes BRCA-proficient cancers to PARP inhibition. Nat Med. 2011;17:875–82. doi: 10.1038/nm.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]