Abstract
A major obstacle to curing chronic myeloid leukaemia (CML) is residual disease maintained by tyrosine kinase inhibitor (TKI)-persistent leukemic stem cells (LSCs). These are BCR-ABL1 kinase-independent (1, 2), refractory to apoptosis (3, 4) and serve as a reservoir to drive relapse or TKI-resistance. We demonstrate that Polycomb Repressive Complex 2 (PRC2) is mis-regulated in chronic phase (CP) CML LSCs. This is associated with extensive reprogramming of H3K27me3 targets in LSCs, thus sensitizing them to apoptosis upon treatment with an EZH2-specific inhibitor (EZH2i). EZH2i does not impair normal hematopoietic stem cell (HSC) survival. Strikingly, treatment of primary CML cells with either EZH2i or TKI alone caused significant up-regulation of H3K27me3 targets, and combined treatment further potentiated these effects and resulted in significant loss of LSCs compared to TKI alone, in vitro, and in long-term bone marrow murine xenografts. Our findings point to a promising epigenetic-based therapeutic strategy to more effectively target LSCs in CML patients receiving TKIs.
Introduction
Chronic myeloid leukemia (CML) is a clonal HSC disorder, arising through the acquisition and expression of the fusion BCR-ABL1 tyrosine kinase which stimulates a number of key survival pathways to drive the disease. Despite the introduction of TKIs which have transformed the clinical course of CML from a fatal to a manageable disease for the vast majority of patients, only ~10% of those who present in chronic phase can discontinue treatment and maintain a therapy-free remission without relapse (5). The remaining 90% of patients require life-long TKIs and can experience serious side effects, compliance issues and high costs, or they develop TKI-resistance (20-30% of cases), and/or progress to advanced disease for which there are no effective drug therapies. The low cure rate in CML is thought to be due to a pool of TKI-persistent and BCR-ABL1 kinase-independent (1) LSCs. LSCs in CML are defined as Philadelphia chromosome-positive (Ph+) CD34+CD38- primitive progenitor cells that show a higher capacity to engraft in immunocompromised mice than bulk CD34+ cells (6), have stem cell properties (quiescence and self-renewal), are prone to genomic instability (7) and resistant to apoptosis (3, 4). Novel therapeutic strategies that target these LSCs must be developed to address an important unmet clinical need for the vast majority of patients currently on TKI therapy.
The epigenetic “writer” complex PRC2 is fundamental for defining stem cell identity where it primarily regulates gene repression using H3K27me3 (8). Whilst its methyltransferase activity can be mediated by either EZH1 or EZH2; aberrant H3K27me3 and EZH2 activity has been implicated in the progression or poor prognosis of solid tumors and hematological malignancies (9, 10). However, little is known about the role of EZH2 in CML and unlike other myeloid malignancies, inactivating mutations of EZH2 in CML are rare, (11, 12) although up-regulation of EZH2 can result in a form of myeloproliferative disease (13).
Results
In comparative mRNA profiling of normal and CP CML primary samples representative of those in our CML biobank (on average ≥ 95% Ph+ in CD34+ or CD34+CD38- cells; see Methods), we observed significant mis-regulation in the levels of several PRC2 components - predominantly between CD34+CD38- HSCs and CD34+CD38- LSCs, and to a lesser extent between CD34+CD38+ leukemic and normal CD34+CD38+ progenitors (LPCs and HPCs respectively) (Fig. 1a; Supplementary Fig. S1a-c). Significant down-regulation of EZH1 in LSCs was found often in combination with EZH2 up-regulation, creating a scenario where the relative levels of EZH2 compared to EZH1 increased 2-16 fold across the ten CML samples we examined (Fig. 1a). Levels of EZH2, as well as other PRC2 components, were significantly down-regulated in LSCs treated with TKI in vitro, indicative of kinase dependent regulation. The levels of EZH1, however, appeared kinase independent, thus maintaining an EZH balance favouring EZH2 in TKI-persistent cells (Supplementary Fig. S2a-c).
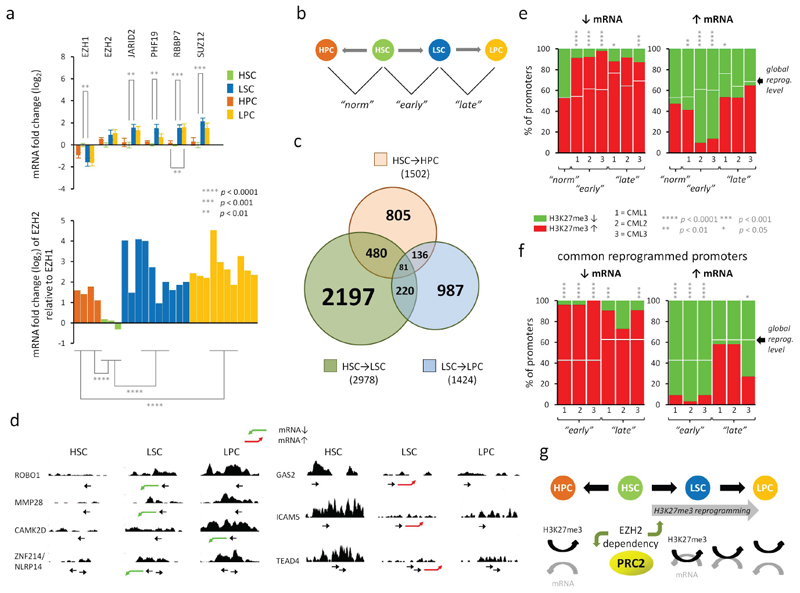
Figure 1. Mis-regulation of PRC2 leads to H3K27me3 reprogramming and increased coupling of H3K27me3 and mRNA expression in CML LSCs.
a. Top; the mean fold changes in mRNA expression for 6 PRC2 components and BCR-ABL1 in normal (HSCs and HPCs) and in CML (LSCs and LPCs) cells as determined by Fluidigm analysis performed in triplicate. Fold changes (log2) are expressed relative to levels found in HSCs. Error bars are standard error measurements (SEMs). Bottom; the mean levels of EZH2 mRNA relative to EZH1 mRNA in each of the bioreplicate samples as above. Significant p values (*; Student’s t-test) between equivalent cell type comparisons (HSC vs LSC, HPC vs LPC) are shown. Bioreplicates are n = 4 (HPC), n = 3 (HSC), n = 10 (LSC), n = 10 (LPC). b. Schematic diagram depicts how changes in H3K27me3 levels between normal and CML cells were determined with respect to three pairwise cell type comparisons: HSCs vs HPCs (“norm”), HSCs vs LSCs (“early”), LSCs vs LPCs (“late”). c. Venn diagram depicts the relationship between the numbers of promoters which show altered levels of H3K27me3 in three pairwise comparisons (HSC→HPC; HSC→LSC; LSC→LPC). d. Examples of H3K27me3 read densities across genes that are up- (red arrow) or down-(green arrow) regulated (mRNA) in CML cells compared to HSCs. Location of transcription start site and direction of transcription are shown by →. Profiles were visualised in the UCSC genome browser (hg18; NCBI build 36.1). e. Bar diagrams show the percentages (%) of all reprogrammed promoters having significant gains (red) or losses (green) of H3K27me3 (p < 0.05; one sample t-tests) when their cognate mRNA levels are down- (↓) or up- (↑) regulated (Affymetrix analysis) in each of three pairwise comparisons for n = 3 bioreplicate CML(1,2,3) and normal samples. f. Bar diagrams show the percentages (%) of all promoters reprogrammed in all three CML samples (common) having significant gains (red) or losses (green) of H3K27me3 (p < 0.05; one sample t-tests) when their cognate mRNA levels are down- (↓) or up- (↑) regulated (Affymetrix analysis) as above. In e and f, expected percentages of H3K27me3 gains associated with global reprogramming of H3K27me3 levels, irrespective of mRNA expression changes, are shown (white bars). Significant p values (*) are shown based on observed vs expected levels of H3K27me3 gains or losses at promoters (Yates Chi-squared or Fischer Exact tests). g. Schematic representation of H3K27me3 reprogramming in CML leading to increased mechanistic coupling of H3K27me3 levels and mRNA expression - consistent with an altered dependency for EZH2 in CML (upward green arrow). HSCs require EZH1(14) but not EZH2(15) (downward green arrow).
To understand the molecular consequences of PRC2 mis-regulation, we performed ChIP-sequencing in normal and CML primary stem and progenitor cell samples (a first in CML LSCs). We identified statistically significant changes in H3K27me3 levels at promoters in three pairwise comparisons (Fig. 1b; Supplementary Fig. S3a). In total, across the three CML samples, changes in H3K27me3 levels were identified at 4101 promoters, 83% of which were unique to CML (Fig. 1c). A quarter of these were common to all three samples and consistent with gains or losses of EZH2 target genes (Supplementary Fig. S3b). A significant majority of these changes (2978; 73%) occurred between the HSC and LSC, indicative of extensive epigenetic reprogramming in the LSC.
Whilst the repressive function of EZH2 and H3K27me3 is well documented, changes in gene expression at EZH2 target genes are not always mechanistically linked to H3K27me3 levels in normal hematopoiesis (14). We therefore asked what impact H3K27me3 reprogramming would have on gene expression in CML. Significant mRNA expression changes between normal and leukemic stem and progenitor cells were determined globally (Supplementary Fig. S4a; Supplementary Table S1) and from these we identified a subset of H3K27me3 reprogrammed target genes, whose mRNAs were either up- or down-regulated (examples shown, Fig. 1d). In agreement with previous observations (14), we found no evidence that H3K27me3 levels were linked to changes in gene expression during normal cell fate decisions (HSC → HPC) (Fig. 1e). However, in the three CML samples, the association between changes in gene expression and H3K27me3 levels was significantly different from that predicted by H3K27me3 reprogramming (Fig. 1e; Supplementary Fig. S4b) and indicative of H3K27me3 having its canonical repressive role. This association was most significant in the “early” changes (HSC vs LSC), particularly amongst promoters reprogrammed in all 3 CML samples (common; Fig. 1f; Supplementary Fig. S4c), and pointed to an altered dependency between H3K27me3 and gene expression primarily in the LSC (Fig. 1g).
Based on the epigenetic evidence, we hypothesized that EZH2 inhibition would represent a potential novel therapeutic strategy to selectively target LSCs given that HSCs require EZH1 (14) for survival but not EZH2 (15). We therefore treated CML and normal primary cells in vitro with a highly specific EZH2i, GSK343 (16) (Fig. 2a; Supplementary Fig. S5a-h). We confirmed a selective dose-dependent decrease, but not complete loss, of H3K27me3 in response to EZH2i in both CML and normal CD34+ cells (Fig. 2b, Supplementary Fig. S5b). Treatment of bulk CML CD34+ cells and LSCs with the EZH2i resulted in modest but significant (≈ 20-40%) reductions in viable cells including quiescent (undivided; CTVmax staining; see Methods) cells (17), along with increases in apoptosis, particularly in undivided cells (Fig. 2c,d; Supplementary Figs. S5a,c, S6a-c). More pronounced decreases (≈ 60-80%) however were seen for total colony forming cell (CFC) and primitive granulocyte/erythroid/macrophage/megakaryocyte (GEMMs) outputs (Fig. 2e; Supplementary Figs. S5a,d, S6a,b,d), and for those of long-term-culture initiating cell (LTC-IC) assays which enrich for primitive LSCs (Fig. 2f; Supplementary Fig. S5e). This suggested that the primitive stem and progenitor cells were being preferentially targeted by EZH2i, supporting our hypothesis. Similar results were seen for CML CD34+ cells treated with other novel EZH2i (Supplementary Fig. S5a). Normal CD34+ cells treated with EZH2i were spared from the deleterious effects seen in CML cells (Fig. 2c-f; Supplementary Fig. S5f-h), thus demonstrating a potential therapeutic window for EZH2is in CML. We observed similar results using shRNAs against EZH2 in a CML cell line (Supplementary Fig. S7a-c) and in CML CD34+ cells (Fig. 2g). On-target effects were confirmed by global mRNA analysis, where CML and normal CD34+ cells treated with EZH2i both showed significant over-representation of up-regulated H3K27me3 targets (Fig. 2h; Supplementary Fig. S8a,b; Supplementary Table S2). However, relatively few EZH2i-deregulated targets were in common between CML and normal and only CML cells showed significant up-regulation of apoptotic pathways (Fig. 2i; Supplementary Table S3). This provides further mechanistic support that global epigenetic reprogramming results in an altered dependency on EZH2 and H3K27me3 in CML cells, and this sensitizes LSCs, but not HSCs, to EZH2 inhibition.
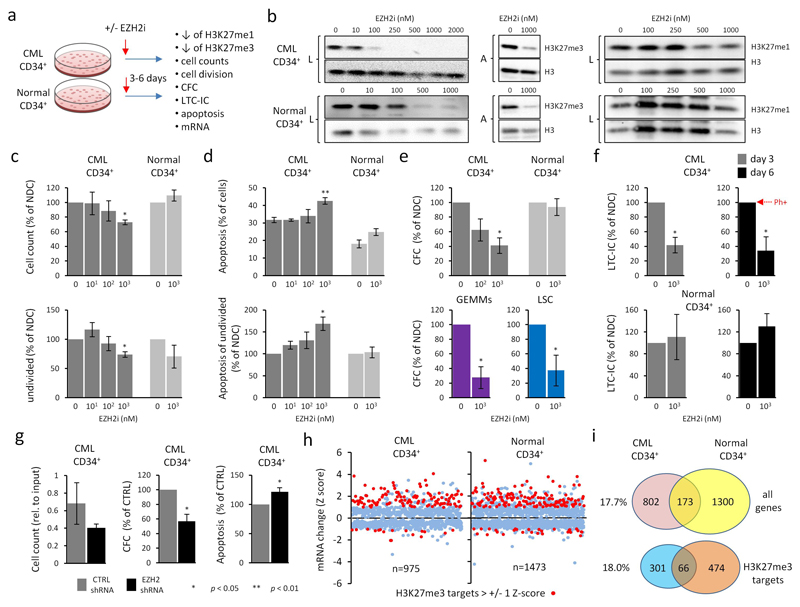
Figure 2. CML cells are sensitive to chemical and genetic inhibition of EZH2.
a. Schematic for EZH2i treatment in primary CML and normal CD34+ cells. n = 3 bioreplicates in all instances described below. b. Representative western analysis of H3K27me3 and H3K27me1 levels in CML and normal CD34+ cells treated for 6 days with increasing concentrations of EZH2i as indicated. L = laemmli buffer; total cellular protein. A = acid extraction; enriched for histones. Levels of bulk histone H3 were used as loading controls. c. Viability of CML and normal CD34+ cells treated with EZH2i for 6 days. Mean total viable cell counts (top); mean viable undivided (CTVmax) CD34+ cells (bottom). d. Levels of apoptosis (Annexin V-positive cells) in CML and normal CD34+ cells treated with EZH2i. Mean total apoptosis (top) and mean apoptosis in undivided (CTVmax) CD34+ cells (bottom). e. Colonogenic (CFC) potential of CML and normal CD34+ cells treated with EZH2i. Mean total CFC outputs (top) and GEMM outputs (bottom) from CD34+ cells; total CFC outputs from LSCs (CD34+CD38-) (bottom). f. Colonogenic potential of CML and normal CD34+ cells from LTC-IC after 3 days or 6 days treatment with EZH2i. % of cells Ph+ (i.e., BCR-ABL+) in NDC (day 6) is shown by the arrowhead (red). g. Effect of EZH2 shRNA knockdown in CML34+ cells. Cell counts (left), CFC output (middle) and apoptosis (Annexin V-positive cells) (right) for CML CD34+ cells transduced with EZH2 or control shRNA for 3 days. Two different EZH2 shRNAs were used to derive data. h. Scatterplot depicting global mRNA expression changes (Z-scores) for 975 and 1473 genes in CML and normal CD34+ cells respectively as a consequence of EZH2i treatment (and where both mRNA and H3K27me3 status is known) (E-MTAB-2893; E-MTAB-3552). H3K27me3 targets (> +/- 1 SD from the mean) are highlighted (red). Significant p values (*; one sample t-tests in c → g) are shown in the key below panel g. EZH2i is GSK343 for data shown in b → f and in h. i. Venn diagrams depict the degree of overlap between genes showing mRNA changes due to EZH2i treatment in CML and normal CD34+ cells; all genes (top), H3K27me3 target genes (bottom). % of overlap with respect to totals for CML CD34+ cells (left).
An important area of unmet clinical need in CML is the eradication of a sup-population of LSCs that are refractory to TKI treatment (18, 19). By analysing global datasets of CML CD34+ cells or LSCs treated with EZH2i or TKI and by using an additive model for combined drug effects, we predicted that a combination of EZH2i and TKI would be more effective at up-regulating H3K27me3 target genes than EZH2i alone (Supplementary Figs. S9a-d, S10a-d). This was, in part, attributable to TKI treatment alone resulting in substantial up-regulation of H3K27me3 targets (Fig. 3a). However, we found little evidence that TKI or EZH2i returned mRNA levels of H3K27me3 targets to those found in normal stem or progenitor cells (Supplementary Figs. S11a-c, S12).
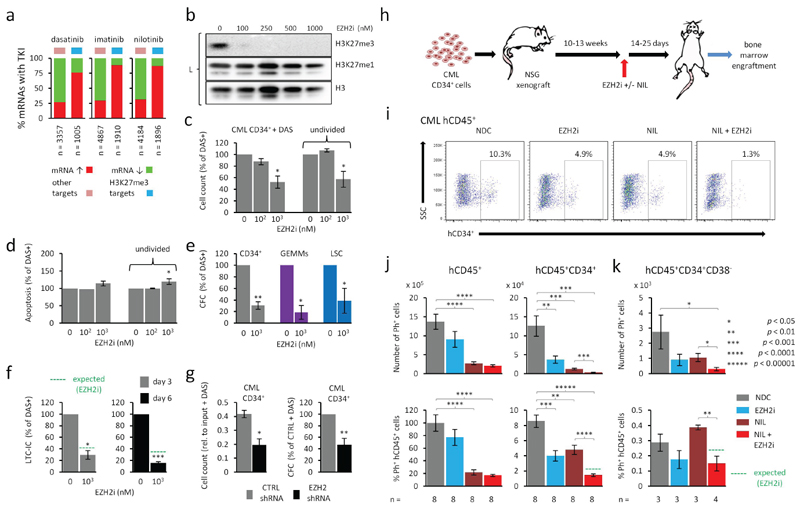
Figure 3. Both TKI and EZH2i up-regulate H3K27me3 targets and combined treatment potentiates loss of LSCs.
a. Summary of global mean mRNA expression changes found in LSCs after treatment with TKI for 7 days in vitro based on Affymetrix analysis (E-MTAB-2594). Percentages of genes up- or down-regulated are shown for H3K27me3 targets and other genes for each TKI treatment (n = 6 bioreplicates in all cases). Total number of mRNAs affected shown along the bottom. b. Representative western analysis (L = laemmli buffer) of H3K27me3 and H3K27me1 levels in CML CD34+ treated with DAS with and without increasing concentrations of EZH2i (GSK343). c. Viability of CML CD34+ cells treated with EZH2i and DAS for 6 days. Mean total viable cell counts (left); mean viable undivided (CTVmax) CD34+ cells (right). d. Levels of apoptosis (Annexin V-positive cells) in CML CD34+ cells treated with EZH2i and DAS for 6 days. Mean total apoptosis (left) and mean apoptosis in undivided (CTVmax) CD34+ cells (right). e. Colonogenic (CFC) potential of CML cells treated with EZH2i and DAS for 6 days. Mean total CFC outputs (left) and GEMM outputs (middle) from CML CD34+ cells and total CFC outputs from LSCs (CD34+CD38-) (right). f. Colonogenic potential of CML CD34+ cells from LTC-IC following 3 or 6 day treatment with EZH2i and DAS (same samples as shown in Fig. 3e). Expected levels (green dotted line) are based on efficacy of EZH2i alone on CD34+ cells (from Fig. 3f). g. Cell counts (left) and CFC output (right) for CML CD34+ cells transduced with EZH2 or control shRNA for 4 days in the presence of DAS. Two different EZH2 shRNAs were used to derive data. Results in c → g are shown as a percentage change from treatment with DAS (150 nM) only, unless indicated. n = 3 bioreplicates in c,d,e,f; n = 4 bioreplicates in g. h. Schematic showing the experimental design for EZH2i and TKI treatment of NSG mice engrafted with CML CD34+ samples. i. Representative FACs scatter plots showing the proportions (%) of human CML CD45+ cells staining positive for CD34+ (hCD34+) in bone marrow from NSG xenografts following in vivo treatment with EZH2i (EPZ-6438) or TKI (nilotinib; NIL) alone and in combination for 25 days. SSC = side scatter. j. Left side, mean absolute number (top) and mean % (bottom) of Ph+ hCD45+ cells from bone marrow of NSG xenografts treated with vehicle (NDC), EZH2i, NIL (nilotinib) and EZH2i + NIL (25 day treatment; n = 2 CML CD34+ samples; n = 8 mice per arm). Right side, mean absolute number (top) and mean % (bottom) Ph+ hCD45+CD34+ cells from the same experiments. k. Mean absolute number (top) and mean % (bottom) of Ph+ hCD45+CD34+CD38- cells from bone marrow of NSG xenografts treated as above (n = 1 CML CD34+ sample; n = 3-4 mice per arm). In j, k, % Ph+ cells are expressed as a % of hCD45+ in NDC. Significant p values (*; one sample t-tests in c → g; Student t-tests in bar diagrams of j, k) are as shown in the key next to panel k.
To provide support for our in silico prediction, we treated CML and normal CD34+ cells and LSCs with EZH2i and dasatinib (DAS), alone and in combination (Fig. 3b-f; Supplementary Fig. S5c-e, S6b-d). EZH2i selectively targeted the loss of H3K27me3 in the presence of DAS (Fig. 3b; Supplementary Fig. S5b). Furthermore, combination treatment with DAS and EZH2i led to a significant reduction in cell viability, even in the undivided ‘TKI-persistent’ cells (Fig. 3c), and was accompanied by increased apoptosis (Fig. 3d) and a pronounced reduction in CFC and CFC/GEMM outputs compared to DAS treatment alone (Fig. 3e). Furthermore, combined EZH2i and DAS treatments reduced LTC-IC outputs to 1% of those of no drug controls (Supplementary Fig. S5e) and the effects of EZH2i in the presence of DAS were more than additive (Fig. 3f). DAS treatment similarly reduced cell viability and the CFC potential of cells in which EZH2 was knocked down by shRNAs (Fig. 3g).
To determine the effect treatment would have on LSCs within a bone marrow microenvironment, human CD34+ cells were engrafted into NOD scid gamma (NSG) mice for 10-13 weeks and then treated in vivo with nilotinib (NIL) and an EZH2i (EPZ-6438) (20) singly and in combination for either 14 or 25 days (Fig. 3h). After 14 days of treatment, we observed significant reductions in levels of leukemic (Ph+) human CD45+ cells, CD45+CD34+ progenitor cells, and primitive CD45+CD34+CD38- stem cells in the bone marrow of mice treated with both NIL and EZH2i compared to NIL treatment alone (Supplementary Fig. S13a-d). Importantly, after 25 days of treatment, we observed near complete elimination of CD45+CD34+ progenitor cells (Fig. 3i,j; Supplementary Fig. S13e,f) and greater than a 70% reduction of CD45+CD34+CD38- cells (Fig. 3k) in combined NIL/EZH2i treatment compared to NIL alone. Again, the effects of EZH2i on CML stem and progenitor cells in the presence of NIL appeared more than additive in vivo, and mirrored our in vitro findings. These results taken together demonstrate a clear rationale for combining TKI treatment with EZH2i, given the efficacy of an EZH2i at targeting primitive CML cells not eradicated by TKI alone.
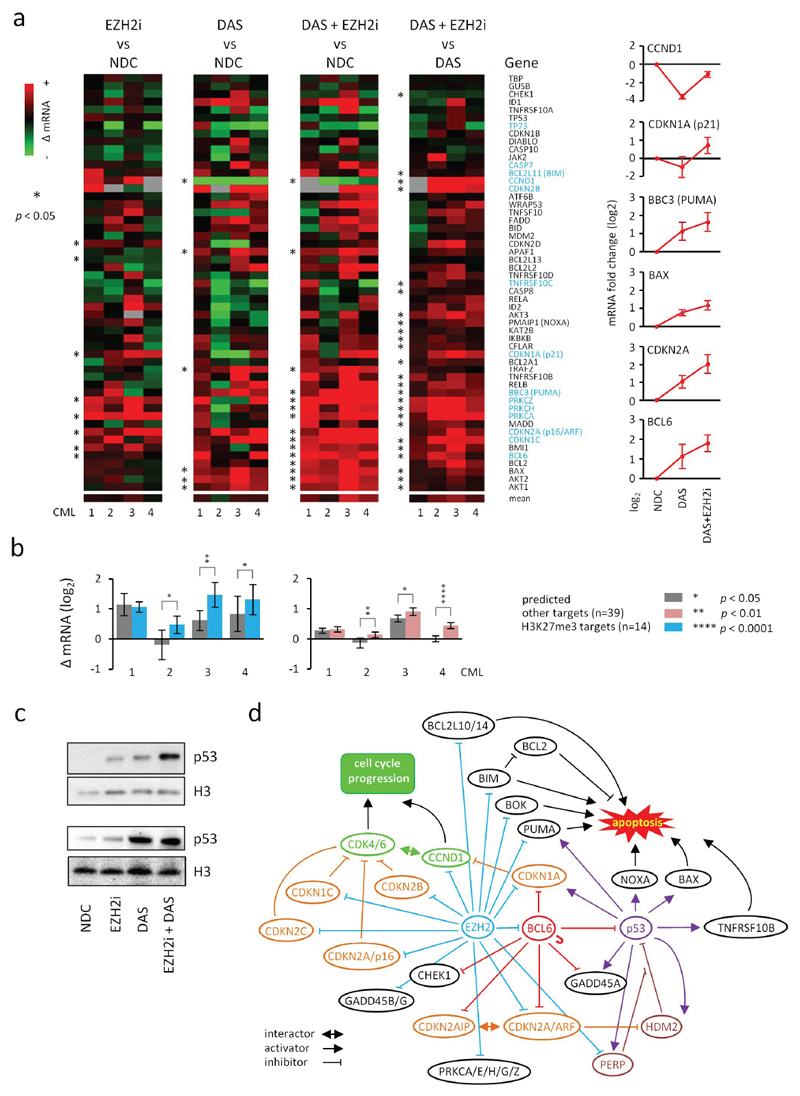
As TKI-persistent cells are refractory to apoptosis (3, 4), we examined the mechanism of action of combined DAS plus EZH2i on the mRNA expression of a set of genes, including 14 H3K27me3 targets from apoptotic pathways (Fig. 4a). BCL6, an H3K27me3 target and a known repressor of p53 and CDKN2A (p16/ARF) in TKI-persistent cells (21, 22) was up-regulated by combined DAS plus EZH2i treatment. Nonetheless, there was a significant up-regulation of apoptotic genes, including p53 transcriptional targets (e.g. CDKN1A, BAX, PUMA, NOXA, TNFRS10B), and an enhanced activation of H3K27me3 targets, including CDKN2A, compared to DAS or EZH2i treatments alone. In keeping with our phenotypic data, the enhanced activation of H3K27me3 targets and apoptotic genes exceeded predicted levels based on additive effects (Fig. 4b). Furthermore, we confirmed increased levels of the p53 protein in CML CD34+ cells treated with EZH2i and in combination with DAS (Fig. 4c). We generated a connectivity map between targets of EZH2 (H3K27me3), BCL6 and p53 in the apoptosis pathway (Fig. 4d) and propose that EZH2 inhibition, when potentiated by TKI, could engage apoptosis downstream of p53 through re-activation of pro-apoptotic targets (e.g. PUMA, BIM), and/or by directly rescuing p53-mediated apoptosis by overcoming BCL6- and EZH2- mediated repression upstream of p53.
Figure 4. Molecular analysis of the effects of EZH2i and TKI on apoptotic-pathways in CML CD34+ cells.
a. Main panel, heat maps showing mean fold changes in the mRNA expression (Fluidigm) of a number of apoptotic pathway genes following treatment of CML CD34+ samples with EZH2i (1000 nM) or DAS (150 nM) alone or in combination (n =4 bioreplicates assayed in triplicate); mean changes in mRNA expression of specific p53 pathway genes and BCL6 are shown (right). b. Bar diagrams showing comparisons of predicted (from individual treatments using an additive model; grey) and actual changes in mean mRNA expression levels of H3K27me3 (blue; left) and other targets (pink; right) following treatment with both EZH2i and DAS in each bioreplicate CML sample (n = 4) shown in a. Error bars are SEMs in all instances. Significant p values (*; one sample t-tests in a, Student t-tests in bar diagrams of b) are as shown in the key next to panel b. c. Representative western analysis (laemmli buffer) of p53 levels in two CML CD34+ samples before and after EZH2i +/- DAS treatment. d. Schematic of a putative interactome map between EZH2/H3K27me3 (blue), BCL6 (red) and p53 (purple) targets which mediate cell cycle arrest and apoptosis in CML CD34+ cells. BCL6 targets were obtained from previously published data(22). BCL6 is self-regulatory as shown. Cell cycle inhibitors and activators are shown in orange and green respectively.
Discussion
Due to increased patient survival, CML will become the most prevalent form of leukemia in the next 20-30 years (23), with TKI-persistent LSCs remaining a significant unmet clinical need for the majority of these patients. Since TKIs are highly effective against the bulk of leukemic cells and are generally well tolerated by most chronic phase CML patients, new therapies that target LSCs must be effective and safe when combined with TKI. Our pre-clinical results, derived from more than 40 primary samples, and hypothesis-driven by global analyses, demonstrate that inhibition of EZH2 in combination with TKI is comparable, if not more effective, than other novel strategies for targeting CML LSCs (Supplementary Table S4); none of which have reached the clinic to supplement or replace TKI as the standard of care. Given that EZH2 can have tumor suppressor or oncogenic roles, further investigations however are required to optimise pre-clinical therapeutic strategies, and to identify biomarkers of EZH2 and H3K27me3 inhibition in CML cells. Nonetheless, novel, orally-administered EZH2 inhibitors currently in trials are showing low toxicity, significant reductions of H3K27me3 levels at clinically achievable doses, and promising objective responses across a range of tumour types. These early trial data, taken together with our results and those of Xie et al. (24), point to a promising new avenue for the development of the first epigenetic therapy for the treatment of CML.
Methods
Primary samples
Primary CML samples (Ph+ in > 90% of CD34+ cells) were obtained from patients with CP CML obtained at point of diagnosis prior to TKI treatment (n = 32 bioreplicates in total, for experiments described in this paper). Normal (i.e., non-CML) control primary samples were obtained by granulocyte-colony stimulating factor (G-CSF) mobilisation from individuals negative for bone marrow involvement upon lymphoma staging or from normal cadaveric bone marrow (BM) samples (normal controls; n = 12 bioreplicates). In all instances, ethical permission was with written informed consent (West of Scotland Research Ethics Service; REC Ref 10/S0704/2 and REC Ref 15/WS/0077). See also Supplementary Table S5. Primary CML samples were randomly chosen for the experiments described below and in the text without prior knowledge of clinical outcome or response to TKI. The investigators were not blinded to the experimental conditions when assessing the outcomes and no datasets were excluded from analysis (for all in vitro assays). Experiments on live vertebrates (NSG mice) were performed as described below.
Primary samples were stored frozen as mononuclear cells (MNCs) isolated by density gradient centrifugation or as CD34+ cells isolated using CliniMACS (Miltenyi Biotech). After thawing, CML cells were recovered by culturing in physiological growth factors (SCF 0.2ng/ml, G-CSF 1ng/ml, GM-CSF 0.2ng/ml, IL-6 1ng/ml, LIF 0.05ng/ml, MIP-α 0.2ng/ml, StemCell Technologies), while normal control stem cells were cultured in 5 growth factors (SCF 100 ng/ml, G-CSF 20 ng.ml, IL-3 20 ng/ml, IL-6 20 ng/ml, Flt3 100 ng/ml). For isolation of HSC/LSC (CD34+CD38-) and HPC/LPC (CD34+CD38+), MNCs or CD34+ cells were stained with CD34-APC, CD38-FITC or CD38-PerCP-Cy™5.5 antibodies (555824, 555259, 551400 respectively; BD Biosciences), before being subject to fluorescence activated cell sorting (FACS) to obtain the appropriate populations using a FACS Aria cell sorter (Beckton Dickinson).
Cell culturing
For experiments involving GSK343, CML and normal primary cells were cultured as above for the treatment regimens described below and in the text with media change every 3 days. For experiments involving treatment of LSCs with TKIs (imatinib (IM)/5µM, dasatinib (DAS)/150 nM and nilotinib (NIL)/5 µM; Bristol-Myers Squibb and Selleckchem) for up to 7 days, cells were grown in serum-free media without growth factors. The KCL22 cell line (CML, blast crisis; obtained from DSMZ cell bank in 2014; mycoplasma-negative; authenticated by DSMZ using RT-PCR for BCR-ABL1 expression and microsatellite repeat typing) was grown in RPMI 1640 (Sigma) supplemented with 10% fetal bovine serum, 2 mM L-glutamine and 1% penicillin-streptomycin. Experiments involving KCL22 cells were performed within 6 months from the date cells were obtained from DSMZ.
Proliferation, differentiation, apoptosis and colony-forming cell assays for EZH2 inhibition with GSK343, GSK126 and EZP-6438
Primary cells from CML patients and normal controls (n = 3 each) (CD34+, CD34+CD38-, or CD34+CD38+) were cultured as above for 6 days with or without EZH2 inhibitors and/or DAS (50-150 nM), with media and drug change every three days. Viable cell numbers (trypan blue exclusion) in culture were measured every three days and fold change determined relative to input. To monitor proliferation/cell division, cells were labelled with cell trace violet (CTV; Invitrogen). Following culture, the cells were co-stained with CD34-APC to assess differentiation status and loss of CD34+. CTV and CD34 levels were analyzed by flow cytometry and the percentage recovery of input for CD34+ cells in each cell division was determined. Colcemid treatment was used to determine gating for undivided cells. To assess levels of apoptosis, cells were incubated with Annexin V-FITC and 7-AAD (556419 and 559925 respectively; BD Biosciences) and analysed by flow cytometry. Apoptotic cells were defined as Annexin V and Annexin V-7AAD positive. For colony forming cells (CFC) assays, cells cultured with or without GSK343 +/- DAS for 6 days were plated in methylcellulose (MethoCult™; StemCell Technologies) and the number and morphology of colonies determined after 12 days. Statistical analysis of differences in proliferation, CFC counts and apoptosis was performed using one sample t-tests.
LTC-IC assays for EZH2 inhibition with GSK343
Primary CD34+ cells from CML patients and normal controls (n = 4 bioreplicates for CML; n = 3 bioreplicates for normal) were cultured as above for 3 or 6 days with or without GSK343 (1000 nM) and DAS (150 nM). M2-10B4 and SL/SL fibroblasts were established as feeder layers and then irradiated at 80 Gy. 5 x 104 cells were then placed in long-term culture-initiating culture (LTC-IC) on irradiated feeders in MyeloCult™ (StemCell Technologies) and maintained for 5 weeks in duplicate in the absence of drugs. Cells were then transferred to CFC assays in duplicate and maintained in culture for 12 days and then scored as above.
Immunodeficient murine xenograft experiments
CML CD34+ cells were transplanted (1×106 cells per mouse) via tail vein injection into female 8-12-week-old sub-lethally irradiated (2.25Gy) NOD scid gamma (NSG) mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ; The Jackson Laboratory). Human cell engraftment was monitored at 8 to 12 weeks post-transplantation by FACS for the presence of human CD45+ cells in the blood. Mice which showed evidence of human cell engraftment were randomized and treated with vehicle/NDC (10% NMP in PEG300), a clinical grade EZH2i (EPZ-6438; Medkoo Biosciences), nilotinib (LC Laboratories), or a combination of EPZ-6438 and nilotinib for 14 or 25 days, commencing at 10-13 weeks post-transplantation. Drugs or vehicle were administered once daily by oral gavage at a dose of 300 mg/kg for EPZ-6438 and 50 mg/kg for nilotinib. Mice were euthanized at the end of treatments and marrow contents of ilia, femurs, and tibia were obtained. The presence of human CD45, CD34 and CD38 on engrafted cells was assessed by FACS. Engraftment data was obtained from n = 3 (14 day treatment) or n = 2 (25 day treatment) CML CD34+ samples derived from different patients. The investigators were blinded to the experimental conditions when assessing the outcomes and no datasets or animals were excluded from analysis. Mice were handled humanely in accordance with regulations under Home Office License PPL. 60/4492.
Fluorescence in situ hybridization (FISH)
Colonies from LTC-IC colonogenic outputs and human CD45+ or CD45+CD34+ cells from NSG xenografts were scored for the presence of the Philadelphia chromosome (Ph+). FISH was performed with the LS1 BCR-ABL Dual Color, Dual Fusion translocation probe according to the manufacturer’s instructions (Abbott Diagnostics). For each LTC-IC, nuclei from at least 10 colonies were scored where possible. Two hundred nuclei were scored from bone marrow from each xenograft.
EZH2 knockdown
EZH2 shRNAs were subcloned from the pLKO.1 puro vector (Sigma Mission shRNA collection) into the NdeI and SpeI sites of the pLKO.1 GFP vector. Sequences for EZH2 shRNA 1 and 3 are CCGGGCTAGGTTAATTGGGACCAAACTCGAGTTTGGTCCCAATTAACCT AGCTTTTTG and CCGGGCTAGGTTAATTGGGACCAAACTCGAGTTTGGTCCCAATTAACCTAGCTTTTTG respectively. A scrambled, non-targeting shRNA was used as a control. 10 μg of each shRNA were transfected into 293 T human embryonic kidney (HEK) cells along with 6.5 μg Hiv1 and 3 μg VSV-G constructs. After 24 hrs, the medium was replaced with DMEM containing 20% serum. KCL22 cells or CML CD34+ cells were transduced in 2 consecutive rounds of 24 and 48 hrs with lentiviral supernatant supplemented with Transdux (Systems Biosciences). Transduction efficiency (% GFP+ cells) was measured by flow cytometry. GFP positive cells were isolated by FACS using a FACS Aria cell sorter and knockdown assessed at the mRNA and protein level.
Western blotting
Total cellular protein was extracted from primary CML and normal samples by direct lysis in laemmli buffer or by acid extraction. Cell lysates were then run out on 4-12% SDS-PAGE gels, transferred to PVDF membrane and probed with the appropriate antibodies. Antibodies used were for H3K27me3 and H3K27me1 (see ChIP-sequencing below), histone H3 (9715; New England Biolabs), EZH2 (5246; New England Biolabs) and p53 (sc-126; Santa Cruz Biotechnology). Validation of specificities of, and citations for, these antibodies can be found on the manufacturers’ websites.
Experimental analysis by Affymetrix GeneChips, Taqman Low Density Array (TLDA) cards and Fluidigm
mRNA was extracted using the RNeasy Mini Kit (Qiagen) and DNase I treated on columns using the ribonuclease (RNase)-Free DNase Set (Qiagen). Purity was determined by the Agilent Bioanalyzer and Agilent RNA 6000 Nano kit (Agilent Technologies). Affymetrix GeneChIP analysis (using HuGene-1_0-st-v1 arrays) was performed using 50 ng of total RNA as starting material according to manufacturer’s instructions. For q-PCR, cDNA was obtained using the High Capacity RNA-to-cDNA kit (Applied Biosystems) or alternatively, 300 cells were sorted and processed with “Cells direct one-step q RT-PCR” (Invitrogen). Real Time qPCR was carried out using FAM-MGB probes for the genes of interest on the TLDA (ABI7900, Applied Biosystems) and Fluidigm 48.48 Dynamic Array IFC platforms (Fluidigm Corporation, San Francisco, CA, USA). Validation of mRNA expression changes determined by Affymetrix GeneChip analysis is shown in Supplementary Fig. S14a-e. All oligonucleotide primer pairs available upon request.
Computational analysis of Affymetrix GeneChips
Affymetrix datasets were processed using one of two different approaches. For CD34+CD38- and CD34+CD38+ normal and CML datasets generated for this study (E-MTAB-2581, Affymetrix HuGe 1.0 ST) or for datasets generated from treatment of CML or normal CD34+ with GSK343 (E-MTAB-2893, E-MTAB-3552, Affymetrix HuGe 1.0 ST), Bioconductor was used for data preprocessing (affy and aroma.affymetrix packages), quality control (simpleaffy package), differential gene expression analysis (limma package) and gene annotation (annotate package). Datasets for CML CD34+ cells treated with IM in vivo (GSE12211, Affymetrix Human Genome U133A 2.0) were normalised using RMA (affy v1.42.3). Datasets from LSCs treated with DAS, IM and NIL (E-MTAB-2594, Affymetrix HuGe 1.0 ST) generated for this study were normalised using RMA (oligo v1.28.2) and subsequently merged and batch corrected using COMBAT (inSilicoDb v2.0.1, inSilicoMerging v1.8.6). For pathway analysis, differentially expressed genes from E-MTAB-2893 and E-MTAB-3552 were analysed for enrichment of Panther Pathways using the DAVID Bioinformatics Database. See also Supplementary Table S3.
ChIP-sequencing
H3K4me3 (Abcam, ab8580), H3K27me3 (Millipore, 07-449) and H3K27me1 (Millipore, 07-448) ChIPs were performed (validated for specificity as previously described (25)) using between 104-106 primary HSCs (CD34+CD38-), LSCs (CD34+CD38-) or LPCs (CD34+CD38+) as starting material with no pre-clearing of chromatin (n=3 for all cell types). The three CML samples used for ChIP-sequencing displayed typical patterns of PRC2 mis-regulation (Supplementary Fig. S15a-c). ChIP-sequencing libraries (see Supplementary Table S6) were prepared according to manufacturer’s protocols (Illumina). HSC, LSC and LPC ChIP-sequencing was performed in singlicate per bioreplicate and the read data obtained was deposited into the European Nucleotide Archive (Access. No. PRJEB8291).
Reads obtained from ChIP-sequencing were aligned to the human genome (hg18; NCBI build 36.1) and uniquely mapped reads were extended to 150 bp and processed as BED or bigWig files for further analysis and visualisation in the UCSC genome browser. H3K27me3 read densities (per 106 reads) at each of 33,626 gene promoters were determined for each CML and normal sample across 2 kilobases (kb) windows flanking the transcriptional start site (TSS) (1 kb up- and down- stream of the TSS). These read densities were quantile normalised to reduce bias and only promoters where all three samples gave read densities greater than 0 at a given promoter were used for further data mining. As a result, read densities for 26,760 promoters (H3K27me3) or 24,292 promoters (H3K27me1) were used for the analyses described in the text.
Bona fide H3K27me3 target genes were defined as follows. k-means clustering of ChIP-sequencing read densities/106 reads at a resolution of 50 bp across 5 kb at each each of 33,626 promoters (2.5 kb bp up- and down- stream of the TSS) was performed by seqMINER (26). Clusters were generated that defined promoters having (i) H3K4me3 only, (ii) both H3K4me3 and H3K27me3, or (iii) neither histone modification (Supplementary Fig. S16a,b). Membership of gene promoters in these three classifications was validated by ChIP qPCR (Supplementary Fig. S17a-c) using SYBR-Green Master (Rox) (Roche) in a Stratagene Mx3000P qPCR system (Agilent Technologies). Oligonucleotide primer pairs available upon request.
Promoters defined as “H3K4me3 only” or having “neither” histone modification were then used as a measure of background noise for H3K27me3 in ChIP-sequencing datasets derived from each patient or normal sample. On a per sample basis, those promoters with H3K27me3 read densities (across 2 kb promoter windows) that were 3 standard deviations above background noise were considered as H3K27me3 targets. In this way, each H3K27me3 ChIP-sequencing dataset had a uniquely defined measure of background (i.e., a threshold) above which bona fide H3K27me3 targets could be identified.
Statistically significant changes in H3K27me3 read densities at promoters of H3K27me3 target genes (defined as above) were determined using either Student t-tests or one-sample t-tests as appropriate. Significance was determined at the level of individual promoters. In addition, promoters shown to be statistically significant were also required to have at least 1.5 fold changes in H3K27me3 levels to be considered as promoters with obvious changes. Promoters ranked as having H3K27me1 levels in the top 25% of all promoters genome-wide were used for the analysis described in Supplementary Fig. S8a,b. Yates Chi-square or Fischer Exact tests were used to determine statistically significant differences between observed vs expected frequencies of changes in H3K27me3 at promoters (gains or losses) with respect to changes in mRNA expression. Expected frequencies were the number of promoters showing gains or losses in H3K27me3 due to global H3K27me3 reprogramming on a per sample basis, or for common H3K27me3 reprogrammed genes. Observed frequencies were the data shown in Supplementary Figure S4b,c. Heatmaps of common H3K27me3 reprogrammed genes (Supplementary Fig. S3b) were generated using Cluster 3.0 (27).
Supplementary Material
Significance.
In CML, TKI-persistent LSCs remain an obstacle to cure and approaches to eradicate them remain a significant unmet clinical need. We demonstrate that EZH2 and H3K27me3 reprogramming is important for LSC survival, but renders them sensitive to the combined effects of EZH2i and TKI. This represents a novel approach to more effectively target LSCs in patients receiving TKI treatment.
Footnotes
Conflict of Interest Statement:
The authors declare no conflict of interest and/or no competing financial interests.
References
- 1.Hamilton A, Helgason GV, Schemionek M, Zhang B, Myssina S, Allan EK, et al. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood. 2012;119:1501–10. doi: 10.1182/blood-2010-12-326843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011;121:396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu S, McDonald T, Lin A, Chakraborty S, Huang Q, Snyder DS, et al. Persistence of leukemia stem cells in chronic myelogenous leukemia patients in prolonged remission with imatinib treatment. Blood. 2011;118:5565–72. doi: 10.1182/blood-2010-12-327437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chomel JC, Bonnet ML, Sorel N, Bertrand A, Meunier MC, Fichelson S, et al. Leukemic stem cell persistence in chronic myeloid leukemia patients with sustained undetectable molecular residual disease. Blood. 2011;118:3657–60. doi: 10.1182/blood-2011-02-335497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross DM, Branford S, Seymour JF, Schwarer AP, Arthur C, Yeung DT, et al. Safety and efficacy of imatinib cessation for CML patients with stable undetectable minimal residual disease: results from the TWISTER study. Blood. 2013;122:515–22. doi: 10.1182/blood-2013-02-483750. [DOI] [PubMed] [Google Scholar]
- 6.Gerber JM, Qin L, Kowalski J, Smith BD, Griffin CA, Vala MS, et al. Characterization of chronic myeloid leukemia stem cells. American journal of hematology. 2011;86:31–7. doi: 10.1002/ajh.21915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nieborowska-Skorska M, Kopinski PK, Ray R, Hoser G, Ngaba D, Flis S, et al. Rac2-MRC-cIII-generated ROS cause genomic instability in chronic myeloid leukemia stem cells and primitive progenitors. Blood. 2012;119:4253–63. doi: 10.1182/blood-2011-10-385658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lund K, Adams PD, Copland M. EZH2 in normal and malignant hematopoiesis. Leukemia. 2014;28:44–9. doi: 10.1038/leu.2013.288. [DOI] [PubMed] [Google Scholar]
- 10.Crea F, Paolicchi E, Marquez VE, Danesi R. Polycomb genes and cancer: Time for clinical application? Critical Reviews in Oncology/Hematology. 2012;83:184–93. doi: 10.1016/j.critrevonc.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 11.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–6. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt M, Rinke J, Schafer V, Schnittger S, Kohlmann A, Obstfelder E, et al. Molecular-defined clonal evolution in patients with chronic myeloid leukemia independent of the BCR-ABL status. Leukemia. 2014;28:2292–9. doi: 10.1038/leu.2014.272. [DOI] [PubMed] [Google Scholar]
- 13.Herrera-Merchan A, Arranz L, Ligos JM, de Molina A, Dominguez O, Gonzalez S. Ectopic expression of the histone methyltransferase Ezh2 in haematopoietic stem cells causes myeloproliferative disease. Nat Commun. 2012;3:623. doi: 10.1038/ncomms1623. [DOI] [PubMed] [Google Scholar]
- 14.Hidalgo I, Herrera-Merchan A, Ligos Jose M, Carramolino L, Nuñez J, Martinez F, et al. Ezh1 Is Required for Hematopoietic Stem Cell Maintenance and Prevents Senescence-like Cell Cycle Arrest. Cell Stem Cell. 2012;11:649–62. doi: 10.1016/j.stem.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Xie H, Xu J, Hsu JH, Nguyen M, Fujiwara Y, Peng C, et al. Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell Stem Cell. 2014;14:68–80. doi: 10.1016/j.stem.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verma SK, Tian X, LaFrance LV, Duquenne C, Suarez DP, Newlander KA, et al. Identification of Potent, Selective, Cell-Active Inhibitors of the Histone Lysine Methyltransferase EZH2. ACS Medicinal Chemistry Letters. 2012;3:1091–6. doi: 10.1021/ml3003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holyoake T, Jiang X, Eaves C, Eaves A. Isolation of a highly quiescent subpopulation of primitive leukemic cells in chronic myeloid leukemia. Blood. 1999;94:2056–64. [PubMed] [Google Scholar]
- 18.Hamad A, Sahli Z, El Sabban M, Mouteirik M, Nasr R. Emerging therapeutic strategies for targeting chronic myeloid leukemia stem cells. Stem Cells Int. 2013;2013 doi: 10.1155/2013/724360. 724360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pellicano F, Sinclair A, Holyoake TL. In search of CML stem cells' deadly weakness. Curr Hematol Malig Rep. 2011;6:82–7. doi: 10.1007/s11899-011-0085-y. [DOI] [PubMed] [Google Scholar]
- 20.Knutson SK, Kawano S, Minoshima Y, Warholic NM, Huang KC, Xiao Y, et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol Cancer Ther. 2014;13:842–54. doi: 10.1158/1535-7163.MCT-13-0773. [DOI] [PubMed] [Google Scholar]
- 21.Pellicano F, Scott MT, Helgason GV, Hopcroft LE, Allan EK, Aspinall-O'Dea M, et al. The Antiproliferative Activity of Kinase Inhibitors in Chronic Myeloid Leukemia Cells Is Mediated by FOXO Transcription Factors. Stem Cells. 2014;32:2324–37. doi: 10.1002/stem.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurtz C, Hatzi K, Cerchietti L, Braig M, Park E, Kim YM, et al. BCL6-mediated repression of p53 is critical for leukemia stem cell survival in chronic myeloid leukemia. J Exp Med. 2011;208:2163–74. doi: 10.1084/jem.20110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X, Cortes J, Kantarjian H. Estimations of the increasing prevalence and plateau prevalence of chronic myeloid leukemia in the era of tyrosine kinase inhibitor therapy. Cancer. 2012;118:3123–7. doi: 10.1002/cncr.26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie H, Peng C, Huang J, Li B, Kim W, Smith E, et al. Chronic myelogenous leukemia initiating cells require Polycomb group protein EZH2. Cancer Discov. 2016 doi: 10.1158/2159-8290.CD-15-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhami P, Saffrey P, Bruce AW, Dillon SC, Chiang K, Bonhoure N, et al. Complex exon-intron marking by histone modifications is not determined solely by nucleosome distribution. PLoS One. 2010;5:e12339. doi: 10.1371/journal.pone.0012339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye T, Krebs AR, Choukrallah MA, Keime C, Plewniak F, Davidson I, et al. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 2011;39:e35. doi: 10.1093/nar/gkq1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.