Abstract
Hepatitis B virus-encoded X protein (HBx) acts as a tumor promoter during hepatocellular carcinoma (HCC) development, probably by regulating the expression of host proteins through protein–protein interaction. A proteomics approach was used to identify HBx-interacting proteins involved in HBx-induced hepatocarcinogenesis. We validated the proteomics findings by co-immunoprecipitation and confocal microscopy. We performed cell proliferation, migration assays and cell cycle analyses in HCC cells. Finally, we confirmed the clinical significance of our findings in samples from patients. We found that cortactin (CTTN) is a novel HBx-interacting protein, and HBx regulates the expression of CTTN in the HCC cell lines MHCC-LM3 and HepG2. Mechanistically, by upregulating the expression of cAMP response element-binding protein (CREB1) and its downstream targets, such as cyclin D1 and MMP-9, the effects of the HBx-CTTN interaction on the enhancement of cellular proliferation and migration were maintained by inhibiting cell cycle arrest. In addition, we demonstrated that the levels of CTTN and CREB1 were closely correlated in clinical samples from HBV-infected patients with HCC. Overall, our data suggests that HBx contributes to cell migration and proliferation of HCC cells by interacting with CTTN and regulating the expression of CTTN and CREB1. Therefore, the HBx/CTTN/CREB1 axis is a potential novel therapeutic target in HCC.
Subject terms: Cancer, Viral hepatitis
Introduction
Hepatocellular carcinoma (HCC) is an aggressive human malignancy1–3. Hepatitis B virus (HBV) infection is the most common risk factor for HCC, and there are ~250 million chronic HBV carriers in the world4–7. Although HBV vaccines have obviously decreased the number of new HBV infection cases, they have limited benefits for previously diagnosed patients with chronic HBV infection8,9. The HBV genome comprises four overlapping open-reading frames: preC/C, preS/S, P, and X10. The X protein (HBx), encoded by the X region, is a multifunctional viral regulator involved in viral pathogenesis and carcinogenesis, and it plays an important role in HBV-related HCC processes, such as autophagy regulation, DNA repair inhibition, post‐transcriptional regulation, and cell cycle arrest9,11–14.
HBx interacts with many host targets, and as such, it is important in viral hepatocarcinogenesis15–19. For example, HBx activates transcription by interacting with transcription factors or complexes, including P53, C/EBP, Sp1, and STAT320–27. HBx also activates various cellular signal transduction pathways related to its transactivation, such as the NF-κB, AMP-activated protein kinase (AMPK), and MEKK1/Jun kinase signaling pathways22,28. Thus, the identification of novel HBx–host interactors and the pathways they are involved with might aid in the development of effective therapies for patients with HCC.
In this study, we looked for HBx-interacting proteins in HCC and explored their mechanism of action. Our data demonstrated the physical association between HBx and CTTN and identified the new HBx/CTTN/CREB1 axis as a crucial switch regulating the proliferation and migration of HCC cells.
Methods
Cell lines, cell culture, and cell transfection
The HCC cell lines HepG2 and MHCC-LM3 were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and China Center for Type Culture Collection (CCTCC, China) and cultured in Dulbecco’s Modified Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS) at a humidified incubator with 5% CO2 at 37 °C.
HBx and CREB1 overexpression was achieved by transfection of HBx (Plasmid #42596, Addgene, MA, USA) or CREB1-overexpressing vectors (GeneCopoecia, Guangzhou, China). The empty vector pcDNA3.1 (vector NC) was used as control; CTTN knockdown was achieved by transfection of small interfering RNAs targeting CTTN (si-CTTN; GeneCopoeia, Maryland, USA). HCC cells (1.5 × 105) grown on six-well plates were transfected with 100 pmol siRNA or 2 µg of overexpression vectors using 6 µL of Lipofectamine 2000 (#11668019, Invitrogen, MA, USA) as described by the manufacturer. The cells were harvested after 48 h. Western blot analyses or other experiments were performed. The siRNA and vector construction primer sequences are listed in Table S1.
Establishment of stably HBx-expressing HepG2 cells
Target HBx sequences of pcDNA3.1-Flag HBx plasmid (Plasmid #42596, Addgene, MA, USA) were amplified by PrimeSTAR GXL DNA polymerase (#R050A, Takara, Dalian, Japan). Next, HBx gene products and pLV-cDNA (No. 632177, Clontech, CA, USA) were amplified via enzyme digestion and purification according to the manufacturer’s instructions. After the enzyme-linked reaction, amplification, and DNA sequencing, we constructed a recombinant pLV-cDNA-HBx plasmid through the recombination of the digested HBx fragment and the purified pLV-cDNA product. According to the lentiviral packaging system protocol (Clontech, CA, USA), we collected lentiviral particles by transient transfection of 293 T cells, and then infected HepG2 cells with lentivirus. A Blasticidin S (#60218ES10, Yeasen, Shanghai, China) concentration of 6 µg/mL selected stably expressing HBx target cells. In this study, the stably modified cells (HBx-HepG2) were only used in the identification of proteins interacting with HBx, verification of the interaction between CTTN and HBx, and determination of CTTN mRNA levels and CTTN protein stability experiments.
Immunoprecipitations
Stably HBx-expressing HepG2 cells were lysed in a co-immunoprecipitation lysis buffer (buffer composed of 20 mM Tris (pH 7.5), 150 mM NaCl, and 1% Triton X-100; P0013, BBI, Shanghai, China) containing a protease inhibitor cocktail for 60 min at 4 °C. After centrifugation, the supernatant was incubated with anti-mouse IgG (#3420, CST, MA, USA) at 4 °C for 60 min and then with 20 μL of protein G agarose beads (#37478, CST, MA, USA); an anti-HBx antibody (M10514, Xiamen Innovax Biotech, Xiamen, China) was then added, and the mixture was incubated at 4 °C overnight. Protein complexes containing the HBx antibody were precipitated with anti-mouse IgG beads, and washed with RIPA lysis buffer (P0013, BBI, Shanghai, China) and phosphate-buffered saline (PBS), sequentially. Finally, the pellet was eluted and subjected to SDS-PAGE analysis.
Mass spectrometry (MS) and database analysis
LC-ESI-LTQ-Orbitrap-MS analysis of proteins was performed as previously described by us29. The protein bands were excised and destained with 100 mM NH4HCO3 in 50% acetonitrile (ACN). The proteins were reduced (10 mM dithiothreitol, 56 °C, 30 min), alkylated (50 mM iodoacetamide, in the dark, 20−25 °C, 30 min), and dried in a vacuum centrifuge. The gel pieces that contained proteins were incubated in a digestion solution (40 mM NH4HCO3, 9% ACN, and 20 μg/mL trypsin) at 37 °C for 18–24 h. The tryptic peptide mixture was purified with a ZipTipC18 microcolumn (cat. no. ZTC18S096; Millipore, Darmstadt, Germany). The purified tryptic peptide mixture was separated onto a PepMap C18 trap column (75 μm, 15 cm) at a column flow rate of 200 nL/min. The MS scan and MS spectra were measured in data-dependent mode with MS/MS analysis of the seven strongest ions in the LTQ. The MS data were analyzed using the Xcalibur software and submitted to the database search via Proteome Discoverer. Search parameters included taxonomy (Homo sapiens), enzyme (trypsin), MS/MS tolerance (±0.5 Da), peptide tolerance (20 ppm), peptide charge (2 + , 3 + , and 4 + ), missed cleavage sites (2), fixed modification (carbamoyl methylation of cysteine), and variable modification (methionine oxidation).
Western blot analysis
Cells were lysed in RIPA lysis buffer (BBI) and separated by electrophoresis, transferred to membranes, and subjected to western blot according to the standard procedure. The primary antibodies used included anti-HBx (dilution 1:1000, M10514, Xiamen Innovax Biotech, Xiamen, China), anti-CTTN (dilution 1:1000, ab81208, Abcam), anti-CREB1 (dilution 1:1000, Cat. #9197, CST), anti-cyclin D1 (dilution 1:1000, D220509, BBI), anti-MMP-9 (dilution 1:1000, ab73734, Abcam), anti-GAPDH (dilution 1:2000, ab8245, Abcam), anti-E-cadherin (Cat. 20874, Proteintech), and anti-vimentin (dilution 1:1000, sc-80975, Santa Cruz). After incubating with goat anti-mouse and goat anti-rabbit secondary antibodies for 1 h at room temperature, immunoreactive bands were visualized with a chemiluminescence system and quantified using Image J software.
Immunofluorescence staining
For the detection of the intracellular distribution of HBx and CTTN, cells (1 × 105 per well) were seeded in six-well glass-bottom plates, fixed in 4% paraformaldehyde for 15 min, and then permeabilized with 0.2% Triton X-100 (PBS) for 10 min. Nonspecific binding sites were blocked with 1% bovine serum albumin in PBS for 1 h. Cells were treated with a primary antibody specific for HBx (1:1000, M10514, Xiamen Innovax Biotech) or CTTN (dilution 1:1000, ab81208, Abcam) overnight at 4 °C. Thereafter, the cells were incubated with eFluor570-F(ab’)2-goat anti-mouse IgG/IgM (H + L) (dilution 1:20, Cat. # 41-4010-82, Invitrogen) and FITC-goat anti-Rabbit IgG (H + L) cross-adsorbed secondary antibody (dilution 1:20, Cat. #F-2765, Invitrogen, Carlsbad, CA, USA). DAPI (Beyotime, Shanghai, China) was used to stain nuclei before capturing images. The images were acquired using a confocal microscope (Zeiss, Oberkochen, Germany). The green fluorescence indicated HBx expression, the red fluorescence indicated CTTN expression, and the blue fluorescence indicated the nuclei.
Protein–protein interaction analysis
The Mentha (http://mentha.uniroma2.it/about.php), BioGrid (https://thebiogrid.org/), IntAct (https://www.ebi.ac.uk/intact/), and BIND (https://www.bindingdb.org/bind/aboutus.jsp) databases were used to analyze the proteins interacting with HBx or CTTN. The candidate proteins were then applicated for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation (http://www.genome.jp/kegg/) to identify the related pathways. The interaction network of these proteins was constructed using String (https://string-db.org/) and visualized with the Cytoscape software (version 3.4) 18.
MTT assays
MTT assays were performed to evaluate cell viability. Twenty-four hours after seeding into 96-well plates (5 × 103 cells/well), HepG2 and MHCC-LM3 cells were transfected with si-CTTN; 48 h after transfection, 20 μL of MTT (at a concentration of 5 mg/mL; Sigma-Aldrich) was added, and the cells were incubated for an additional 4 h in a humidified incubator with 5% CO2 at 37 °C. DMSO (200 μL) was added after the supernatant was discarded to dissolve the formazan crystals, and finally absorbance at 490 nm was measured (PerkinElmer). The viability of non-treated cells (control) was defined as 100%, and the viability of the other groups of cells was calculated accordingly.
5-Bromo-2-deoxyuridine assays
5-Bromo-2-deoxyuridine (BrdU) assays were conducted 24 and 48 h after cells were co-transfected with si-CTTN and pcDNA3.1( + )-HBx-overexpressing vectors. Cells were seeded in 96-well culture plates at a density of 2 × 103 cells/well, cultured for 48 h, and then incubated with a final concentration of 10 μM BrdU (BD Pharmingen, San Diego, CA, USA) for 2 h. Cells were fixed for 30 min at room temperature, incubated with peroxidase-coupled BrdU antibody (Sigma-Aldrich) for 60 min at room temperature, washed three times with PBS, incubated with peroxidase substrate (tetramethylbenzidine) for 30 min, and the 450-nm absorbance values were measured for each well. Background BrdU immunofluorescence was determined in cells not exposed to BrdU and stained with the BrdU antibody.
Transwell assays
Cells were co-transfected with si-CTTN and pcDNA3.1( + )-HBx-overexpressing vectors, and seeded on the top side of polycarbonate transwell filters. The cells were suspended in a medium without serum, while the bottom chamber was filled with medium with serum. After 24 h, the non-migrating cells in the top chambers were removed with cotton swabs. The migrated cells on the lower membrane surface were fixed in 100% methanol for 10 min, air-dried, stained with crystal violet (BBI, Shanghai, China), and counted under a microscope.
Cell cycle analysis
Cells (5 × 105) were silenced for CTTN or transfected with a plasmid CREB1 knockdown for 48 h. The cells were washed with PBS and collected into a 1.5-mL centrifuge tube. Subsequently, the cells were fixed with 70% ethanol for 24 h and then incubated with a propidium iodide solution (C1052, BBI, Shanghai, China). Flow cytometry was used to study the cell cycle distribution after HBx overexpression and CREB1 knockdown, according to the manufacturer’s instructions (Millipore Guava easyCyte, ON, Canada).
Quantitative PCR (qPCR) and tissue specimens
The total RNA was isolated from stably HBx-expressing HepG2 cells and HBx ( + )/(−) HCC tissues by using TRIzol (Invitrogen) following the manufacturer’s protocol. A SYBR green PCR Master Mix (Qiagen) was used for mRNA expression detection following the protocol of the manufacturer. The GAPDH expression was used as an endogenous control. The 2−ΔΔCT method30 was used to calculate the relative fold changes.
Thirty HBx ( + ) and 22 HBx (−) HCC tissues were collected from tumor surgical resection in the Xiangya Hospital (Central South University, Changsha, China) with the approval of the Medical Ethics Committee of Xiangya hospital at Central South University. Informed consent was obtained from all patients enrolled.
Statistical analysis
The data were processed using SPSS 17.0 statistical software and presented as the mean ± SD of at least three independent experiments. A Student's t test was used for statistical comparison between means, where applicable. Differences between more than two groups were estimated using one-way ANOVA. *P < 0.05; **P < 0.01.
Results
Identification of proteins interacting with HBx in HepG2 cells
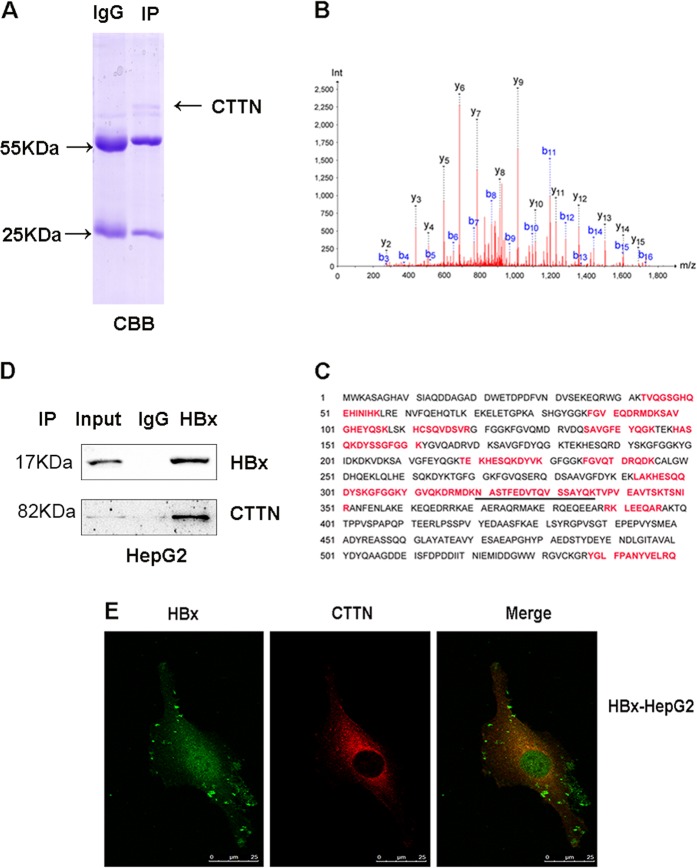
To identify proteins that interact with HBx, we used a CoIP-MS proteomic analysis of the HBx complexes isolated from HepG2 stably expressing HBx (HBx-HepG2) cells using a HBx antibody; nonimmune IgGs were used as a negative control. The complexes were eluted and separated on 12% SDS-PAGE (Fig. 1a). Proteins in the gels were detected by Coomassie brilliant blue staining (CBB), in-gel trypsin digested, and subjected to MS analysis. Fifteen proteins were identified in the anti-HBx-treated cells and not in the negative control: their mass spectra are shown as supplementary data (Table S2). Among them, CTTN was identified from a specific band that originated the highest score and 18 unique peptides. We confirmed the protein sequence of CTTN (Fig. 1b–c; the matched peptides are labeled in red bold letters).
Fig. 1. CTTN interacts with HBx in HepG2 HCC cells.
a Analysis of HBx-associated proteins by CoIP-MS in HepG2 cells overexpressing HBx (HBx-HepG2). b Representative MS/MS spectrum of CTTN; the amino acid sequence of a doubly charged peptide with m/z 937.9402 was identified as NASTFEDVTQVSSAYQK from mass differences in the y and b fragment ions series. c Protein sequence of CTTN: the matched peptides are labeled in red bold letters. d CoIP to validate the HBx-CTTN interaction. CTTN was immunoprecipitated by an anti-HBx but not by IgG control. e Expression of HBx and CTTN in the cytoplasm HBx-HepG2 by confocal microscopy
Verification of the interaction between CTTN and HBx
We confirmed the interaction between CTTN and HBx in co-immunoprecipitation assays. HBx-HepG2 cell lysates were immunoprecipitated with an HBx antibody or IgG; immunocomplexes were separated on a gel and subjected to western blot with a CTTN antibody. As shown in Fig. 1d, CTTN was detected in anti-HBx immunoprecipitates but not in the control samples, suggesting the interaction between CTTN and HBx in vitro. Subsequently, we investigated CTTN and HBx subcellular localization in HBx-HepG2 cells to determine where they interacted with each other. Immunofluorescence staining indicated that HBx enhanced CTTN fluorescence intensity (Fig. 1d; Supplementary Fig. S1). Moreover, CTTN and HBx colocalized in the cytoplasm (Fig. 1e).
Roles of HBx/CTTN interaction in enhancing cell proliferation and migration
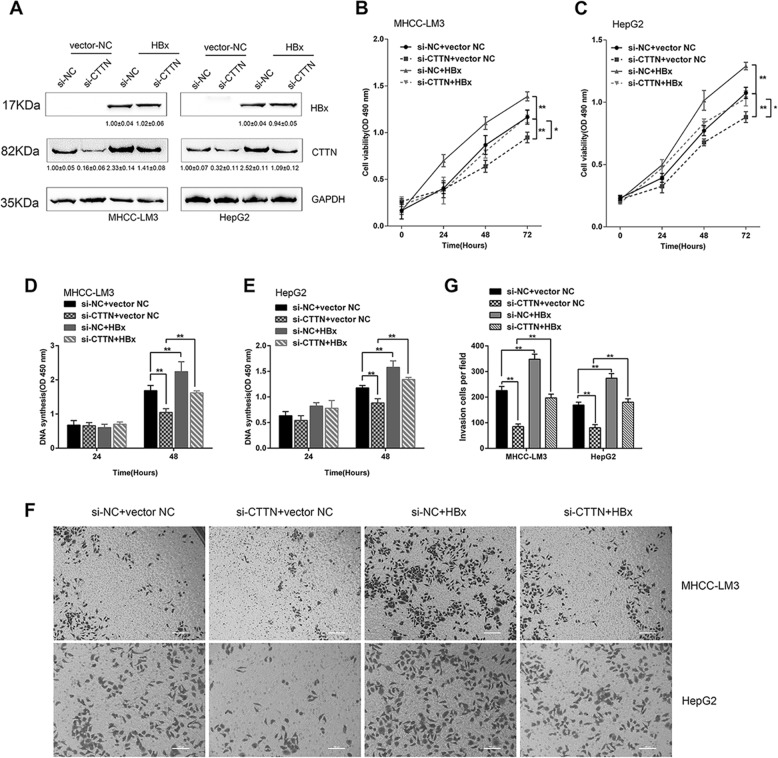
To investigate whether the HBx/CTTN interaction had an impact on cellular processes, we conducted MTT, BrdU, and transwell assays. We found that HBx significantly increased the CTTN in HepG2 and MHCC-LM3 cells. While CTTN silencing could partially reverse the effect of HBx on CTTN protein expression (Fig. 2a). The upregulation of CTTN expression by HBx might be associated with the increase of protein stability (Supplementary Fig. S2). MTT and BrdU assays showed that the viability and DNA synthesis ability of HepG2 and MHCC-LM3 cells increased upon HBx overexpression and decreased upon CTTN silencing, and CTTN silencing reversed the effects of HBx overexpression in cells that overexpressed HBx and were silenced for CTTN (Fig. 2b–e). Similarly, transwell assays revealed that cell migration was significantly suppressed by CTTN knockdown and promoted by HBx overexpression (Fig. 2f, g). These findings indicate that HBx promotes cell proliferation and migration of HCC cells by interacting with CTTN.
Fig. 2. Effects of the HBx-CTTN interaction on cell proliferation and migration.
a HepG2 and MHCC-LM3 cells were co-transfected with si-CTTN and pcDNA3.1( + )-HBx-overexpressing vectors. Protein expressions of HBx and CTTN were measured by western blot. b, c HepG2 and MHCC-LM3 cells were co-transfected with si-CTTN and pcDNA3.1( + )-HBx-overexpressing vectors and the cell viability was examined using MTT assays. d, e DNA synthesis was examined in BrdU assays; the data are presented as the mean ± SD of three independent experiments. f, g Migration of the indicated groups of cells examined using transwell assays on a light microscope. The statistical analysis was shown in the right panel. *P < 0.05, **P < 0.01
HBx-CTTN interaction modulates CREB1 and its downstream target genes
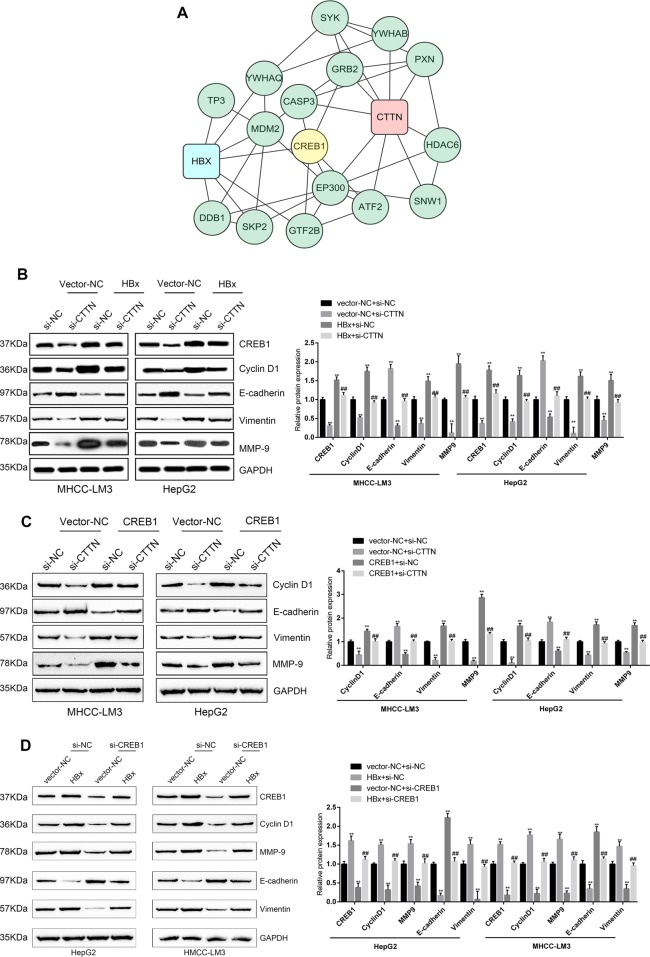
To investigate how HBx-CTTN modulates cell proliferation and migration, we performed bioinformatics analyses and analyzed the signaling pathways that are involved (Fig. S3). Proteins that might be related to HBx and CTTN were analyzed using online databases. A total of 124 human proteins were predicted to interact with CTTN (Table S3), and a total of 20 proteins were predicted to bind to HBx (Table S4). KEGG pathway annotation (Tables S5 and S6) showed that 16 proteins (nine for CTTN and seven for HBx) were involved in viral carcinogenesis (hsa05023); these proteins were selected and visualized using Cytoscape. We found that, among them (Fig. 3a), a cellular transcription factor was reported to participate in HBx-induced dysregulation of lipogenesis in HCC cells31. Previous studies have shown that CREB1 knockdown inhibits the expression of its downstream targets: cyclin D1, Bcl-2, and MMP-9 in gastric cancer2. Based on its essential role in HCC, CREB1 was chosen as a potential target. Therefore, we decided to focus on CREB1 as the possible target of HBx-CTTN.
Fig. 3. The HBx-CTTN interaction modulates CREB1 and its downstream target genes.
a Analysis of HBx and CTTN protein–protein network using online databases. The network nodes represent proteins, and the connecting lines represent protein–protein interactions. b CREB1, cyclin D1, MMP-9, E-cadherin, and vimentin were detected by western blot in HepG2 and MHCC-LM3 cells. HepG2 and MHCC-LM3 cells were transfected with si-CTTN or si-NC and pcDNA3.1( + )-HBx or vector NC. si-CTTN or si-NC and pcDNA3.1( + )-HBx or vector NC were compared with the si-NC + vector-NC group. **P < 0.01. si-CTTN and pcDNA3.1( + )-HBx group were compared with HBx + si-NC group. ##P < 0.01. c The expression of EMT-associated proteins was detected in HepG2 and MHCC-LM3 cells transfected with si-CTTN or si-NC and CREB1 or vector NC. si-CTTN or si-NC and CREB1 or vector NC were compared with the si-NC + vector-NC group. **P < 0.01. si-CTTN and the CREB1 group were compared with the CREB1 + si-NC group. ##P < 0.01. d The expression of CTTN, HBx, CREB1, cyclin D1, MMP-9, E-cadherin, and vimentin was determined by western blot when HepG2 and MHCC-LM3 cells were transfected with si-CREB1 or si-NC and pcDNA3.1( + )-HBx or vector NC compared with the si-NC + vector-NC group. **P < 0.01. si-CREB1 and pcDNA3.1( + )-HBx group were compared with the HBx + si-NC group. ##P < 0.01
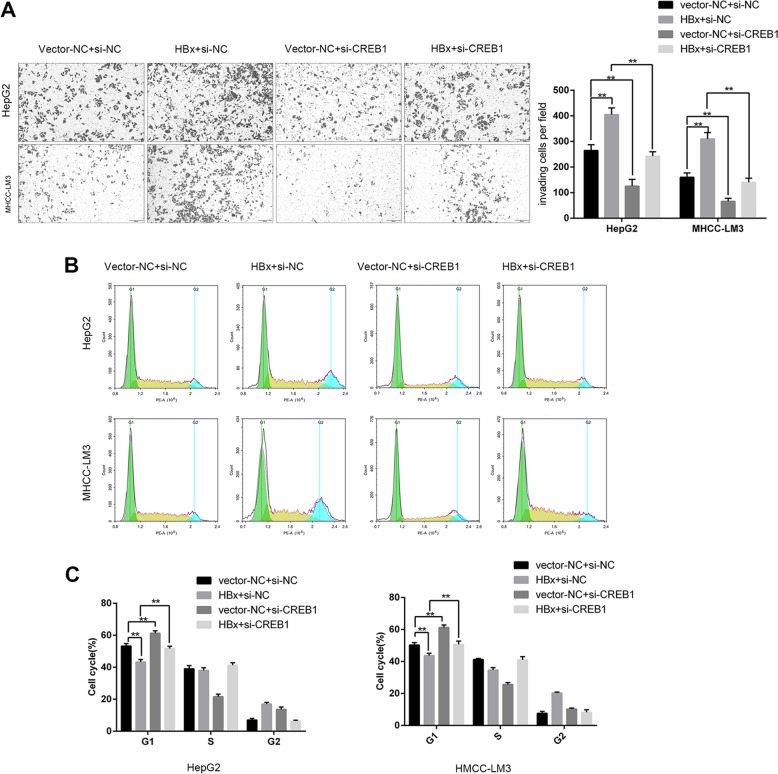
To evaluate the combined effects of HBx and CTTN on CREB1 and its downstream target genes, MHCC-LM3 and HepG2 cells were silenced for CTTN, and the protein levels of CREB1, cyclin D1, E-cadherin, vimentin, and MMP-9 were examined. We found that CTTN knockdown remarkably increased E-cadherin protein levels and reduced the levels of CREB1, cyclin D1, vimentin, and MMP-9, while HBx overexpression decreased E-cadherin expression and increased the levels of CREB1, cyclin D1, vimentin, and MMP-9, and the effect of HBx overexpression was partially attenuated by CTTN knockdown (Fig. 3b). The epithelial–mesenchymal transition (EMT) is directly linked to tumor initiation and invasion32. Therefore, we monitored the expression of EMT markers and found that CREB1 overexpression significantly reduced E-cadherin expression and increased vimentin and MMP-9 levels in MHCC-LM3 and HepG2 cells, and this effect was reversed by CTTN knockdown (Fig. 3c). We further assessed the relationship between HBx-CTTN and CREB1. The western blot results showed that CREB1 knockdown increased E-cadherin expression and reduced MMP-9 and vimentin expression;these effects were reversed by HBx overexpression (Fig. 3d). We found that CREB1 knockdown inhibited cell migration and induced cell cycle arrest at G1, while these effects could be reversed by HBx overexpression (Fig. 4a–c). Therefore, our results suggested that CREB1 and its downstream targets can be regulated by the HBx-CTTN complex.
Fig. 4. HBx-CTTN regulates cell proliferation and migration via CREB1.
HepG2 and MHCC-LM3 cells were transfected with si-CREB1 or si-NC and pcDNA3.1( + )-HBx or vector NC. a Migration of the indicated groups of cells was examined using transwell assays. b, c Cell cycle analysis by flow cytometry in HepG2 and MHCC-LM3 cells. The results show the percentage of cells in each phase of the cell cycle. *P < 0.05, **P < 0.01
Positive correlation between the high expression of CTTN and CREB1 in HBV-associated patients
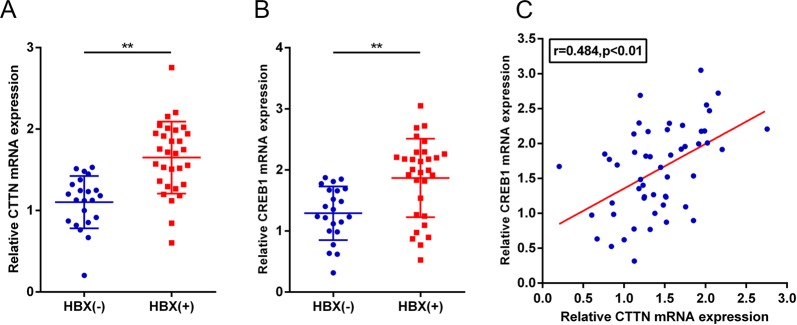
To investigate the clinical significance of our findings, the mRNA levels of CTTN and CREB1 were evaluated by qPCR in HCC tissues of patients. As shown in Fig. 5, the levels of CTTN in the HBx ( + ) group were significantly upregulated compared with the HBx(−) group (P < 0.01), and similar results were obtained with CREB1 (P < 0.01). In addition, CTTN and CREB1 expression were positively correlated in tissue samples.
Fig. 5. Upregulation of CTTN and CREB1 in clinical specimens.
a, b CTTN and CREB1 expression in 30 HBx ( + ) and 22 HBx (−) HCC tissues from patients, as assessed by qPCR. The data are presented as mean ± SD of three independent experiments. **P < 0.01. c The correlation between CTTN and CREB1 expression in tissue samples was analyzed using Spearman’s rank correlation analysis
Discussion
HBx viral protein contributes to the development of HCC mainly through HBx–host interactions, and it is involved in numerous cellular activities, including transcription activation, transactivation, DNA damage repair, and cell transformation inhibition7,9,13,23,33. For example, HBx inhibits DNA repair by interacting with PARP1 and SIRT6, resulting in dissociation of SIRT6 and PPAP19. HBx also inhibits cell apoptosis by regulating the endoplasmic reticulum (ER) stress response: GRP78 interacts with HBx in the ER, and this interaction leads to the interruption of ER stress response and host cell apoptosis17. Many studies have shown that HBx participates in the regulation of viral or host proteins by protein–protein interactions, providing potent evidence for HBV-related tumorigenesis. However, how HBx protein induces HCC is still unclarified. In this study, we undertook a proteomics approach to identify HBx-interacting proteins and found that CTTN, a Src substrate, was a novel HBx-interacting protein.
Previous studies have shown that Src protein and RNA expression levels are significantly upregulated in most HBV+–HCC specimens and HBx transgenic mice, and the overexpression of Src promotes cell viability, migration, and cell colony formation34–36. The Src substrate CTTN has been found to be overexpressed in various cancers, including human colorectal cancer, esophageal tumors, and non-small-cell lung cancer, and HCC and CTTN overexpression closely correlates with tumor aggressiveness, cell adhesion, and cell motility37–42. We demonstrated that HBx contributed to cell migration and proliferation of HCC by binding CTTN in the cytoplasm and upregulating its expression. It has been reported that the phosphorylation and activation of Src are increased in HCC and result in the phosphorylation of CTTN at Y42136,43,44. This modification is closely associated with CTTN localization and function through the regulation of its interaction with other host proteins45. We believe that the adjacent region between Y421 and the SH3 domain of CTTN (aa 490–550) in the C terminus contributes to the phosphorylation of CTTN at Y421 upon HBx-CTTN interaction, and affects the structure and biological functions of CTTN. This hypothesis needs further investigation. Furthermore, we found that HBx regulates CTTN protein expression at the post-transcriptional level, and HBx increases CTTN protein stability (Figure S2). Given that Zhao J. et al. reported that CTTN protein degradation was conducted by ubiquitin-independent proteasome system, we assumed that HBx could increase the stability of CTTN through attenuation of CTTN’s ubiquitination46.
We also performed bioinformatics analyses to identify the potential targets of HBx and CTTN. Based on its essential role in HCC, CREB1 was chosen as a potential target. We hypothesized that the HBx-CTTN interaction may affect CREB1 and its downstream targets. CREB1 is part of leucine zipper family of proteins, and its expression is essential for various cell functions, including apoptosis, cell cycle, and DNA repair47–49. Once activated, CREB1 regulates its downstream target genes, including the apoptosis suppressor gene Bcl2, a metalloproteinase involved in extracellular matrix remodeling, MMP9, and the cell cycle-related genes, cyclin D1, A1, and B12,50. Previous studies have revealed that HBx can induce CREB-mediated activation of miR-3188 and Notch signaling, resulting in the development of HBV-related HCC51–53. Based on our data, we illustrate a critical oncogenic role of CREB1, which is overexpressed and correlates with CTTN in HBV+ HCC. Furthermore, by targeting CREB1, the HBx-CTTN interaction forms a critical oncogenic axis, which regulates cell proliferation, migration, and oncogenic signaling in HCC. Bioinformatics analysis has shown that SKP2, a vital component of the SCF ubiquitin E3 ligase, is involved in HBV-related tumorigenesis54. CTTN also plays a role in SKP2 expression55,56. Therefore, the HBx-CTTN interaction has a possible effect on SKP2. This hypothesis might be investigated in the future.
Based on this work, we provided a novel mechanism of how HBx contributes to the progression of HCC: HBx interacts with CTTN in the cytoplasm, resulting in the upregulation of CREB1, which promotes the proliferation and migration of HCC. The crosstalk between CTTN and HBx may also provide a potential therapeutic strategy for HCC therapy.
Supplementary information
Acknowledgements
This work was supported by a grant from International Scientific and Technology Cooperation Program of China (No. 2015DFA31490), grants from National Natural Science Foundation of China (No. 81602572, 81873574, and 81402623), and the Natural Science Foundation of Hunan Province (2017JJ3496 and 2018ZX10732202).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by G. Giannelli
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Fang Peng, Email: pengfang@csu.edu.cn.
Xue-gong Fan, Email: xgfan@hotmail.com.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-019-1650-x).
References
- 1.Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA: a Cancer Journal for Clinicians. 2012;62:394–399. doi: 10.3322/caac.21161. [DOI] [PubMed] [Google Scholar]
- 2.Rao M, Zhu Y, Cong X, Li Q. Knockdown of CREB1 inhibits tumor growth of human gastric cancer in vitro and in vivo. Oncol. Rep. 2017;37:3361–3368. doi: 10.3892/or.2017.5636. [DOI] [PubMed] [Google Scholar]
- 3.Luo J, et al. MicroRNA-146a-5p Enhances Radiosensitivity in Hepatocellular Carcinoma through Replication Protein A3 Induced Activation of the DNA Repair Pathway. Am. J. Physiol. Cell Physiol. 2018;316:C299–C311. doi: 10.1152/ajpcell.00189.2018. [DOI] [PubMed] [Google Scholar]
- 4.Chen RC, et al. Integrated analysis of microRNA and mRNA expression profiles in HBx-expressing hepatic cells. World J. Gastroenterol. 2017;23:1787–1795. doi: 10.3748/wjg.v23.i10.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi J, et al. Risk of hepatocellular carcinoma in patients treated with entecavir vs tenofovir for chronic hepatitis B: A Korean Nationwide Cohort Study. JAMA Oncol. 2018;5:30–36. doi: 10.1001/jamaoncol.2018.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu S, et al. Detection of HBV DNA and antigens in HBsAg-positive patients with primary hepatocellular carcinoma. Clin. Res. Hepatol. Gas. 2017;41:415–423. doi: 10.1016/j.clinre.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Duan M, et al. Diverse modes of clonal evolution in HBV-related hepatocellular carcinoma revealed by single-cell genome sequencing. Cell Res. 2018;28:359–373. doi: 10.1038/cr.2018.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan L, et al. Optimized HepaRG is a suitable cell source to generate the human liver chimeric mouse model for the chronic hepatitis B virus infection. Emerg Microbes Infect. 2018;7:144. doi: 10.1038/s41426-018-0143-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Na TY, et al. Interaction of hepatitis B virus X protein with PARP1 results in inhibition of DNA repair in hepatocellular carcinoma. Oncogene. 2016;35:5435–5445. doi: 10.1038/onc.2016.82. [DOI] [PubMed] [Google Scholar]
- 10.Bai W, et al. Re-designed recombinant hepatitis B virus vectors enable efficient delivery of versatile cargo genes to hepatocytes with improved safety. Viruses. 2016;8:129. doi: 10.3390/v8050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An P, Xu J, Yu Y, Winkler CA. Host and viral genetic variation in HBV-related hepatocellular carcinoma. Front. Genet. 2018;9:261. doi: 10.3389/fgene.2018.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu S, et al. Crosstalk between hepatitis B virus X and high-mobility group box 1 facilitates autophagy in hepatocytes. Mol. Oncol. 2018;12:322–338. doi: 10.1002/1878-0261.12165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen SL, et al. HBx-mediated decrease of AIM2 contributes to hepatocellular carcinoma metastasis. Mol. Oncol. 2017;11:1225–1240. doi: 10.1002/1878-0261.12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian Y, et al. HBx promotes cell proliferation by disturbing the cross-talk between miR-181a and PTEN. Sci. Rep. 2017;7:40089. doi: 10.1038/srep40089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sitterlin D, et al. Transy C. Interaction of the UV-damaged DNA-binding protein with hepatitis B virus X protein is conserved among mammalian hepadnaviruses and restricted to transactivation-proficient X-insertion mutants. J. Virol. 1997;71:6194–6199. doi: 10.1128/jvi.71.8.6194-6199.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei C, et al. The hepatitis B virus X protein disrupts innate immunity by downregulating mitochondrial antiviral signaling protein. J. Immunol. 2010;185:1158–1168. doi: 10.4049/jimmunol.0903874. [DOI] [PubMed] [Google Scholar]
- 17.Li J, et al. Hepatitis B virus X protein inhibits apoptosis by modulating endoplasmic reticulum stress response. Oncotarget. 2017;8:96027–96034. doi: 10.18632/oncotarget.21630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh A, Kim HS, Lim SO, Yu DY, Jung G. Hepatitis B viral X protein interacts with tumor suppressor adenomatous polyposis coli to activate Wnt/beta-catenin signaling. Cancer Lett. 2011;300:162–172. doi: 10.1016/j.canlet.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 19.Abdul F, et al. Smc5/6 antagonism by HBx is an evolutionarily conserved function of hepatitis B virus infection in mammals. J Virol. 2018;92:e00769–18. doi: 10.1128/JVI.00769-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi BH, Park GT, Rho HM. Interaction of hepatitis B viral X protein and CCAAT/ enhancer-binding protein alpha synergistically activates the hepatitis B viral enhancer II/pregenomic promoter. J. Biol. Chem. 1999;274:2858–2865. doi: 10.1074/jbc.274.5.2858. [DOI] [PubMed] [Google Scholar]
- 21.Chan C, et al. Global re-wiring of p53 transcription regulation by the hepatitis B virus X protein. Mol. Oncol. 2016;10:1183–1195. doi: 10.1016/j.molonc.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shukla R, et al. Proinflammatory cytokine TNF-alpha increases the stability of hepatitis B virus X protein through NF-kappaB signaling. Carcinogenesis. 2011;32:978–985. doi: 10.1093/carcin/bgr057. [DOI] [PubMed] [Google Scholar]
- 23.Jiang T, Liu M, Wu J, Shi Y. Structural and biochemical analysis of Bcl-2 interaction with the hepatitis B virus protein HBx. Proc. Natl Acad. Sci. USA. 2016;113:2074–2079. doi: 10.1073/pnas.1525616113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang WG, et al. Targeted proteomic analysis of 14-3-3sigma in nasopharyngeal carcinoma. Int. J. Biochem. Cell Biol. 2010;42:137–147. doi: 10.1016/j.biocel.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Zhang T, et al. An integrated proteomics and bioinformatics analyses of hepatitis B virus X interacting proteins and identification of a novel interactor apoA-I. J. proteomics. 2013;84:92–105. doi: 10.1016/j.jprot.2013.03.028. [DOI] [PubMed] [Google Scholar]
- 26.Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J. Hepatol. 2016;64(1 Suppl):S84–S101. doi: 10.1016/j.jhep.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 27.Al-Anazi MR, Nazir N, Colak D, Al-Ahdal MN, Al-Qahtani AA. Deletion and functional analysis of hepatitis B virus X protein: evidence for an effect on cell cycle regulators. Cell. Physiol. Bioch.: Int. J. Exp. Cell. Physiol., Bioch., And Pharmacol. 2018;49:1987–1998. doi: 10.1159/000493670. [DOI] [PubMed] [Google Scholar]
- 28.Bagga S, Rawat S, Ajenjo M, Bouchard MJ. Hepatitis B virus (HBV) X protein-mediated regulation of hepatocyte metabolic pathways affects viral replication. Virology. 2016;498:9–22. doi: 10.1016/j.virol.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Peng F, et al. Nitroproteins in human astrocytomas discovered by gel electrophoresis and tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2015;26:2062–2076. doi: 10.1007/s13361-015-1270-3. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.You X, et al. Hepatitis B virus X protein upregulates oncogene Rab18 to result in the dysregulation of lipogenesis and proliferation of hepatoma cells. Carcinogenesis. 2013;34:1644–1652. doi: 10.1093/carcin/bgt089. [DOI] [PubMed] [Google Scholar]
- 32.von Olshausen G, et al. Hepatitis B virus promotes beta-catenin-signalling and disassembly of adherens junctions in a Src kinase dependent fashion. Oncotarget. 2018;9:33947–33960. doi: 10.18632/oncotarget.26103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mouzannar K, et al. Farnesoid X receptor-alpha is a proviral host factor for hepatitis B virus that is inhibited by ligands in vitro and in vivo. FASEB J. 2018;33:2472–2483. doi: 10.1096/fj.201801181R. [DOI] [PubMed] [Google Scholar]
- 34.Rothschild BL, et al. Cortactin overexpression regulates actin-related protein 2/3 complex activity, motility, and invasion in carcinomas with chromosome 11q13 amplification. Cancer Res. 2006;66:8017–8025. doi: 10.1158/0008-5472.CAN-05-4490. [DOI] [PubMed] [Google Scholar]
- 35.MacGrath SM, Koleske AJ. Cortactin in cell migration and cancer at a glance. J. Cell Sci. 2012;125(Pt 7):1621–1626. doi: 10.1242/jcs.093781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu JW, et al. Identification of the common regulators for hepatocellular carcinoma induced by hepatitis B virus X antigen in a mouse model. Carcinogenesis. 2012;33:209–219. doi: 10.1093/carcin/bgr224. [DOI] [PubMed] [Google Scholar]
- 37.Hong CC, et al. miR326 maturation is crucial for VEGF-C-driven cortactin expression and esophageal cancer progression. Cancer Res. 2014;74:6280–6290. doi: 10.1158/0008-5472.CAN-14-0524. [DOI] [PubMed] [Google Scholar]
- 38.Jing X, et al. Cortactin promotes cell migration and invasion through upregulation of the dedicator of cytokinesis 1 expression in human colorectal cancer. Oncology Rep. 2016;36:1946–1952. doi: 10.3892/or.2016.5058. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, et al. miR-182 suppresses invadopodia formation and metastasis in non-small cell lung cancer by targeting cortactin gene. J. Exp. Clin. Cancer Res. 2018;37:141. doi: 10.1186/s13046-018-0824-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jia D, et al. Amplification of MPZL1/PZR promotes tumor cell migration through Src-mediated phosphorylation of cortactin in hepatocellular carcinoma. Cell Res. 2014;24:204–217. doi: 10.1038/cr.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chuma M, et al. Overexpression of cortactin is involved in motility and metastasis of hepatocellular carcinoma. J. Hepatol. 2004;41:629–636. doi: 10.1016/j.jhep.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 42.He J, et al. Fra-1 is upregulated in gastric cancer tissues and affects the PI3K/Akt and p53 signaling pathway in gastric cancer. Int. J. Oncol. 2015;47:1725–1734. doi: 10.3892/ijo.2015.3146. [DOI] [PubMed] [Google Scholar]
- 43.Martinez-Quiles N, Ho HY, Kirschner MW, Ramesh N, Geha RS. Erk/Src phosphorylation of cortactin acts as a switch on-switch off mechanism that controls its ability to activate N-WASP. Mol. Cell. Biol. 2004;24:5269–5280. doi: 10.1128/MCB.24.12.5269-5280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chellaiah MA, Schaller MD. Activation of Src kinase by protein-tyrosine phosphatase-PEST in osteoclasts: comparative analysis of the effects of bisphosphonate and protein-tyrosine phosphatase inhibitor on Src activation in vitro. J. Cell. Physiol. 2009;220:382–393. doi: 10.1002/jcp.21777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martini V, Gattazzo C, Frezzato F. Cortactin, a Lyn substrate, is a checkpoint molecule at the intersection of BCR and CXCR4 signalling pathway in chronic lymphocytic leukaemia cells. Br J Haematol. 2017;178:81–93. doi: 10.1111/bjh.14642. [DOI] [PubMed] [Google Scholar]
- 46.Zhao J, et al. Extracellular signal-regulated kinase (ERK) regulates cortactin ubiquitination and degradation in lung epithelial cells. J. Biol. Chem. 2012;287:19105–19114. doi: 10.1074/jbc.M112.339507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mukherjee S, et al. CDK5 inhibition resolves PKA/cAMP-independent activation of CREB1 signaling in glioma stem cells. Cell Rep. 2018;23:1651–1664. doi: 10.1016/j.celrep.2018.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang P, et al. Knockdown of CREB1 promotes apoptosis and decreases estradiol synthesis in mouse granulosa cells. Biomed. Pharmacother. 2018;105:1141–1146. doi: 10.1016/j.biopha.2018.06.101. [DOI] [PubMed] [Google Scholar]
- 49.Walia MK, Taylor S, Ho PWM, Martin TJ, Walkley CR. Tolerance to sustained activation of the cAMP/Creb pathway activity in osteoblastic cells is enabled by loss ofp53. Cell Death Dis. 2018;9:844. doi: 10.1038/s41419-018-0944-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Esparza SD, et al. CREB regulates Meis1 expression in normal and malignant hematopoietic cells. Leukemia. 2008;22:665–667. doi: 10.1038/sj.leu.2404933. [DOI] [PubMed] [Google Scholar]
- 51.Zhou SJ, Deng YL, Liang HF, Jaoude JC, Liu FY. Hepatitis B virus X protein promotes CREB-mediated activation of miR-3188 and Notch signaling in hepatocellular carcinoma. Cell Death Differ. 2017;24:1577–1587. doi: 10.1038/cdd.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu FY, et al. MiR-216b is involved in pathogenesis and progression of hepatocellular carcinoma through HBx-miR-216b-IGF2BP2 signaling pathway. Cell Death & Dis. 2015;6:e1670. doi: 10.1038/cddis.2015.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao J, et al. Hepatitis B virus X protein activates Notch signaling by its effects on Notch1 and Notch4 in human hepatocellular carcinoma. Int. J. Oncol. 2016;48:329–337. doi: 10.3892/ijo.2015.3221. [DOI] [PubMed] [Google Scholar]
- 54.Wei X, et al. SKP2 promotes hepatocellular carcinoma progression through nuclear AMPK-SKP2-CARM1 signaling transcriptionally regulating nutrient-deprived autophagy induction. Cell. Physiol. Biochem.: Int. J. Exp. Cell. Physiol., Biochem., Pharmacol. 2018;47:2484–2497. doi: 10.1159/000491622. [DOI] [PubMed] [Google Scholar]
- 55.Croucher DR, Rickwood D, Tactacan CM, Musgrove EA, Daly RJ. Cortactin modulates RhoA activation and expression of Cip/Kip cyclin-dependent kinase inhibitors to promote cell cycle progression in 11q13-amplified head and neck squamous cell carcinoma cells. Mol. Cell. Biol. 2010;30:5057–5070. doi: 10.1128/MCB.00249-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee S, Kim W, Ko C, Ryu WS. Hepatitis B virus X protein enhances Myc stability by inhibiting SCF(Skp2) ubiquitin E3 ligase-mediated Myc ubiquitination and contributes to oncogenesis. Oncogene. 2016;35:1857–1867. doi: 10.1038/onc.2015.251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.