Abstract
Twist1, a key transcription factor regulating epithelial–mesenchymal transition and cancer metastasis, is highly expressed in invasive cancers in contrast to the loss of BTG2/TIS21 expression. Based on our observation that forced expression of BTG2/TIS21 downregulated Twist1 protein expression without altering mRNA level, we investigated molecular mechanisms of the BTG2/TIS21-inhibited Twist1 translation in the triple negative breast cancer (TNBC) cells and in vivo BTG2/TIS21-knockout (KO) mice and human breast cancer tissues. (1) C-terminal domain of Twist1 and Box B of BTG2/TIS21 interacted with each other, which abrogated Twist1 activity. (2) BTG2/TIS21 inhibited translational initiation by depleting eIF4E availability via inhibiting 4EBP1 phosphorylation. (3) Expression of BTG2/TIS21 maintained p-eIF2α that downregulates initiation of protein translation, confirmed by eIF2α-AA mutant expression and BTG2/TIS21 knockdown in MEF cells. (4) cDNA microarray analysis revealed significantly higher expression of initiation factors-eIF2A, eIF3A, and eIF4G2-in the BTG2/TIS21-KO mouse than that in the wild type. (5) BTG2/TIS21-inhibited translation initiation lead to the collapse of polysome formation and the huge peak of 80s monomer in the BTG2/TIS21 expresser, but not in the control. (6) mRNAs and protein expressions of elongation factors were also downregulated by BTG2/TIS21 expression in TNBC cells, but much higher in both TIS21-KO mice and lymph node-positive human breast cancers. (7) BTG2/TIS21-mediated Twist1 loss was not due to the protein degradation by ubiquitination and autophagy activation. (8) Twist1 protein level was significantly higher in various organs of TIS21-KO mice compared with that in the control, indicating the in vivo role of BTG2/TIS21 gene in the regulation of Twist1 protein level. Altogether, the present study support our hypothesis that BTG2/TIS21 is a promising target to combat with metastatic cancers with high level of Twist1 without BTG2/TIS21 expression.
Subject terms: Breast cancer, Tumour-suppressor proteins
Introduction
Epithelial–mesenchymal transition (EMT) is a process trans-differentiating epithelial cells to mesenchymal cells, which results in the increase of motility, migration, and invasion of cancer cells into extracellular matrix1. Metastasis of epithelial tumors takes place when the cancer cells lose cell–cell contact inhibition in contrast to obtaining migration capacity along with EMT process2. Twist1, a basic helix-loop-helix transcription factor, plays an important role in the embryonal development as a master regulator of morphogenesis3 and in EMT process during tumor invasion and metastasis4. Twist1 along with Bmi suppress the expression of E-cadherin5 in contrast to increasing N-cadherin expression by β1-integrin6, which promotes metastatic process. Twist1 expression induced by sunitinib accelerates vasculogenic mimicry and induction of cancer stem cell marker in triple negative breast cancer (TNBC) cells7.
In pathological context, initiation of cancer invasion requires actin polymerization by Dia1 and myosin recruitment to the fully matured focal adhesion site, which impose cells to get invasive motility rather than planar motility8. Among the several tumor suppressors, BTG2/TIS21 gene was primarily focused due to its targeting activity on actin nucleator, Dia19. BTG210 is a human ortholog of mouse TIS2111 and rat PC312, which belongs to antiproliferative gene family. Expression of BTG2/TIS21 is frequently lost early in carcinogenesis13–19 by epigenetic change and miRNA regulation20–22. BTG2 helps CCR4-cNOT complex that degrades mRNAs and thereby inhibits target gene expression23,24. Overexpression of BTG2/TIS21 has been significantly associated with G2/M arrest by interacting with cyclin B1-Cdc2 complex25, and with cell death by inducing MnSOD expression via nuclear factor-κB activation26,27. BTG2/TIS21 high expression in breast cancer increases survival rate28, whereas loss of BTG2 expression induces breast tumor progression that responds to ErbB/HER inhibitor lapatinib29. Indeed, overexpression of BTG2/TIS21 inhibits invadopodia formation in the TNBC, MDA-MB-231, cells30 and reduces cancer invasion to muscle layer and lymph nodes of human bladder and breast cancers9,20. Constitutive expression of BTG2/TIS21 in the ductal carcinoma in situ (DCIS), but not in the infiltrating ductal carcinoma, strongly suggests that BTG2 might be an important barrier to block cancer progression from DCIS to infiltrating cancers31, and the notion is well supported by the report that p53 deficiency-enhanced metastatic potential of breast cancer is linked to BTG2/TIS21 loss in the primary and metastatic sites of TNBC-PDX model32.
In contrast to BTG2, Twist1 expression promotes invadopodia formation by inducing PDGFRα expression33 and exhibits poor prognosis in human breast cancer through Src co-expression34. We have screened in vitro protein interaction network of BTG2/TIS21 and observed Twist1 as one of the interacting partners of BTG2. Thus, we could hypothesize that BTG2/TIS21-inhibited cancer invasion might be through the downregulation of Twist1 activity in human breast cancers. To validate our hypothesis, adenoviral transduction of BTG2/TIS21 gene was employed in the TNBC cells and found the significant reduction of Twist1 translation, but not mRNA transcription, in the BTG2/TIS21 expresser along with the collapse of polysome formation on the Twist1 mRNAs. In the present study, we report potential mechanisms of the translational inhibition of Twist1 gene by BTG2/TIS21 gene. To our best knowledge, this is the first report that BTG2/TIS21, a tumor suppressor, acts as a potential candidate to block EMT phenomenon by inhibiting Twist1 translation in TNBC cells.
Results
Downregulation of Twist1 protein expression by BTG2/TIS21 gene
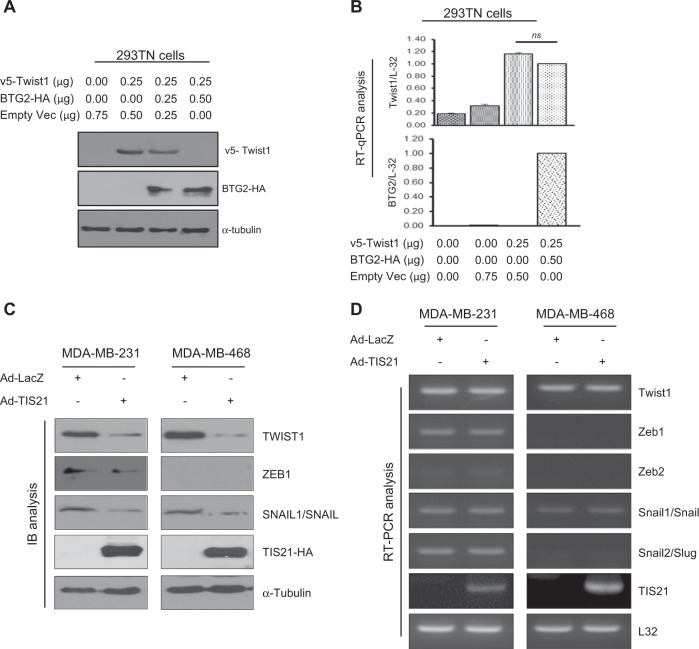
To explore regulation of Twist1 expression by BTG2/TIS21 gene, transfection analysis was performed in 293TN cells for 48 h with v5-Twist1 and BTG2-HA gene, and the cells were analyzed by immunoblotting. Twist1 protein expression was significantly downregulated by BTG2/TIS21 expression (Fig. 1a) without any significant change in the Twist1 mRNA level (Fig. 1b). Initially, we screened various breast cancer cells with and without invasiveness to identify if BTG2 and Twist1 expressions are mutually exclusive. As shown in Supplementary Fig. S1A, we found that MCF-7 and ZR-75-1 cells expressed endogenous BTG2 without TWIST1 expression. The finding was opposite in the TNBC cells with high TWIST1, but no BTG2, expression. To explore whether the amount of exogenous BTG2/TIS21 expressed in the TNBC cells by adenoviral transduction is relevant to physiological level or not, immunoblot analysis was performed and confirmed that the exogenous BTG2/TIS21 level was about physiologically relevant (Supplementary Fig. S1B). To further examine whether endogenous BTG2 protein can also regulate Twist1 expression or not, MCF-7 cells were employed and the endogenous BTG2 expression was knocked down by siBTG2 transfection. As expected, the transfection significantly induced TWIST1 protein level, but not Twist1 mRNA, in the MCF-7 cells (Supplementary Fig. S1C). In addition to Twist1, Zeb1 and Snail protein expressions were also regulated by BTG2/TIS21 expression (Fig. 1c), whereas their mRNA expressions were not altered (Fig. 1d). BTG2/TIS21-regulated TWIST1 protein level was dependent on the concentration of BTG2/TIS21 gene (Supplementary Fig. S2A). As BTG2/TIS21 downregulates interleukin (IL)-6 mRNA expression35, IL-6 transcription was monitored as a control experiment to prove our observation. As expected, it was significantly reduced in both TNBC cells with BTG2/TIS21 expression (Supplementary Fig. S2B). To validate the phenomenon in the MDA-MB-231 cells depleted with serum for 22 h, fetal bovine serum (FBS; 0–10%) was supplemented for 6 h before immunoblot analysis. As expected, Twist1 biosynthesis was able to be induced in the LacZ expresser by serum stimulation in the concentration- and the time-dependent manners, whereas it was not in the BTG2/TIS21 expresser (Supplementary Fig. S2C and S2D). To screen whether BTG2/TIS21 downregulated poly(A) tail length of Twist1 mRNA or not, poly(A) tail assay (PAT) was performed. Twist1 mRNA and poly(A) tail length unchanged by transduction of MDA-MB-231 cells with Ad-BTG2/TIS21 (Supplementary Fig. S3). The data support that the BTG2/TIS21-mediated downregulation of EMT factors might be at the protein, but not mRNA, levels.
Fig. 1. Expression of BTG2/TIS21 accompanies with loss of Twist1 protein, not mRNA, level.
a When 293TN cells were transfected with v5-Twist1 for 48 h, its expression was significantly reduced in the dose-dependent manner of BTG2-HA. Anti-v5, anti-HA, and anti-α-tubulin antibodies were applied for immunoblot analysis. b To examine regulation of v5-Twist1 mRNA expression by BTG2-HA, real-time PCR analysis was performed at 48 h. L32 was used for loading control. Note the absence of significant difference in the Twist1 mRNA expression by BTG2-HA (c) MDA-MB-231 and MDA-MB-468 cells transduced with either Ad-LacZ (100 moi) or Ad-TIS21 (100 moi) for 48 h were subjected to immunoblot analysis and the significant loss of Twist1 expression along with ZEB and Snail was observed in the BTG2/TIS21 expresser. d The TNBC cells were subjected to RT-PCR analysis and mRNAs levels were measured. Note absence of significant changes in the levels of Twist1, Zeb1, and Snail between the BTG2/TIS21 and the LacZ expressers. Immunoblots (a, c, d) are representative of three independent experiments. Data are expressed as mean ± SD from the more than two independent experiments
Inhibition of Twist1 activity by BTG2/TIS21 gene in the TNBC cells
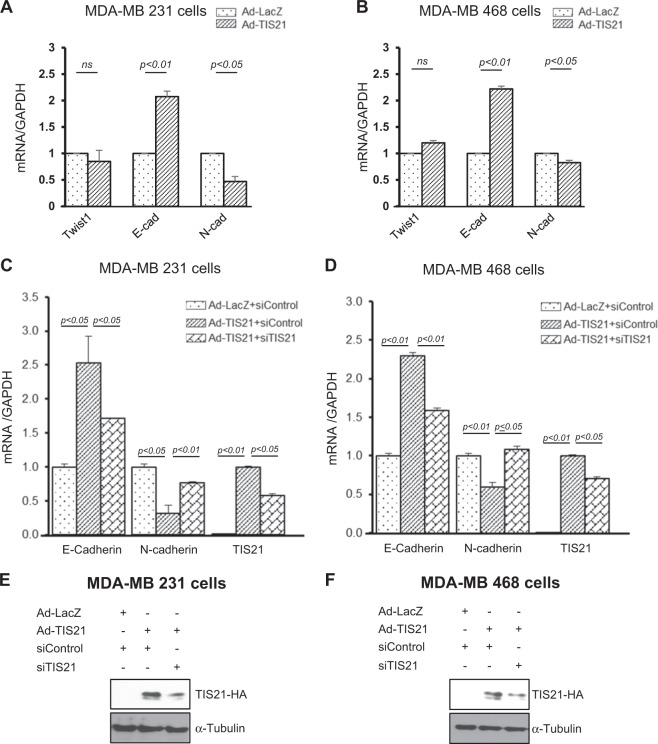
To further examine whether the BTG2/TIS21-mediated Twist1 protein loss downregulates expression of Twist1 targets genes, E-cadherin and N-cadherin levels were measured by reverse transcriptase-quantitative PCR (RT-qPCR). Despite no significant change in the Twist1 mRNA expression, E-cadherin and N-cadherin expressions were significantly altered in the BTG2/TIS21 expresser compared with those in the LacZ control (Fig. 2a, b). To further evaluate specificity of the regulation, expression of BTG2/TIS21 was knocked down by siTIS21 transfection and the recovery of the target gene expressions was observed in both TNBC cells (Fig. 2c, d). Efficiency of short interfering RNA (siRNA) knockdown was confirmed by immunoblot analysis with anti-hemagglutinin (HA) and anti-α-Tubulin antibodies (Fig. 2e, f).
Fig. 2. Knockdown of BTG2/TIS21 expression recovers Twist1 activity.
a Regulation of Twist1 expression by BTG2/TIS21 gene was re-examined via measuring its target gene expression by real-time PCR after forced expression of BTG2/TIS21 gene in MDA-MB-231 cells. Note the reciprocal regulation of E-cadherin and N-cadherin expressions without significant change of Twist1 mRNA expression by BTG2/TIS21 gene. GAPDH served as a control for amplification. b The same experiment was performed in MDA-MB-468 cells and similar regulation was observed in the BTG2/TIS21 expresser. c To confirm the activity of BTG2/TIS21 gene in the regulation of Twist1 target genes, MDA-MB-231 cells were transfected with siControl or siTIS21 (100 nM) for 24 h, and then transduced with either Ad-LacZ or Ad-TIS21 (100 moi). Total cellular RNAs were isolated 24 h later and subjected to real-time PCR analysis, and the expressions of E-cadherin, N-cadherin, and BTG2/TIS21 were measured along with GAPDH as an internal control. p < 0.05 was considered as statistically significant. Note the significant changes of E-cadherin and N-cadherin expressions by the level of BTG2TIS21 gene expression. d The same experiment was performed in MDA-MB-468 cells and the similar regulation of Twist1 target gene expression by BTG2TIS21 was also found in the cells. Immunoblot analyses reveal the knockdown efficiency of siTIS21 in the MDA-MB-231 (e) and MDA-MB-468 (f) cells. Anti-HA and anti-α-tubulin antibodies were applied for the experiment (n = 2). All data are expressed as mean ± SD after two independent experiments
In vitro interaction of BTG2-box B with C-terminal region of Twist1
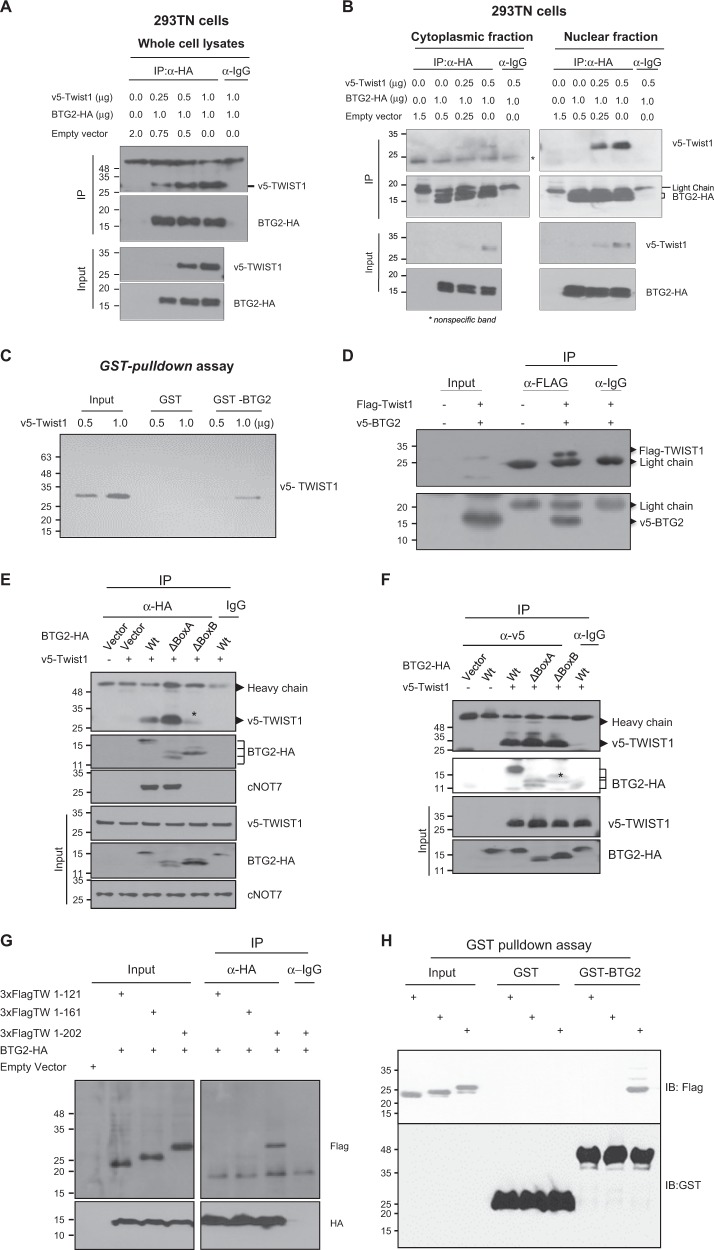
BTG2 exerts its activity by interacting with biologically significant proteins such as Hoxb936, rPICK137, cNOT7/823,24, PRMT138, and Pin-139. By using 19K-protein chip analysis, various proteins interacting with BTG2/TIS21 protein could be isolated by Ho Chul Kang at Ajou University, BTG2-HA interacted with v5-Twist1 dose dependently (Fig. 3a), and the interaction occurred mainly in the nucleus rather than the cytoplasmic fraction (Fig. 3b). It has been known that cNOT7 binds to BTG223,24; thus, competition assay was performed with constant amounts of BTG2-HA and v5-Twist1 along with the increasing amount of Flag-cNOT7 construct. Immunoprecipitation (IP) with anti-BTG2-HA antibody revealed strong interaction of BTG2-HA with v5-TWIST1, independent of the cNOT7 in the whole-cell lysates of 293TN cells (Supplementary Fig. S4A). When fractionation was applied, cNOT7 was expressed in the cytoplasm, but TWIST1 was in the nuclear fractions (Supplementary Fig. S4B). GST-pulldown analysis also exhibited in vitro interaction of v5-Twist1 with GST-BTG2 (Fig. 3c). To exclude the effect of protein tag on their interaction, the reciprocal IP was performed and observed the interaction of Flag-Twist1 with v5-BTG2 in 293TN cells (Fig. 3d). As intracellular location of Twist1 expression has been reported as variable in different cancer cells40,41, HeLa cells were employed and the cytoplasmic expression of Twist1 was found (Supplementary Fig. S4C); the phenomenon was further confirmed by confocal analysis (Supplementary Fig. S4D). When competition assay was performed in HeLa cells, the expression of Flag-cNOT7 significantly reduced v5-Twist1 binding to BTG2-HA concentration dependently (Supplementary Fig. S4E). To identify BTG2 domain interacting with Twist1, reciprocal IP analyses were performed. The Box B-deleted BTG2 mutant significantly abrogated BTG2 binding with v5-Twist1 and with cNOT7 (Fig. 3e, f), indicating the interaction of BTG2-Box B with TWIST1 and cNOT7. For mapping the domain of Twist1 protein interacting with BTG2, 3xFlag-tagged Twist1 mutants (1–121 and 1–161) and its wild-type (Wt) construct were co-transfected with BTG2-HA and then evaluated by IP analysis. Fig. 3g showed that only the full-length Twist1, but not the deletion mutants, could strongly bind to BTG2-HA. The data were confirmed by in vitro GST-pulldown assay with the clear interaction of BTG2 only with the full-length Twist1 (Fig. 3h). The data were further supported by the prediction of intrinsically unstructured protein analysis that exhibits only the C-terminal region (121–202 amino acid) of Twist1 forms ordered structure (Supplementary Fig. S4F), whereas the whole regions of BTG2 and TIS21 proteins form the globular structure (Supplementary Fig. S4G and S4H). To further investigate whether endogenous BTG2 also interacts with TWIST1, v5-Twist1 was transfected into MCF-7 cells and subjected to IP with anti-BTG2 antibody. Endogenous BTG2 protein clearly interacted with v5-TWIST1 and with cNOT7 as shown in Supplementary Fig. S5A. To analyze localization of the proteins in MCF-7 cells, fractionation was performed and we observed cNOT7 only in the cytoplasm; however, BTG2 and v5-TWIST1 proteins were more in the nuclear fraction than in the cytoplasm (Supplementary Fig. S5B).
Fig. 3. C-terminal region of TWIST1 interacts with BTG2 box B domain.
a In vitro immunoprecipitation (IP) analysis was performed in 293TN cells with anti-HA antibody. Pulldown of BTG2-HA using 1.0 µg of anti-HA antibody showed its interaction with v5-Twist1, depending on its concentration. Normal IgG was employed as a negative control of IP assay. Immunoblotting is the representative of three independent experiments. b Fractionation of the cytoplasm and nucleus of 293TN cells transfected with v5-Twist1 and BTG2-HA was performed, and the interaction of Twist1 and BTG2 was further examined by IP with 1.0 µg of anti-HA antibody. The data suggest that interaction was mainly observed in the nuclear fraction than in the cytoplasm. Star indicates nonspecific band. c In vitro GST-pulldown assay. GST and GST-BTG2 beads were incubated with 293TN cell lysates transfected with v5-Twist1 at 4 °C overnight. The beads were pulled down and subjected to immunoblot analysis with anti-v5 antibody. Note the direct interaction of BTG2 with v5-Twist1. d IP analysis was performed in 293TN cells after transfection with Flag-Twist1 (1.0 μg) and v5-BTG2 (1.0 μg) genes. In vitro interaction of Flag-Twist1 and v5-BTG2 was able to be shown by IP with 1.0 µg of anti-Flag antibody. Immunoblotting is the representative of three independent experiments. e For mapping BTG2 domain bound to Twist1, 293TN cells were transfected with either BTG2-HA wild type or deletion mutants (ΔBox-A, ΔBox B) along with v5-Twist1 gene. IP analysis performed with anti-HA antibody found that overexpression of ΔBox-B mutant significantly reduced BTG2-Twist1 interaction, indicating that box B in BTG2-HA interacted with v5-Twist1 protein. *The reduced interaction between BTG2-HA and v5-Twist1. BTG2 with ΔBox-B also abrogated interaction with endogenous cNOT7. f IP analysis with anti-v5 antibody. Procedure was the same as described in the e. Note the star revealing significant loss of the interaction. g To investigate the domain in Twist1 interacting with BTG2-HA, 293TN cells were transfected with either Twist1 wild type (1–202) and its deletion constructs (1–121, 1–161) along with BTG2-HA. IP performed with anti-HA antibody showed that the wild-type Twist1, but not the deletion mutants, could interact with BTG2-HA. h In vitro interaction analysis by GST-pulldown assay. GST and GST-BTG2 beads were incubated with 293TN cell lysates transfected with either wild type or the deletion constructs of Twist1 gene at 4 °C overnight. When the beads were subjected to immunoblot analysis with anti-Flag antibody, the interaction of BTG2 with wild-type Twist1, but not deletion mutants, was observed. All data clearly indicate the in vitro interactions of C-terminal region of Twist1 with BTG2 box B domain. Blots (b, c, and e–g) are representative of two independent experiments
Proteasome- and lysosome-independent loss of TWIST1 protein by BTG2/TIS21 gene
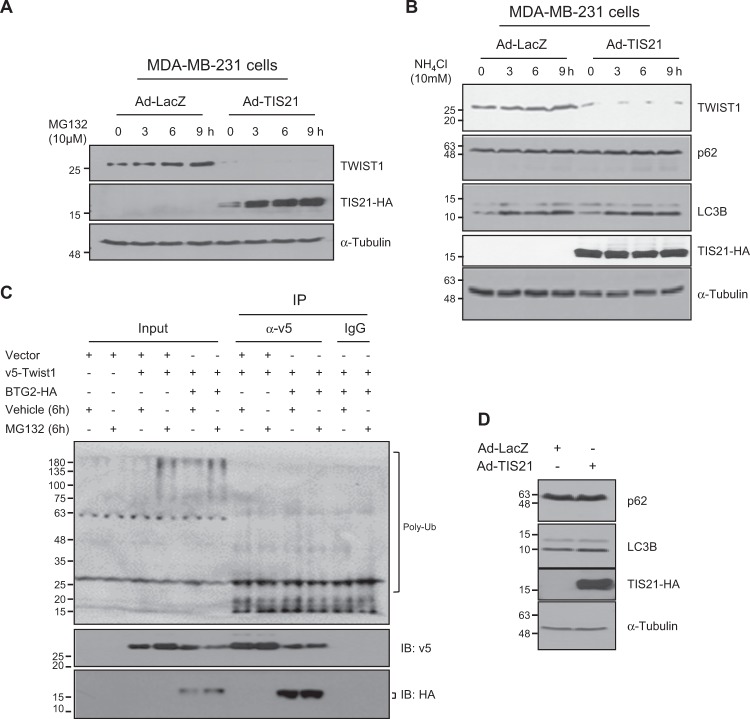
MDA-MB-231 cells with BTG2/TIS21 or LacZ overexpression were incubated with either MG132 (proteasome inhibitor) or ammonium chloride (lysosome inhibitor) for 9 h and the change of TWIST1 protein was measured at every 3 h along with the TIS21-HA and LC3B accumulation as the positive controls; the inhibitions of proteasome and lysosome activity failed to block the BTG2/TIS21-mediated Twist1 loss (Fig. 4a, b). To examine whether Twist1 ubiquitination is enhanced by BTG2/TIS21, MDA-MB-231 cells transfected with 3xFlag-Twist1 with or without BTG2-HA were examined by IP and immunoblot analysis; ubiquitination of Twist1 protein was not enhanced in the BTG2/TIS21 expresser (Supplementary Fig. S6A), and that was similar in the 293TN cells transfected with v5-Twist1 with or without BTG2-HA under MG132 treatment (Fig. 4c). We have recently reported that ABI-2 is targeted to proteasome-mediated degradation by BTG242. Hence, ABI-2 was employed as a positive control of proteasome activation and confirmed that BTG2/TIS21 expression slightly increased ubiquitination of ABI-2 protein along with its interaction (Supplementary Fig. S6B). When the onset of autophagy program upon Ad-BTG2/TIS21 transduction was examined, the expression of p62 and LC3B conversion were not significantly changed (Fig. 4d).
Fig. 4. BTG2/TIS21-induced loss of TWIST1 expression is not due to protein degradation.
a MG132 treatment failed to protect the BTG2/TIS21-induced Twist1 loss. MDA-MB-231 (3 × 105/60 mm dish) cells were transduced with either Ad-LacZ (100 moi) or Ad-TIS21(100 moi) virus for 48 h and then treated with MG132 (10 µM) for various time points to block proteasome activity. Twist1 protein was accumulated by the treatment in the LacZ expresser, whereas it was failed in the BTG2/TIS21 expresser as opposed to accumulation of BTG2/TIS21 expression that served as a positive control. α-Tubulin served as a loading control. b BTG2/TIS21-induced Twist1 loss cannot be protected by NH4Cl treatment. MDA-MB-231 (3 × 105 cells) cells transduced with either Ad-LacZ or Ad-TIS21 were treated with 10 mM of NH4Cl to block lysosome activity before immunoblot analysis at the time points. Twist1 protein was accumulated in the LacZ expresser by the treatment; however, it was lost in the BTG2/TIS21 expresser. NH4Cl treatment also failed to alter the expression of p62. NH4Cl increased LC3B, the autophagosome-associated lipid form of LC3, which accumulates if their lysosomal degradation is inhibited, which served as a positive control. α-Tubulin served as a loading control. c Ubiquitination analysis. To investigate whether BTG2/TIS21-induced TWIST1 loss is regulated by ubiquitination or not, 293TN cells were transfected with either v5-Twist1 (1.0 μg) and/or BTG2-HA (1.0 μg) before IP analysis with anti-v5 antibody and immunoblot assay with anti-ubiquitin antibody. v5-Twist1 expression was slightly increased in the vector expresser by MG132 treatment (compare lane 4 vs. lane 3 in the v5-panel); however, it was rather decreased in the BTG2/TIS21 expresser (compare lane 6 vs. lane 5 in the v5 panel). The interaction between v5-Twist1 and BTG2/TIS21-HA was almost similar and there was no difference in the ubiquitination after MG132 treatment. d No alteration of autophagy signal in the BTG2/TIS21 expresser vs. LacZ control. MDA-MB-231 cells transduced with either Ad-LacZ or Ad-TIS21 were subjected to immunoblot analysis, to examine p62 and LC3B conversion. α-Tubulin as loading control. Blots are a representative of two independent experiment
BTG2/TIS21-mediated Twist1 loss via downregulating translational initiation of protein biosynthesis
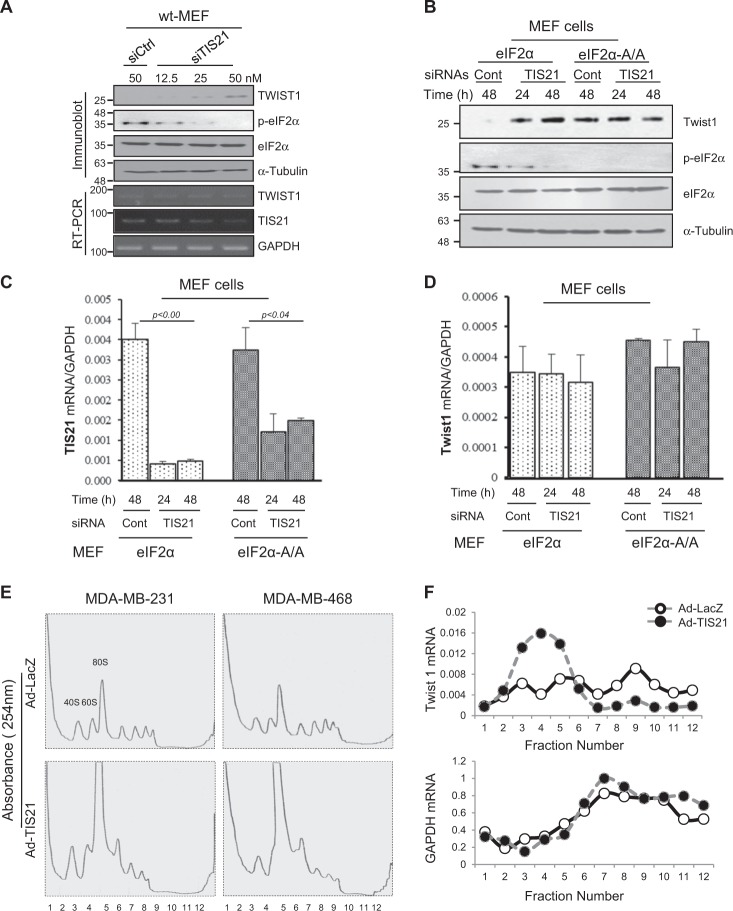
To further investigate whether the BTG2/TIS21-mediated Twist1 loss is ascribed to the translational defect, endogenous BTG2/TIS21 expression was knocked down by siRNAs to TIS21 (siTIS21, 0–50 nM) in the Wt MEF cells, and found the induction of TWIST1 expression by siTIS21 transfection along with the loss of p-eIF2α without eIF2α protein change (Fig. 5a). When the endogenous TIS21 expression was knocked down, TWIST1 protein level was clearly increased without change in its mRNA levels (Fig. 5a). To explore whether BTG2/TIS21 regulates Twist1 translation at initiation phase, MEF cells were overexpressed with either Wt eIF2α or mutant eIF2α (eIF2α-Α/Α) for 48 h. Knockdown of TIS21 expression significantly induced TWIST1 protein along with the loss of p-eIF2α (Fig. 5b, lanes 1–3). In contrast to wt-MEF, mutant MEF with eIF2α-A/A cannot be phosphorylated as shown in Fig. 5b. The data suggest that BTG2/TIS21 might inhibit TWIST1 biosynthesis through maintaining eIF2α phosphorylation, whereas Twist1 mRNA expression was not changed by knockdown of BTG2/TIS21 expression in the MEF cells (Fig. 5c, d), implying that BTG2/TIS21 downregulates Twist1 translation at the initiation phase by inhibiting eIF2α activity via inducing its phosphorylation in normal cells. The notion can be supported by the reports that homozygous mutant eIF2α-Α/Α fails to attenuate protein synthesis under stress condition43 and phosphorylation of eIF2α on S51 residue cannot form ternary complex to pre-initiate protein translation44. In TNBC cells, there was no physical interaction between BTG2/TIS21 and eIF2α molecules (Supplementary Fig. S7A and S7B). Moreover, transduction of TNBC cells with Ad-BTG2/TIS21 significantly inhibited phosphorylation of 4EBP1 at S65 and at T37/46, but not T70, residues (Supplementary Fig. S7C and S7D), which is the essential step to activate 4EBP145,46. Phosphorylation of T37 and T46 has been reported as a priming event that permits phosphorylation of 4EBP1 at S65 residue that promotes dissociation of eIF4E from p-4EBP1 and permits formation of a functional eIF4F complex45. In addition, our cDNA microarray data (NCBI Gene Expression Omnibus as GSE 105772) exhibited significantly elevated expression of the initiation factors in the TIS21-KO mice than those in the wt mice (Supplementary Fig. S8). To validate the notion in detail, we performed polysome profiling assay in the TNBC cells and observed that BTG2/TIS21 expresser revealed collapse of the 80s polysome formation (fraction 6–10) along with the huge single peak of 80S monosome (fraction 5, Fig. 5e). To further confirm the phenomenon, abundance of Twist1 mRNA on the ribosomes was determined by real-time PCR analysis. Twist1 mRNA level was much lower in the BTG2/TIS21 expresser than that in the LacZ control (upper panel, Fig. 5f), whereas GAPDH mRNA level was not different in the two expressers (lower panel, Fig. 5f). As mTORc1 activity can be inhibited by BTG2/TIS21 expression31 and Twist1 mRNA contains 5′TOP motif (Supplementary Fig. S9A), we treated the MDA-MB-231 cells with Rapamycin, Torin, and PP242 for 6–12 h and observed that TWIST1 protein was significantly reduced by mTORc inhibition (Supplementary Fig. S9B). Collectively, the expression of BTG2/TIS21 downregulated initiation of Twist1 translation in the TNBC cells.
Fig. 5. BTG2/TIS21-induced Twist1 loss is associated with the failure of polysome formation.
a Twist1 expression was significantly induced by knockdown of endogenous BTG2/TIS21 expression. To confirm the effect of BTG2/TIS21 on the Twist1 protein loss, TIS21/BTG2 expression in the wild-type MEF was removed by transfection with siTIS21 (12.5~50 nM) for 2 days and then analyzed by immunoblotting with anti-Twist1 antibody. The induction of Twist1 expression appeared in the siTIS21-dependent manner, compared with that in the control. In the same cells, p-eIF2α level maintained in the siControl-transfected cells was significantly reduced by knockdown of BTG2/TIS21 expression. Total eIF2α levels remained unaltered. α-Tubulin served as a loading control. RT-PCR analysis showed that Twist1 mRNA level was unchanged. TIS21 knockdown was analyzed by RT-PCR and GAPDH served as a control. b Wild-type MEF cells overexpressed with either eIF2α or eIF2α-A/A (S51A/S51A) mutant were transfected with siTIS21-RNAs (50 nM) and siControl-RNAs (50 nM), and the cells were subjected to immunoblot assay in 24 and 48 h of the transfection. Knockdown of BTG2/TIS21 expression by siTIS21 significantly induced Twist1 expression compared with that in the siControl (lanes 2 and 3 vs. lane 1); however, Twist1 expression was consistently high in the MEF cells with eIF2α-A/A mutant expresser (lanes 5 and 6 vs. lane 4). The phosphorylation of eIF2α completely disappeared by the knockdown of BTG2/TIS21 expression for 48 h, suggesting that BTG2/TIS21 expression might maintain the p-eIF2α level that inhibits translation of Twist1. c, d Real-time qPCR analyses revealing mRNA expressions of BTG2/TIS21 (c) and Twist1 (d) in the MEF cells transfected with wild-type and mutant eIF2α. Transfection of MEF with siTIS21 markedly reduced BTG2/TIS21 expression; however, Twist1 transcription was not significantly changed by BTG2/TIS21 knockdown. Relative expressions were presented based on that of GAPDH. e Polysome profiling assay; to assess a general translational control by BTG2/TIS21 gene, TNBC cells transduced with either Ad-LacZ or Ad-TIS21 for 48 h were subjected to polysome profiling analysis. Polysome formation in the BTG2/TIS21 expresser was collapsed after a huge peak of the 80S monomer, whereas the profile was well maintained in the LacZ control. Numbers in X axis (1–12) indicate fractions collected up to 500 μL per each tube. f Real-time PCR analysis; to analyze abundance of the Twist1 mRNAs in each fraction, real-time PCR analysis was performed. The amount of Twist1 mRNA was higher in the 2–5 fractions of the BTG2/TIS21 expresser; however, it was much less in the 6–12 fractions compared with those in the LacZ expresser. In contrast, levels of GAPDH mRNA were similar between the LacZ and BTG2/TIS21 expressers in each fraction. Blots are representative of three independent experiments. All data are expressed as mean ± SD after two independent experiments
Attenuated expression of eukaryotic elongation factors by BTG2/TIS21 gene
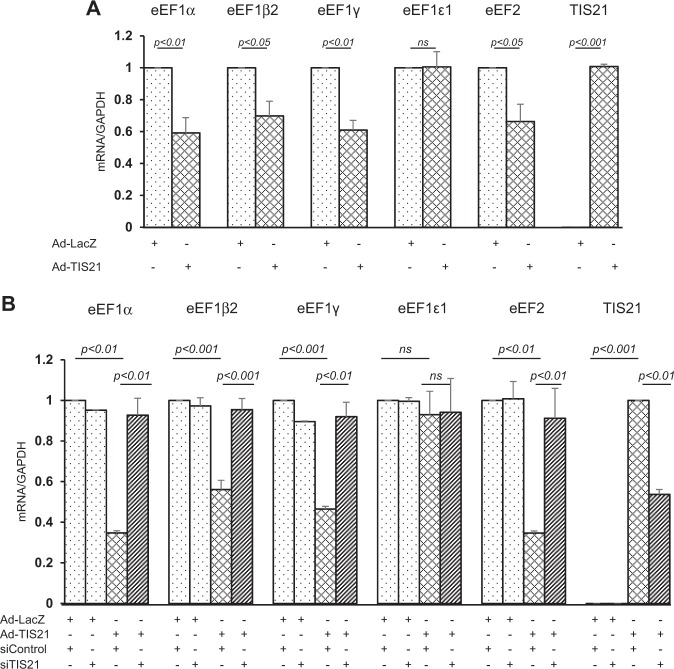
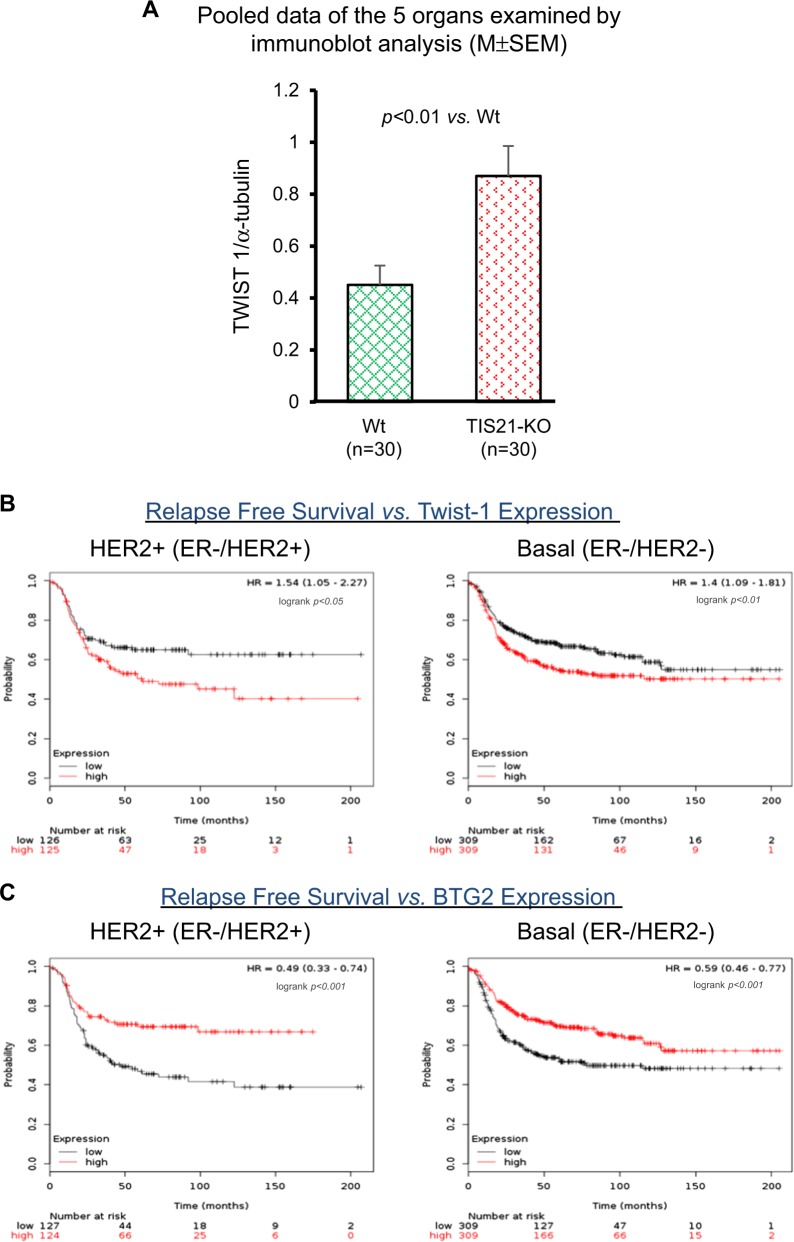
As polysome formation of Twist1 mRNA was significantly reduced by BTG2/TIS21, expression of elongation factors (eEFs) was measured in the TNBC cells by real-time PCR analysis and observed the significant reduction of eEF1α, eEF1β2, eEF1γ, and eEF2, except eEF1ε1 mRNAs in the BTG2/TIS21 expresser (Fig. 6a). However, knockdown of BTG2/TIS21 expression by siTIS21 transfection significantly recovered eEFs mRNA levels (Fig. 6b) and transduction with Ad-BTG2/TIS21 abolished eEF2 protein expression (Supplementary Fig. S7D). In addition, the expressions of eEF1γ and eEF2 were higher in the liver of the TIS21-KO mice, and eEF1α, eEF1β2, and eEF1γ were higher in the lung and spleen of the TIS21-KO mice than those in the wt mice (Supplementary Fig. S10C–S10E) without any significant alterations in the body and organ weights of the mice (Supplementary Fig. S10A, S10B). In vivo Twist1 mRNA expressions between the TIS21-KO and the wt mice were not statistically different (Supplementary Fig. S10F and S10G); however, in vivo Twist1 protein level in the lung and spleen was higher in the TIS21-KO mice compared with that in the Wt mice (Supplementary Fig. S11). When the expression ratio of Twist1/α-tubulin was pooled from the 30 organs dissected from six mice, Twist1 protein level was significantly higher in the TIS21-KO mice than that in the Wt evaluated by Mann–Whitney U-test (Fig. 7a). Above data strongly supported in vivo effect of BTG2/TIS21 gene on the translation of Twist1 mRNA. To further estimate the role of BTG2/TIS21 playing in human breast cancer tissues, lymph node-negative and -positive tumor samples and the matched surrounding normal tissues were analyzed and found the active expressions of TWIST1, SNAIL, and eEF2 in the node-positive tumors compared with the node-negative tissues. BTG2 expression was detectable only in the normal tissues (Supplementary Fig. S12). The data can be supported by the report that high expression of eEF2, regulating the rate of peptide chain elongation during protein translation, was significantly associated with node positivity and tumor size in breast cancers47. In addition, relapse-free survival rates of breast cancers (both HER2+ and HER2−) were negatively correlated with the level of Twist1, but positively correlated with BTG2 expression by open data analysis (Fig. 7b, c).
Fig. 6. Downregulation of elongation factor expression by BTG2/TIS21 gene in MDA-MB-231 cells.
a When the cells transduced with either Ad-LacZ or Ad-TIS21 were analyzed by real-time PCR, mRNA expressions of eEF1α, eEF1β2, eEF1γ, and eEF2 were significantly reduced in the BTG2/TIS21 expresser than those in the LacZ control. GAPDH served as an internal control of transcription. All data are expressed as mean ± SD after three independent experiments. b To validate the activity of BTG2/TIS21 gene regulating the transcription of elongation factors, knockdown of the exogenously expressed TIS21 gene was performed by transfection with either siControl or siTIS21 (100 nM) for 24 h after adenovirus transduction for 24 h. Real-time PCR analysis showed significant recovery of the BTG2/TIS21-induced transcriptional repression by TIS21 knockdown in the cells. GAPDH served as an internal control. p < 0.05 was considered as statistically significant. All data are expressed as mean ± SD after two independent experiments
Fig. 7. In vivo activity of BTG2/TIS21 gene in the TIS21-knockout mice and human breast cancers.
a To analyze in vivo regulation of Twist1 expression by BTG2/TIS21 gene, five organs were extracted from each TIS21-KO (six heads) and the WT (six heads) female mice, and then subjected to immunoblot analyses with 40 µg/lane in each sample. When the ratios of Twist1/α-tubulin expression were examined by Image J software, the level was significantly higher in the TIS21-KO mice than that in the WT (n = 30 organs in each group). b Relapse-free survival vs. Twist1 expression was analyzed from open data in the HER2+ and Basal-type breast cancers. Higher expression of Twist1 significantly decreased the relapse free survival of HER2+ (p < 0.05) and Basal-type (p < 0.01) breast cancers. c Relapse-free survival vs. BTG2 expression was analyzed from open data in the HER2+ and Basal-type breast cancers. Higher expression of BTG2 significantly increased the relapse free survival of the HER2+ (p < 0.001) and Basal-type (p < 0.001) breast cancer patients
Discussion
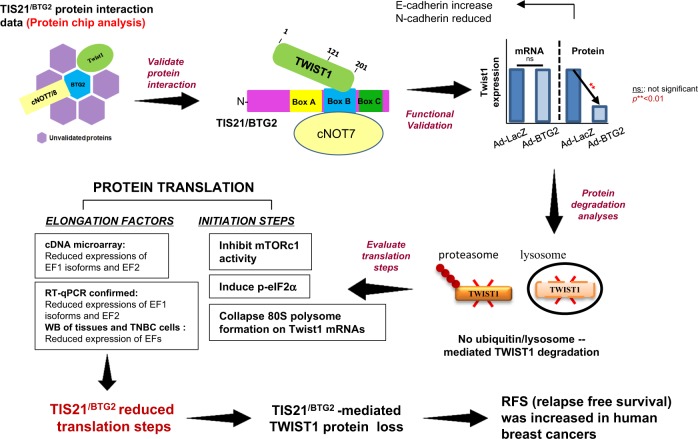
We hereby presented the new role of BTG2/TIS21 gene as an inhibitor of Twist1 translation in the TNBC cells and the phenomenon was evidenced in tissues of the BTG2/TIS21-KO mice and human breast cancers (Fig. 8). To the best of our knowledge, this is the first report revealing the BTG2/TIS21-mediated translational downregulation of Twist1. Consequences of the BTG2/TIS21-mediated Twist1 loss in the TNBC cells was evidenced by the inverse regulations of N-cadherin and E-cadherin expression, which resulted in the inhibition of EMT phenomenon. EMT-inducing transcription factors, Twist1, Snail, and Zeb, are responsible for regulating oncogenesis48.
Fig. 8. Summary depicting the effects of BTG2/TIS21 gene on the downregulation of Twist1 translation.
Reciprocal immunoprecipitation analysis confirmed the interaction of C-terminal of Twist1 to BTG2/TIS21 box B domain, which had been screened by protein chip analysis. In the present assay, cNOT7 binding to BTG2/TIS21 was employed as the interaction control. Adenoviral transfer of BTG2/TIS21 gene into TNBC cells significantly reduced Twist1 protein, but not mRNA, expression and exhibited inhibition of Twist1 activity regulating E-cadherin and N-cadherin expressions. BTG2/TIS21-mediated Twist1 protein loss was due to the failures of protein translation by inhibiting cap-dependent initiation via reduced mTORc1 activity and p-eIF2α maintenance, and by collapse in 80S polysome formation. That could lead to failure of protein translation at the initiation step. In addition, BTG2/TIS21 reduced in vivo and in vitro expressions of eukaryotic elongation factor 1 (eEF1) isoforms and eEF2. All the mechanisms could be confirmed by cDNA microarray data, RT-qPCR, and immunoblotting analyses in the TNBC cells, BTG2/TIS21-KO mice, and human breast cancer tissues. Taken altogether, BTG2/TIS21 gene inhibited the initiation and elongation steps of Twist1 translation. Thus, cancer progression including EMT phenomenon can be reduced in the cells with BTG2/TIS21 high expresser. In conclusion, the BTG2/TIS21-mediated Twist1 protein loss exhibited better prognosis in the relapse-free survival of malignant breast cancers than that in the lower BTG2/TIS21 and higher Twist1 expressers
Deletion of 161–202 residues of TWIST1 abrogated its interaction with BTG2
Reciprocal interaction of BTG2/TIS21 and C-terminal region of Twist1 was proved by in vitro IP, GST-pulldown, and deletion construct analyses (Fig. 3). The observation reminded us that Twist and p53 reciprocally regulate target genes via direct interaction49. Although cNOT7, a representative protein bound to BTG2 protein, and Twist1 bind to the same Box-B domain of BTG2/TIS21, their interaction was mutually exclusive in 293TN cells due to the differential localizations of cNOT7 and Twist1 (Supplementary Fig. S4A and S4B). Flag-cNOT7 blot for input in the Supplementary Fig. S4A shows no increase, and that may have contributed to the failure of the competitive inhibition assay in 293TN cells. However, it was competitive in HeLa cells due to their expression in the cytoplasm (Supplementary Fig. S4C–S4E). All data indicate that both Twist1 and cNOT7 bind to the same domain of BTG2/TIS21 protein.
BTG2/TIS21 inhibited translational initiation of Twist1 mRNA
The mRNA-specific translational control can be driven by RNA sequences and/or structures in the 5′/3′-untranslated region (UTR) of the transcript44,50. Cap structure of mRNAs is circularized by binding to poly(A) tail via interaction of eIF4E–eIF4G–polyA-binding-protein (PABP). When the 43S preinitiation complex scans mRNAs in a 5′→3′ direction until it meets the initiation codon, the 60S ribosome subunit joins to form 80S. On the other hand, BTG2/TIS21 interacts with RNA-binding domain of PABPC151, which can bind to N-terminal of eIF4G in the translation initiation complex52. The significant collapse of the polysome formation in the BTG2/TIS21 expresser and the loss of Twist1, but not GAPDH, mRNAs in the ribosomes (Fig. 5e, f) clearly support the inhibition of Twist1 translation by BTG2/TIS21 gene. Moreover, BTG2 inhibits mTORC1 activity in cancer cells31 and TIS21-KO MEF exhibits constitutive activation of mTORc153.
In addition to failure of the cap structure formation, the BTG2/TIS21-mediated Twist1 loss shown in the MEF cells (Fig. 5a–d) can be regulated by maintaining eIF2α phosphorylation, the stress signal from endoplasmic reticulum54. When the endogenous BTG2 in MCF-7 cells was knocked down, phosphorylation of PERK was lost (evident by the band shift, please compare Lane 3 vs. Lane 1 in Supplementary Fig. S1C) and followed by eIF2α dephosphorylation. In contrast to MEF cells, BTG2/TIS21 expression could not alter the eIF2α phosphorylation in the TNBC cells. The discrepancy can be supported by the previous reports that phosphorylation of eIF2α is higher in human breast cancers and cells than that in the normal tissues and mammary epithelial cells55,56. In yeast, eIF2A functions as a suppressor of internal ribosome entry site (IRES)-mediated translation57,58 and regulates non-canonical translation initiation events near cognate UUG and CUG codons independent of GTP59. When we examined on the website RegRNA2.060, there was no IRES sequences on the Twist1 mRNAs and no ribosome-binding sites on the 5′-UTR; therefore, BTG2/TIS21-mediated Twist1 loss might be due to the general repression of protein translation. Although further studies are required to identify more translational targets of BTG2, we here elucidated the mechanism of TWIST1 regulation by BTG2.
The BTG2/TIS21-mediated Twist1 loss was not due to the protein degradation by ubiquitination and autophagy reaction; in contrast, the inhibition of proteasome and lysosome rather aggravated Twist1 loss by the BTG2/TIS21 accumulation (Fig. 4). The data can be supported by the report that ubiquitination of BTG2/TIS21 by Skp261. In vivo Twist1 expression was significantly higher in the TIS21-KO mice than the Wt mice (Fig. 7a, p < 0.01 vs. Wt), indicating the in vivo role of BTG2/TIS21 gene in the downregulating Twist1 translation. Considering the previous reports23,24 that BTG2/TIS21 plays in the poly(A) deadenylation by interacting with hCAF1, and that short poly(A) tails (20–50 A) repress translation as opposed to enhancement by the long tails (80–500 A)62, suggesting that translation regulating factors might be the potential targets of the BTG2/TIS21-mediated deadenylation reaction, although further evidences are required.
In eukaryotes, peptide chain elongation is mediated by the elongation factors, EF1 isoforms, and EF263–65. mRNA expressions of Twist1 and the EFs were evidenced in the TNBC cells and tissues (Fig. 6, Supplementary Fig. S10). In vivo Twist1 protein expression was higher in both TIS21-KO mice and the lymph node-positive human breast cancers (Supplementary Figs. S11 and S12). Another evidence about the strong association between BTG2 expression and visceral metastatic disease in breast cancer patients (p < 0.001, 95% confidence interval 1.43–4.27) has been reported and it was independent of other clinico-pathologic features66. The findings imply the significant role of BTG2 that can prevent cancer progression via inhibition of cancer cell invasion and metastasis into surrounding tissues.
Although further analysis is needed, the present study can support our hypothesis that BTG2/TIS21 is a promising target to treat metastatic cancers with high TWIST1 expression by employing inhibitors to relieve epigenetic silencing of BTG2, which can prevent cancer progression from DCIS to infiltrating carcinoma.
Materials and methods
Cell culture
MCF-7, ZR-75-1, MDA-MB-231, MDA-MB-468, MDA-MB-453, and Hs578T cells were maintained in RPMI medium-1640 containing 10% FBS, 1% antibiotic–antimycotic solution (GIBCO, Life Technologies, Grand Island, NY) at 37 °C with 5% CO2 air. 293TN, HeLa, Wt MEF, and eIF2αA/A mutant MEF cells were maintained in Dulbecco’s modified Eagle medium with high glucose containing 10% FBS, 1% antibiotic–antimycotic solution (GIBCO, Life Technologies, Grand Island, NY) at 37 °C with 5% CO2 air. Cell lines used in our study were authenticated and tested Mycoplasma free. Short tandem repeat (STR) profiling and mycoplasma contamination of the cells used for our experiments were evaluated by AmpFlSTR PCR reaction analysis (Thermo Fisher Scientific, Waltham, MA) and e-MycoTM plus Mycoplasma PCR detection kit (iNtRON Biotech. South Korea).
Immunoblotting
Immunoblot analysis was performed as described previously18. For protein expression analysis, cell lysates were separated by SDS-polyacrylamide gel electrophoresis (PAGE). Resolved proteins were transferred to nitrocellulose membrane and incubated with primary antibodies overnight at 4 °C, followed by washing and incubation with secondary antibodies. Membranes were then washed and incubated with horseradish peroxidase conjugated with anti-mouse or anti-rabbit antibodies, and the proteins were visualized by means of the enhanced chemiluminescence kit (AbClon, Inc., Seoul, Republic of Korea). Anti-Twist1, E-cadherin, N-cadherin, HA, ZEB1, BTG2 (Q-22), GAPDH, and α-tubulin antibodies were obtained from Santacruz (Dallas, TX); anti-v5 was obtained from Invitrogen (Carlsbad, CA); anti-eIF2α, Snail, eEF2, and PERK antibodies were from Cell Signaling (Danvers, MA), and anti-Flag antibody was from Sigma (St Louis, MO).
RNA isolation and reverse transcription
For RNA isolation, samples were collected in 1.0 mL of TRIzol (Invitrogen, Carlsbad, CA) and 1.0 µg of the purified total RNAs were subjected to reverse transcription for cDNA preparation using Prime-Script reverse transcriptase (Takara, Inc., Kyoto, Japan). Extracted RNAs were stored at −80 °C and the prepared cDNAs were diluted tenfold before PCR analysis.
Real-time PCR analysis
Real-time PCR analysis was performed to analyze the mRNA expression of various genes. Real-time quantitative RT-PCR analysis was performed in a 20 µl total volume with 2 µl diluted cDNA, 10 µl of 2× qPCR premix (RealHelixTM qPCR kit, NanoHelix, Daejeon, Korea) containing hot-start polymerase. Reaction mixtures were amplified for one cycle at 95 °C for 15 min, 50 cycles at 95 °C for 20 s and 60 °C for 40 s, and then the temperature was increased to 95 °C in 0.5 °C increments every 0.05 s along with plate reading. GAPDH and L32 were used as an internal control.
Transduction and transfection experiments
Adenovirus carrying β-galactosidase (Ad-lacZ) or TIS21-HA (Ad-TIS21) were transduced for 48 h into MDA-MB-231 and MDA-MB-468 cells. We have used exogenous expression of TIS21 (a mouse ortholog of human BTG2) in human cancer cells. BTG2-HA and Ad-TIS21-HA plasmids contain the tag at the C-terminal end, and the constructs contain two translational start sites that make double bands in the higher percentage gels (>15%) and long separation time (~3–3.5 h). Exogenous expression was confirmed with immunoblot analysis with HA antibody. α-Tubulin served as a loading control. For siRNA transfection, siControl, or siTIS21, siBTG2 were transfected into cells using Lipofectamine 2000 reagent according to the manufacturer’s protocol. TIS21 knockdown was confirmed using real-time PCR analysis. Forced expression and knockdown experiments, transfection with siRNAs were performed 24 h before transducing the cells with adenoviruses carrying with either Ad-LacZ or Ad-TIS21. For IP analysis, 293TN cells were transfected for 24 h with plasmids either BTG2-HA, v5-Twist1, Flag-Twist1, v5-BTG2, various deletion mutants of BTG2, or Twist1. The cells were subjected to IP and immunoblot analyses.
Serum starvation and FBS addition
To examine the regulation of Twist1 biosynthesis by the serum stimulation, MDA-MB-231 cells transduced with Ad-LacZ/Ad-TIS21 for 24 h were depleted with FBS for 18 h and then supplemented with FBS up to 1–10% for 6 h before immunoblot analysis. To further confirm the induction of Twist 1 protein biosynthesis in response to serum stimulation, the treatment time-dependent induction of Twist1 biosynthesis was tested in the MDA-MB-231 cells depleted with FBS for 18 h, and then 10% FBS was added up to 6 h before analyzed by immunoblotting.
Subcellular fractionation
Fractionation of the 293TN and HeLa cells was performed as the adapted method from Abcam protocol (http://www.abcam.com/protocols/subcellular-fractionation-protocol).
IP analysis
Cell lysates (1.0 mg) collected and sonicated in E1A buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 0.1% NP-40, 5 mM EDTA, protease inhibitors) were incubated overnight at 4 °C with either anti-v5, anti-HA, or anti-Flag antibodies for IP analysis, and then protein G beads were added and incubated at 4 °C for 2 h before centrifugation at 1000 r.p.m. for 1 min. The beads were washed three times thoroughly and the proteins bound were eluted with 2× SDS loading buffer to examine by immunoblot analysis. Normal IgG was used as a negative control. For IP analysis, subcellular fractionation was followed and 1 mg of cytoplasmic/nuclear fractions were pulled down with 1 μg of anti-HA antibody overnight at 4 °C. Following steps were similar as mentioned above.
Polysome profiling analysis
MDA-MB-231 and MDA-MB-468 cells transduced with Ad-LacZ/Ad-TIS21 for 48 h were treated with cycloheximide (10 µg/mL) for 10 min at room temperature, washed with cold PBS, and then lysed at cold room with 1 mL of polysome lysis buffer [20 mM HEPES pH 7.6, 5 mM MgCl2, 125 mM KCl, 1% NP-40, 2 mM dithiothreitol] supplemented with 100 µg/mL cycloheximide (Sigma), protease inhibitor cocktail (EDTA-free; Pierce), and RNAsin (Ambion). The cell lysates were tumbled for 15 min at 4 °C and centrifuged at 13,000 r.p.m. for 15 min. The supernatants were fractionated in 17.5–50% linear sucrose gradients by centrifugation (35,000 r.p.m. for 2 h 40 min) in a Beckman ultracentrifuge using SW40-Ti rotor. Gradients were eluted with a gradient fractionator (Brandel) and monitored with a UA-5 detector (ISCO). RNAs from each fraction were isolated and reverse transcribed as mentioned above. cDNAs were amplified using real-time PCR analysis for polysome abundance on Twist1 and GAPDH mRNAs.
Poly(A) tail length assay
PAT assay was performed and adapted from the previous report67. Total RNAs were isolated from the MDA-MB-231 cells transduced with Ad-LacZ/Ad-TIS21 for 48 h. To abolish the higher ordered secondary structure at the 3′-end, RNA samples were heated at 65 °C for 5 min and immediately placed on ice. The samples were incubated for 2 h at 37 °C with 10× PAP buffer, 1 μl of PAP (1 U/μL), 1 mM MnCl2, and 1 mM GTP (Thermo Fisher Scientific, Waltham, MA) in a 20 µL reaction mixture. PAP buffer, PAP, and MnCl2 (BioVision, Milpitas, CA). Ten microliters of Guanylated RNA was used for cDNA reaction (20 μL). The first-strand cDNA synthesis was primed by oligo(dCT). As specificity control, oligo(dT) was used. The RT reaction was stopped by denaturation at 95 °C (3 min) and the cDNA product was applied to PCR amplification using Gene-specific forward and reverse primers (38 cycles) and examined by agarose gel electrophoresis.
Isolation of mouse organs
TIS21 Wt and TIS21-KO mice under the C57BL/6 background were maintained at the Ajou University Animal Care Center under the specific pathogen-free condition with constant temperature and constant humidity. All of the animal procedures were followed by the Ajou University Institutional Review Board. Mouse organs—liver, lung, spleen, kidney, and pancreas—isolated from the TIS21-KO mice (15w–17w old) and the Wt mice (17w–20w old) were immediately snap frozen in the liquid nitrogen. Tissue samples were homogenized using a Dounce homogenizer under cold conditions (4 °C) and the samples were sonicated in RIPA buffer. Proteins (40 µg) were separated by SDS-PAGE and the resolved proteins were transferred onto nitrocellulose membrane to examine the expressions of Twist1, eEF2, and α-tubulin. For mRNA expression analysis, tissues were pulverized with liquid nitrogen and RNA extraction was performed by using TRIzol reagent. cDNAs were amplified using primers specific for eEF1α, eEF1β2, eEF1γ, eEF1ε1, and eEF2. GAPDH served as an internal control.
Analysis of human samples
Clinical data about the breast cancer patients were collected from the Ajou University Hospital after informed consent. Tumor tissues were divided based on the degree of lymph node invasion and the matched normal tissues were obtained according to the regulations of Institutional Review Board at the Ajou University Hospital (AJIRB-GEN-SMP-11-066).
Statistical analysis
The values presented as bar graphs are means ± SD. Statistical differences were analyzed either by Student’s t-test or Mann–Whitney U-test and the p-values < 0.05 were considered as statistically significant.
Supplementary information
Acknowledgements
This study was funded by National R&D Program for Cancer Control (131280) by Ministry for Health and Welfare, and by the National Research Foundation (No. 2016R1A2B4006466) funded by the Korean government (MSIP) to I.K.L., National Research Foundation (No. 2018R1D1A1B07049128) funded by the Korean government to P.D., and National Research Foundation (No. 2018R1D1A1B07044478) funded by the Korean government (MSIP) to M.S.R.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by A. Willis
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-019-1640-z).
References
- 1.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trimboli AJ, et al. Direct evidence for epithelial-mesenchymal transitions in breast cancer. Cancer Res. 2008;68:937–945. doi: 10.1158/0008-5472.CAN-07-2148. [DOI] [PubMed] [Google Scholar]
- 3.Chen ZF, Behringer RR. Twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 1995;9:686–699. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- 4.Yang J, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Yang MH, et al. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat. Cell Biol. 2010;12:982–992. doi: 10.1038/ncb2099. [DOI] [PubMed] [Google Scholar]
- 6.Alexander NR, et al. N-cadherin gene expression in prostate carcinoma is modulated by integrin-dependent nuclear translocation of Twist1. Cancer Res. 2006;66:3365–3369. doi: 10.1158/0008-5472.CAN-05-3401. [DOI] [PubMed] [Google Scholar]
- 7.Zhang D, et al. Twist1 expression induced by sunitinib accelerates tumor cell vasculogenic mimicry by increasing the population of CD133+ cells in triple-negative breast cancer. Mol. Cancer. 2014;13:207. doi: 10.1186/1476-4598-13-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fessenden TB, et al. Dia1-dependent adhesions are required by epithelial tissues to initiate invasion. J. Cell Biol. 2018;217:1485–1502. doi: 10.1083/jcb.201703145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi JA, Jung YS, Kim JY, Kim HM, Lim IK. Inhibition of breast cancer invasion by TIS21/BTG2/Pc3-Akt1-Sp1-Nox4 pathway targeting actin nucleators, mDia genes. Oncogene. 2016;35:83–93. doi: 10.1038/onc.2015.64. [DOI] [PubMed] [Google Scholar]
- 10.Rouault JP, et al. Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat. Genet. 1996;14:482–486. doi: 10.1038/ng1296-482. [DOI] [PubMed] [Google Scholar]
- 11.Fletcher BS, et al. Structure and expression of TIS21, a primary response gene induced by growth factors and tumor promoters. J. Biol. Chem. 1991;266:14511–14518. [PubMed] [Google Scholar]
- 12.Bradbury A, Possenti R, Shooter EM, Tirone F. Molecular cloning of PC3, a putatively secreted protein whose mRNA is induced by nerve growth factor and depolarization. Proc. Natl Acad. Sci. USA. 1991;88:3353–3357. doi: 10.1073/pnas.88.8.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim IK, et al. Differential expression of TIS21 and TIS1 genes in the various organs of Balb/c mice, thymic carcinoma tissues and human cancer cell lines. J. Cancer Res. Clin. Oncol. 1995;121:279–284. doi: 10.1007/BF01209594. [DOI] [PubMed] [Google Scholar]
- 14.Ficazzola MA, et al. Antiproliferative B cell translocation gene 2 protein is down-regulated post-transcriptionally as an early event in prostate carcinogenesis. Carcinogenesis. 2001;22:1271–1279. doi: 10.1093/carcin/22.8.1271. [DOI] [PubMed] [Google Scholar]
- 15.Struckmann K, et al. Impaired expression of the cell cycle regulator BTG2 is common in clear cell renal cell carcinoma. Cancer Res. 2004;64:1632–1638. doi: 10.1158/0008-5472.CAN-03-1687. [DOI] [PubMed] [Google Scholar]
- 16.Kawakubo H, et al. Expression of the NF-kappaB-responsive gene BTG2 is aberrantly regulated in breast cancer. Oncogene. 2004;23:8310–8319. doi: 10.1038/sj.onc.1208008. [DOI] [PubMed] [Google Scholar]
- 17.Farioli-Vecchioli S, et al. Inhibition of medulloblastoma tumorigenesis by the antiproliferative and pro-differentiative gene PC3. FASEB J. 2007;21:2215–2225. doi: 10.1096/fj.06-7548com. [DOI] [PubMed] [Google Scholar]
- 18.Park TJ, et al. TIS21 negatively regulates hepatocarcinogenesis by disruption of cyclin B1-Forkhead box M1 regulation loop. Hepatology. 2008;47:1533–1543. doi: 10.1002/hep.22212. [DOI] [PubMed] [Google Scholar]
- 19.Liu M, et al. Regulation of the cell cycle gene, BTG2, by miR-21 in human laryngeal carcinoma. Cell Res. 2009;19:828–837. doi: 10.1038/cr.2009.72. [DOI] [PubMed] [Google Scholar]
- 20.Devanand P, et al. Inhibition of bladder cancer invasion by Sp1-mediated BTG2 expression via inhibition of DNA methyltransferase 1. FEBS J. 2014;281:5581–5601. doi: 10.1111/febs.13099. [DOI] [PubMed] [Google Scholar]
- 21.Frampton AE, et al. Integrated molecular analysis to investigate the role of microRNAs in pancreatic tumour growth and progression. Lancet. 2015;385(Suppl 1):S37. doi: 10.1016/S0140-6736(15)60352-X. [DOI] [PubMed] [Google Scholar]
- 22.Tajima K, et al. SETD1A modulates cell cycle progression through a miRNA network that regulates p53 target genes. Nat. Commun. 2015;6:8257. doi: 10.1038/ncomms9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mauxion F, Faux C, Seraphin B. The BTG2 protein is a general activator of mRNA deadenylation. EMBO J. 2008;27:1039–1048. doi: 10.1038/emboj.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doidge R, Mittal S, Aslam A, Winkler GS. The anti-proliferative activity of BTG/TOB proteins is mediated via the Caf1a (CNOT7) and Caf1b (CNOT8) deadenylase subunits of the Ccr4-not complex. PLoS ONE. 2012;7:e51331. doi: 10.1371/journal.pone.0051331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryu Min Sook, Lee Myung Soog, Hong Jong Wook, Hahn Tae-Ryong, Moon Eunpyo, Lim In Kyoung. TIS21/BTG2/PC3 is expressed through PKC-δ pathway and inhibits binding of cyclin B1-Cdc2 and its activity, independent of p53 expression. Experimental Cell Research. 2004;299(1):159–170. doi: 10.1016/j.yexcr.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 26.Lim YB, Park TJ, Lim IK. B cell translocation gene 2 enhances susceptibility of HeLa cells to doxorubicin-induced oxidative damage. J. Biol. Chem. 2008;283:33110–33118. doi: 10.1074/jbc.M804255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sundaramoorthy S, Ryu MS, Lim IK. B-cell translocation gene 2 mediates crosstalk between PI3K/Akt1 and NFkappaB pathways which enhances transcription of MnSOD by accelerating IkappaBalpha degradation in normal and cancer cells. Cell Commun. Signal. 2013;11:69. doi: 10.1186/1478-811X-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mollerstrom E, et al. Up-regulation of cell cycle arrest protein BTG2 correlates with increased overall survival in breast cancer, as detected by immunohistochemistry using tissue microarray. BMC Cancer. 2010;10:296. doi: 10.1186/1471-2407-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi F, et al. Breast tumor progression induced by loss of BTG2 expression is inhibited by targeted therapy with the ErbB/HER inhibitor lapatinib. Oncogene. 2011;30:3084–3095. doi: 10.1038/onc.2011.24. [DOI] [PubMed] [Google Scholar]
- 30.Choi JA, Lim IK. TIS21/BTG2 inhibits invadopodia formation by downregulating reactive oxygen species level in MDA-MB-231 cells. J. Cancer Res. Clin. Oncol. 2013;139:1657–1665. doi: 10.1007/s00432-013-1484-3. [DOI] [PubMed] [Google Scholar]
- 31.Sundaramoorthy S, et al. TIS21(/BTG2) inhibits breast cancer growth and progression by differential regulation of mTORc1 and mTORc2-AKT1-NFAT1-PHLPP2 signaling axis. J. Cancer Res. Clin. Oncol. 2018;144:1445–1462. doi: 10.1007/s00432-018-2677-6. [DOI] [PubMed] [Google Scholar]
- 32.Powell E, et al. p53 deficiency linked to B cell translocation gene 2 (BTG2) loss enhances metastatic potential by promoting tumor growth in primary and metastatic sites in patient-derived xenograft (PDX) models of triple-negative breast cancer. Breast Cancer Res. 2016;18:13. doi: 10.1186/s13058-016-0673-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eckert MA, et al. Twist1-induced invadopodia formation promotes tumor metastasis. Cancer Cell. 2011;19:372–386. doi: 10.1016/j.ccr.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y, et al. SRC-1 and Twist1 expression positively correlates with a poor prognosis in human breast cancer. Int. J. Biol. Sci. 2014;10:396–403. doi: 10.7150/ijbs.8193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quy LN, et al. TIS21(/BTG2/PC3) inhibits interleukin-6 expression via downregulation of STAT3 pathway. Cell. Signal. 2013;25:2391–2399. doi: 10.1016/j.cellsig.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 36.Prevot D, et al. The leukemia-associated protein Btg1 and the p53-regulated protein Btg2 interact with the homeoprotein Hoxb9 and enhance its transcriptional activation. J. Biol. Chem. 2000;275:147–153. doi: 10.1074/jbc.275.1.147. [DOI] [PubMed] [Google Scholar]
- 37.Lin WJ, Chang YF, Wang WL, Huang CY. Mitogen-stimulated TIS21 protein interacts with a protein-kinase-Calpha-binding protein rPICK1. Biochem. J. 2001;354:635–643. doi: 10.1042/bj3540635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- 39.Hong JW, Ryu MS, Lim IK. Phosphorylation of serine 147 of tis21/BTG2/pc3 by p-Erk1/2 induces Pin-1 binding in cytoplasm and cell death. J. Biol. Chem. 2005;280:21256–21263. doi: 10.1074/jbc.M500318200. [DOI] [PubMed] [Google Scholar]
- 40.Pallier K, et al. TWIST1 a new determinant of epithelial to mesenchymal transition in EGFR mutated lung adenocarcinoma. PLoS ONE. 2012;7:e29954. doi: 10.1371/journal.pone.0029954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J, et al. The Aurora-A-Twist1 axis promotes highly aggressive phenotypes in pancreatic carcinoma. J. Cell Sci. 2017;130:1078–1093. doi: 10.1242/jcs.196790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim IK, et al. TIS21(/BTG2) inhibits doxorubicin-induced stress fiber-vimentin networks via Nox4-ROS-ABI2-DRF-linked signal cascade. Cell. Signal. 2017;30:179–190. doi: 10.1016/j.cellsig.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Han J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell. Bio. 2013;15:481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 45.Gingras AC, et al. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomoo K, Abiko F, Miyagawa H, Kitamura K, Ishida T. Effect of N-terminal region of eIF4E and Ser65-phosphorylation of 4E-BP1 on interaction between eIF4E and 4E-BP1 fragment peptide. J. Biochem. 2006;140:237–246. doi: 10.1093/jb/mvj143. [DOI] [PubMed] [Google Scholar]
- 47.Meric-Bernstam F, et al. Aberrations in translational regulation are associated with poor prognosis in hormone receptor-positive breast cancer. Breast Cancer Res. 2012;14:R138. doi: 10.1186/bcr3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat. Cell Biol. 2014;16:488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 49.Shiota M, et al. Twist and p53 reciprocally regulate target genes via direct interaction. Oncogene. 2008;27:5543–5553. doi: 10.1038/onc.2008.176. [DOI] [PubMed] [Google Scholar]
- 50.Merrick WC. Mechanism and regulation of eukaryotic protein synthesis. Microbiol. Rev. 1992;56:291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stupfler B, Birck C, Seraphin B, Mauxion F. BTG2 bridges PABPC1 RNA-binding domains and CAF1 deadenylase to control cell proliferation. Nat. Commun. 2016;7:10811. doi: 10.1038/ncomms10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim BC, Ryu MS, Oh SP, Lim IK. TIS21/(BTG2) negatively regulates estradiol-stimulated expansion of hematopoietic stem cells by derepressing Akt phosphorylation and inhibiting mTOR signal transduction. Stem Cells. 2008;26:2339–2348. doi: 10.1634/stemcells.2008-0327. [DOI] [PubMed] [Google Scholar]
- 54.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 55.Guo L, et al. Phosphorylated eIF2alpha predicts disease-free survival in triple-negative breast cancer patients. Sci. Rep. 2017;7:44674. doi: 10.1038/srep44674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim SH, Forman AP, Mathews MB, Gunnery S. Human breast cancer cells contain elevated levels and activity of the protein kinase, PKR. Oncogene. 2000;19:3086–3094. doi: 10.1038/sj.onc.1203632. [DOI] [PubMed] [Google Scholar]
- 57.Zoll WL, Horton LE, Komar AA, Hensold JO, Merrick WC. Characterization of mammalian eIF2A and identification of the yeast homolog. J. Biol. Chem. 2002;277:37079–37087. doi: 10.1074/jbc.M207109200. [DOI] [PubMed] [Google Scholar]
- 58.Komar AA, et al. Novel characteristics of the biological properties of the yeast Saccharomyces cerevisiae eukaryotic initiation factor 2A. J. Biol. Chem. 2005;280:15601–15611. doi: 10.1074/jbc.M413728200. [DOI] [PubMed] [Google Scholar]
- 59.Golovko A, et al. The eIF2A knockout mouse. Cell Cycle. 2016;15:3115–3120. doi: 10.1080/15384101.2016.1237324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang TH, et al. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics. 2013;14(Suppl 2):S4. doi: 10.1186/1471-2105-14-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park TJ, Kim JY, Park SH, Kim HS, Lim IK. Skp2 enhances polyubiquitination and degradation of TIS21/BTG2/PC3, tumor suppressor protein, at the downstream of FoxM1. Exp. Cell Res. 2009;315:3152–3162. doi: 10.1016/j.yexcr.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 62.de Moor CH, Meijer H, Lissenden S. Mechanisms of translational control by the 3′ UTR in development and differentiation. Semin. Cell Dev. Biol. 2005;16:49–58. doi: 10.1016/j.semcdb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 63.Riis B, Rattan SI, Clark BF, Merrick WC. Eukaryotic protein elongation factors. Trends Biochem. Sci. 1990;15:420–424. doi: 10.1016/0968-0004(90)90279-K. [DOI] [PubMed] [Google Scholar]
- 64.Sasikumar AN, Perez WB, Kinzy TG. The many roles of the eukaryotic elongation factor 1 complex. Wiley Interdisciplinary Rev. RNA. 2012;3:543–555. doi: 10.1002/wrna.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaul G, Pattan G, Rafeequi T. Eukaryotic elongation factor-2 (eEF2): its regulation and peptide chain elongation. Cell Biochem. Funct. 2011;29:227–234. doi: 10.1002/cbf.1740. [DOI] [PubMed] [Google Scholar]
- 66.Savci-Heijink CD, Halfwerk H, Koster J, Horlings HM, van de Vijver MJ. A specific gene expression signature for visceral organ metastasis in breast cancer. BMC Cancer. 2019;19:333. doi: 10.1186/s12885-019-5554-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kusov YY, Shatirishvili G, Dzagurov G, Gauss-Muller V. A new G-tailing method for the determination of the poly(A) tail length applied to hepatitis A virus RNA. Nucleic Acids Res. 2001;29:E57–E57. doi: 10.1093/nar/29.12.e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.