Abstract
Abstract
Mentha haplocalyx (Mentha canadensis) is widely used as a medicinal plant in traditional Chinese medicine, and the extracts of its aerial parts are found to significantly inhibit the activity of α-glucosidase with an IC50 value of 21.0 μg/mL. Bioactivity-guided isolation of the extracts afforded two new compounds (1 and 2), together with 23 known ones (3–25). Their structures were established by extensive spectroscopic analyses (1D and 2D NMR, MS, IR and UV). Compounds 1–17 and 21–25 were evaluated for their α-glucosidase inhibitory activities. Compound 11 was the most active ones with an IC50 values of 83.4 μM. These results verify the α-glucosidase inhibitory activity of M. haplocalyx (M. canadensis) and specify its active compounds for the first time.
Graphical Abstract
Electronic supplementary material
The online version of this article (10.1007/s13659-019-0207-0) contains supplementary material, which is available to authorized users.
Keywords: Mentha haplocalyx Briq. (Mentha canadensis L.), Lamiaceae, α-Glucosidase inhibitor
Introduction
Diabetes mellitus is a chronic disease caused by inherited or acquired deficiency in insulin secretion and by decreased responsiveness of the organs to secreted insulin [1]. Such a deficiency results in an increased blood glucose level and in turn damages body systems including blood vessels and nerves [2]. It is one of the most serious diseases worldwide and developing with an increase in obesity and ageing [3]. An efficiently therapeutic approach is to retard absorption of glucose through the inhibition of carbohydrate-hydrolysing enzymes such as α-amylase and α-glucosidase in the digestive organs [4]. Clinically, acarbose and voglibose have been used as effective α-glucosidase inhibitors to delay glucose absorption [5].
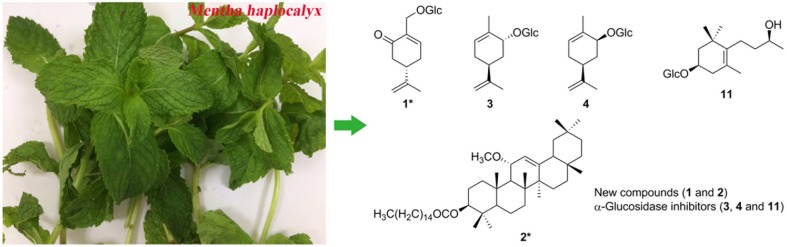
Mentha haplocalyx Briq. (Mentha canadensis L.), a perennial herbaceous plant of the family Lamiaceae, is widely distributed in southwest of China and popularly used in food, cosmetics and medicines. As a traditional Chinese medicine, it is clinically used to treat diseases in the nerve center, breath, procreation and digestive systems [6]. Pharmacological studies of M. haplocalyx (M. canadensis) revealed various biological activities, such as antimicrobial, anti-inflammatory, antioxidant, antitumor, gastrointestinal protective, and hepatoprotective activities [7]. A large number of volatile compounds were reported from M. haplocalyx (M. canadensis), as well as a few polyphenolic acids, flavonoids, monoterpenoids, and glycosides, which might contribute to the medicinal benefits of this plant [8].
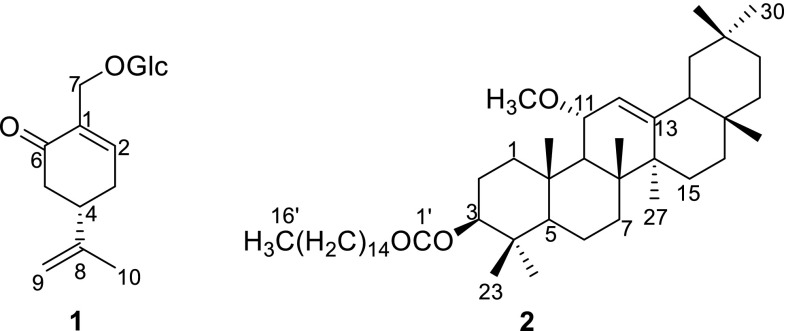
Several findings have depicted the potential antidiabetic capability of genus Mentha. M. piperita could alleviate hyperglycemia induced by streptozotocin–nicotinamide-induced type 2 diabetes in rats, and cause a reduction of glycemia in human [9, 10]. However, no report has referred to the active compounds of Mentha responsible for its antidiabetic capability. Our preliminary bioassay revealed that extracts of the aerial parts of M. haplocalyx (M. canadensis) exhibited significant α-glucosidase inhibitory activity with an IC50 value of 21.0 μg/mL. Subsequently, two new compounds (1 and 2) (Fig. 1) and 23 known ones (3‒25) were isolated and identified through bioactivity-guided fractionation. This paper described the isolation, identification, and α-glucosidase inhibitory activity evaluation of these compounds.
Fig. 1.
The structures of new compounds 1 and 2 from M. haplocalyx
Results and Discussion
Structural Identification
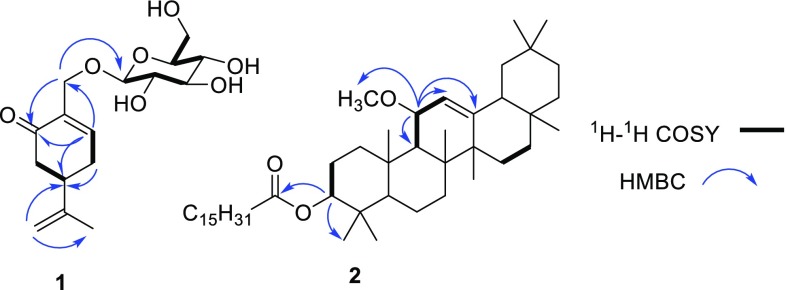
Compound 1 was obtained as colorless caramelized solid, with the molecular formula of C16H24O7 from its HRESIMS at m/z 351.1404 [M+Na]+. The IR absorptions suggested the existence of a hydroxyl group (3427 cm−1) and an α,β-unsaturated carbonyl group (1635 cm−1). The 13C NMR (DEPT) data (Table 1) displayed sixteen carbon signals, including one methyl, five methylenes, seven methines, and three quaternary carbons. Among them, a glucose unit (δC 104.1, 75.2, 78.2, 71.7, 78.2, 62.8) and an α,β-unsaturated carbonyl group (δC 136.6, 149.4, 201.0) were observed. 1H NMR data (Table 1) exhibited a singlet methyl proton (δH 1.78, H-10), a trisubstituted olefinic proton (δH 7.23, H-2), and one terminal double bond proton (δH 4.82, H-9a; 4.80, H-9b). The glucose unit protons [δH 4.30 (d, J = 7.8 Hz, H-1ʹ); 3.19–3.85 (6H, H-2ʹ‒H-6ʹ)] were also present. All the above information suggested that compound 1 was a menthane-monoterpene glycoside. Apart from the glucose unit, the NMR data of 1 closely resembled those of the reported compound (R)-7-hydroxycarvone [11]. Whereas the major differences between their NMR signals at C-7 position [δH 4.49, 4.28 and δC 67.0 for 1; δH 4.27 and δC 61.8 for (R)-7-hydroxycarvone] suggested that 7-OH in 1 might be glycosylated, which was further confirmed by the HMBC correlation of H-7 (δH 4.99 and 4.28) with C-1ʹ (δC 104.1) (Fig. 2). The specific rotation of 1 ( − 22.0) was opposite to (4R)-7-hydroxyisopiperitenone7-O-β-d-glucopyranoside ( + 53.9°), and similar to the (4S)-7-hydroxyisopiperitenone 7-O-β-d-glucopyranoside ( − 14.4) [12], tentatively determining its (4S)-configuration. Therefore, compound 1 was elucidated as (4S)-7-hydroxy-carvone 7-O-β-d-glucopyranoside.
Table 1.
1H NMR (600 MHz, CD3OD) and 13C NMR (150 MHz, CD3OD) data for compound 1
| No | δ H | δ C | 1H–1H COSY | HMBC |
|---|---|---|---|---|
| 1 | 136.6 s | |||
| 2 | 7.23 m | 149.4 d | H-3a, 3b, 7a, 7b | C-1, 3, 4, 6, 7 |
| 3a | 2.67 m | 32.2 t | H-2, 3b | C-1, 2, 4, 5 |
| 3b | 2.41 ddq (18.0, 10.2, 1.8) | H-2, 3a | C-1, 2, 4, 5 | |
| 4 | 2.74 ddd (16.2, 10.2, 4.8) | 43.6 d | H-5a, 5b | C-3, 5, 6, 8, 9, 10 |
| 5a | 2.50 ddd (16.2, 5.4, 1.2) | 44.2 t | H-4 | C-3, 4, 6, 8 |
| 5b | 2.48 m | H-4 | C-3, 4, 6, 8 | |
| 6 | 201.0 s | |||
| 7a | 4.49 dq (13.2, 1.2) | 67.0 t | H-7b | C-1, 2, 6, 1ʹ |
| 7b | 4.28 dd (13.2, 1.2) | H-7a | C-1, 2, 6, 1ʹ | |
| 8 | 148.3 s | |||
| 9a | 4.82 m | 111.3 s | H-10 | C-4, 8, 10 |
| 9b | 4.80 m | C-4, 8, 10 | ||
| 10 | 1.78 s | 20.7 q | H-9a | C-4, 8, 9 |
| 1′ | 4.30 d (7.8) | 104.1 d | H-2ʹ | C-7, 2ʹ, 3ʹ, 5ʹ |
| 2′ | 3.19 dd (9.6, 8.4) | 75.2 d | H-1ʹ, 3ʹ | C-3ʹ |
| 3′ | 3.34 t (9.0) | 78.2 d | H-2ʹ | C-2ʹ, 4ʹ |
| 4′ | 3.24 m | 71.7 d | H-5ʹ | C-5ʹ, 6ʹ |
| 5′ | 3.28 t (9.6) | 78.2 d | H-4ʹ, 6ʹa, 6ʹb | C-1ʹ, 4ʹ |
| 6′a | 3.85 dd (12.0, 1.8) | 62.8 t | H-5ʹ, 6ʹb | C-4ʹ, 5ʹ |
| 6′b | 3.66 dd (12.0, 5.4) | H-5ʹ, 6ʹa | C-4ʹ, 5ʹ |
Fig. 2.
Key 1H–1H COSY and HMBC correlations of compounds 1 and 2
Compound 2, white powder, had a molecular formula of C47H82O3 from its HREIMS (m/z 694.6258 [M]+). The 13C NMR (DEPT) data (Table 2) displayed 47 carbon signals, including ten methyls, 23 methylenes, six methines and eight quaternary carbons. Among them, a methoxyl (δC 53.4), two oxy-methines (δC 80.4, C-3; 75.8, C-11), a trisubstituted double bond (δC 121.8, 149.7) were showed. A palmitoyl group (δC 173.7, C-1′; 34.7, C-2′; 25.2, C-3′; 29.2–29.7, C-4′–C-13′; 31.9, C-14′; 22.7, C-15′; 14.1, C-16′) was also present. The 1H NMR data (Table 2) exhibited eight singlet methyls (δH 0.85–1.21), a triplet methyl (δH 0.88, t, J = 7.8 Hz), a methoxyl (δH 3.25), and a trisubstituted olefinic proton (δH 5.32, d, J = 3.0 Hz, H-12). These NMR data suggested that 2 was an oleanane-type triterpene fused with a palmitoyl group, and closely resembled those of the reported compound (3β,11α)-11-hydroxy-olean-12-en-3-yl palmitate [13]. The major difference was that the NMR signals at C-11 of the known compound (δH 4.50; δC 81.6) were upfield shifted in 2 (δH 3.89; δC 75.8) and a methoxyl group (δH 3.25; δC 53.4) further appeared, indicating the hydroxyl group at C-11 was methylated in 2, which was further confirmed by the HMBC (Fig. 2) correlation of OMe (δH 3.25) with C-11 (δC 75.8). Finally, compound 2 was established as (3β,11α)-3-hydroxy-11α-methoxy-olean-12-en-3-yl palmitate.
Table 2.
1H NMR (600 MHz, CDCl3) and 13C NMR (150 MHz, CDCl3) data for compound 2
| No. | δ H | δ C | No. | δ H | δ C |
|---|---|---|---|---|---|
| 1a | 1.96 m | 39.1 t | 20 | 31.1 s | |
| 1b | 1.32 m | 21a | 1.32 m | 34.9 t | |
| 2a | 1.62 m | 23.8 t | 21b | 1.10 m | |
| 2b | 1.23 m | 22a | 1.44 m | 37.0 t | |
| 3 | 4.51 dd (7.8, 9.0) | 80.4 d | 22b | 1.23 m | |
| 4 | 38.0 s | 23 | 0.85 s | 28.2 q | |
| 5 | 0.88 m | 55.3 d | 24 | 0.90 s | 16. 8 q |
| 6a | 1.52 m | 18.3 t | 25 | 1.07 s | 16.8 q |
| 6b | 1.26 m | 26 | 1.00 s | 18.2 q | |
| 7a | 1.50 m | 33.2 t | 27 | 1.21 s | 25.1 q |
| 7b | 1.30 m | 28 | 0.83 s | 28.5 q | |
| 8 | 43.1 s | 29 | 0.89 s | 33.2 q | |
| 9 | 1.73 m | 51.1 d | 30 | 0.88 s | 23.6 q |
| 10 | 38.1 s | OCH3 | 3.25 s | 53.4 q | |
| 11 | 3.89 dd (3.0, 9.0) | 75.8 d | 1′ | 173.7 s | |
| 12 | 5.32 d (3.0) | 121.8 d | 2′ | 2.29 t (7.8) | 34.7 t |
| 13 | 149.7 s | 3′ | 1.64 m | 25.2 t | |
| 14 | 41.7 s | 4′ | 29.2 t | ||
| 15a | 2.04 m | 26.3 t | 5′ | 29.3 t | |
| 15b | 0.84 m | 6′ | 29.4 t | ||
| 16a | 1.65 m | 26.8 t | 7′ ~ 13′ | 29.6 ~ 29.7 t | |
| 16b | 1.00 m | 14′ | 1.26 m | 31.9 t | |
| 17 | 32.3 s | 15′ | 1.27 m | 22.7 t | |
| 18 | 2.01 dd (13.8, 4.2) | 47.0 d | 16′ | 0.88 t (7.8) | 14.1 q |
| 19a | 1.66 dd (13.8, 4.2) | 46.5 t | |||
| 19b | 1.08 m |
By comparing their physical and spectroscopic data with those reported in the literatures, the known compounds (Figure S1) were elucidated as (4R,6R)-carveol β-d-glucoside (3) [14], (4R,6S)-carveol β-d-glucoside (4) [14], (+)-neodihydrocarvy β-d-glucoside (5) [15], (−)-dihydrocarvy β-d-glucoside (6) [15], uroterpenol β-d-glucoside (7) [15], spicatoside A (8) [16], spicatoside B (9) [16], (3S,6S)-cis-linalool-3,7-oxide (10) [17], (3R,9S)-megastigman-5-en-3,9-diol 3-O-β-d-glucopyranoside (11) [18], linarionoside A (12) [19], 1,1,5-trimethyl- 6-(3-hydroxyl) cyclohexene-5-yl-1-β-d-pyranoglucoside (13) [20], linarionoside B (14) [21], (9S)-linarionoside B (15) [21], (+)-jasmololone glycoside (16) [14], (‒)-5′-(β-d-glucopyranosyloxy) jasmonic acid (17) [22], maniladiol (18) [23], 3β,28-dihydroxy-olean-12-enyl palmitate (19) [24], olean-12-ene-28-arboxy-3-palmitate (20) [25], ursolic acid (21) [26], 1-(β-d-ribofuranosyl)-1H-1,2,4-triazone (22) [27], naphthisoxazol A (23) [28], menthalactone (24) [29], 6-amino-9-[1-(3,4-dihydroxy phenyl)ethyl]-9H-purine (25) [30], respectively.
α-Glucosidase Inhibitory Activity
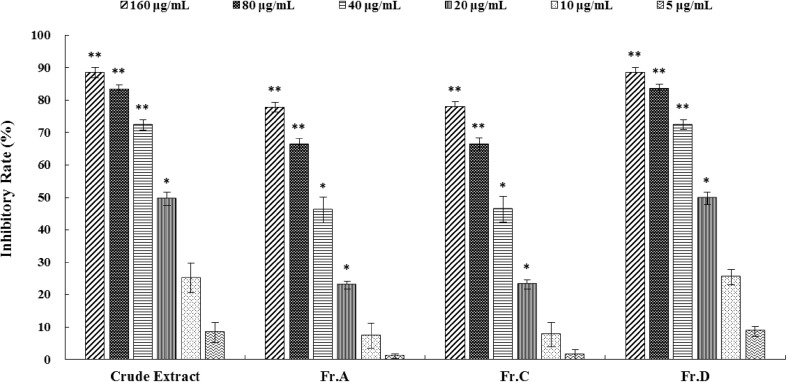
In the preliminary bioassay, the crude extracts, Fr.A, Fr.C and Fr.D all exhibited significantly inhibitory activity against α-glucosidase at concentrations (> 20 μg/mL). These inhibitory effects were dose-dependent (Fig. 3). Their IC50 values were measured as 21.0, 36.7, 37.2, and 20.3 μg/mL, respectively (Table 3). Bioactivity-guided isolation further afforded 25 compounds, while compounds 1–17 and 21–25 were measured their α-glucosidase inhibitory activity. Compound 11 possessed the most significant activity with an IC50 values of 83.4 μM, while compounds 3 and 4 showed moderate inhibitory activity against α-glucosidase with IC50 values of 516.0 and 919.0 μM, respectively (Table 3). Other compounds had no significant inhibitory activity.
Fig. 3.
α-Glucosidase inhibitory activities of crude extract, Fr.A, Fr.C and Fr.D at concentrations of 160, 80, 40, 20, 10 and 5 μg/mL, respectively. Values are presented as inhibitory rate compared to the blank control. Means significantly lower than the controls are indicated with one asterisk (*) (Dunnett’s one-sided t test; p < 0.05) or two asterisks (**) (p < 0.01). Error bars are one standard error of the mean. N = 5
Table 3.
α-Glucosidase inhibitory activities of extracts and compounds from M. haplocalyx
| Extractsa | IC50 ± SD | Compoundsb | IC50 ± SD |
|---|---|---|---|
| Crude extract | 21.0 ± 1.9 | 3 | 516.0 ± 2.1 |
| Fr.A | 36.7 ± 2.6 | 4 | 919.0 ± 37.3 |
| Fr.C | 37.2 ± 5.5 | 11 | 83.4 ± 1.3 |
| Fr.D | 20.3 ± 2.8 | ||
| Acarbosec | 25.8 ± 3.3 |
aIC50 values in μg/mL are mean ± SD from three independent experiments
bIC50 values in μM are mean ± SD from three independent experiments
cIC50 values in nM are mean ± SD from three independent experiments
Comparing the chemical structures and activity of these compounds, it can be found that triterpenoids (2 and 21), jasmonoid glucosides (16 and 17) and N-containing compounds (22–25) were not responsible for the α-glucosidase inhibitory activity of the extracts from M. haplocalyx (M. canadensis). Among the monoterpene glucosides (1 and 3–10), only compounds 3 and 4 manifested moderate inhibitory activity against α-glucosidase, while 5 and 6 did not. Thus, it might be assumed that the absence of cyclic double bond in 3 and 4 markedly decreased their inhibitory activity.
Experimental
General Experimental Instruments and Procedures
LC–MS analyses were performed on a UFLC/MS-IT-TOF apparatus and the analytical conditions were set as previously reported [31]. Mass spectra were measured through a Waters AutoSpec Premier P776 (Waters, USA) mass spectrometer. Optical rotations were measured through a Jasco model 1020 digital polarimeter (Horiba, Tokyo, Japan). UV and IR (KBr) spectra were recorded on a Shimadzu UV2401PC spectrophotometer (Shimadzu, Kyoto, Japan) and a Bio-Rad FTS-135 spectrometer (Hercules, California, USA), respectively. NMR spectra were recorded on the DRX-500 or AdvanceIII-600 NMR (Bruker, Bremerhaven, Germany) spectrometers with TMS as an internal standard. Thin-layer chromatography (TLC) analyses were carried out with silica gel GF254 (Merck, Chemical Co. Ltd., Shanghai, China), and spots were detected under UV light or by heating after spraying with 10% H2SO4 in C2H5OH (v/v). Preparative TLC (PTLC) was purchased from Yantai Jiangyou silicon development Company (Yantai, China). Silica gel (200–300 mesh, Linyi Haixiang Co., Ltd; Linyi, China), Sephadex LH-20 (Amersham Bioscience, Sweden), and D101 macroporous adsorption resin (Tianjin guangfu fine chemical co. LTD, Tianjin, China) were used for column chromatography. MPLC separations were conducted on a Dr-Flash II apparatus using a MCI gel CPH 20P (75-150 u, Mitsubishi Chemical Corporation, Tokyo, Japan) column. Semipreparative HPLC purifications were performed on a Shimadzu LC-CBM-20 system (Shimadzu, Kyoto, Japan) with the XDB-C18 column.
Plant Material
The aerial parts of fresh M. haplocalyx Briq. (M. canadensis L.) were bought from market, Kunming, Yunnan Province, China, in March 2013, and were identified by Prof. Li-gong Lei, Kunming Institute of Botany. A voucher specimen (No. 2013032401) was deposited at the Laboratory of Anti-virus and Natural Medicinal Chemistry, Kunming Institute of Botany, CAS.
Extraction and Isolation
The aerial parts of fresh M. haplocalyx (M. canadensis) (20 kg) were powdered and extracted with 50% ethanol for three times at room temperature (each 200 L). The combined extraction was concentrated in vacuo to yield a residue. The residue was then suspended in water and extracted with ethyl acetate (Fr.A, 130 g). The aqueous phase was subjected to D101 macroporous adsorption resin using a step gradient elution with C2H5OH-H2O (0:100, 50:50 and 90:10, v/v) as the mobile phase to give three fractions (Fr.B–Fr.D).
The Fr.A (130 g) and Fr.D (10 g) were combined (Fr.E, 140 g). Fr.E (140 g) was fractionated on silica gel CC using a step gradient elution with MeOH–CHCl3 (2:98, 5:95, 10:90, v/v) and H2O–MeOH–CHCl3 (2:20:80, 3:30:70. 4:40:60, v/v) as the mobile phase, and following washed with MeOH to give seven fractions (Fr.E-1–Fr.E-7). Fr.E-3 (34 g) was subjected to silica gel CC eluted with EtOAc-petroleum ether (from 6:94 to 50:50) to give five fractions (Fr.E-3-1–Fr.E-3-5). Fr.E-3-1 (8.8 g) was submitted to silica gel CC eluted with Me2CO-petroleum ether (5:95), then applied to Sephadex LH-20 CC eluted with CHCl3–MeOH (50:50), and finally purified by silica gel CC with EtOAc-petroleum ether (40:60) to yield 2 (10 mg), 19 (13 mg) and 20 (12 mg). Compound 21 (2 g) was got from Fr.E-3-2 (9.1 g) that was separately separated on MCI gel CHP 20P CC (MeOH–H2O, 30:70, 70:30, 90:10) and Sephadex LH-20 CC (MeOH–CHCl3, 50:50). Fr.E-3-5 (5.6 g) was subjected on MCI gel CHP 20P CC using MeOH–H2O (60:40, 90:10), subsequently compound 24 (10 mg) was yielded from Sephadex LH-20 CC (MeOH–CHCl3, 50:50). Fr.E-5 (5.8 g) was subjected to MCI gel CHP 20P CC using a step gradient elution with MeOH–H2O (40:60, 80:20) to give two fractions (Fr.A-5-1 and Fr.A-5-2). Fr.A-5-1 (2.7 g) was fractionated on silica gel CC (MeOH–CHCl3, 10:90) to give three fractions (from Fr.E-5-1-1 to Fr.E-5-1-3). Fr.E-5-1-1 (327 mg) was preferred on Sephadex LH-20 eluted with MeOH–CHCl3 (50:50), and following purified by semipreparative HPLC (MeCN–H2O, 25:75, v/v, 3.0 mL/min) over an XDB-C18 column (9.4 × 250 mm, 5 μm) to yield compounds 3 (5 mg), 5 (4 mg), 6 (5 mg) and 25 (8 mg). Fr.E-5-1-2 (1.3 g) was separated on Sephadex LH-20 CC (MeOH–CHCl3, 50:50), and later fractionated on silica gel CC (MeOH–CHCl3, 90:10), therefore, compounds 1 (4 mg), 7 (14 mg), 10 (2 mg), the mixture of 12 and 13 (8 mg), the mixture of 14 and 15 (6 mg) were obtained by semipreparative HPLC (MeCN-H2O, 40:60, v/v, 3.0 mL/min) over an XDB-C18 column (9.4 × 250 mm, 5 μm).
Fr.C (50 g) was subjected to silica gel CC and then Al2O3 CC with H2O–MeOH–CHCl3 (3:30:70) to give four fractions (from Fr.C-1 to Fr.C-4). Fr.C-1 (10.4 g) was fractionated on MCI gel CHP 20P CC with MeOH–H2O (40:60, 80:20), then applied to Sephadex LH-20 CC eluted with MeOH–CHCl3 (50:50), and later purified by silica gel CC eluted with MeOH–EtOAc (2:98) to yield 4 (120 mg), 8 (800 mg), 9 (311 mg), 11 (200 mg) and 16 (20 mg). Compounds 17 (201 mg), 22 (46 mg) and 23 (17 mg) were afforded from Fr.C-2 (20.0 g) by repeated silica gel CC (MeOH–CHCl3, 15:85, 20:80, 25:75) and Sephadex LH-20 (MeOH–CHCl3, 50:50).
Spectroscopy Data of Compounds
Compound 1
Colorless caramelized solid, C16H24O7, − 22.0 (c, 0.20, MeOH); UV (MeOH) λmax (log ε): 230 (3.65) nm; IR (KBr) νmax: 3427, 1635, 1385, 1046, 902 cm−1; 1H NMR (600 MHz, CD3OD) and 13C NMR (DEPT, 150 MHz, CD3OD) see Table 1; HRESIMS m/z: 351.1404 [M+Na]+ (calc. 351.1414 for C16H24O7Na).
Compound 2
Colorless powder, C47H82O3; 1H NMR (600 MHz, CDCl3) and 13C NMR (DEPT, 150 MHz, CDCl3) see Table 2; HREIMS m/z: 694.6258 [M]+ (calc. 694.6264 for C47H82O3).
Inhibitory Assay of α-Glucosidase
The α-glucosidase inhibitory activity was measured in a 96-well microtiter plate based on p-nitrophenyl-α-d-glucopyranoside (PNPG, Yuanye Biosciences Co. Ltd., Shanghai, China) as a substrate following the reported method with slight modifications [32]. In brief, 5.0 mM PNPG (20 μL) and 20 μL tested compounds of dissolved in 10 μL DMSO and 990 μL phosphate buffer (PB, 0.1 M, pH = 6.8) were sequentially added to a 96-well plate to be mixed. The mixture was incubated at 37 °C for 5 min. Reactions were initiated by addition of 2.0 U/mL α-glucosidase (20 μL, Yuanye Biosciences Co. Ltd., Shanghai, China) in PB. The reaction mixture was incubated at 37 °C for 15 min. Then, the incubation solution was stopped the reaction by adding 0.2 M Na2CO3 (40 μL). The absorbance was recorded at 405 nm by a Bio-Rad 680 microplate reader (Hercules, CA, USA). The negative control was set by adding PB instead of the sample using the same procedure for the tests. Acarbose (Bayer) dissolved in PB was utilized as the positive control. The blank was set by adding phosphate buffer instead of the α-glucosidase using the same method. Inhibition rate (%) = [(ODnegative control − ODblank) − (ODtest − ODtest blank)]/(ODnegative blank − ODblank) × 100%. All data were subjected to an analysis of variance using SPSS 18.0. The significant differences in inhibition rates between the treatment and blank control were calculated using one-way analysis of variance (ANOVA).
Conclusion
In this study, the extracts of the aerial parts of M. haplocalyx (M. canadensis) were firstly found to exhibit significantly inhibitory activity against α-glucosidase. Two new compounds (1 and 2) and 23 known ones were isolated and identified through bioactivity-guided fractionation. Among them, compounds 3–9 and 24 were reported from M. haplocalyx (M. canadensis) for the first time, while compounds 10–20, 22–23 and 25 were firstly isolated from the genus Mentha. Bioactivity assay further traced the active compounds (3, 4 and 11), whose inhibitory activity against α-glucosidase had not been reported before. It was noted that the monoterpene glucosides and the ionone glycosides endowed this plant with the α-glucosidase inhibitory activity.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (DOC 1878 kb). HRMS, IR, UV, 1D and 2D NMR spectra of compounds 1 and 2; Structures of the known compounds 3–25.
Acknowledgements
This work was supported by the CAS Hundred Talents Program, the Youth Innovation Promotion Association of the Chinese Academy of Sciences (CN), and Southeast Asia Biodiversity Research Institute, CAS (2017CASSEABRIQG003).
Compliance with Ethical Standards
Conflict of interests
No potential conflict of interest was reported by the authors.
References
- 1.Bhandari MR, Jong-Anurakkun N, Hong G, Kawabata J. Food Chem. 2008;106:247–252. doi: 10.1016/j.foodchem.2007.05.077. [DOI] [Google Scholar]
- 2.Wang H, Du YJ, Song HC. Food Chem. 2010;123:6–13. doi: 10.1016/j.foodchem.2010.03.088. [DOI] [Google Scholar]
- 3.Kim KY, Nam KA, Kurihara H, Kim SM. Phytochemistry. 2008;69:2820–2825. doi: 10.1016/j.phytochem.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Kim KY, Wang MH, Rhee HI. Carbohydr. Res. 2004;339:715–717. doi: 10.1016/j.carres.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Li YQ, Zhou FC, Gao F, Bian JS, Shan F. J. Agric. Food Chem. 2009;57:11463–11468. doi: 10.1021/jf903083h. [DOI] [PubMed] [Google Scholar]
- 6.Zhao BT, Kim TI, Kim YH, Kang JS, Min BS, Son JK, Woo MH. Nat. Prod. Res. 2018;32:239–242. doi: 10.1080/14786419.2017.1343325. [DOI] [PubMed] [Google Scholar]
- 7.Cao G, Shan Q, Li X, Cong X, Zhang Y, Cai H, Cai B. Analyst. 2011;136:4653–4661. doi: 10.1039/c1an15616k. [DOI] [PubMed] [Google Scholar]
- 8.She GM, Xu C, Liu B, Shi RB. Helv. Chim. Acta. 2010;93:2495–2498. doi: 10.1002/hlca.201000143. [DOI] [Google Scholar]
- 9.Abdellatief SA, Beheiry RR, Sam EM. Biomed. Pharmacother. 2017;95:990–999. doi: 10.1016/j.biopha.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 10.Barbalho SM, Machado FMVF, Oshiiwa M, Abreu M, Guiger EL, Tomazela P, Goulart RA. Food Sci. Technol. 2011;31:584–588. doi: 10.1590/S0101-20612011000300006. [DOI] [Google Scholar]
- 11.Lakshmi R, Bateman TD, McIntosh MC. J. Org. Chem. 2005;70:5313–5315. doi: 10.1021/jo050217h. [DOI] [PubMed] [Google Scholar]
- 12.Park SH, Kim KS, Suzuki Y, Kim SU. Phytochemistry. 1997;44:623–626. doi: 10.1016/S0031-9422(96)00584-5. [DOI] [Google Scholar]
- 13.Ragasa CY, Espineli DL, Shen CC. Nat. Prod. Res. 2012;26:1869–1875. doi: 10.1080/14786419.2011.619187. [DOI] [PubMed] [Google Scholar]
- 14.Yamamura S, Ozawa K, Ohtani K, Kasai R, Yamasaki K. Phytochemistry. 1998;48:131–136. doi: 10.1016/S0031-9422(97)01112-6. [DOI] [Google Scholar]
- 15.Shimizu S. J. Essent. Oil Res. 1990;2:81–86. doi: 10.1080/10412905.1990.9697828. [DOI] [Google Scholar]
- 16.Zheng J, Wu LJ, Zheng L, Wu B, Song AH. J. Asian Nat. Prod. Res. 2003;5:69–73. doi: 10.1080/1028602031000080496. [DOI] [PubMed] [Google Scholar]
- 17.Jiang LH, Kojima H, Yamada K, Kobayashi A, Kubota K. J. Agri. Food Chem. 2001;49:5888–5894. doi: 10.1021/jf0104937. [DOI] [PubMed] [Google Scholar]
- 18.Nakanishi T, Iida N, Inatomi Y, Murata H, Inada A, Murata J, Lang FA, Iinuma M, Sakagami Y. Chem. Pharm. Bull. 2005;53:783–787. doi: 10.1248/cpb.53.783. [DOI] [PubMed] [Google Scholar]
- 19.Yan LH, Xu LZ, Wang ZM, Zhang QW, Yang SL. Acta Pharm. Sin. 2010;45:1527–1532. [PubMed] [Google Scholar]
- 20.Li GQ, Jia ZJ, Zhang SS. Chem. Res. Chinese Univ. 2003;19:422–424. [Google Scholar]
- 21.Lin S, Zhang YL, Liu MT, Zi JC, Gan ML, Song WX, Fan XN, Wang SJ, Yang YC, Shi JG. China J. Chin. Mater Med. 2015;40:2602–2611. [PubMed] [Google Scholar]
- 22.Fujita T, Terato K, Nakayama M. Biosci. Biotechnol. Biochem. 1996;60:732–735. doi: 10.1271/bbb.60.732. [DOI] [Google Scholar]
- 23.Lin S, Zhang ZX, Shen HH, Li HL, Shan L, Liu RH, Xu XK, Zhang WD. China J Chin Mater Med. 2010;35:1137–1141. doi: 10.4268/cjcmm20100911. [DOI] [PubMed] [Google Scholar]
- 24.Barreiros ML, David JM, de Pereira PAP, Guedes MLS, David JP. J. Brazil. Chem. Soc. 2002;13:669–673. doi: 10.1590/S0103-50532002000500021. [DOI] [Google Scholar]
- 25.Barbosa L, Mathias FL, Braz-Filho R, Vieira IJC. J. Brazil. Chem. Soc. 2010;21:1434–1438. doi: 10.1590/S0103-50532010000800004. [DOI] [Google Scholar]
- 26.Silva MGV, Vieira ÍGP, Mendes FNP, Albuquerque IL, dos Santos RN, Silva FO, Morais SM. Molecules. 2008;13:2483–2487. doi: 10.3390/molecules13102482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang RM, Zhou XF, Peng Y, Yang XW, Xu TH, Liu YH. Chem. Nat. Compd. 2011;46:1010–1011. doi: 10.1007/s10600-011-9815-6. [DOI] [Google Scholar]
- 28.Li GQ, Zhang YB, Guan HS. Fitoterapia. 2008;79:238–239. doi: 10.1016/j.fitote.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 29.Villaseñor LIM, Sanchez AC. Z Naturforsch C: J. Biosciences. 2009;64:809–812. doi: 10.1515/znc-2009-11-1209. [DOI] [PubMed] [Google Scholar]
- 30.Ma HY, Sun Y, Zhou YZ, Hong M, Pei YH. Molecules. 2008;13:267–271. doi: 10.3390/molecules13020267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geng CA, Chen H, Chen XL, Zhang XM, Lei LG, Chen JJ, Int I. J. Mass Spectrom. 2014;361:9–22. doi: 10.1016/j.ijms.2014.01.021. [DOI] [Google Scholar]
- 32.Luo Y, Xu QL, Dong LM, Zhou ZY, Chen YC, Zhang WM, Tan JW. Phytochem. Lett. 2015;11:127–131. doi: 10.1016/j.phytol.2014.12.005. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 (DOC 1878 kb). HRMS, IR, UV, 1D and 2D NMR spectra of compounds 1 and 2; Structures of the known compounds 3–25.