Abstract
Abstract
Two new C21 steroidal glycosides, paniculatumosides H and I, together with four known ones were isolated from the roots of Cynanchum paniculatum (Bge.) Kitag. Their structures were identified by spectroscopic methods including extensive 1D and 2D NMR techniques. All compounds were subjected to detect the anti-tobacco mosaic virus (TMV) activities and their cytotoxities against three human tumor cell lines (SMMC-7721, MDA-MB-231 and A549). The results showed that compounds 1 and 5 exhibited potent protective activities against TMV, while 2, 4 and 6 had moderate effects on the SMMC-7721 cancer cells viability.
Graphical Abstract
Keywords: Cynanchum paniculatum, Steroidal glycosides, Bioactivities, NMR data
Introduction
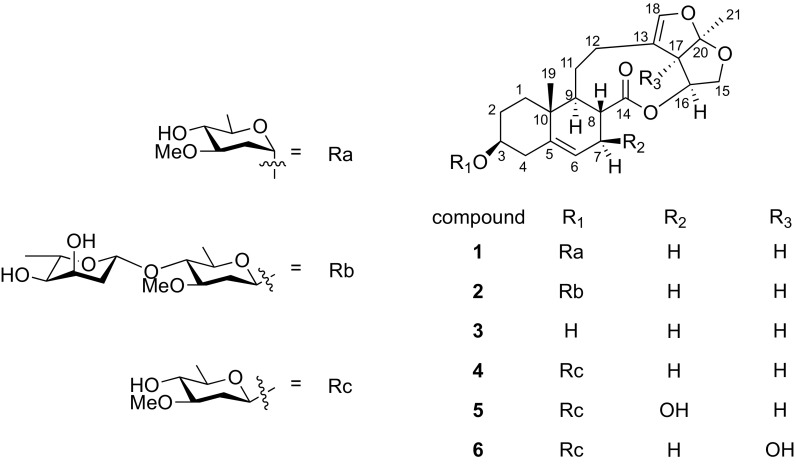
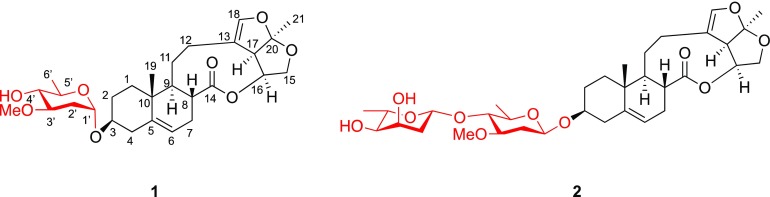
Cynanchum paniculatum (Bunge) Kitag is a vivacious herb broadly distributed in China, Japan and Korea, whose dried roots have been used as a Chinese herbal medicine for the treatment of rheumatic arthralgia, epigastric pain, toothache, lumbago, traumatic injuries, and eczema [1–3]. In the previous work, it has been confirmed that C. paniculatum contains C21 steroidal glycosides [4, 5]. Pregnanes and their glycosides have shown many aspect activities, such as antitumor, antifungal, antiviral and cytotoxic activities [6, 7]. Our previous works found that pregnanes and their glycosides could inhibit tobacco mosaic virus through suppressing the expression of viral subgenomic RNA but without affecting the accumulation of viral genomic RNA [6]. Therefore, in order to find the structurally unique natural products in C. paniculatum and explore their biological activities, we investigated the dichloromethane extract of C. paniculatum, and two new steroidal glycosides paniculatumosides H (1) and I (2), together with four known ones glaucogenin C (3) [8], cynatratoside A (4) [8], cynapanoside A (5) [9] and neocynapanogenin F 3-O-β-d-oleandropyranoside (6) [10] were isolated. Furthermore, we tested the anti-TMV activities and cytotoxicity of these compounds (Fig. 1).
Fig. 1.
Chemical structures of compounds 1–6
Result and Discussion
Structure Elucidation
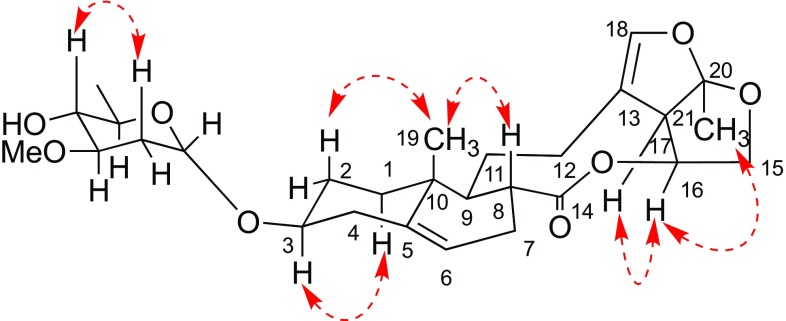
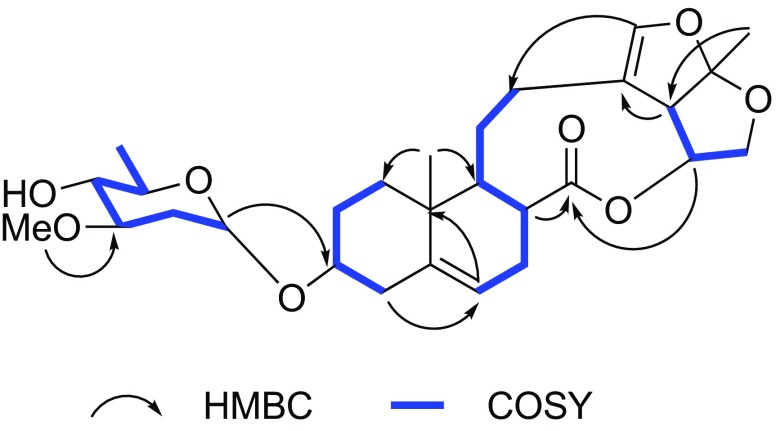
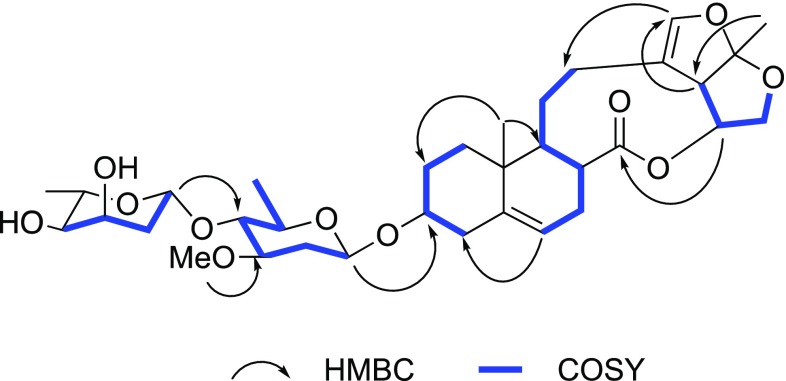
Paniculatumoside H (1) was obtained as a yellow powder. Its molecular formula, C28H40O8, was determined by analysis the peak at m/z 527.2629 [M + Na]+ in the positive HRESI-MS (calcd for C28H40O8Na 527.2615). The IR spectrum displayed absorption bands for hydroxyl (3443 cm−1), carbonyl (1736 cm−1), olefinic (1635 cm−1) and carbon–oxygen bond (1081 cm−1). The 1H NMR spectrum of compound 1 showed the presence of two tertiary methyl groups at δH 0.92 (3H, s, H-19) and δH 1.53 (3H, s, H-21), one olefinic proton at δH 5.38 (d, J = 5.3 Hz, H-6), one olefinic deshielded proton at δH 6.24 (s, H-18), two oxygen-substituted methine protons at δH 3.44 (m, H-3) and δH 5.29 (td, 9.5, 7.1, H-16) and two methylene protons at δH 3.84 (t, J = 8.6 Hz, H-15a) and δH 4.15 (td, J = 8.6, 1.4 Hz, H-15b). By comparison of its 1H and 13C NMR spectra (Table 1) to those of glaucogenin C (3) indicated that the aglycone of 1 is glaucogenin C, which also confirmed by correlations of H-19/H-2b, H-19/H-8, H-1b/H-3, H-21/H-16 and H-16/H-17 in the ROESY experiment (Fig. 2). The main difference was an extra sugar unit present at 1, which could be further verified by the glycosidation shifts at C-2 (− 3.8), C-3 (+ 4.2) and C-4 (− 2.2). The sugar carbon signals (δC 95.2, 78.2, 76.8, 67.5, 56.5, 34.5, 17.8) illustrated that the sugar belonged to 2,6-deoxy sugar because it contained four methines, one methylene, a terminal methyl group and a methoxyl group. In the sugar spin systems, the large coupling constants JH-3′/H-4′ (9.1 Hz) and JH-4′/H-5′ (9.3 Hz) disclosed their trans-diaxial relationship, which suggested that the sugar was oleandropyranose [8]. Moreover, the anomeric proton of d-oleandrose was α-orientation based on the small coupling constants of H-1′ (3JH-1,H-2 = 3.5 Hz) [11]. Furthermore, the sugar carbon and proton signals were identical with α-d-oleandropyranosyl unit by comparison with reported literature [12], and the ROESY correlations of H-2′b/H-4′ supported this configuration (Fig. 2). Thus, compound 1 could be glycosylated at the C-3 with α-linkage by d-oleandropyranosyl unit, which was also concluded from the HMBC correlation of δH 5.04 (d, J = 3.5 Hz, H-1′) to δC 75.8 (C-3) (Fig. 3). The structure of 1 was finally established as glaucogenin C 3-O-α-d-oleandropyranoside.
Table 1.
1H NMR (500 MHz) and 13C NMR (125 MHz) spectroscopic data for compounds 1, 2 and 3 in CDCl3
| No. | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δ C | δ H | δ C | δ H | δ C | δ H | |
|
1a 1b |
36.2 |
1.96 (m) 1.02 (td, 14.4, 3.8) |
36.4 |
1.96 (m) 1.05 (td, 14.6, 3.9) |
36.4 |
1.96 (m) 1.07 (td, 13.7, 3.3) |
|
2a 2b |
27.5 |
1.91 (m) 1.45 (m) |
29.4 |
1.97 (m) 1.60 (m) |
31.3 |
1.86 (m) 1.51 (m) |
| 3 | 75.8 | 3.44 (m) | 77.9 | 3.53 (m) | 71.6 | 3.53 (m) |
|
4a 4b |
39.7 |
2.30 (m) 2.24 (m) |
38.6 |
2.33 (m) 2.16 (m) |
41.9 | 2.32 (dd, 13.0, 2.5), 2.17 (dd, 13.0, 11.9) |
| 5 | 140.3 | – | 140.2 | – | 140.2 | – |
| 6 | 120.2 | 5.38 (d, 5.3) | 120.3 | 5.39 (d, 5.2) | 120.2 | 5.39 (d, 5.3) |
|
7a 7b |
28.0 |
2.43 (m) 2.05 (m) |
28.0 |
2.42 (m) 2.04 (m) |
27.9 |
2.43 (m) 2.02 (m) |
| 8 | 40.4 | 2.42 (m) | 40.4 | 2.42 (m) | 40.4 | 2.43 (m) |
| 9 | 52.8 | 1.19 (overlapped) | 52.9 | 1.19 (overlapped) | 52.8 | 1.20 (t, 9.7) |
| 10 | 38.5 | – | 38.6 | – | 38.3 | – |
|
11a 11b |
23.6 |
2.56 (m) 1.29 (m) |
23.6 |
2.56 (m) 1.28 (m) |
23.6 |
2.57 (m) 1.27 (m) |
|
12a 12b |
29.6 |
2.06 (m) 1.33 (m) |
29.6 |
2.07 (m) 1.33 (m) |
29.6 |
2.07 (m) 1.33 (m) |
| 13 | 118.1 | – | 118.1 | – | 118.1 | – |
| 14 | 175.6 | – | 175.6 | – | 175.6 | – |
|
15a 15b |
67.6 |
4.15 (td, 8.6, 1.4) 3.84 (t, 8.6) |
67.6 |
4.15 (td, 7.8, 1.6), 3.84 (t, 7.8) |
67.5 |
4.16 (td, 8.1, 1.6), 3.85 (t, 8.1) |
| 16 | 75.1 | 5.29 (td, 9.5,7.1) | 75.1 | 5.29 (td, 9.7, 7.3) | 75.0 | 5.29 (td, 9.7, 7.8) |
| 17 | 55.7 | 3.43 (overlapped) | 55.7 | 3.43 (overlapped) | 55.7 | 3.43 (d, 7.8) |
| 18 | 143.3 | 6.24 (s) | 143.3 | 6.25 (s) | 143.3 | 6.25 (s) |
| 19 | 18.0 | 0.92 (s) | 18.0 | 0.91 (s) | 18.0 | 0.91 (s) |
| 20 | 114.0 | – | 113.9 | – | 113.9 | – |
| 21 | 24.6 | 1.53 (s) | 24.6 | 1.53 (s) | 24.5 | 1.53 (s) |
| Sugar | ||||||
| 1′ | 95.2 | 5.04 (d, 3.5) | 97.8 | 4.53 (d, 9.6) | ||
|
2′a 2′b |
34.5 |
2.22 (m) 1.50 (m) |
36.5 |
2.26 (m) 1.53 (m) |
||
| 3′a | 78.2 | 3.52 (ddd, 9.1, 8.8, 4.9) | 79.1 | 3.38 (m) | ||
| 4′ | 76.8 | 3.15 (dd, 9.3, 9.1) | 82.5 | 3.19 (dd, 9.0, 8.8) | ||
| 5′ | 67.5 | 3.73 (dq, 9.3, 6.3) | 71.1 | 3.29 (m) | ||
| 6′ | 17.8 | 1.28 (d, 6.3) | 18.2 | 1.30 (d, 6.3) | ||
| 3′-OCH3 | 56.5 | 3.39 (s) | 56.4 | 3.40 (s) | ||
| 1′′ | 98.5 | 5.02 (d, 9.7) | ||||
| 2′′ | 38.2 |
2.12 (m) 1.72 (m) |
||||
| 3′′ | 68.3 | 4.11 (m) | ||||
| 4′′ | 72.9 | 3.31 (m) | ||||
| 5′′ | 69.6 | 3.74 (dq, 9.7, 6.3) | ||||
| 6′′ | 18.4 | 1.31 (d, 6.3) | ||||
Fig. 2.
The key ROESY correlations of 1
Fig. 3.
The selected HMBC and COSY correlations of 1
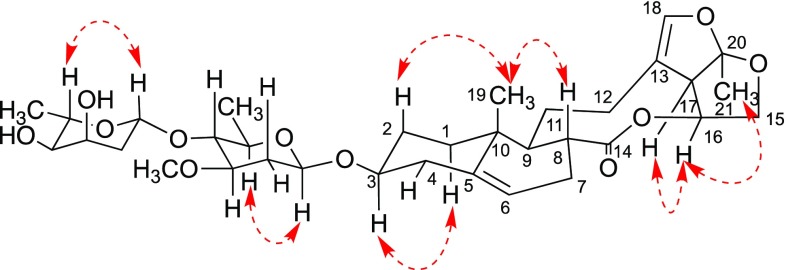
Paniculatumoside I (2) was obtained as a colourless powder. The molecular formula of 2, C34H50O11, was deduced by HRESI-MS (m/z 657.3258 [M + Na]+, calcd for C34H50O11Na 657.3245). The IR spectrum also suggested the presence of hydroxyl (3433 cm−1), carbonyl (1736 cm−1), olefinic (1632 cm−1) and carbon–oxygen bond (1070 cm−1). Compounds 2 and 1 possess the same aglycone by analyzing their 1H and 13C NMR spectra (Table 1), which was confirmed to be glaucogenin C by detailed analysis of their 2D NMR spectra. Two anomeric proton signals δH 4.53 (d, J = 9.6 Hz, H-1′) and δH 5.02 (d, J = 9.7 Hz, H-1′′), which correlated to the corresponding anomeric carbon signals at δC 97.8 (C-1′) and 98.5 (C-1′′) in the HSQC spectrum, respectively. It showed that the sugar moiety of 2 was made up of two monosaccharides. Meanwhile, from the HSQC, HSQC-TOCSY, and 1H, 1H-COSY experiments, the spin system of each monosaccharide could be established. Two methylenes (δC 36.5 and 38.2) and methyl groups (δH 1.30 and 1.31) suggested both monosaccharides of 2 were deoxysugars. The splitting patterns and the coupling constants of H-1′ (3JH-1,H-2 = 9.6 Hz) and H-1″ (3JH-1,H-2 = 9.7 Hz) illustrated that the anomeric protons of the two sugar were β-oriented, which was also proved by the ROESY collations between δH 4.53 (d, J = 9.6 Hz, H-1′) and δH 3.29 (m, H-5′), δH 5.02 (d, J = 9.7 Hz, H-1′′) and δH 3.74 (dq, H-5′′) (Fig. 4). The oligosaccharide moiety was identified as β-d-oleandropyranoside and β-l-digitoxopytanoside by analyzing from comparing the 1H and 13C signals of 2 with those in the literatures [6, 13]. Compound 2 was glycosylated at the C-3, which was concluded from the HMBC correlation of δH 4.53 (H-1′) to δC 77.9 (C-3), and the HMBC correlation of δH 5.02 (H-1′′) of δC 82.5 (C-4′) suggested digitoxopytanose connected to C-4′ of oleandropyranose (Fig. 5). Therefore, compound 2 was established to be glaucogenin C 3-O-β-l-digitoxopytanosyl-(1 → 4)-β-d-oleandropyranoside.
Fig. 4.
The key ROESY correlations of 2
Fig. 5.
The selected HMBC and COSY correlations of 2
Bioactivities
Anti-TMV Activities
All compounds were tested for anti-TMV activity using the half-leaf method. The results revealed that compounds 1 and 5 exhibited protective activities at concentration of 200 μM, with the antiviral inhibition rates of 62.7% and 59.5%, respectively. Ningnanmycin, which was used as a positive control, showed inhibition rates of 57.3%.
Cytotoxic Activities
In order to evaluate whether these compounds have any biological functions on cancer cells, we tested them on SMMC-7721, MDA-MB-231 and A549 human cancer cell lines for their impact on tumor cell growth by MTT method. All the compounds have no inhibitory effect on the MDA-MB-231 and A549 cell lines. But compounds 2, 4 and 6 have moderate effects on the SMMC-7721 cancer cells viability with the IC50 values of 27.4 ± 0.24 μM, 22.2 ± 0.11 μM, 27.2 ± 0.13 μM, respectively. Cisplatin was used as a positive control and its IC50 for the SMMC-7721 cancer cells was 13.07 ± 0.27 μM.
Experimental
General Experimental Procedures
UV spectra were measured with a Shimadzu UV-2401A spectrophotometer. Optical rotations were determined on a Jasco P-1020 polarimeter. Infrared spectroscopy (IR) spectra were measured on a Bio-Rad FTS-135 spectrometer with KBr pellets. HRESIMS data were collected on a triple quadrupole mass spectrometer. 1D and 2D NMR spectra were recorded on a Bruker spectrometer with tetramethylsilane as the internal standard. Preparative HPLC separations were carried out using an Agilent 1200 liquid chromatograph with a Waters X-select CSH Prep RP C18 (19 × 150 mm) column and the flowing rate is 8 mL/min. Semipreparation HPLC separations were performed on an Agilent 1100 liquid chromatograph using a YMC-Pack CDS-A (10 × 250 mm) column with flowing rate of 3 mL/min. Sephadex LH-20 (40–70 mm, Amersham Pharmacia Biotech AB, Uppsala, Sweden) and Silica gel (100–200 mesh and 300–400 mesh, Qingdao Marine Chemical, Inc., Qingdao, P. R. China) were used for column chromatography.
Plant Material
The roots of C. paniculatum were purchased from a medicinal market (Kunming luosiwan Chinese herbal medicine market) in August 2017 and identified by prof. Hua Peng of Kumming Institute of Botany, Chinese Acdemy of Sciences (CAS).
Extraction and Isolation
The roots of C. paniculatum (100.0 kg) were powdered and extracted three times with MeOH at room temperature to afford 7.2 kg of crude extract. The extract was partitioned between CH2Cl2 and aqueous solution portions which yielded 4.4 kg crude CH2Cl2 extract. This extract was subjected to normal-phase silica gel column chromatography eluted with a gradient of petroleum ether–acetone (from 1:0 to 1:2) and CH2Cl2–MeOH (10:1–0:1) to obtain eight major fractions (Fr.1-8). Fr.6 (182.3 g) was separated by reversed-phase separation (CH3OH–H2O, 4:6–9:1) to get twelve subfractions (Fr.6a-6l). Fr.6g (2.1 g) was purified by Sephadex LH-20 eluting with MeOH to yield three fractions. Fr.6g-2 (161.4 mg) was applied to a silica gel column using CH2Cl2–MeOH (200:1–0:1) to obtain 3 (7.1 mg). Fr.6g-3 (92.5 mg) was subject to a normal-phase column chromatography and further purified by semiprepative HPLC (68% CH3CN in water) to yield 6 (5.0 mg, tR = 15.0 min). Fr6I (4.3 g) was separated into four subfractions by Sephadex LH-20 eluting with CH2Cl2–MeOH (1:1). Fr.6I-2 (1.9 g) was chromatographed on a silica gel column eluting with CH2Cl2–MeOH (200:1–0:1) to get Fr.6I-2c (109.2 mg) and Fr.6I-2e (79.3 mg). Fr6I-2c was purified by semiprepative HPLC (50% CH3CN in water) to obtain 1 (5 mg, tR = 27.5 min) and 4 (92.0 mg, tR = 29.0 min). Fr.6I-2d also used semiprepative HPLC (46% CH3CN in water) to yield 5 (10.2 mg, tR = 35.0 min). Fr.6 k (3.6 g) was fractioned by a silica gel column eluting with petroleum ether-acetone (80:1–0:1) to afford the Fr6 k-3 (39.3 mg), which was purified by Sephadex LH-20 (MeOH) and semiprepative HPLC (49% CH3CN in water) in sequence to yield 2 (8.2 mg, tR = 31.5 min).
Paniculatumoside H (1)
White amorphous powder; [α]20D−21.5 (c 0.11, MeOH); IR (KBr) νmax 3443 (OH), 2919, 1736, 1635, 1384, 1081 cm−1; 1H and 13C NMR data, see Table 1; ESIMS m/z 527 [M + Na]+; HRESIMS m/z 527.2629 [M + Na]+ (calcd for C28H40O8Na, 527.2615).
Paniculatumoside I (2)
White amorphous powder; [α]20D−52.0 (c 0.09, MeOH); IR (KBr) νmax 3433 (OH), 2922, 1736, 1632, 1383, 1070 cm−1; 1H and 13C NMR data, see Table 1; ESIMS m/z 657 [M + Na]+; HRESIMS m/z 657.3258 [M + Na]+ (calcd for C34H50O11Na, 657.3245).
Biological Activity Assays
Anti-TMV Assays
The half-leaf method was used to evaluate the anti-TMV activities as literatures reported [14]. Ningnanmycin, a Chinese commercial product for plant disease, obtained from Heilongjiang Qiang’er Biochemical Technology Development Company, was administered as a positive control.
Cytotoxicity Assay
The cytotoxicity of each compound on three cultured human cancer cell lines was tested by MTT assay. The cell lines used were SMMC-7721 (human hepatoma cells), MDA-MB-231 (triple-negative breast cancer cells), A549 (human lung cancer cell). Cell growth inhibition assay was performed as reported literatures [15]. Cisplatin was used as a positive control.
Acknowledgements
This project was supported financially by grants from the National Natural Science Foundation of China (Nos. 31770389, 81703393). We thank Professor Hua Peng, Kunming Institute of Botany (KIB), CAS, for identifying the plant material; Analysis and Test Center, KIB, CAS, for the technical support.
Compliance with Ethical Standards
Conflicts of interest
The authors declare that they have no conflict of interest.
Contributor Information
Xiao Ding, Phone: +86-18487336458, Email: dingxiao@mail.kib.ac.cn.
Shun-Lin Li, Phone: +86-871-65223263, Email: lisl@mail.kib.ac.cn.
References
- 1.Zhao D, Feng B, Chen S, Chen G, Li Z, Lu X, Sang X, An X, Wang H, Pei Y. Fitoterapia. 2016;113:51–57. doi: 10.1016/j.fitote.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Jiang SP. Chin J. Chin. Mater. Med. 1994;5:311–314. [PubMed] [Google Scholar]
- 3.Dou J, Li P, Bi ZM, Zhou JL. Chin. Chem. Lett. 2007;18:300–302. doi: 10.1016/j.cclet.2007.01.013. [DOI] [Google Scholar]
- 4.Mitsuhashi H, Hayashi K, Nomura T. Chem. Pharm. Bull. 1966;14:779–783. doi: 10.1248/cpb.14.779. [DOI] [PubMed] [Google Scholar]
- 5.Sugama KO, Hayashi K, Mitsuhashi H, Kaneko KO. Chem. Pharm. Bull. 1986;34:4500–4507. doi: 10.1248/cpb.34.4500. [DOI] [PubMed] [Google Scholar]
- 6.Yan Y, Zhang JX, Liu KX, Huang T, Yan C, Huang LJ, Liu S, Mu SZ, Hao XJ. Fitoterapia. 2014;97:50–63. doi: 10.1016/j.fitote.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 7.Liu H, Xiong Z, Li F, Qu G, Kobayashi H, Yao X. Chem. Pharm. Bull. 2003;51:1089–1091. doi: 10.1248/cpb.51.1089. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Z, Zhou J, Hayashi K, Mitsuhashi H. Chem. Pharm. Bull. 1985;33:1507–1514. doi: 10.1248/cpb.33.1507. [DOI] [PubMed] [Google Scholar]
- 9.Sugama K, Hayashi K, Mitsuhashi H, Kaneko K. Chem. Pharm. Bull. 1986;34:4500–4507. doi: 10.1248/cpb.34.4500. [DOI] [PubMed] [Google Scholar]
- 10.Dou J, Bi ZM, Zhang YQ, Li P. Chin. J. Nat. Med. 2006;4:192–194. [Google Scholar]
- 11.Simpson JH. Organic Structure Determination Using 2-D NMR Spectroscopy. 2. Cambridge: Academic Press; 2011. p. 111. [Google Scholar]
- 12.Huang X, Tan AM, Yang SB, Zhang AY, Zhang H. Helv. Chim. Acta. 2009;92:937–943. doi: 10.1002/hlca.200800314. [DOI] [Google Scholar]
- 13.Gui C, Zhang S, Zhu X, Ding W, Huang H, Gu YC, Duan Y, Ju J. J. Nat. Prod. 2017;80:1594–1603. doi: 10.1021/acs.jnatprod.7b00176. [DOI] [PubMed] [Google Scholar]
- 14.Hu ZX, Zhao LH, Tang HY, Aisa HA, Zhang Y, Hao XJ. Fitoterapia. 2018;128:73–78. doi: 10.1016/j.fitote.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Yan C, Zhang WQ, Sun M, Liang W, Wang TY, Zhang YD, Ding X. Tetrahedron Lett. 2018;59:4063–4066. doi: 10.1016/j.tetlet.2018.09.067. [DOI] [Google Scholar]