Abstract
Background
The MADS-box transcription factors mainly function in floral organ organogenesis and identity specification. Few research on their roles in vegetative growth has been reported.
Results
Here we investigated the functions of OsMADS57 in plant vegetative growth in rice (Oryza sativa). Knockdown of OsMADS57 reduced the plant height, internode elongation and panicle exsertion in rice plants. Further study showed that the cell length was remarkably reduced in the uppermost internode in OsMADS57 knockdown plants at maturity. Moreover, OsMADS57 knockdown plants were more sensitive to gibberellic acid (GA3), and contained less bioactive GA3 than wild-type plants, which implied that OsMADS57 is involved in gibberellin (GA) pathway. Expectedly, the transcript levels of OsGA2ox3, encoding GAs deactivated enzyme, were significantly enhanced in OsMADS57 knockdown plants. The level of EUI1 transcripts involved in GA deactivation was also increased in OsMADS57 knockdown plants. More importantly, dual-luciferase reporter assay and electrophoretic mobility shift assay showed that OsMADS57 directly regulates the transcription of OsGA2ox3 as well as EUI1 through binding to the CArG-box motifs in their promoter regions. In addition, OsMADS57 also modulated the expression of multiple genes involved in GA metabolism or GA signaling pathway, indicating the key and complex regulatory role of OsMADS57 in GA pathway in rice.
Conclusions
These results indicated that OsMADS57 acts as an important transcriptional regulator that regulates stem elongation and panicle exsertion in rice via GA-mediated regulatory pathway.
Electronic supplementary material
The online version of this article (10.1186/s12284-019-0298-6) contains supplementary material, which is available to authorized users.
Keywords: OsMADS57, Plant height, GA catabolism, Panicle exsertion, Rice
Background
Plant height, determined by the number of elongated internodes as well as the length of internodes, is an important agronomic trait that directly affects yield potential (Yang and Hwa 2008). In “Green Revolution”, high-yielding cultivars have been created that have suitable height in rice and wheat (Khush 2001; Thomas et al. 2005). Furthermore, both the improved lodging resistance and increased total biomass result from their short stature (Sakamoto and Matsuoka 2004). Therefore, it is of great significance to clarify the signal network regulating plant height.
Gibberellin (GA) is an important phytohormone in the regulation of seed germination, flowering, leaf shape and stem elongation (Richards et al. 2001; Swain and Singh 2005). Generally, plants with a dwarf or semi-dwarf stature are deficient in GA accumulation. In the GA biosynthetic pathway, geranylgeranyl diphosphate is converted to ent-kaurene, which is catalyzed by ent-copalyl diphosphate synthase (CPS) and ent-kaurene synthase (KS) (Helmut Aach et al. 1997). With the catalysis of two cytochrome P450 enzymes, ent-kaurene oxidase (KO) and ent-kaurenoic acid oxidase (KAO), ent-kaurene is converted to GA12 (Helliwell et al. 2001). Then GA12 is converted to the bioactive GA1 and GA4 form through the 13-hydroxylation and non-13-hydroxylation pathway, respectively (Yamaguchi 2008). CYP714B1 and CYP714B2, two cytochrome P450 genes, encode GA 13-oxidase that is responsible for converting GA12 to GA53, thus determining the ratio of GA1 and GA4 in rice (Magome et al. 2013). In GA homeostasis, GA intermediates can be converted into bioactive forms by GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox), while GA 2-oxidase (GA2ox) has the opposite effect (Yamaguchi 2008; Hedden and Phillips 2000). ELONGATED UPPERMOST INTERNODE1 (EUI1) encodes a putative cytochrome P450 monooxygenase that deactivates GAs through GA 16α, 17-epoxidation (Zhu et al. 2006; Luo et al. 2006; Hedden and Thomas 2012).
GA governing plant growth requires signal transduction (Xu et al. 2014). GIBBERELLIN INSENSITIVE DWARF1 (GID1) encodes a soluble receptor for perceiving GA (Ueguchi-Tanaka et al. 2005). Binding of GA to GID1 causes GID1 conformation change, stimulating the interaction of GID1 and SLENDER1 (SLR1), a DELLA family protein acting as the GA signaling repressor (Tong et al. 2014; Hedden and Sponsel 2015). The GA-GID1-DELLA complex is recognized and polyubiquitylated by E3 ubiquitin ligase complex, and subsequently degraded through the 26S proteasome pathway, thereby stimulating GA response (Daviere and Achard 2016; Feng et al. 2008; Fleet and Sun 2005). Mutations in the components involved in GA metabolism as well as GA signaling pathway significantly affect plant growth (Thomas et al. 2005). d18 and sd1 are loss-of-function mutants that result from mutations in OsGA3ox2 and OsGA20ox2, respectively, and exhibit remarkable dwarf or semi-dwarf phenotype (Sakamoto et al. 2004). In addition, overexpression of the EUI1 greatly reduces plant height (Zhang et al. 2008). Accordingly, rice slender-type mutant (slr1) contains a loss-of-function mutation in the SLR1 gene, and thereby shows a significant increase in plant height (Ikeda et al. 2001).
A number of transcription factors are involved in the development of plant height through regulating GA metabolism. In rice, OsNAC2 negatively regulates plant height by suppressing the expression of KO2 that is involved in GA biosynthesis (Chen et al. 2015). The YABBY family members such as OsYABBY1 and OsYABBY4 can reduce bioactive GA levels by repressing the expression of OsGA3ox2 and OsGA20ox2, respectively, thus reducing plant height. OsWOX3A directly represses the transcription of KAO that encodes GA biosynthetic enzyme. Thus overexpression of OsWOX3A leads to severe dwarfism in rice plants (Cho et al. 2016). Homeodomain-leucine zipper (HD-ZIP) transcription factor HOX12 activates the expression of EUI1 by binding to its promoter sequence, thus reducing plant height and panicle exsertion (Gao et al. 2016); overexpression of rice HOMEOBOX4 (Oshox4) also leads to a semi-dwarf phenotype by inhibiting cell elongation in stems (Dai et al. 2008; Zhou et al. 2014). Additionally, OsEATB, an APETALA2 (AP2)/Ethylene-Responsive Element Binding Factor (ERF), can reduce rice plant height by down-regulating the transcription of OsCPS2 that encodes GA biosynthetic enzyme (Qi et al. 2011). These transcription factors form a regulatory system to participate in the development of plant height.
MADS-box proteins are an important class of transcription factors that can bind to CArG motifs in the promoter regions of the target genes (Tang and Perry 2003; Huang et al. 1996), which has a vital significance in plant growth and development. In Arabidopsis thaliana, AGL15 (AGAMOUS-Like 15) increases production of somatic embryos via activating the expression of AtGA2ox6 (Wang et al. 2004). In rice, suppression of short vegetative phase (SVP)-group MADS-box genes such as OsMADS22, OsMADS47 and OsMADS55 that play the negative regulatory role in BR signaling, results in increased lamina joint inclination (Lee et al. 2008; Duan et al. 2006). OsMADS3 is involved in regulating flower meristem determinacy and pistil development (Yasui et al. 2017). In addition, OsMADS6 regulates flower morphogenesis by activating the expression of FACTOR OF DNA METHYLATION LIKE 1 (OsFDML1) that is involved in maintaining normal floral organ identity (Tao et al. 2018). OsMADS57 mainly expresses in leaves (Puig et al. 2013), and the physical interaction between OsTB1 and OsMADS57 can alleviate the transcriptional inhibition of OsMADS57 on D14, thereby inhibiting the production of axillary buds and reducing the number of tillers (Yao et al. 2016; Arite et al. 2009; Guo et al. 2013a). In addition, only under the cold stress, OsMADS57 can activate the expression of OsWRKY94 by directly binding to the promoter region of OsWRKY94 to enhance the chilling tolerance of rice plants (Chen et al. 2018). Although MADS-box proteins play a variety of functions in plant growth and development, little is known about the function of OsMADS57 regulating plant height in rice. Here, we found osmads57 mutants show a semi-dwarf phenotype that can be rescued by application of GA3. Further research demonstrated that OsMADS57 binds to the promoter regions of OsGA2ox3 as well as EUI1, respectively, thus inhibiting their expression. These results show that OsMADS57 affects plant growth by regulating gene expression of GA metabolism, which reveals a molecular mechanism that OsMADS57 controls plant height in rice.
Results
osmads57 mutants display a semi-dwarf phenotype
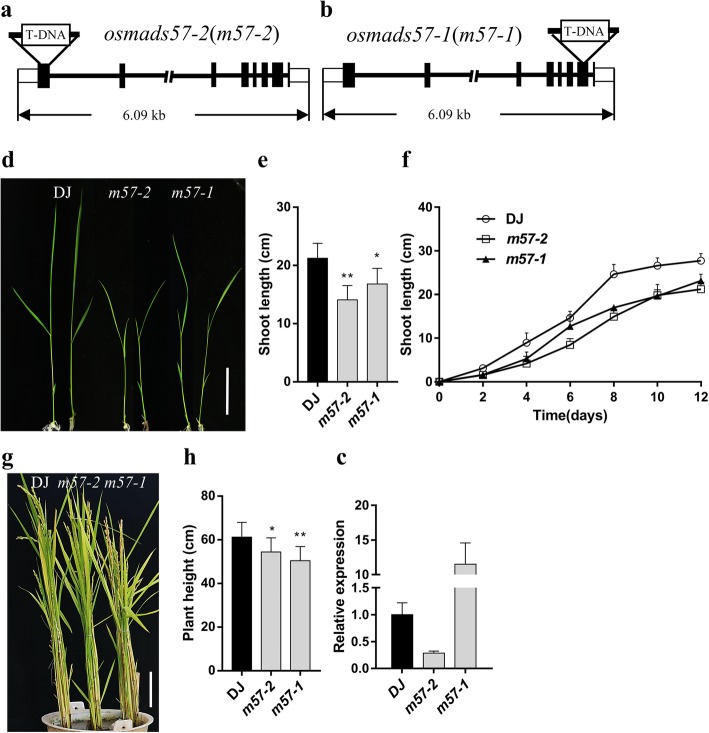
The rice gain-of-function mutant osmads57–1 (m57–1) shows increased tiller numbers and improved chilling tolerance (Guo et al. 2013a; Chen et al. 2018), whereas the loss-of-function mutant osmads57–2 (m57–2) exhibits weakened chilling tolerance (Chen et al. 2018). The m57–2 line is a knockdown mutant in which the T-DNA was inserted in the 5′ terminus of OsMADS57, + 61 from ATG (Fig. 1a, Additional file 1: Figure S1a-c). In m57–1 line, the T-DNA was inserted in 3′ terminus of OsMADS57 (Fig. 1b), forming truncated protein (Guo et al. 2013a). Further, quantitative PCR (Q-PCR) analysis exhibited the transcript levels of OsMADS57 were significantly reduced in m57–2, whereas remarkably increased in m57–1 (Fig. 1c). Intriguingly, both m57–2 and m57–1 showed a semi-dwarf phenotype in rice plants at seedling stage as well as at maturity (Fig. 1d and g). Compared to that in wild type, the shoot length was significantly reduced in the mutant seedlings, with 34% less in m57–2 and 21% less in m57–1 than wild type, respectively (Fig. 1d-f). Expectedly, the plant height of m57–2 and m57–1 was also significantly reduced compared to that of wild type at maturity, with 12% less in m57–2 and 18% less in m57–1 than wild type, respectively (Fig. 1g and h). At maturity, m57–1 plants exhibited more tillers (Fig. 1g), which was consistent with the previous report (Guo et al. 2013a). GA is also involved in flowering and seed germination (Guo et al. 2013b; Olszewski et al. 2002). We found that m57–2 plants showed the phenotype of late flowering (Additional file 1: Figure S1d and e). In addition, m57–2 also exhibited delayed seed germination compared to that of wild type, although almost all seeds germinated on day 5 (Additional file 1: Figure S1f). These investigations indicate that abnormal expression of OsMADS57 affects plant height, which might be caused by GA deficiency and/or defect in GA response.
Fig. 1.
Phenotypes of the m57–2 and m57–1 mutants. a and b Schematic diagram indicating the T–DNA insertion site in genomic region in m57–2 (a) and m57–1 (b). c Transcript levels of OsMADS57 in mutants and wild type lines by Q–PCR analysis. d The 10-day-old wild type and osmads57 plants grown in standard 1/2 MS medium. Bars = 5 cm. e Shoot length of 10-day-old wild type and osmads57 seedlings. f Shoot length of wild type and osmads57 plants grown in standard 1/2 MS medium for different days. g Morphology of wild type and osmads57 plants at maturity. Bars = 10 cm. h Plant height of wild type and osmads57 plants at maturity. DJ, wild type; m57–2, osmads57–2; m57–1, osmads57–1. Three independent experiments were conducted with similar results. The data are means± SD (n = 30). Error bars indicate SD. The statistical significance of the measurements was determined by Student’s t-test. Asterisks indicate the significant difference between osmads57 and wild type (t–test, * P < 0.05, ** P < 0.01 or *** P < 0.001)
Abnormal expression of OsMADS57 affects internode cell length
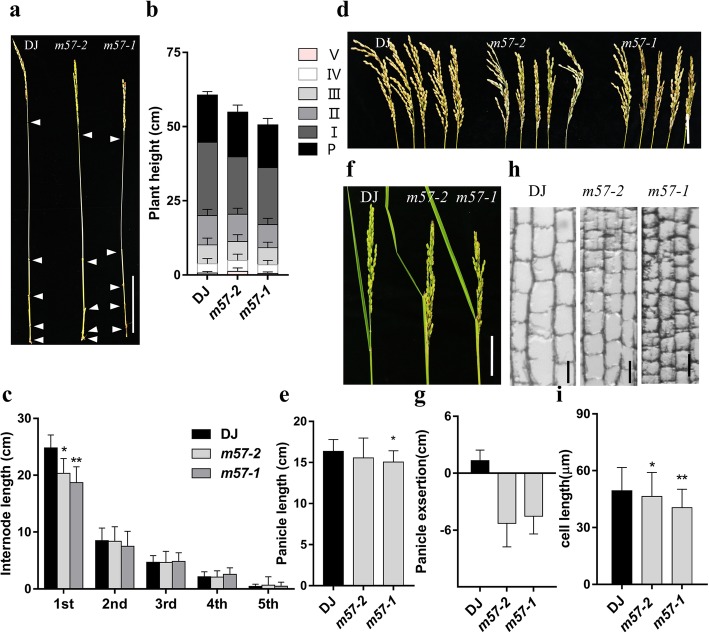
The plant height is determined by the length of each internode and the total number of internodes (Wang and Li 2005; Yang and Hwa 2008). Then we compared the internode length of mutants with that of wild-type plants. Statistical analysis showed that the uppermost internode length of m57–2 and m57–1 was drastically reduced compared to that of the wild type (Fig. 2a-c). We also found that the panicle length of m57–1 was greatly reduced than that in wild type, but the panicle length of m57–2 was not significantly altered (Fig. 2d and e). Thus, our results indicate that the semi-dwarf phenotype of two mutants is caused by the reduced internode length. In addition, there was no great significance in the flag leaf length between mutants and wild type (Additional file 2: Figure S2a and b). It is notable that, different from panicles completely out of the leaf sheath in wild type, panicles of mutants were only partly out of the leaf sheath and even m57–2 plants displayed severer panicle enclosure phenotype than m57–1 plants at heading stage (Fig. 2f and g). In addition, we found the seed setting rate in m57–2 and m57–1 was also reduced (Additional file 2: Figure S2c), with many shrunken grains (Additional file 2: Figure S2d). Nevertheless, the grain size between mutants and wild type was not significantly different (Additional file 2: Figure S2d). Together, our data show that the mutants are defective in the elongation of the uppermost internode. Internode length is mostly influenced by the cell length and cell number (Dai et al. 2007; Gao et al. 2016). To explore whether the reduction of internode length in mutant lines is caused by abnormal cell elongation or/and cell proliferation, we compared the longitudinal sections of the elongation zone in the uppermost internode of wild type, m57–2 and m57–1 plants. Microscopic observation revealed that the cell length in the uppermost internode of m57–2 and m57–1 significantly reduced compared to that of the wild type (Fig. 2h and i), respectively. These data indicate that the abnormal expression of OsMADS57 impairs the cell elongation of the uppermost internode.
Fig. 2.
Characterization of osmads57 plants. a Plant height of wild type and mutants. Bars = 10 cm. b Plant height and the length of its components of wild type and mutants. P, panicle; I, the uppermost internode; II, III, IV, the second, third, and fourth internodes counted from the uppermost internode, respectively. c Individual internode lengths of wild type and mutants. d The phenotype of panicles. Bars = 5 cm. e Panicle length of wild type and mutants at maturity. f Panicle exsertion of wild type and mutants. Bars = 2 cm. g Measurement of panicle exsertion of wild type and mutants. h Longitudinal sections of the uppermost internodes of wild type and mutants. Bars = 50 μm. i Quantitation of cell length of the uppermost internode of wild type and mutants. DJ, wild type; m57–2, osmads57–2; m57–1, osmads57–1. Three independent experiments were conducted with similar results. The data are means± SD (n = 15). Error bars indicate SD. The statistical significance of the measurements was determined by Student’s t-test. Asterisks indicate the significant difference between osmads57 and wild type (t–test, * P < 0.05, ** P < 0.01 or *** P < 0.001)
Exogenous GA eliminates the semi-dwarf stature of osmads57 mutants
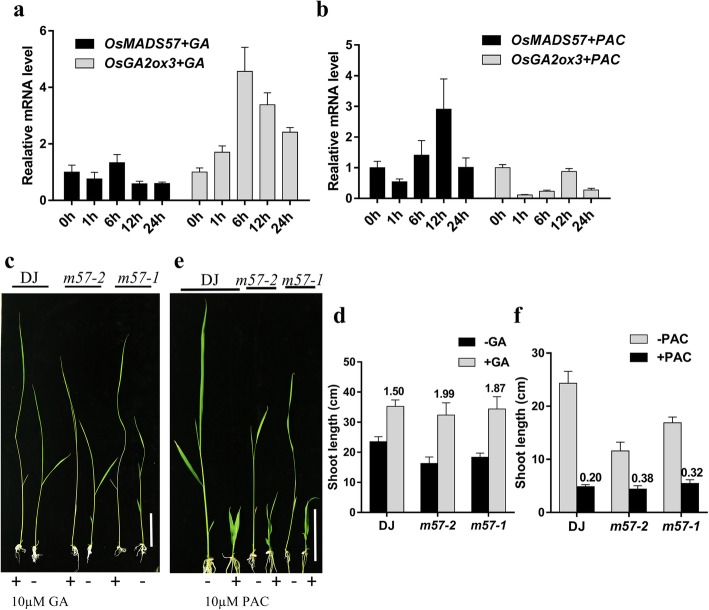
Phytohormones play an important role in regulating the growth and development of plants. GA, one of the important plant hormones, generally regulates cell elongation and determines plant height (Thomas et al. 2005). To explore whether GA modulates the expression of OsMADS57, the wild-type plants were treated with GA3 and paclobutrazol (PAC), a GA biosynthesis inhibitor, for transcriptional analysis. OsGA2ox3, up-regulated by GA3 and down-regulated by PAC (Boden et al. 2014; Sakai et al. 2003; Puig et al. 2013; Magome et al. 2013), was used as a positive control for the experiment. Q-PCR showed that the expression of OsMADS57 was suppressed by GA3, whereas up-regulated by PAC (Fig. 3a and b). GA, acting as a signal molecule, has a major function in signal transduction, and the malfunction in biosynthesis or signaling pathway of GA can result in typical dwarf or semi-dwarf phenotype (Thomas et al. 2005). To investigate whether the semi-dwarf phenotype of mutants is caused by GA deficiency or defects in GA signaling, we investigated the response of m57–2 and m57–1 to exogenous GA3. The application of exogenous GA greatly promoted the growth of two mutant seedlings, which exhibited a slender phenotype, and the shoot length of m57–2 and m57–1 was fully rescued compared to that of wild type without GA application (Fig. 3c), although the shoot length of two mutant plants was slightly shorter than that of wild-type plants under GA treatment (Additional file 3: Figure S3a). For comparison, we calculated the elongation ratio of seedlings under 10 μМ GA3 treatment compared with the untreated plants. Both m57–2 and m57–1 had the elongation ratio of 1.99 and 1.87, respectively. However, the ratio for the wild type was 1.50, which was lower than that of the two mutants (Fig. 3d). Thus m57–2 and m57–1 mutants were more sensitive to GA3 than wild type. Furthermore, we also treated seedlings with PAC. We found that the growth of mutants and wild type was all inhibited, with thick root and wide leaf phenotypes (Fig. 3e, Additional file 3: Figure S3b). The elongation ratio of seedlings under 10 μМ PAC treatment compared with the untreated plants was calculated. We found m57–2 and m57–1 had the higher elongation ratio compared with wild type, with the ratio of 0.38:0.20 and 0.32:0.20 (Fig. 3f), respectively. Together, these data indicate that both m57–2 and m57–1 have enhanced GA3 sensitivity and reduced PAC sensitivity, which implies that GA signaling pathway in osmads57 mutant plants is not impaired.
Fig. 3.
Response of m57–2 and m57–1 mutants to GA and PAC. a and b 50 μМ GA3 (a) or 10 μМ paclobutrazol (b) treatment suppressed and enhanced the expression of OsMADS57, respectively. OsGA2ox3 was used as a positive control. c Semi-dwarf phenotype of m57–2 and m57–1 mutants can be rescued by GA3. The mutants were treated with exogenous 10 μМ GA3 for 10 d. Bars = 5 cm. d Shoot length of seedlings before and after 10 μМ GA3 treatment for 10 d. e Seedling morphology of mutants and wild-type plants grown with or without 10 μМ PAC for 10 d. Bars = 5 cm. f Shoot length of seedlings before and after 10 μМ PAC treatment for 10 d. Numbers above the bars indicate the elongation ratio of seedlings after treatment (the shoot length after treatment divided by the shoot length before treatment). DJ, wild type; m57–2, osmads57–2; m57–1, osmads57–1. Three independent experiments were conducted with similar results. The data are means± SD (n = 15). Error bars indicate SD
OsMADS57 alters the transcription of genes involved in GA metabolism
To determine whether the inhibition of internode elongation in m57–2 and m57–1 plants is caused by reduced contents of bioactive GAs, we firstly investigated the expression of genes encoding GA metabolic enzymes by Q-PCR analysis. We found that the transcript levels of genes involved in GA biosynthesis such as OsGA20ox1, OsGA20ox2, CPS1 and KO2 were down-regulated in m57–2 and m57–1 plants compared with that in wild type (Fig. 4a). Next we compared the expression of genes encoding GA catabolic enzymes in mutants and wild type (Fig. 4b). We found that both m57–2 and m57–1 exhibited reduced transcription levels of OsGA2ox1 and OsGA2ox6. In contrast, the mRNA levels of OsGA2ox3 as well as EUI1 were increased in m57–2 plants but reduced in m57–1 plants.
Fig. 4.
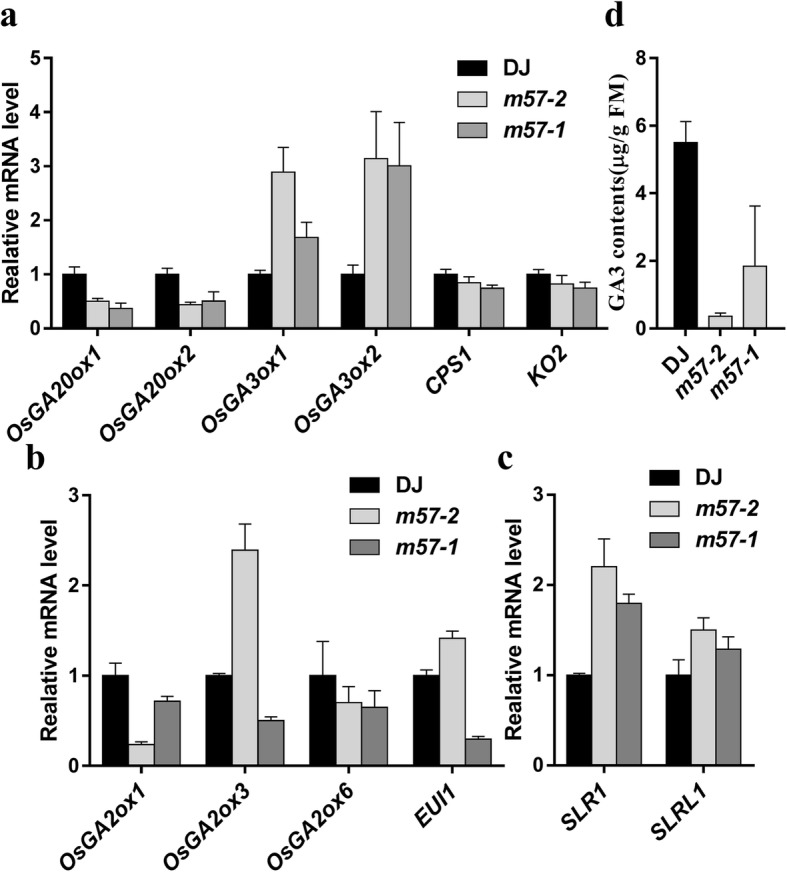
OsMADS57 regulate the expression of GA metabolic genes. a-c Altered expression of genes involved in GA metabolism (a and b) and signaling (c). Expression of genes involved in GA metabolism and signaling was analysed by Q-PCR in wild type and mutants lines. The PCR signals were normalized with that from actin1 transcripts. Transcript levels from wild type were set at 1. d Quantification of GA3 in four-leaf stage seedlings. DJ, wild type; m57–2, osmads57–2; m57–1, osmads57–1. Three independent experiments were conducted with similar results. Data are means ± SE (n = 10). Error bars indicate SE
The response of plants to GA is not only determined by GA accumulation, but by GA signaling (Ueguchi-Tanaka et al. 2005; Sakamoto et al. 2004). Altered the expression of genes involved in GA signaling pathway also affects the GA response (Ikeda et al. 2001). SLR1 and SLRL1, as the central suppressors of GA signaling pathway, play an important role in GA response (Fukao and Bailey-Serres 2008). Overexpression of SLRL1, homologous to SLR1, induces a dwarf phenotype in rice (Itoh et al. 2005). Thus, we examined the transcript levels of SLR1 and SLRL1 in wild type, m57–2 and m57–1. Intriguingly, the results showed that the transcription of SLR1 and SLRL1 was up-regulated in two mutant plants (Fig. 4c). We speculated that the down-regulated expression of genes involved in GA biosynthesis led to reduced accumulation in GA levels in m57–2 and m57–1. To confirm this hypothesis, endogenous levels of bioactive GA were measured. We found that the GA3 content in m57–1 plants decreased by more than half compared with that of the wild type, while the GA3 content in m57–2 plants decreased more severely (Fig. 4d). Taken together, the above data show that defect in the stem elongation in m57–2 and m57–1 is caused by the reduced GA levels.
OsMADS57 regulates the transcription of OsGA2ox3 and EUI1 by interacting with their promoters
DELLA protein is the repressor of GA signaling, and inhibition of the function of DELLA protein triggers GA responses (Itoh 2002; Hirano et al. 2012; Ikeda et al. 2001). DELLA, lacking a DNA-binding domain, regulates downstream genes via interacting with other proteins (Yoshida et al. 2014; Fukazawa et al. 2014; Chen et al. 2017). To investigate whether OsMADS57 interacts with SLR1, we conducted yeast two-hybrid assay. We found that there was no direct physical interaction between OsMADS57 and SLR1 (Additional file 4: Figure S4).
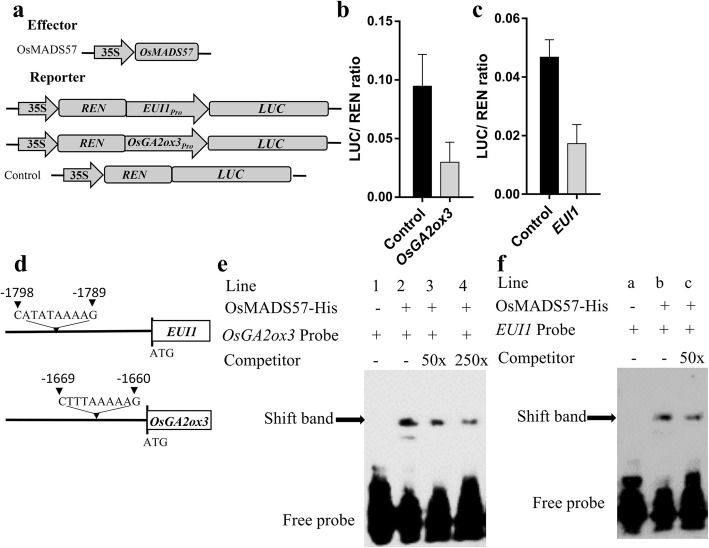
It was shown that the eui1 mutant shows the elongated uppermost internodes and elevated levels of bioactive GA compared with wild type (Luo et al. 2006; Zhang et al. 2008; Zhu et al. 2006); while overexpression of EUI1 caused extreme dwarfism and reduced the content of bioactive GA (Hedden and Thomas 2012). Q-PCR analysis showed that the transcript levels of OsGA2ox3 as well as EUI1 were increased in m57–2 plants but reduced in m57–1 plants (Fig. 4b). As a transcription factor, OsMADS57 binds to CArG-box motifs in the promoter regions to regulate the transcription of target genes (Chen et al. 2018; Wang et al. 2004). Thus, to investigate whether OsMADS57 regulates the transcription of OsGA2ox3 and EUI1, we performed a dual-luciferase reporter assay. The promoter sequence containing the CArG motif was constructed into the reporter vector, which contains 35SPro:REN as an internal control, and the firefly luciferase reporter driven by the OsGA2ox3 or EUI1 promoter. OsMADS57 was expressed in the effector vector (Fig. 5a). The reporter and effector were co-transformed into rice protoplasts. After transiently expressed in rice protoplasts, the LUC and REN activities were then measured and the LUC activity was normalized to REN activity. We found that co-expression of 35S:OsMADS57 and OsGA2ox3Pro:LUC significantly reduced the LUC:REN ratio, and the LUC activity was 32% of that in the control (Fig. 5b). As to EUI1, the LUC activity was also repressed by co-transformation of 35S:OsMADS57 and EUI1Pro:LUC, and the LUC activity was 37% of that in the control (Fig. 5c). LUC activity analysis showed that OsMADS57 inhibited the expression of OsGA2ox3 and EUI1, indicating that OsMADS57 is a negative regulator of OsGA2ox3 and EUI1. To further investigate whether OsMADS57 can bind to DNA motifs in the promoter region of EUI1 or OsGA2ox3, electrophoretic mobility shift assays (EMSAs) were used with His-OsMADS57 protein. We identified CArG motifs by surveying the promoter regions of EUI1 and OsGA2ox3. As shown in Fig. 5d, the potential OsMADS57-binding CArG-box sites, CATATAAAAG at − 1789 to − 1798 bp from ATG position of EUI1 and CTTTAAAAAG at − 1660 to − 1669 bp from ATG position of OsGA2ox3, were found. As shown in Fig. 5e, shifted band was observed when probes containing CTTTAAAAAG in the OsGA2ox3 promoter region were incubated with OsMADS57 protein (lane 2). By contrast, no shift band was detected when the sample contains the probe alone (lane 1). The binding of OsMADS57 protein to the labeled probe was competed by unlabeled probe (lane 3; 4). As we expected, shifted band was also observed when probes containing CATATAAAAG in the EUI1 promoter region were incubated with OsMADS57 protein (lane b) (Fig. 5f). On the contrary, no shift band was detected when the sample only contains the probe (lane a). As expected, the binding band can be competed by unlabeled probe (lane c). Thus, our results indicate that OsMADS57 is an upstream transcriptional regulator of OsGA2ox3 as well as EUI1.
Fig. 5.
OsMADS57 directly regulates the expression of OsGA2ox3 and EUI1 by binding to the promoter regions of OsGA2ox3 and EUI1, respectively. a Schematic diagrams of the effector and reporter constructs used in the dual-luciferase reporter assays. b and c The transient transactivation assay in rice protoplasts. d Schematic diagram of OsGA2ox3 and EUI1 promoter regions showing the CArG-box. e and f EMSA showed that OsMADS57 bound to CArG-box in the promoter regions of OsGA2ox3 (e) and EUI1 (f). Three independent experiments were conducted with similar results. Data are means± SD (n = 9). Error bars indicate SD
mRNA expression analysis of mutant plants
Based on the semi-dwarf phenotype of mutants, we conducted RNA sequencing (RNA-seq) using shoots of m57–2 and wild type to analyze the genes involved in plant growth and development. The transcriptome analysis showed that 7013 genes were up-regulated and 2440 genes were down-regulated in m57–2 compared with that in wild type (Fig. 6a, Additional file 6: Data Set S1). Among the changed genes, we found that the expression of genes involved in GA metabolism and cell wall loosening had changed greatly (Table 1). The expression of HOX12 was significantly induced in m57–2 plants. Accordingly, reducing the expression of HOX12 promotes panicle exsertion in rice plants (Gao et al. 2016). The transcript levels of OsYABBY4 in m57–2 plants were 6.40-fold greater than that in wild-type plants. It was shown that overexpression of OsYABBY4 reduces the plant height through repressing the expression of OsGA20ox2 (Yang et al. 2016). In addition, cell wall loosening-related genes (EXPs), which influence plant height through wall extension (Shcherban et al. 1995), were also altered greatly. The promoter region of OsEXPA4 that is involved in expansin synthesis contains GA-responsive elements (GARE), and OsEXPA4 can be induced by GA (Lee et al. 2001). However, the expression of OsEXPA4 was found to be 2.02-fold less in m57–2 plants than that in wild-type plants. To further verify the gene expression was changed, we performed Q-PCR analysis. The results showed that expansin genes were down-regulated in mutant plants (Fig. 6b). Increased transcription levels of HOX12 and OsYABBY4 were detected in m57–2 plants, which was consistent with the results of RNA-seq (Table 1). Our results suggest that OsMADS57 affects plant height by regulating the expression of genes involved in GA metabolism and extension of the plant cell wall.
Fig. 6.
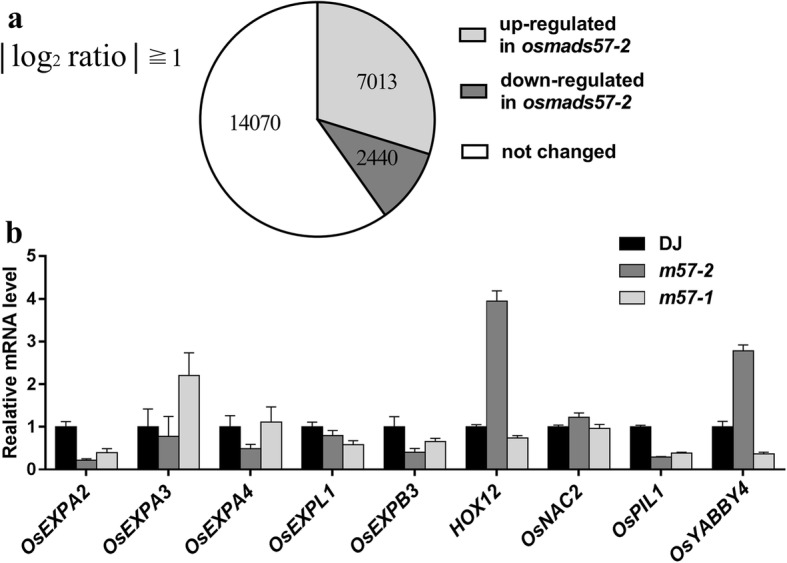
Expression analysis of genes involved in GA metabolism and cell wall loosening. a Expression changes of genes in m57–2 seedlings from the RNA-seq analysis. b Expression of genes involved in GA metabolism and cell wall loosening in mutants and wild-type lines was analysed by Q-PCR. DJ, wild type; m57–2, osmads57–2; m57–1, osmads57–1. Three independent experiments were conducted with similar results. Data are means± SE (n = 12). Error bars indicate SE
Table 1.
Expression of genes involved in GA metabolism and cell wall loosening in osmads57–2 plants
| gene | Change(Log2 ratio) | Annotation |
|---|---|---|
| Os04g0228400 | −4.75 | OsEXPA1 |
| Os01g0823100 | −2.06 | OsEXPA2 |
| Os05g0477600 | −2.02 | OsEXPA4 |
| Os06g0725300 | −1.69 | OsEXPL4 |
| Os10g0555900 | −1.94 | OsEXPB3 |
| Os04g0552200 | −3.00 | OsEXPB5 |
| Os04g0598300 | 2.58 | APO2 |
| Os04g0610400 | 2.16 | OsAP2–39 |
| Os03g0782500 | −3.20 | OsPIL1 |
| Os03g0198600 | 7.45 | HOX12 |
| Os02g0643200 | 6.40 | OsYABBY4 |
| Os04g0460600 | 2.16 | OsNAC2 |
| Os06g0275000 | −2.88 | OsHd1 |
Changes in gene expression based on the RNA-seq data
Discussion
OsMADS57 controls plant height via GA homeostasis
Rice plant height, an important agronomic trait, largely affects crop yields (Yang and Hwa 2008). Many transcription factors are involved in regulating plant height (Chen et al. 2015; Todaka et al. 2012; Qi et al. 2011). Increased expression of SHORT and SOLID CULM, the rice homologue of Arabidopsis LFY, drastically reduces the plant height (Wang et al. 2017). In this work, we demonstrated that the MADS-box transcription factor OsMADS57 directly suppressed the expression of OsGA2ox3 and EUI1, resulting in reduced levels of bioactive GA and plant height in m57–2. The expression of OsMADS57 was induced by PAC but repressed by GA3 (Fig. 3a and b), which was consistent with previous research (Puig et al. 2013). Both the loss-of-function mutant m57–2 and gain-of-function mutant m57–1 displayed semi-dwarf phenotype in seedlings, and the semi-dwarf phenotype can be rescued by applying GA3 (Figs. 1 and 3). These results indicate that OsMADS57 is associated with GA-mediated regulatory pathway. Endogenous GA levels are modulated by the expression of genes involved in GA biosynthesis and deactivation, and also fine-tuned by feedback control of GA metabolism (Yamaguchi 2008; Hedden and Thomas 2012; Fukazawa et al. 2017; Boden et al. 2014; Thomas et al. 2005; Fukazawa et al. 2011). The transcript levels of GA biosynthesis genes were reduced in m57–2 and m57–1 plants, resulting in less bioactive GA accumulation than wild type (Fig. 4a and d). Interestingly, the transcription of OsGA3ox1 and OsGA3ox2 was up-regulated in both mutant plants, which may account for a feedback mechanism due to the reduced levels of bioactive GA in both mutants. Thus, it was likely that two mutants showed reduced transcription of OsGA2ox1 and OsGA2ox6. In contrast, the increase in OsGA2ox3 transcripts observed in m57–2 and decrease in OsGA2ox3 transcripts observed in m57–1 were not due to feedback mechanism (Fig. 4b). By scanning the promoter region of OsGA2ox3, we found that a cis-element was the same as the CArG-box in D14 promoter region, bound by OsMADS57 protein (Guo et al. 2013a). Data from EMSA and dual-luciferase reporter assays in rice protoplasts proved that OsGA2ox3 is a direct target of OsMADS57 (Fig. 5e). Deficiency of OsMADS57 protein in m57–2 relieved the inhibition of OsMADS57 on OsGA2ox3, resulting in reduced bioactive GA levels, followed by the reduced plant height of m57–2. Surprisingly, m57–1, containing truncated OsMADS57 protein lacking an intact C-terminus (Guo et al. 2013a), also showed semi-dwarf phenotype (Fig. 1), which may result from the down-regulation of OsGA20ox2 by other regulators because mutation in OsGA20ox2 led to reduced GA levels and semi-dwarf phenotype (Spielmeyer et al. 2002; Sasaki et al. 2002). In addition, m57–2 and m57–1 had increased response to GA compared to wild type (Fig. 3c and d). It is intriguing that, as the suppressor of GA signaling pathway, the transcription of SLR1 and SLRL1 was promoted in two mutants (Fig. 4c). Although accumulation of SLR1 and SLRL1 limits GA responsiveness, post-transcriptional modification may also determine protein levels (Badodi et al. 2015). SLR1 and SLRL1 protein accumulation in vivo need to be further verified. These results prove that semi-dwarf phenotype in osmads57 mutants result from GA deficiency but not malfunction in GA signaling.
OsMADS57 directly represses EUI1 transcription to regulate internode elongation
GA signaling and metabolism are involved in the regulation of internode elongation (Fleet and Sun 2005; Vriezen et al. 2003). GA levels are determined by GA3ox and GA20ox that are involved in GAs biosynthesis (Hedden and Thomas 2012), but also GA2ox that deactivates GAs (Thomas et al. 1999). EUI1 is also involved in GA deactivation reaction by catalyzing 16α, 17-epoxidation reaction of GA4, GA9 and GA12 (Zhu et al. 2006; Ma et al. 2006). In eui1 mutant, the uppermost internode is significantly elongated with enhanced panicle exsertion, and accumulates higher levels of GA than wild type. By contrast, the lines overexpressing EUI1 exhibited significantly dwarf phenotype (Luo et al. 2006; Zhu et al. 2006). Data showed that both m57–2 and m57–1 had reduced elongation in the uppermost internode and defected in panicle exsertion compared to that of wild type (Fig. 2). In addition, the expression of EUI1 was enhanced in m57–2 but reduced in m57–1 (Fig. 4b). MADS-box proteins function by regulating the expression of target genes via binding to CArG-box motifs (Gramzow and Theissen 2010; Tang and Perry 2003). Through browsing the promoter region of EUI1, a potential binding site of OsMADS57 was found. Analysis of LUC activity and EMSA confirmed the hypothesis that OsMADS57 directly regulated the expression of EUI1. In m57–2, the suppression of OsMADS57 on EUI1 was abolished, which caused higher transcript levels of EUI1 and thus reducing bioactive GA levels. Hence, internode elongation and panicle exsertion were hindered in m57–2 plants. In addition, HOX12 can directly promote EUI1 expression (Gao et al. 2016). Expectedly, the expression of HOX12 was also greatly enhanced in m57–2 plants. Therefore, panicle exsertion progress may not be regulated by OsMADS57 alone, but other protein factors synergisticly regulated this development process in combination with OsMADS57. However, m57–1 plants also exhibited semi-dwarf phenotype with reduced uppermost internode elongation and defected panicle exsertion (Figs. 1 and 2). Previous research reported that overexpression of CYP714B1 or CYP714B2, which are highly expressed in the uppermost internode of adult plants, causes semi-dwarf phenotype by reducing GA activity; but cyp714b1cyp714b2 double mutant shows a longer uppermost internode, thus exposing longer internode from the flag leaf sheath (Magome et al. 2013). EUI1 (CYP714D1), CYP714B1 and CYP714B2, belonging to CYP714 subfamilies, likely co-modulate GA-mediated growth and development. We suppose that, as a compensation mechanism, inhibition of EUI1 expression may cause the up-regulation of homologous genes, which led to shorter internode and panicle enclosure. In addition, the m57–1 plants also showed increased tiller numbers (Fig. 1), which was consistent with previous study (Guo et al. 2013a). Generally, rice plants with more tillers tends to exhibit dwarf phenotype (Ishikawa et al. 2005; Qi et al. 2011; Dai et al. 2018). In rice, overexpressing genes encoding GA2-oxidase increases tiller numbers but inhibits stem elongation, which is coupled with GA deficiency (Lo et al. 2008). Accordingly, overexpression of OsYABBY1 leads to reduced GA levels in rice plants, and overexpression lines exhibit semi-dwarf phenotype and more tillers (Dai et al. 2007). Also, MOC1 overexpression lines display increased tiller numbers and reduced plant height (Li et al. 2003). Although there is a negative correlation between plant height and tiller numbers, the specific molecular mechanisms is not clear. It is worth noting that expansins regulate cell size/elongation via cell wall expansion (Magneschi et al. 2009; Vriezen et al. 2003). OsEXPA4 overexpressors have enhanced stem elongation via affecting cell sizes (Choi et al. 2003). We speculate that reduced stem length in m57–2 and m57–1 plants is associated with down-regulated expression of expansin genes.
Conclusions
In this work, OsMADS57 was shown to regulate internode elongation and plant height by directly repressing OsGA2ox3 and EUI1 expression. Deficiency of OsMADS57 protein promoted the expression of OsGA2ox3 and EUI1, which enhanced the conversion of bioactive GA to deactivated GA, thus resulting in panicle enclosure and semi-dwarf phenotype (Fig. 7). The role of OsMADS57 in regulating plant growth provides an opportunity to improve grain yield by genetic manipulation to modulate stem elongation.
Fig. 7.
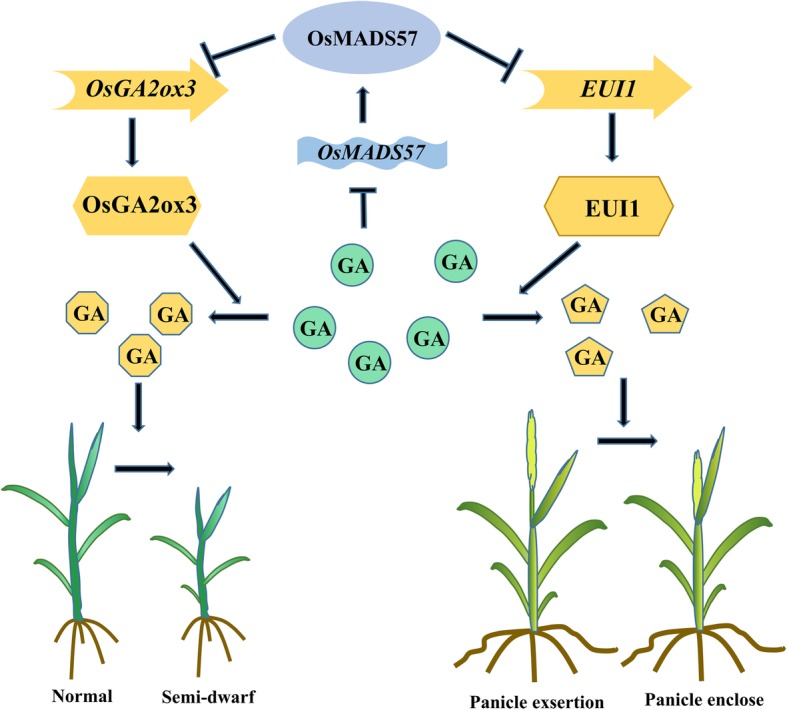
Model of OsMADS57 regulatory network through GA catabolic genes. The expression of OsMADS57 is inhibited by GA3. And OsMADS57 suppresses the transcription of OsGA2ox3 and EUI1 that are involved in GA deactivation. Deficiency of OsMADS57 protein enhances the expression of OsGA2ox3 and EUI1, which results in converting from bioactive GA to deactivated GA, thus reducing bioactive GA levels. Plants deficiency in bioactive GA shows panicle enclosure and semi-dwarf phenotype
Methods
Plant materials and hormone treatment
The rice (Oryza sativa) mutants PFG_3A-15,619.R (osmads57–2) and PFG_3A-05432.L (osmads57–1) were obtained from RiceGE (the Rice Functional Genomics Express Database), in Pohang city, Korea. The mutants and their origin cultivar, Oryza sativa ssp. japonica cv Dongjin, were grown in a chamber with controlled temperature about 30 °C or in the fields during the suitable seasons. All the field management abides normal agricultural action. At maturity, the length of internode was recorded. For the phenotype analysis, the seeds of mutants and wild type were sterilized with 0.2% HgCl2 solution for 10 min, then washed five times with sterile distilled water and immersed in water for 2–3 d. These seeds were sown in half-strength Murashige and Skoog (1/2 MS) medium supplemented with 10 μМ GA3 or PAC at 30 °C. The shoot length of the seeding was measured. For OsMADS57 expression analysis, 7-day-old wild-type seedlings were treated with 50 μМ GA3 or 10 μМ PAC, and harvested at different time points for Q-PCR analysis. For the seed germination assays, the sterilized seeds were placed on sterile filter paper moistened with double sterilized H2O and germinated in the dark. The germination rates were recorded each day.
Total RNA extraction and quantitative PCR analysis
Total RNA was extracted from shoots of mutants and wild-type plants using TRIzol reagent (TaKaRa). RNA was used to synthesize cDNA using a PrimeScript™ RT reagent Kit from TaKaRa (code: RR047A). Q-PCR was performed on Bio-Rad CFX96 instrument with GoTaq qPCR Master Mix reagent (Promega) according to manufacturer’s instructions. The gene Actin1 as rice housekeeping gene was used for an internal reference. Each analysis includes three biological repeats and three technical replicates. Primers used for Q-PCR are listed in Additional file 5: Table S1.
Dual-luciferase reporter assays
For dual-luciferase reporter assay, the coding sequence (CDS) of OsMADS57 was amplified by PCR and inserted into the pGreenII 62-SK vector for generating the effector. To generate reporter constructs, the promoter fragments of EUI1 or OsGA2ox3 were inserted into the pGreenII 0800 vector, respectively. The Renilla luciferase (REN) gene under the control of the 35S promoter in the pGreenII 0800 vector was used as an internal control (Hellens et al. 2005). Empty pGreenII 0800 vector was used as the control. Protoplasts were prepared from rice shoots and transfected using a polyethylene glycolcalcium-mediated method followed by a 20-h incubation to allow transient expression (Zhang et al. 2011). Firefly LUC and REN activities were measured with a dual-luciferase reporter assay kit (Promega). The relative ratio of LUC to REN was calculated to represent the expression of reporter genes. Primers used for these constructs are listed in Additional file 5: Table S1.
Electrophoretic mobility shift assays (EMSAs)
To test the binding activity of OsMADS57 protein, the full-length coding sequence of OsMADS57 was cloned into pET32a (+) and transformed into BL21 (DE3). The OsMADS57 recombinant protein was purified using His60 Ni Superflow Resin. Oligonucleotide probes containing CArG motifs were synthesized and labeled with using a Biotin 3′End DNA Labeling Kit (Thermo Scientific). Unlabeled probe was used for competitive reactions. EMSA was performed using a LightShift® Chemiluminescent EMSA Kit (Thermo). Probe sequences are shown in Additional file 5: Table S1.
Yeast two-hybrid
For yeast two-hybrid (Y2H) analysis, the CDS for OsMADS57 and SLR1 were cloned into the pGBKT7 and pGADT7 vectors, respectively. Detecting the interaction between OsMADS57 and SLR1 in yeast was performed. The yeast two-hybrid assay was conducted following the manufacturer’s instructions (Clontech). Primers used for these constructs are listed in Additional file 5: Table S1.
Quantification of endogenous GA
For GA quantification, the shoots of two-week-old wild type and mutants plants were harvested and used for measurement of GA3 by a liquid chromatography-MS system (UPLC/Quattro Premier XE; Waters) according to the protocol (Kojima et al. 2009).
Accession numbers
Sequence data from this article can be found in RICEGE or GenBank/EMBL databases under the following accession number: OsMADS57 (Os02g0731200), EUI1 (Os05g0482400), OsGA2ox3 (Os01g0757200), SLR1 (Os03g0707600), SLRL1 (Os01g0646300), HOX12 (Os03g0198600), OsNAC2 (Os04g0460600), OsYABBY4 (Os02g0643200), OsPIL1 (Os03g0782500), OsGA20ox1 (Os03g0856700), OsGA20ox2 (Os01g0883800), OsGA2ox6 (Os04g0522500), CPS1 (Os02g0278700), KO2 (Os06g0570100), OsGA2ox1 (Os05g0158600), OsGA3ox1 (Os05g0178100), OsEXPA2 (Os01g0823100), OsGA3ox2 (Os01g0177400), OsEXPA1 (Os04g0228400), OsEXPA3 (Os05g0276500), OsEXPA4 (Os05g0477600), OsEXPL1 (Os03g0132200) and OsEXPB3 (Os10g0555900).
Additional files
Figure S1. m57–2 and m57–1 mutants exhibit delayed flowering phenotype. a Schematic diagram indicating the T-DNA insertion site in genomic region in m57–2. b PCR analyse the genotype of m57–2 T2 seedlings. c Sequencing result for identifying of insertion site in the genomic region of m57–2. d Comparison of flowering between mutants and wild type. Arrows indicate flowering panicles. Bars = 10 cm. e Magnification of the boxed region in (d). Bars = 5 cm. f Quantitation of the seed germination rate of wild type and mutants lines. DJ, wild type; m57–2, osmads57–2; m57–1, osmads57–1. Three independent experiments were conducted with similar results. (TIF 6740 kb)
Figure S2. m57–2 and m57–1 displayed reduced seed setting rate. a Flag leaf of wild type and mutants. Bars = 5 cm. b Flag leaf length of wild type and mutants at maturity. c Quantification of seed setting rate in wild type and mutants. d Comparison of grains between wild type and mutants, indicating blight grain rate increased in mutants. Bars = 2 cm. DJ, wild type; m57–2, osmads57–2; m57–1, osmads57–1. Three independent experiments were conducted with similar results. The data are means± SD (n = 10). Error bars indicate SD. The statistical significance of the measurements was determined by Student’s t-test. Asterisks indicate the significant difference between osmads57 and wild type. (t-test, * P < 0.05, ** P < 0.01 or *** P < 0.001). (TIF 5617 kb)
Figure S3. Shoot length of m57–2, m57–1 and wild type under GA or PAC treatment at different time point. a Shoot length of wild type and mutants with GA treatment. b Shoot length of wild type and mutants with PAC treatment. DJ, wild type; m57–2, osmads57–2; m57–1, osmads57–1. Three independent experiments were conducted with similar results. The data are means± SD (n = 20). Error bars indicate SD. (TIF 938 kb)
Figure S4. Physical interaction analysis between OsMADS57 and SLR1 in yeast. a Schematic representation of various combination between different constructs. b Yeast two-hybrid analysis the interaction of SLR1 and OsMADS57, and no interaction was observed between SLR1 and OsMADS57. Positive control, pGADT7-T (SV40 large T antigen)/pGBKT7–53 (murine p53). (TIF 5207 kb)
Table S1. Primer and probe sequences used in this study. (XLSX 12 kb)
Data Set S1. The up-regulated or down-regulated genes with│log2 ratio│≧1 in osmads57–2 plants from the RNA-seq data. (XLSX 558 kb)
Acknowledgements
Not applicable.
Abbreviations
- CPS
Ent-copalyl diphosphate synthase
- EMSAs
Electrophoretic mobility shift assays
- EUI1
ELONGATED UPPERMOST INTERNODE1
- GA
Gibberellin
- GA20ox
GA 20-oxidase
- GA2ox
GA 2-oxidase
- GA3
Gibberellic acid
- GA3ox
GA 3-oxidase
- GARE
GA-responsive elements
- GID1
GIBBERELLIN INSENSITIVE DWARF1
- HD-ZIP
Homeodomain-leucine zipper
- KAO
Ent-kaurenoic acid oxidase
- KO
Ent-kaurene oxidase
- KS
Ent-kaurene synthase
- MS
Murashige and Skoog
- PAC
Paclobutrazol
- Q-PCR
Quantitative PCR
- RNA-seq
RNA sequencing
- SLR1
SLENDER1
- WT
Wild type
- Y2H
Yeast two-hybrid
Authors’ contributions
H. J. and C. Y. designed the experiments; C.Y. performed experiments, data analysis and wrote the manuscript; X. N., Y. B. and W. Q. contributed to assist in performing part of the experiments; L. X. and C. R. contributed to assist in analyzing and discussing the data; and H. J. supervised the project and revised the manuscript. All authors read and approved the final manuscript.
Funding
Funding for this work was provided by National Natural Science Foundation of China (31771747, 31271685).
Availability of data and materials
The datasets used or analysed in this study are included in the article and its additional files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 2009;50:1416–1424. doi: 10.1093/pcp/pcp091. [DOI] [PubMed] [Google Scholar]
- Badodi S, Baruffaldi F, Ganassi M, Battini R, Molinari S. Phosphorylation-dependent degradation of MEF2C contributes to regulate G2/M transition. Cell Cycle. 2015;14:1517–1528. doi: 10.1080/15384101.2015.1026519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden SA, Weiss D, Ross JJ, Davies NW, Trevaskis B, Chandler PM, Swain SM. EARLY FLOWERING3 regulates FLOWERING in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression. Plant Cell. 2014;26:1557–1569. doi: 10.1105/tpc.114.123794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Xiang S, Chen Y, Li D, Yu D. Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol Plant. 2017;10:1174–1189. doi: 10.1016/j.molp.2017.07.008. [DOI] [PubMed] [Google Scholar]
- Chen L, Zhao Y, Xu S, Zhang Z, Xu Y, Zhang J, Chong K. OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytol. 2018;218:219–231. doi: 10.1111/nph.14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Lu S, Wang Y, Zhang X, Lv B, Luo L, Xi D, Shen J, Ma H, Ming F. OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberellic acid pathway in rice. Plant J. 2015;82:302–314. doi: 10.1111/tpj.12819. [DOI] [PubMed] [Google Scholar]
- Cho SH, Kang K, Lee SH, Lee IJ, Paek NC. OsWOX3A is involved in negative feedback regulation of the gibberellic acid biosynthetic pathway in rice (Oryza sativa) J Exp Bot. 2016;67:1677–1687. doi: 10.1093/jxb/erv559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D, Lee Y, Cho HT, Kende H. Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell. 2003;15:1386–1398. doi: 10.1105/tpc.011965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Hu Y, Ma Q, Zhao Y, Zhou DX. Functional analysis of rice HOMEOBOX4 (Oshox4) gene reveals a negative function in gibberellin responses. Plant Mol Biol. 2008;66:289–301. doi: 10.1007/s11103-007-9270-8. [DOI] [PubMed] [Google Scholar]
- Dai M, Zhao Y, Ma Q, Hu Y, Hedden P, Zhang Q, Zhou DX. The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol. 2007;144:121–133. doi: 10.1104/pp.107.096586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Wang J, Yang X, Lu H, Miao X, Shi Z. Modulation of plant architecture by the miR156f-OsSPL7-OsGH3.8 pathway in rice. J Exp Bot. 2018;69:5117–5130. doi: 10.1093/jxb/ery273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daviere JM, Achard P. A pivotal role of DELLAs in regulating multiple hormone signals. Mol Plant. 2016;9:10–20. doi: 10.1016/j.molp.2015.09.011. [DOI] [PubMed] [Google Scholar]
- Duan K, Li L, Hu P, Xu SP, Xu ZH, Xue HW. A brassinolide-suppressed rice MADS-box transcription factor, OsMDP1, has a negative regulatory role in BR signaling. Plant J. 2006;47:519–531. doi: 10.1111/j.1365-313X.2006.02804.x. [DOI] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, Schafer E, Fu X, Fan LM, Deng XW. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet CM, Sun TP. A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr Opin Plant Biol. 2005;8:77–85. doi: 10.1016/j.pbi.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Fukao T, Bailey-Serres J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc Natl Acad Sci U S A. 2008;105:16814–16819. doi: 10.1073/pnas.0807821105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J, Mori M, Watanabe S, Miyamoto C, Ito T, Takahashi Y. DELLA-GAF1 complex is a Main component in gibberellin feedback regulation of GA20 oxidase 2. Plant Physiol. 2017;175:1395–1406. doi: 10.1104/pp.17.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J, Nakata M, Ito T, Matsushita A, Yamaguchi S, Takahashi Y. bZIP transcription factor RSG controls the feedback regulation of NtGA20ox1 via intracellular localization and epigenetic mechanism. Plant Signal Behav. 2011;6:26–28. doi: 10.4161/psb.6.1.14114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J, Teramura H, Murakoshi S, Nasuno K, Nishida N, Ito T, Yoshida M, Kamiya Y, Yamaguchi S, Takahashi Y. DELLAs function as coactivators of GAI-ASSOCIATED FACTOR1 in regulation of gibberellin homeostasis and signaling in Arabidopsis. Plant Cell. 2014;26:2920–2938. doi: 10.1105/tpc.114.125690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Fang J, Xu F, Wang W, Chu C. Rice HOX12 regulates panicle Exsertion by directly modulating the expression of ELONGATED UPPERMOST INTERNODE1. Plant Cell. 2016;28:680–695. doi: 10.1105/tpc.15.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramzow L, Theissen G. A hitchhiker's guide to the MADS world of plants. Genome Biol. 2010;11:214. doi: 10.1186/gb-2010-11-6-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Xu Y, Liu H, Mao Z, Zhang C, Ma Y, Zhang Q, Meng Z, Chong K. The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat Commun. 2013;4:1566. doi: 10.1038/ncomms2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Hou X, Fang J, Wei P, Xu B, Chen M, Feng Y, Chu C. The rice GERMINATION DEFECTIVE 1, encoding a B3 domain transcriptional repressor, regulates seed germination and seedling development by integrating GA and carbohydrate metabolism. Plant J. 2013;75:403–416. doi: 10.1111/tpj.12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Phillips AL. Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 2000;5:523–530. doi: 10.1016/S1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- Hedden P, Sponsel V. A century of gibberellin research. J Plant Growth Regul. 2015;34:740–760. doi: 10.1007/s00344-015-9546-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Thomas SG. Gibberellin biosynthesis and its regulation. Biochem J. 2012;444:11–25. doi: 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1:13. doi: 10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Chandler PM, Poole A, Dennis ES, Peacock WJ. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc Natl Acad Sci U S A. 2001;98:2065–2070. doi: 10.1073/pnas.98.4.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmut Aach, Heike Bode, G. D, Robinson JE. Graebe ent-Kaurene synthase is located in proplastids of meristematic shoot tissues. Planta. 1997;202:211–219. doi: 10.1007/s004250050121. [DOI] [Google Scholar]
- Hirano K, Kouketu E, Katoh H, Aya K, Ueguchi-Tanaka M, Matsuoka M. The suppressive function of the rice DELLA protein SLR1 is dependent on its transcriptional activation activity. Plant J. 2012;71:443–453. doi: 10.1111/j.1365-313X.2012.05000.x. [DOI] [PubMed] [Google Scholar]
- Huang H, Tudor M, Su T, Zhang Y, Hu Y, Ma H. DNA binding properties of two Arabidopsis MADS domain proteins: binding consensus and dimer formation. Plant Cell. 1996;8:81–94. doi: 10.1105/tpc.8.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J. Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell. 2001;13:999–1010. doi: 10.1105/tpc.13.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J. Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol. 2005;46:79–86. doi: 10.1093/pcp/pci022. [DOI] [PubMed] [Google Scholar]
- Itoh H. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. The Plant Cell Online. 2002;14:57–70. doi: 10.1105/tpc.010319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Shimada A, Ueguchi-Tanaka M, Kamiya N, Hasegawa Y, Ashikari M, Matsuoka M. Overexpression of a GRAS protein lacking the DELLA domain confers altered gibberellin responses in rice. Plant J. 2005;44:669–679. doi: 10.1111/j.1365-313X.2005.02562.x. [DOI] [PubMed] [Google Scholar]
- Khush GS. Green revolution: the way forward. Nat Rev Genet. 2001;2:815–822. doi: 10.1038/35093585. [DOI] [PubMed] [Google Scholar]
- Kojima M, Kamada-Nobusada T, Komatsu H, Takei K, Kuroha T, Mizutani M, Ashikari M, Ueguchi-Tanaka M, Matsuoka M, Suzuki K, Sakakibara H. Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol. 2009;50:1201–1214. doi: 10.1093/pcp/pcp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Choi SC, An G. Rice SVP-group MADS-box proteins, OsMADS22 and OsMADS55, are negative regulators of brassinosteroid responses. Plant J. 2008;54:93–105. doi: 10.1111/j.1365-313X.2008.03406.x. [DOI] [PubMed] [Google Scholar]
- Lee Y, Choi D, Kende H. Expansins: ever-expanding numbers and functions. Curr Opin Plant Biol. 2001;4:527–532. doi: 10.1016/S1369-5266(00)00211-9. [DOI] [PubMed] [Google Scholar]
- Li X, Qian Q, Fu Z, Wang Y, Xiong G, Zeng D, Wang X, Liu X, Teng S, Hiroshi F, Yuan M, Luo D, Han B, Li J. Control of tillering in rice. Nature. 2003;422:618–621. doi: 10.1038/nature01518. [DOI] [PubMed] [Google Scholar]
- Lo SF, Yang SY, Chen KT, Hsing YI, Zeevaart JA, Chen LJ, Yu SM. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell. 2008;20:2603–2618. doi: 10.1105/tpc.108.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo A, Qian Q, Yin H, Liu X, Yin C, Lan Y, Tang J, Tang Z, Cao S, Wang X, Xia K, Fu X, Luo D, Chu C. EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol. 2006;47:181–191. doi: 10.1093/pcp/pci233. [DOI] [PubMed] [Google Scholar]
- Ma H, Zhang S, Ji L, Zhu H, Yang S, Fang X, Yang R. Fine mapping and in silico isolation of the EUI1 gene controlling upper internode elongation in rice. Plant Mol Biol. 2006;60:87–94. doi: 10.1007/s11103-005-2762-5. [DOI] [PubMed] [Google Scholar]
- Magneschi L, Kudahettige RL, Alpi A, Perata P. Expansin gene expression and anoxic coleoptile elongation in rice cultivars. J Plant Physiol. 2009;166:1576–1580. doi: 10.1016/j.jplph.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Magome H, Nomura T, Hanada A, Takeda-Kamiya N, Ohnishi T, Shinma Y, Katsumata T, Kawaide H, Kamiya Y, Yamaguchi S. CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc Natl Acad Sci U S A. 2013;110:1947–1952. doi: 10.1073/pnas.1215788110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N, T-p S, Gubler F. Gibberellin Signaling. Plant Cell. 2002;14:S61–S80. doi: 10.1105/tpc.010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig J, Meynard D, Khong GN, Pauluzzi G, Guiderdoni E, Gantet P. Analysis of the expression of the AGL17-like clade of MADS-box transcription factors in rice. Gene Expr Patterns. 2013;13:160–170. doi: 10.1016/j.gep.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Qi W, Sun F, Wang Q, Chen M, Huang Y, Feng YQ, Luo X, Yang J. Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol. 2011;157:216–228. doi: 10.1104/pp.111.179945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards DE, King KE, Ait-Ali T, Harberd NP. HOW GIBBERELLIN REGULATES PLANT GROWTH AND DEVELOPMENT: a molecular genetic analysis of gibberellin signaling. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:67–88. doi: 10.1146/annurev.arplant.52.1.67. [DOI] [PubMed] [Google Scholar]
- Sakai M, Sakamoto T, Saito T, Matsuoka M, Tanaka H, Kobayashi M. Expression of novel rice gibberellin 2-oxidase gene is under homeostatic regulation by biologically active gibberellins. J Plant Res. 2003;116:161–164. doi: 10.1007/s10265-003-0080-z. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Matsuoka M. Generating high-yielding varieties by genetic manipulation of plant architecture. Curr Opin Biotechnol. 2004;15:144–147. doi: 10.1016/j.copbio.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, Miyao A, Hirochika H, Kitano H, Ashikari M, Matsuoka M. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004;134:1642–1653. doi: 10.1104/pp.103.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, Kitano H, Matsuoka M. Green revolution: a mutant gibberellin-synthesis gene in rice. Nature. 2002;416:701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- Shcherban TY, Shi J, Durachko DM, Guiltinan MJ, McQueen-Mason SJ, Shieh M, Cosgrove DJ. Molecular cloning and sequence analysis of expansins--a highly conserved, multigene family of proteins that mediate cell wall extension in plants. Proc Natl Acad Sci U S A. 1995;92:9245–9249. doi: 10.1073/pnas.92.20.9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmeyer W, Ellis MH, Chandler PM. Semidwarf (sd-1), "green revolution" rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci U S A. 2002;99:9043–9048. doi: 10.1073/pnas.132266399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SM, Singh DP. Tall tales from sly dwarves: novel functions of gibberellins in plant development. Trends Plant Sci. 2005;10:123–129. doi: 10.1016/j.tplants.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Tang W, Perry SE. Binding site selection for the plant MADS domain protein AGL15: an in vitro and in vivo study. J Biol Chem. 2003;278:28154–28159. doi: 10.1074/jbc.M212976200. [DOI] [PubMed] [Google Scholar]
- Tao J, Liang W, An G, Zhang D. OsMADS6 controls flower development by activating Rice FACTOR OF DNA METHYLATION LIKE1. Plant Physiol. 2018;177:713–727. doi: 10.1104/pp.18.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SG, Phillips AL, Hedden P. Molecular cloning and functional expression of gibberellin 2- oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci U S A. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SG, Rieu I, Steber CM. Gibberellin metabolism and signaling. Vitam Horm. 2005;72:289–338. doi: 10.1016/S0083-6729(05)72009-4. [DOI] [PubMed] [Google Scholar]
- Todaka D, Nakashima K, Maruyama K, Kidokoro S, Osakabe Y, Ito Y, Matsukura S, Fujita Y, Yoshiwara K, Ohme-Takagi M, Kojima M, Sakakibara H, Shinozaki K, Yamaguchi-Shinozaki K. Rice phytochrome-interacting factor-like protein OsPIL1 functions as a key regulator of internode elongation and induces a morphological response to drought stress. Proc Natl Acad Sci U S A. 2012;109:15947–15952. doi: 10.1073/pnas.1207324109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H, Xiao Y, Liu D, Gao S, Liu L, Yin Y, Jin Y, Qian Q, Chu C. Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell. 2014;26:4376–4393. doi: 10.1105/tpc.114.132092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, Matsuoka M. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437:693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- VRIEZEN W. H. Regulation of Submergence-induced Enhanced Shoot Elongation in Oryza sativa L. Annals of Botany. 2003;91(2):263–270. doi: 10.1093/aob/mcf121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Caruso LV, Downie AB, Perry SE. The embryo MADS domain protein AGAMOUS-like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. Plant Cell. 2004;16:1206–1219. doi: 10.1105/tpc.021261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang R, Wang Y, Zhang L, Zhang L, Xu Y, Yao S. Short and Solid culm/RFL/APO2 for culm development in rice. Plant J. 2017;91:85–96. doi: 10.1111/tpj.13548. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li J. The plant architecture of rice (Oryza sativa) Plant Mol Biol. 2005;59:75–84. doi: 10.1007/s11103-004-4038-x. [DOI] [PubMed] [Google Scholar]
- Xu H, Liu Q, Yao T, Fu X. Shedding light on integrative GA signaling. Curr Opin Plant Biol. 2014;21:89–95. doi: 10.1016/j.pbi.2014.06.010. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. Gibberellin metabolism and its regulation. Annu Rev Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- Yang C, Ma Y, Li J. The rice YABBY4 gene regulates plant growth and development through modulating the gibberellin pathway. J Exp Bot. 2016;67:5545–5556. doi: 10.1093/jxb/erw319. [DOI] [PubMed] [Google Scholar]
- Yang XC, Hwa CM. Genetic modification of plant architecture and variety improvement in rice. Heredity (Edinb) 2008;101:396–404. doi: 10.1038/hdy.2008.90. [DOI] [PubMed] [Google Scholar]
- Yao R, Ming Z, Yan L, Li S, Wang F, Ma S, Yu C, Yang M, Chen L, Chen L, Li Y, Yan C, Miao D, Sun Z, Yan J, Sun Y, Wang L, Chu J, Fan S, He W, Deng H, Nan F, Li J, Rao Z, Lou Z, Xie D. DWARF14 is a non-canonical hormone receptor for strigolactone. Nature. 2016;536:469–473. doi: 10.1038/nature19073. [DOI] [PubMed] [Google Scholar]
- Yasui Y, Tanaka W, Sakamoto T, Kurata T, Hirano HY. Genetic enhancer analysis reveals that FLORAL ORGAN NUMBER2 and OsMADS3 co-operatively regulate maintenance and determinacy of the flower meristem in Rice. Plant Cell Physiol. 2017;58:893–903. doi: 10.1093/pcp/pcx038. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Hirano K, Sato T, Mitsuda N, Nomoto M, Maeo K, Koketsu E, Mitani R, Kawamura M, Ishiguro S, Tada Y, Ohme-Takagi M, Matsuoka M, Ueguchi-Tanaka M. DELLA protein functions as a transcriptional activator through the DNA binding of the indeterminate domain family proteins. Proc Natl Acad Sci U S A. 2014;111:7861–7866. doi: 10.1073/pnas.1321669111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, Wang J, Wang H. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods. 2011;7:30. doi: 10.1186/1746-4811-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhu Y, Peng Y, Yan D, Li Q, Wang J, Wang L, He Z. Gibberellin homeostasis and plant height control by EUI and a role for gibberellin in root gravity responses in rice. Cell Res. 2008;18:412–421. doi: 10.1038/cr.2008.28. [DOI] [PubMed] [Google Scholar]
- Zhou W, Malabanan PB, Abrigo E. OsHox4 regulates GA signaling by interacting with DELLA-like genes and GA oxidase genes in rice. Euphytica. 2014;201:97–107. doi: 10.1007/s10681-014-1191-4. [DOI] [Google Scholar]
- Zhu Y, Nomura T, Xu Y, Zhang Y, Peng Y, Mao B, Hanada A, Zhou H, Wang R, Li P, Zhu X, Mander LN, Kamiya Y, Yamaguchi S, He Z. ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell. 2006;18:442–456. doi: 10.1105/tpc.105.038455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. m57–2 and m57–1 mutants exhibit delayed flowering phenotype. a Schematic diagram indicating the T-DNA insertion site in genomic region in m57–2. b PCR analyse the genotype of m57–2 T2 seedlings. c Sequencing result for identifying of insertion site in the genomic region of m57–2. d Comparison of flowering between mutants and wild type. Arrows indicate flowering panicles. Bars = 10 cm. e Magnification of the boxed region in (d). Bars = 5 cm. f Quantitation of the seed germination rate of wild type and mutants lines. DJ, wild type; m57–2, osmads57–2; m57–1, osmads57–1. Three independent experiments were conducted with similar results. (TIF 6740 kb)
Figure S2. m57–2 and m57–1 displayed reduced seed setting rate. a Flag leaf of wild type and mutants. Bars = 5 cm. b Flag leaf length of wild type and mutants at maturity. c Quantification of seed setting rate in wild type and mutants. d Comparison of grains between wild type and mutants, indicating blight grain rate increased in mutants. Bars = 2 cm. DJ, wild type; m57–2, osmads57–2; m57–1, osmads57–1. Three independent experiments were conducted with similar results. The data are means± SD (n = 10). Error bars indicate SD. The statistical significance of the measurements was determined by Student’s t-test. Asterisks indicate the significant difference between osmads57 and wild type. (t-test, * P < 0.05, ** P < 0.01 or *** P < 0.001). (TIF 5617 kb)
Figure S3. Shoot length of m57–2, m57–1 and wild type under GA or PAC treatment at different time point. a Shoot length of wild type and mutants with GA treatment. b Shoot length of wild type and mutants with PAC treatment. DJ, wild type; m57–2, osmads57–2; m57–1, osmads57–1. Three independent experiments were conducted with similar results. The data are means± SD (n = 20). Error bars indicate SD. (TIF 938 kb)
Figure S4. Physical interaction analysis between OsMADS57 and SLR1 in yeast. a Schematic representation of various combination between different constructs. b Yeast two-hybrid analysis the interaction of SLR1 and OsMADS57, and no interaction was observed between SLR1 and OsMADS57. Positive control, pGADT7-T (SV40 large T antigen)/pGBKT7–53 (murine p53). (TIF 5207 kb)
Table S1. Primer and probe sequences used in this study. (XLSX 12 kb)
Data Set S1. The up-regulated or down-regulated genes with│log2 ratio│≧1 in osmads57–2 plants from the RNA-seq data. (XLSX 558 kb)
Data Availability Statement
The datasets used or analysed in this study are included in the article and its additional files.