Abstract
Technological advances over the past decade have unraveled the remarkable complexity of RNA. The identification of small peptides encoded by long non-coding RNAs (lncRNAs) as well as regulatory functions mediated by non-coding regions of mRNAs have further complicated our understanding of the multifaceted functions of RNA. In this review, we summarize current evidence pointing to dual roles of RNA molecules defined by their coding and non-coding potentials. We also discuss how the emerging roles of RNA transform our understanding of gene expression and evolution.
Keywords: messenger RNA, long noncoding RNA, coding potential, ribosome profiling, micropeptide
Introduction
Benefiting from the advances in science and technology, our understanding of the complexity of organisms is constantly increasing. The “central dogma” of molecular biology states that genetic information is typically processed from DNA to RNA to protein, and this decides cellular and organismal phenotype (Crick, 1970). In the past, RNAs, except for infrastructural RNAs (such as rRNAs and tRNAs), were commonly considered as an intermediate between DNA and proteins. However, over recent decades, the rapid development of high-throughput sequencing technologies has revealed the pervasive transcription of eukaryotic genomes (Okazaki et al., 2002; Carninci et al., 2005; Kapranov et al., 2007; Lander, 2011), thus revealing RNA-mediated gene regulation. The fact that most regulatory RNAs function without involvement in protein translation led us to re-examine the roles of RNAs in the development and evolution of higher organisms.
In higher organisms, only a small fraction of genetic transcripts (less than 3%) have the capability to encode proteins, despite pervasive transcription across genomes. This raises the question of whether the remaining non-protein-coding transcripts are transcriptional “noise” or contain more genetic information. Large-scale projects for the systematic annotation and functional characterization of genes (such as ENCODE and FANTOM) have reported that at least 80% of mammalian genomic DNA is actively transcribed and elaborately regulated, with the vast majority of this considered to be noncoding RNA (ncRNA) genes (Consortium, 2012; Hon et al., 2017). The numbers of ncRNA genes vary between species, and interestingly, the complexity of an organism is highly associated with the abundance of ncRNA genes but not protein-coding genes, implying the potential importance of ncRNAs (Rubin et al., 2000; Stover et al., 2000; Mattick, 2001; Venter et al., 2001; Kapusta and Feschotte, 2014). Among these, lncRNAs that are defined as transcripts longer than 200 nucleotides with low/no protein-coding potential, represent a considerable proportion.
Long non-coding RNAs can regulate gene expression in various ways, including epigenetic, transcriptional, post-transcriptional, translational and protein location effects. Corresponding to functional diversity, the modes of action of lncRNAs are also quite varied. lncRNAs can recruit epigenetic factors to modify chromatin state (Rinn and Chang, 2012), assemble transcriptional machinery to trigger the initiation of transcription (Bonasio and Shiekhattar, 2014), or act as a structural organizer to participate in the formation of subcellular organelles (Naganuma and Hirose, 2013). Additionally, lncRNAs can complementarily bind with other forms of RNA molecules to modulate gene expression at transcriptional, post-transcriptional and translational levels, for example as a moderator of mRNA activity or a decoy/sponge for miRNA (Poliseno et al., 2010; Gong and Maquat, 2011; Bonasio and Shiekhattar, 2014; Tay et al., 2014; Yoon et al., 2014). Moreover, lncRNAs couple with proteins through particular structures to act as a location transferor, or to modulate enzyme activities (Wang and Chang, 2011).
Based on the “noncoding” definition, the modes of action of lncRNAs mentioned above are exerted primarily through ncRNAs. Intriguingly, recent bioinformatics analyses of large-scale data from ribosome-protected RNA fragments (ribosome profiling or ribo-profiling) have revealed that a considerably large part of these transcripts tends to contain sORFs and binds with ribosomes (Aspden et al., 2014; Ruiz-Orera et al., 2014; Anderson et al., 2015; Mackowiak et al., 2015; Olexiouk et al., 2016), suggesting that the coding potential of lncRNAs has been vastly underestimated. Several functional experiments have demonstrated that some lncRNAs can encode small peptides (named “micropeptides” with a length less than 100 aa) that are involved in various biological processes, although this is rare (Hubé and Francastel, 2018). In addition, certain coding transcripts, such as TP53 mRNA, could also function as RNA, without translation to proteins, to regulate significant biological processes (Candeias, 2011; Kloc et al., 2011). Therefore, it seems reasonable to presume that the demarcation of RNA depending on its coding or noncoding status is somewhat blurred, and partially intertwined. That is, RNA roles are likely not tightly constrained (such as RNA functioning only as mRNA or ncRNA), but rather converge and overlap: lncRNAs can function by encoding small peptides, while mRNAs can use their special structural features, such as the 3′ UTR or 5′ UTR, to function (Figure 1).
FIGURE 1.
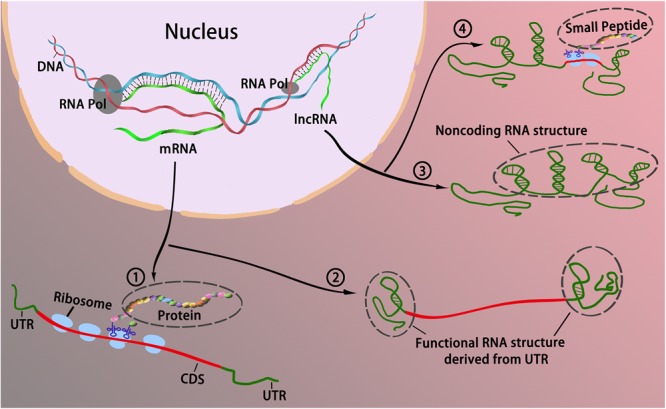
The interchangeable roles between coding and long noncoding RNAs. Traditionally, RNAs could be divided into two categories in accordance with their coding potential, that is, coding RNAs and noncoding RNAs. Coding RNAs generally refers to mRNA that encodes protein ① to act as various components including enzymes, cell structures, and signal transductors. Noncoding RNAs act as cellular regulators without encoding proteins ③. However, it appears that the boundaries blur between coding RNA and noncoding RNA as some coding mRNAs can function without translating to protein via the formation of RNA secondary structure primarily derived from the UTR ②; some lncRNAs can bind with ribosomes, and encode peptides to modulate cellular activities ④.
In the present article, we will review current studies of the bilateral functionality of lncRNAs and mRNAs in terms of their coding potential, as well as the advancement of high-throughput techniques that would facilitate a deeper recognition of functional diversity of RNAs. This review will highlight the cases that illuminate the contrapositive roles between lncRNAs and mRNAs, and briefly discuss the biological significance of these discoveries for gene expression and evolution.
Long Noncoding Rnas Encode Small Peptides/Proteins With Regulatory Functions
Peptides/Proteins Encoded by Regular Long Noncoding RNAs
The original definition of lncRNAs concerns their low/non-coding potential. However, with accumulating evidence from bioinformatics and ribosome transcriptome profiling, lncRNAs have been shown to display strong ribosomal associations in many species, varying from plant to animal, indicating a potential coding capacity in lncRNA sORFs (Kageyama et al., 2011; Nam et al., 2016; Yeasmin et al., 2018). In recent years, several micropeptides derived from lncRNAs have been shown to be functional. We have summarized these micropeptides in Table 1.
Table 1.
Peptides encoded by lncRNAs in plants and animals.
| Origin | Micropeptides | Gene | Function | Size (AAs) | References |
|---|---|---|---|---|---|
| Plants | Early nodulin 40 (Enod 40) | Enod 40 | Nodule organogenesis | 12; 24 | Rohrig et al., 2002 |
| Plants | POLARIS (PLS) | POLARIS | Leaf morphogenesis | 36 | Chilley et al., 2006 |
| Plants | ROTUNDIFOLIA (ROT4) | ROT4 | Leaf morphogenesis | 53 | Narita et al., 2004 |
| Plants | ROT18/ DLV1 | DEVIL1 (DVL1) | Plant organogenesis | 51 | Wen et al., 2004 |
| Plants | Kiss of death (KOD) | Kiss of death (KOD) | Programmed cell death regulation | 25 | Blanvillain et al., 2011 |
| Plants and animals | Brick1 (Brk) | Brick1 (Brk) | Leaf morphogenesis | 84 | Frank and Smith, 2002 |
| Poaceae | Zm401p10; Zm908p11 | Zm401 | Pollen development | 89; 97 | Ma et al., 2008 |
| Mammals | DWORF | LOC100507537 | Enhance muscle performance | 34 | Nelson et al., 2016 |
| Vertebrates | Toddler | LOC100506013 | Promotes cell migration; activator of APJ/Apelin receptor signaling | 54 | Pauli et al., 2014 |
| Vertebrates | Myomixer | LOC101929726 | Functionally involve in controlling muscle performance | 84 | Bi et al., 2017 |
| Fruit fly | Pri | polished rice (pri) | Epidermal morphogenesis in embryogenesis | 11; 32 | Kondo et al., 2007 |
| Fruit fly | MOTS-c | 12s rRNA | Insulin sensitivity and metabolic Homeostasis. | 16 | Lee et al., 2015 |
| Fruit fly | Pgc polypeptide | pgc | Positivephosphorylation of transcription elongation factor b (P-TEFb) | 71 | Hanyu-Nakamura et al., 2008 |
| Fruit fly | sarcolamban (Scl) | pncr003:2L | Calcium transport and muscle contraction, | 28; 29 | Magny et al., 2013 |
| Fruit Fly | Tarsal-less/tal | LP10384 | Morphogenesis, including tissue morphogenesis and pattern formation | 11 | Galindo et al., 2007 |
| Mammals | PINT87aa | LINC-PINT | Inhibit the transcriptional elongation of multiple oncogenes | 87 | Zhang et al., 2018b |
| Mammals | Mitoregulin (Mtln) | LINC00116 | Mitochondrial respiration, ROS, and Ca2+ retention capacity | 56 | Stein et al., 2018 |
| Mammals | MRI-2 | C7orf49 | Non-homologous end-joining DNA repair | 69 | Slavoff et al., 2014 |
| From humans to zebrafish | AGD3 | AGU1 | Human stem Cell differentiation | 63 | Kikuchi et al., 2009 |
| Mammals | NoBody | LINC01420/ LOC550643 | mRNA turnover and nonsense-mediated decay (NMD) | 67 | D’Lima et al., 2017 |
| Mammals | Minion | LOC101929726 | Muscle development | 84 | Zhang et al., 2017 |
| Human and mouse | SPAR | LINC00961 | Regulate muscle regeneration | 90 | Matsumoto et al., 2017 |
| Primates | HOXB-AS3 peptide | HOXB-AS3 | Suppresses colon cancer growth, PKM splicing and subsequent metabolic reprogramming | 53 | Huang et al., 2017 |
| Different species | Humanin | 16s rRNA | Program cell death | 24 | Hashimoto et al., 2001 |
| Human | SRAP | SRA | Diverse roles in both normal biological processes and pathological changes | 224; 236 | Emberley et al., 2003; Chooniedass-Kothari et al., 2006 |
| Mouse and human | Myoregulin (MLN) | LINC00948 in human; 2310015B20Rik in mouse | Inhibit the SERCA activity and regulate Ca2+ flow in muscle | 46 | Anderson et al., 2015 |
Steroid receptor RNA activator is a prototypic example of lncRNAs with both coding and noncoding products (Lanz et al., 1999; Mattick, 2003; Hubé et al., 2006, 2011). SRA was initially identified as a noncoding gene with multiple RNA isoforms, which is critical in many biological processes, such as acting as a co-activator of nuclear receptors and a regulator of steroid receptor-dependent gene expression (Hubé et al., 2006, 2011; Cooper et al., 2011). Interestingly, SRA can also encode for a conserved SRAP, which, in turn, represses the transcriptional regulatory activity of the SRA1 gene by interacting with a specific SRA stem-loop (Emberley et al., 2003; Chooniedass-Kothari et al., 2006; Hubé et al., 2011). The transmissible functionalities between the coding and noncoding SRA gene are caused by alternative splicing (AS) of introns/extrons (Colley and Leedman, 2011), suggesting the significance of AS events in the generation of bifunctional RNA.
Of note, among the small number of already-known functional micropeptides, a few are muscle-specific, and have been implicated in the regulation of the activities of SERCA (Anderson et al., 2016; Nelson et al., 2016; Matsumoto et al., 2017). For example, MLN, a 46-aa micropeptide specifically expressed in skeletal-muscle, is encoded by a lncRNA (LINC00948 in human and 2310015B20Rik in mouse); it can directly interact with SERCA to decrease the affinity of this ATPase for Ca2+ and inhibit Ca2+ entry into the SR (Anderson et al., 2015). The Scl micropeptide is encoded by the noncoding pncr003:2L gene, and can affect Ca2+ traffic in cardiac muscle in the fly; the mutation of this gene triggers an arrhythmic phenotype (Magny et al., 2013). The MOTS-C micropeptide can regulate insulin sensitivity and metabolic homeostasis in the mitochondria of muscle cells, and derives from mitochondrial 12S rRNA (Slavoff et al., 2014; Lee et al., 2015). In the above example, the Scl peptides and their respective regulatory functions in the heart are quite conserved between species, including the fly and humans (Magny et al., 2013). These results indicate that several sORFs embedded in the noncoding region of the genome seem to undergo a relatively stricter natural selection than adjacent sequences, raising the question of whether these sORFs have a capability to sprout into a new gene in situ or to be integrated as a component into new genes elsewhere during evolution.
The tal gene in Drosophila is of vital importance in tarsal morphogenesis in the fly leg, and stage-and position-specific expression have been reported in embryonic development. Although tal is regarded as a noncoding gene as none of its ORFs are over 100 aa, deeper analysis has found that the functionality of tal is predominantly dependent on the ORF regions (Manak et al., 2006). There are five ORFs in the tal gene, four of which contain a similar and conserved 7 aa motif that determines the functionality of the gene, with the shortest peptide of only 11 aa. Phylogenetic analysis revealed that these tal-like peptides are conserved in metazoans and represent a new class of eukaryotic genes. The discovery of these mini-peptides further expands the possible scope and function of lncRNA-encoded peptides that are hidden in currently sequenced genomes and the transcriptome (Galindo et al., 2007).
Peptides/Proteins Encoded by Circular RNAs
Circular RNAs pertain to a sub-category of specialized lncRNAs, which are primarily produced by backsplicing the 3′ end to the 5′ end of exons in the same transcript (often a coding gene) via the spliceosome, thereby forming lncRNAs in a circular shape (Ashwal-Fluss et al., 2014; Zhang X.O. et al., 2014; Starke et al., 2015). Through bioinformatics analysis and high-throughput sequencing, many circRNAs have been identified in multiple species (Sanger et al., 1976; Capel et al., 1993; Danan et al., 2012; Memczak et al., 2013; Jeck and Sharpless, 2014; Wang et al., 2014). However, understanding of circRNA function is still very limited. The reported biological activities of circRNAs include acting as a sponge for microRNAs (Hansen et al., 2013; Memczak et al., 2013), as a competitor during pre-mRNA splicing (Ashwal-Fluss et al., 2014), and as a transcriptional regulator in the nucleus (Li et al., 2015). The majority of circRNAs are chimeric lncRNAs derived from mRNA transcripts and likely in part encompass the exons of protein-coding genes. This poses the question of whether circRNAs have protein coding capabilities. In fact, many studies have demonstrated that circRNAs have coding capabilities both in vitro and in vivo in terms of cap-independent translation (Chen and Sarnow, 1995; Li and Lytton, 1999; Guo et al., 2014; Jeck and Sharpless, 2014; Abe et al., 2015; Wang and Wang, 2015; Pamudurti et al., 2017). Moreover, some functional protein products are encoded by circRNAs (such as circ-FBXW7; circ-Mbl, circ-ZNF609 and circ-SHPRH) (Rybak-Wolf et al., 2015; Pamudurti et al., 2017; Yang et al., 2018; Zhang et al., 2018a).
circ-ZNF609 was initially screened out in a functional genetic screen, and is differentially expressed during myogenesis (Legnini et al., 2017). This circRNA contains an ORF covering almost all ORF regions of the host gene, but has a small variation at the splice junction. Its protein product lacks the zinc-finger domain compared with its linear counterpart, with an obvious impact on myoblast proliferation. Interestingly, heat shock could significantly activate the translation of circ-ZNF609; suggesting a possible regulatory role of circRNA translation under specific stimuli (Legnini et al., 2017).
circ-Mbl was first detected in the lodge of the second exon of the splicing factor muscleblind (MBL/MBNL1) in flies and humans, with a function of competing with pre-mRNA splicing (Ashwal-Fluss et al., 2014). Recently, through a bioinformatics analysis of ribosome foot-printing datasets, Pamudurti and co-workers revealed that circ-Mbl could encode a peptide in the fly head, as detected through MS. Both circ-Mbl1 RNA and its protein-related product reside in the synaptosome and can be regulated by the 4E-BP and the transcription factor in forkhead family – FOXO, suggesting that this circRNA translation might be distinctively important in the brain (Pamudurti et al., 2017).
The observation that circRNAs generate proteins can be traced back to much earlier studies in Archaea, where circularized introns produce a site-specific endonuclease (Dalgaard et al., 1993). However, to date, direct experimental evidence for circRNA translation to peptides is still scarce; as a result, it is even tougher to understand the function of their translated products. Considering that most circRNAs stem from coding transcripts and contain complete exons, it is possibly assumed that the circRNAs and their coding-products might provide uncharacterized modes of regulation of gene and protein expression (Pamudurti et al., 2017). Therefore, it is important to further investigate the possible functions associated with circRNA coding.
Large-Scale Approaches for the Identification of Potential sORFs
To date, hundreds of thousands of lncRNAs have been discovered in various species, and there is a desire to study their relevant functional mechanisms (Okazaki et al., 2002; Liu et al., 2005; Kapranov et al., 2007; Ponting et al., 2009; Ulitsky and Bartel, 2013; Volders et al., 2013). However, it is unpractical to identify lncRNAs and predict their functions using only traditional technical approaches, irrespective of the requirement for intensive validation of the exact mechanisms underlying lncRNA activities. The same is true for the identification of lncRNA coding capacity. Therefore, new large-scale technologic approaches based on computational analysis of transcriptome data and proteomics data have been developed, all of which are mutually reinforcing and cross-validated.
A ribo-seq technique has been recently developed and is widely used to measure the full coding potential of RNA transcripts on a genome scale through deep sequencing of ribosome-protected RNA fragments (Ingolia et al., 2009). By identifying the precise ribosomal positions of RNAs, ribo-seq can plot the potential on-going events of translation in the cytosol, which is useful in identifying potentially functional micropeptides (Ingolia et al., 2011; Ingolia, 2016). With the advent of Ribo-Seq, thousands of translated sORFs were discovered in lncRNAs (Ingolia et al., 2011; Bazzini et al., 2014; Ruiz-Orera et al., 2014; Ji et al., 2015), with a few functional peptides, such as MLN (Anderson et al., 2015) and HOXB-AS3 (Huang et al., 2017). However, the proportion of coding lncRNAs estimated by various ribosome-profiling studies differ widely (Guttman et al., 2013; Ingolia et al., 2014), resulting from false positive and distinct prediction thresholds. Therefore, MS has emerged as a complementary method.
Mass spectrometry demonstrates excellent performance in detecting and characterizing the products of proteins/peptides in a complex biological sample. The detection of lncRNA-encoded peptides is the most direct evidence for lncRNA coding potential. However, to date, the proportion of coding lncRNAs detected by MS-based proteomes is small compared with that in ribo-seq results (Verheggen et al., 2017). The main weakness attributed to this approach is that MS-based proteomics is obviously impacted by the length and concentration of the detected samples. Therefore, specialized methods have been developed to circumvent these detection limitations. Short translation products at low abundance can surmount the threshold of MS detection through the use of peptidomics approaches (Schulz-Knappe et al., 2005) and enrichment protocols (Mustafa et al., 2015).
Both of the above techniques have their respective advantages and shortcomings; therefore, “proteogenomics” has been developed (Nesvizhskii, 2014; Menschaert and Fenyö, 2017; Ruggles et al., 2017). In proteogenomics, proteomics data are systematically integrated and analyzed with genomics and transcriptomic data generated from DNA-sequencing, RNA-sequencing and ribosome-profiling. The predicted sequences of proteins/peptides are tracked back to the genome and transcripts to identify the gene expression patterns and actual translational events. The significance of proteogenomics studies lies in improving genome annotation, and reasonably applying multi-omics data to explore complex and profound mechanisms in biological activities and complex diseases (Zhang B. et al., 2014; Zhang et al., 2016; Mertins et al., 2016).
Noncoding RNA Regulatory Functions Embedded in mRNAs
3′ UTR Regulatory Roles of mRNAs
Based on current research results, the noncoding regulatory functions discovered in mRNAs are mainly present in the 3′ UTRs, which were previously supposed to be the vital regulative elements for mRNA self-stability and location. Compared with highly conserved coding regions that have to undergo strictly selected pressure, the 3′ UTR displays more flexibility and plasticity between species. Its size varies from a few to hundreds of nucleotides, and likely has a close relationship with biological complexity (Chen et al., 2012; Mayr, 2016). Moreover, for an RNA molecule, other than the impact on base pairing, the changes in sequence are most likely to induce corresponding changes in structure, resulting in information transmitted from RNA to protein (Berkovits and Mayr, 2015).
By comprehensively estimating up-to-date cases where mRNAs regulate biologic activities without translating to protein, we found that the 3′ UTR of mRNA plays a large role as an effectors. Increasing evidence has demonstrated that the 3′ UTRs of mRNAs are actively involved in repressing the occurrence and progression of cancer cells, such as the 3′ UTRs of α-tropomyosin mRNA, prohibitin mRNA and ribonucleotide reductase mRNA (Rastinejad et al., 1993; Fan et al., 1996; Manjeshwar et al., 2003). These studies demonstrate that the 3′ UTRs of some mRNAs can antagonize tumor development, likely through RNA interactions with regulatory factors involved in cellular growth in a post-transcriptional pattern. Indeed, the 3′ UTR can recruit RNA-binding proteins, as in the case of CD47 mRNA. CD47 mRNA has two isoforms of the 3′ UTR, long (CD47-LU) and short (CD47-SU), and only the CD47-LU, which is AU-rich, can interact with the RNA-binding protein TIS11B to form a membraneless organelle with a specific biochemical and biophysical environment which is separate from the cytosol (Ma and Mayr, 2018). However, the most prevalent mode of action of the 3′ UTR is as ceRNAs, such as in the cases of CCR2 mRNA and Ube3a1 RNA, which confer to the function of lncRNAs (Valluy et al., 2015; Hu et al., 2017).
Noncoding Regulatory Roles of mRNA Not Involving the 3′ UTR
Other than the 3′ UTR, the 5′ UTR and ORF can also be involved in RNA-mediated regulatory function, although recent reports of this phenomenon are scare. Two mRNAs, TP53 mRNA and HIST1H1C mRNA, are recognized as being involved in ORF-mediated regulation. TP53 protein is a tumor suppressor implicated in many processes during tumor occurrence and development. However, a triple synonymous mutant (TriMp53) in codons led to a misshapen structure, resulting in loss of the IRES activity of p47 (one isoform of p53) and an abrogated affinity of hnRNPC, but with better binding to Mdm2, which is an E3 ubiquitin-protein ligase in mediating p53/TP53 ubiquitination, and an augmented ability of p53 to activate apoptosis. These facts indicate that TP53 has intricate regulatory roles at both the RNA and protein levels, suggesting that the functions of the RNA and protein molecules are closely intertwined (Candeias, 2011). HIST1H1C mRNA participates in regulating telomere length homeostasis. Aside from the protein-related product, a 15-nt long region in the ORF region (nt334–nt348) is attributed to HIST1H1C-mRNA-mediated biological activity, through complementation with the terminal stem-loop sequence of the P6b region of hTR, in a base-pairing pattern. These results extend the functional potency of mRNA ORF regions in a non-traditional and noncoding direction (Ivanyi-Nagy et al., 2018).
In terms of the 5′ UTR, there are only two examples. VEGF is a key regulator of angiogenesis during embryonal and cancerous development, and this regulatory function is closely correlated with the 5′ UTR. vegf mRNA has an unusually long 5′ UTR of 1,038 nucleotides, and contains two IRES, resulting in an intricate regulation of VEGF expression. In addition, the presence of the 5′ UTR of vegf mRNA alone in tumor cells could promote the expression of anti-apoptotic genes but repress pro-apoptotic genes, suggesting an anti-apoptotic role of the vegf 5′ UTR, and demonstrating its potential as a target for cancer treatment. To the best of our knowledge, the 5′ UTR of vegf mRNA represents the only example of an mRNA UTR which can promote tumor progression (Akiri et al., 1998; Huez et al., 1998; Masuda et al., 2008).
The c-myc P0 transcript is an isoform transcript from the promoter 0 (P0) of the c-myc gene, which has an extra ˜639-nucleotide extension of the 5′ UTR when compared with two major isoforms (P1 and P2) of c-myc mRNA. Ectopic expression of the 5′ UTR of the c-myc P0 transcript alone in HeLa cells results in significantly increased expression of the c-Myc1 (p67) and c-Myc2 (p64) proteins as well as incremental apoptosis sensitivity, but decreased tumorigenicity, all of which are likely attributable to competitive regulation of gene expression in the c-myc locus. These results demonstrate that the 5′ UTR potentially functions in trans to perform gene regulation (Blume et al., 2003).
Perspective
In recent years, researchers have begun to pay close attention to the development of bifunctional RNAs, and have discussed the evolved roles of RNAs with multiple functions (Dinger et al., 2011; Ulveling et al., 2011; Kageyama et al., 2011; Hubé and Francastel, 2018). In the early stages of such research, researchers discovered individual gene on a case-by-case basis. However, in the last ten years, rapid advances in large-scale detection and identification techniques (such as ribo-seq and MS-based proteomics) have facilitated multi-faceted investigations of genomes and vital processes, thus shedding light on the complex activities of various RNA molecules. Bifunctional RNAs raise questions about the concept of a gene, in terms of whether RNA, both coding and noncoding, is an independent gene type or a convergence of coding and noncoding genes which occurred during evolution. In this review, we intend to not only investigate the current status of bifunctional RNAs as reported in recent years, but also discuss the potential pervasiveness of bifunctional RNAs from a global perspective in terms of large-scale data.
With recent estimates of ribosome profiling, small peptides encoded by lncRNAs have significantly expanded the extent and diversity of the proteome, and predictions suggested that a large fraction of the annotated lncRNAs in various eukaryotic organisms would be translated with sORFs (Aspden et al., 2014; Ruiz-Orera et al., 2014; Mackowiak et al., 2015; Olexiouk et al., 2016). Proteogenomic evidence has confirmed that many small peptides which stem from regions of lncRNA genes are expressed differentially in different cell types and during different developmental/disease stages, although their functions are somewhat enigmatic (Nesvizhskii, 2014; Zhu et al., 2018). However, other studies have revealed that mRNAs could also be involved in cellular regulatory processes in a coding-independent manner (Nam et al., 2016). The results from a large-scale RNA structure analysis revealed that the secondary structures of mRNA have an essential regulatory effect on its maturation and stability, even for the evolutionarily conserved RNA silencing pathways of eukaryotes, suggesting that mRNAs partially retain the functionality of structure that exists in many RNA molecules (Katz and Burge, 2003; Li et al., 2012; Taggart et al., 2012). All these facts indicate that the coding potential and biological roles of mRNAs and lncRNAs could be switched in some cases, implying a conceptual blurring between coding and noncoding genes.
Many lncRNAs share similar features with classical mRNAs, such as transcription by polymerase II with a 5′-cap and 3′-polyadenylated tail, and frequent accumulation in the cytoplasm (van Heesch et al., 2014). Therefore, when associated with ribosomes, sORFs embedded in lncRNAs have a significant chance to be translated to peptides. The peptides derived from lncRNAs have a relatively shorter chain length and weaker conservation across different species, and this is consistent with the original lncRNAs which often have few introns, a low expression level and weak phylogenetic conservation (Cabili et al., 2011; Derrien et al., 2012; Kutter et al., 2012; Necsulea et al., 2014). From the perspective of proteins driving evolution, these peptides are likely considered to be an important source for new protein (Ruiz-Orera et al., 2014). Previously reported experimental evidence indicates that noncoding RNAs expressed at low levels could contribute to the birth of novel protein coding genes (Levine et al., 2006; Cai et al., 2008; Reinhardt et al., 2013). Given that several lncRNA-derived peptides have been demonstrated to play essential roles in many biological activities, it is worth investigating the putative significance of the generation of these lncRNA-derived peptides in gene evolution, expression and regulation.
However, in view of the huge quantity, diversified mechanisms of action, and intricate functions of lncRNAs, it is inappropriate to regard lncRNAs just as a pool for evolved peptides. In terms of RNA alone, its roles are diverse, including the potential to be retro-transcribed into DNA, or to act as an enzyme to participate in complex biochemical processes (Cech, 1986). Moreover, random RNA sequences can inoculate structurally complex and highly active RNA ligases, suggesting that randomness can produce functionality (Ekland et al., 1995). Therefore, it is very likely that RNA molecules alone comprise abundant genetic information, such as particular structural features and ultra-conservative sequence elements, which could regulate the timing and place of gene expression during cellular differentiation and development.
In recent decades, because of the addition of the huge family of noncoding genes, RNAs have provoked great interest for their mysterious roles in organisms. lncRNA-encoded peptides expand the horizon of functional mechanisms for these bio-macromolecules. To date, thousands of peptide products have been identified in human cells, with limited understanding of their function. The current review has summarized the recently discovered micropeptides implicated in various biological processes. We also discussed the potential noncoding roles of mRNAs as a regulator. The continued discovery and functional characterization of bifunctional RNAs will provide new insights into important cellular processes and organismal evolution.
Author Contributions
Both authors have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- 4E-BP
Eukaryotic translation initiation factor 4E (eIF4E)-binding protein
- aa
amino acid
- CCR2
C-C chemokine receptor type 2
- ceRNAs
competing endogenous RNAs
- circRNAs
circular RNAs
- FBXW7
F-Box and WD repeat domain containing 7
- Foxo
Forkhead box-o
- HIST1H1C
histone cluster 1 H1 family member C
- hnRNPC
heterogeneous nuclear ribonucleoprotein C
- hTR
human telomerase RNA
- IRES
internal ribosome entry site
- lncRNA
long non-coding RNA
- Mbl
Mannose-binding lectin
- Mdm2
mouse double minute 2
- MLN
myoregulin
- MOTS-C
mitochondrial open reading frame of the 12S rRNA-c
- MS
mass spectrometry
- nt
nucleotide
- ribo-seq
ribosome profiling sequencing
- Scl
sarcolamb
- SERCA
sarco/endoplasmic reticulum Ca2+-ATPase
- sORF
small open reading frame
- SR
sarcoplasmic reticulum
- SRA
steroid receptor RNA activator
- SRAP
steroid receptor RNA activator protein
- Ube3a1
ubiquitin-protein ligase E3A
- UTRs
untranslated regions
- VEGF
vascular endothelial growth factor
- ZNF609
zinc Finger Protein 609
Footnotes
Funding. This work was supported through the following grants by the National Natural Science Foundation of China (Grant Nos. 31471220 and 91440113), the Start-up Fund from Xishuangbanna Tropical Botanical Garden, and the “Top Talents Program in Science and Technology” from Yunnan Province.
References
- Abe N., Matsumoto K., Nishihara M., Nakano Y., Shibata A., Maruyama H., et al. (2015). Rolling circle translation of circular RNA in living human cells. Sci. Rep. 5:16435. 10.1038/srep16435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiri G., Nahari D., Finkelstein Y., Le S. Y., Elroy-Stein O., Levi B. Z. (1998). Regulation of vascular endothelial growth factor (VEGF) expression is mediated by internal initiation of translation and alternative initiation of transcription. Oncogene 17 227–236. 10.1038/sj.onc.1202019 [DOI] [PubMed] [Google Scholar]
- Anderson D. M., Anderson K. M., Chang C. L., Makarewich C. A., Nelson B. R., McAnally J. R., et al. (2015). A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160 595–606. 10.1016/j.cell.2015.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. M., Makarewich C. A., Anderson K. M., Shelton J. M., Bezprozvannaya S., Bassel-Duby R., et al. (2016). Widespread control of calcium signaling by a family of SERCA-inhibiting micropeptides. Sci. Signal. 9:ra119. 10.1126/scisignal.aaj1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwal-Fluss R., Meyer M., Pamudurti N. R., Ivanov A., Bartok O., Hanan M., et al. (2014). circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56 55–66. 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- Aspden J. L., Eyre-Walker Y. C., Phillips R. J., Amin U., Mumtaz M. A., Brocard M., et al. (2014). Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. eLife 3:e03528. 10.7554/eLife.03528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini A. A., Johnstone T. G., Christiano R., Mackowiak S. D., Obermayer B., Fleming E. S., et al. (2014). Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 33 981–993. 10.1002/embj.201488411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovits B. D., Mayr C. (2015). Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature 522 363–367. 10.1002/embj.201488411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi P., Ramirez-Martinez A., Li H., Cannavino J., McAnally J. R., Shelton J. M., et al. (2017). Control of muscle formation by the fusogenic micropeptide myomixer. Science 356 323–327. 10.1126/science.aam9361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanvillain R., Young B., Cai Y. M., Hecht V., Varoquaux F., Delorme V., et al. (2011). The Arabidopsis peptide kiss of death is an inducer of programmed cell death. EMBO J. 30 1173–1183. 10.1038/emboj.2011.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume S. W., Miller D. M., Guarcello V., Shrestha K., Meng Z., Snyder R. C., et al. (2003). Inhibition of tumorigenicity by the 5′-untranslated RNA of the human c-myc P0 transcript. Exp. Cell Res. 288 131–142. 10.1016/S0014-4827(03)00182-4 [DOI] [PubMed] [Google Scholar]
- Bonasio R., Shiekhattar R. (2014). Regulation of transcription by long noncoding RNAs. Annu. Rev. Genet. 48 433–455. 10.1146/annurev-genet-120213-092323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili M. N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., et al. (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25 1915–1927. 10.1101/gad.17446611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Zhao R., Jiang H., Wang W. (2008). De novo origination of a new protein-coding gene in Saccharomyces cerevisiae. Genetics 179 487–496. 10.1534/genetics.107.084491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candeias M. M. (2011). The can and can’t dos of p53 RNA. Biochimie 93 1962–1965. 10.1016/j.biochi.2011.06.010 [DOI] [PubMed] [Google Scholar]
- Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., et al. (1993). Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73 1019–1030. 10.1016/0092-8674(93)90279-Y [DOI] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M. C., Maeda N., et al. (2005). The transcriptional landscape of the mammalian genome. Science 309 1559–1563. 10.1126/science.1112014 [DOI] [PubMed] [Google Scholar]
- Cech T. R. (1986). A model for the RNA-catalyzed replication of RNA. Proc. Natl. Acad. Sci. U.S.A. 83 4360–4363. 10.1073/pnas.83.12.4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Y., Chen S. T., Juan H. F., Huang H. C. (2012). Lengthening of 3′UTR increases with morphological complexity in animal evolution. Bioinformatics 28 3178–3181. 10.1093/bioinformatics/bts623 [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Sarnow P. (1995). Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268 415–417. 10.1126/science.7536344 [DOI] [PubMed] [Google Scholar]
- Chilley P. M., Casson S. A., Tarkowski P., Hawkins N., Wang K. L., Hussey P. J., et al. (2006). The POLARIS peptide of Arabidopsis regulates auxin transport and root growth via effects on ethylene signaling. Plant Cell 18 3058–3072. 10.1105/tpc.106.040790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chooniedass-Kothari S., Hamedani M. K., Troup S., Hube F., Leygue E. (2006). The steroid receptor RNA activator protein is expressed in breast tumor tissues. Int. J. Cancer 118 1054–1059. 10.1002/ijc.21425 [DOI] [PubMed] [Google Scholar]
- Colley S. M., Leedman P. J. (2011). Steroid Receptor RNA Activator - A nuclear receptor coregulator with multiple partners: insights and challenges. Biochimie 93 1966–1972. 10.1016/j.biochi.2011.07.004 [DOI] [PubMed] [Google Scholar]
- Consortium E. P. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper C., Vincett D., Yan Y., Hamedani M. K., Myal Y., Leygue E. (2011). Steroid Receptor RNA Activator bi-faceted genetic system: heads or Tails? Biochimie 93 1973–1980. 10.1016/j.biochi.2011.07.002 [DOI] [PubMed] [Google Scholar]
- Crick F. (1970). Central dogma of molecular biology. Nature 227 561–563. 10.1038/227561a0 [DOI] [PubMed] [Google Scholar]
- Dalgaard J. Z., Garrett R. A., Belfort M. (1993). A site-specific endonuclease encoded by a typical archaeal intron. Proc. Natl. Acad. Sci. U.S.A. 90 5414–5417. 10.1073/pnas.90.12.5414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danan M., Schwartz S., Edelheit S., Sorek R. (2012). Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 40 3131–3142. 10.1093/nar/gkr1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22 1775–1789. 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger M. E., Gascoigne D. K., Mattick J. S. (2011). The evolution of RNAs with multiple functions. Biochimie 93 2013–2018. 10.1016/j.biochi.2011.07.018 [DOI] [PubMed] [Google Scholar]
- D’Lima N. G., Ma J., Winkler L., Chu Q., Loh K. H., Corpuz E. O., et al. (2017). A human microprotein that interacts with the mRNA decapping complex. Nat. Chem. Biol. 13 174–180. 10.1038/nchembio.2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekland E. H., Szostak J. W., Bartel D. P. (1995). Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 269 364–370. 10.1126/science.7618102 [DOI] [PubMed] [Google Scholar]
- Emberley E., Huang G. J., Hamedani M. K., Czosnek A., Ali D., Grolla A., et al. (2003). Identification of new human coding steroid receptor RNA activator isoforms. Biochem. Biophys. Res. Commun. 301 509–515. 10.1016/s0006-291x(02)03070-x [DOI] [PubMed] [Google Scholar]
- Fan H., Villegas C., Huang A., Wright J. A. (1996). Suppression of malignancy by the 3′ untranslated regions of ribonucleotide reductase R1 and R2 messenger RNAs. Cancer Res. 56 4366–4369. [PubMed] [Google Scholar]
- Frank M. J., Smith L. G. (2002). A small, novel protein highly conserved in plants and animals promotes the polarized growth and division of maize leaf epidermal cells. Curr. Biol. 12 849–853. 10.1016/s0960-9822(02)00819-9 [DOI] [PubMed] [Google Scholar]
- Galindo M. I., Pueyo J. I., Fouix S., Bishop S. A., Couso J. P. (2007). Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 5:e106. 10.1371/journal.pbio.0050106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C., Maquat L. E. (2011). lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470 284–288. 10.1038/nature09701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. U., Agarwal V., Guo H., Bartel D. P. (2014). Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 15:409. 10.1186/s13059-014-0409-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Russell P., Ingolia N. T., Weissman J. S., Lander E. S. (2013). Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154 240–251. 10.1016/j.cell.2013.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T. B., Jensen T. I., Clausen B. H., Bramsen J. B., Finsen B., Damgaard C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495 384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Hanyu-Nakamura K., Sonobe-Nojima H., Tanigawa A., Lasko P., Nakamura A. (2008). Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature 451 730–733. 10.1038/nature06498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y., Niikura T., Tajima H., Yasukawa T., Sudo H., Ito Y., et al. (2001). A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer’s disease genes and Abeta. Proc. Natl. Acad. Sci. U.S.A. 98 6336–6341. 10.1073/pnas.101133498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon C. C., Ramilowski J. A., Harshbarger J., Bertin N., Rackham O. J., Gough J., et al. (2017). An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543 199–204. 10.1038/nature21374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Li X., Guo X., Guo Q., Xiang C., Zhang Z., et al. (2017). The CCR2 3′UTR functions as a competing endogenous RNA to inhibit breast cancer metastasis. J. Cell Sci. 130 3399–3413. 10.1242/jcs.202127 [DOI] [PubMed] [Google Scholar]
- Huang J. Z., Chen M., Chen X., Gao C., Zhu S., Huang H., et al. (2017). A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol. Cell 68 171–184.e6. 10.1016/j.molcel.2017.09.015 [DOI] [PubMed] [Google Scholar]
- Hubé F., Francastel C. (2018). Coding and Non-coding RNAs, the frontier has never been so blurred. Front. Genet. 9:140. 10.3389/fgene.2018.00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubé F., Guo J., Chooniedass-Kothari S., Cooper C., Hamedani M. K., Dibrov A. A., et al. (2006). Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol. 25 418–428. 10.1089/dna.2006.25.418 [DOI] [PubMed] [Google Scholar]
- Hubé F., Velasco G., Rollin J., Furling D., Francastel C. (2011). Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Res. 39 513–525. 10.1093/nar/gkq833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huez I., Creancier L., Audigier S., Gensac M. C., Prats A. C., Prats H. (1998). Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol. Cell Biol. 18 6178–6190. 10.1128/mcb.18.11.6178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N. T. (2016). Ribosome footprint profiling of translation throughout the genome. Cell 165 22–33. 10.1016/j.cell.2016.02.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N. T., Brar G. A., Stern-Ginossar N., Harris M. S., Talhouarne G. J., Jackson S. E., et al. (2014). Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 8 1365–1379. 10.1016/j.celrep.2014.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N. T., Ghaemmaghami S., Newman J. R., Weissman J. S. (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324 218–223. 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N. T., Lareau L. F., Weissman J. S. (2011). Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147 789–802. 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanyi-Nagy R., Ahmed S. M., Peter S., Ramani P. D., Ong P. F., Dreesen O., et al. (2018). The RNA interactome of human telomerase RNA reveals a coding-independent role for a histone mRNA in telomere homeostasis. eLife 7:e40037. 10.7554/eLife.40037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W. R., Sharpless N. E. (2014). Detecting and characterizing circular RNAs. Nat. Biotechnol. 32 453–461. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z., Song R., Regev A., Struhl K. (2015). Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. eLife 4:e08890. 10.7554/eLife.08890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y., Kondo T., Hashimoto Y. (2011). Coding vs non-coding: translatability of short ORFs found in putative non-coding transcripts. Biochimie 93 1981–1986. 10.1016/j.biochi.2011.06.024 [DOI] [PubMed] [Google Scholar]
- Kapranov P., Cheng J., Dike S., Nix D. A., Duttagupta R., Willingham A. T., et al. (2007). RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316 1484–1488. 10.1126/science.1138341 [DOI] [PubMed] [Google Scholar]
- Kapusta A., Feschotte C. (2014). Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Genet. 30 439–452. 10.1016/j.tig.2014.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L., Burge C. B. (2003). Widespread selection for local RNA secondary structure in coding regions of bacterial genes. Genome Res. 13 2042–2051. 10.1101/gr.1257503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Fukuda M., Ito T., Inoue M., Yokoi T., Chiku S., et al. (2009). Transcripts of unknown function in multiple-signaling pathways involved in human stem cell differentiation. Nucleic Acids Res. 37 4987–5000. 10.1093/nar/gkp426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M., Foreman V., Reddy S. A. (2011). Binary function of mRNA. Biochimie 93 1955–1961. 10.1016/j.biochi.2011.07.008 [DOI] [PubMed] [Google Scholar]
- Kondo T., Hashimoto Y., Kato K., Inagaki S., Hayashi S., Kageyama Y. (2007). Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat. Cell Biol. 9 660–665. 10.1038/ncb1595 [DOI] [PubMed] [Google Scholar]
- Kutter C., Watt S., Stefflova K., Wilson M. D., Goncalves A., Ponting C. P., et al. (2012). Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 8:e1002841. 10.1371/journal.pgen.1002841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S. (2011). Initial impact of the sequencing of the human genome. Nature 470 187–197. 10.1038/nature09792 [DOI] [PubMed] [Google Scholar]
- Lanz R. B., McKenna N. J., Onate S. A., Albrecht U., Wong J., Tsai S. Y., et al. (1999). A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 97 17–27. 10.1016/S0092-8674(00)80711-4 [DOI] [PubMed] [Google Scholar]
- Lee C., Zeng J., Drew B. G., Sallam T., Martin-Montalvo A., Wan J., et al. (2015). The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 21 443–454. 10.1016/j.cmet.2015.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., et al. (2017). Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 66 22–37.e9. 10.1016/j.molcel.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. T., Jones C. D., Kern A. D., Lindfors H. A., Begun D. J. (2006). Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc. Natl. Acad. Sci. U.S.A. 103 9935–9939. 10.1073/pnas.0509809103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Zheng Q., Vandivier L. E., Willmann M. R., Chen Y., Gregory B. D. (2012). Regulatory impact of RNA secondary structure across the Arabidopsis transcriptome. Plant Cell 24 4346–4359. 10.1105/tpc.112.104232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. F., Lytton J. (1999). A circularized sodium-calcium exchanger exon 2 transcript. J. Biol. Chem. 274 8153–8160. 10.1074/jbc.274.12.8153 [DOI] [PubMed] [Google Scholar]
- Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., et al. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22 256–264. 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- Liu C., Bai B., Skogerbo G., Cai L., Deng W., Zhang Y., et al. (2005). NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 33 D112–D115. 10.1093/nar/gki041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Yan B., Qu Y., Qin F., Yang Y., Hao X., et al. (2008). Zm401, a short-open reading-frame mRNA or noncoding RNA, is essential for tapetum and microspore development and can regulate the floret formation in maize. J. Cell. Biochem. 105 136–146. 10.1002/jcb.21807 [DOI] [PubMed] [Google Scholar]
- Ma W., Mayr C. (2018). A membraneless organelle associated with the endoplasmic reticulum enables 3′UTR-mediated protein-protein interactions. Cell 175 1492–1506.e19. 10.1016/j.cell.2018.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackowiak S. D., Zauber H., Bielow C., Thiel D., Kutz K., Calviello L., et al. (2015). Extensive identification and analysis of conserved small ORFs in animals. Genome Biol. 16:179. 10.1186/s13059-015-0742-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magny E. G., Pueyo J. I., Pearl F. M., Cespedes M. A., Niven J. E., Bishop S. A., et al. (2013). Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 341 1116–1120. 10.1126/science.1238802 [DOI] [PubMed] [Google Scholar]
- Manak J. R., Dike S., Sementchenko V., Kapranov P., Biemar F., Long J., et al. (2006). Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat. Genet. 38 1151–1158. 10.1038/ng1875 [DOI] [PubMed] [Google Scholar]
- Manjeshwar S., Branam D. E., Lerner M. R., Brackett D. J., Jupe E. R. (2003). Tumor suppression by the prohibitin gene 3′untranslated region RNA in human breast cancer. Cancer Res. 63 5251–5256. [PubMed] [Google Scholar]
- Masuda K., Teshima-Kondo S., Mukaijo M., Yamagishi N., Nishikawa Y., Nishida K., et al. (2008). A novel tumor-promoting function residing in the 5′ non-coding region of vascular endothelial growth factor mRNA. PLoS Med. 5:e94. 10.1371/journal.pmed.0050094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A., Pasut A., Matsumoto M., Yamashita R., Fung J., Monteleone E., et al. (2017). mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 541 228–232. 10.1038/nature21034 [DOI] [PubMed] [Google Scholar]
- Mattick J. S. (2001). Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2 986–991. 10.1093/embo-reports/kve230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. S. (2003). Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays 25 930–939. 10.1002/bies.10332 [DOI] [PubMed] [Google Scholar]
- Mayr C. (2016). Evolution and biological roles of alternative 3′UTRs. Trends Cell Biol. 26 227–237. 10.1016/j.tcb.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495 333–338. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- Menschaert G., Fenyö D. (2017). Proteogenomics from a bioinformatics angle: a growing field. Mass Spectrom. Rev. 36 584–599. 10.1002/mas.21483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P., Mani D. R., Ruggles K. V., Gillette M. A., Clauser K. R., Wang P., et al. (2016). Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534 55–62. 10.1038/nature18003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa G. M., Larry D., Petersen J. R., Elferink C. J. (2015). Targeted proteomics for biomarker discovery and validation of hepatocellular carcinoma in hepatitis C infected patients. World J. Hepatol. 7 1312–1324. 10.4254/wjh.v7.i10.1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganuma T., Hirose T. (2013). Paraspeckle formation during the biogenesis of long noncoding RNAs. RNA Biol. 10 456–461. 10.4161/rna.23547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J. W., Choi S. W., You B. H. (2016). Incredible RNA: dual functions of coding and noncoding. Mol. Cells 39 367–374. 10.14348/molcells.2016.0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita N. N., Moore S., Horiguchi G., Kubo M., Demura T., Fukuda H., et al. (2004). Overexpression of a novel small peptide ROTUNDIFOLIA4 decreases cell proliferation and alters leaf shape in Arabidopsis thaliana. Plant J. 38 699–713. 10.1111/j.1365-313X.2004.02078.x [DOI] [PubMed] [Google Scholar]
- Necsulea A., Soumillon M., Warnefors M., Liechti A., Daish T., Zeller U., et al. (2014). The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505 635–640. 10.1038/nature12943 [DOI] [PubMed] [Google Scholar]
- Nelson B. R., Makarewich C. A., Anderson D. M., Winders B. R., Troupes C. D., Wu F., et al. (2016). A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351 271–275. 10.1126/science.aad4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii A. I. (2014). Proteogenomics: concepts, applications and computational strategies. Nat. Methods 11 1114–1125. 10.1038/nmeth.3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., et al. (2002). Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420 563–573. 10.1038/nature01266 [DOI] [PubMed] [Google Scholar]
- Olexiouk V., Crappe J., Verbruggen S., Verhegen K., Martens L., Menschaert G. (2016). sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 44 D324–D329. 10.1093/nar/gkv1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamudurti N. R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., et al. (2017). Translation of CircRNAs. Mol. Cell 66 9–21.e7. 10.1016/j.molcel.2017.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A., Norris M. L., Valen E., Chew G. L., Gagnon J. A., Zimmerman S., et al. (2014). Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science 343:1248636. 10.1126/science.1248636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L., Salmena L., Zhang J., Carver B., Haveman W. J., Pandolfi P. P. (2010). A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465 1033–1038. 10.1038/nature09144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C. P., Oliver P. L., Reik W. (2009). Evolution and functions of long noncoding RNAs. Cell 136 629–641. 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- Rastinejad F., Conboy M. J., Rando T. A., Blau H. M. (1993). Tumor suppression by RNA from the 3′ untranslated region of alpha-tropomyosin. Cell 75 1107–1117. 10.1016/0092-8674(93)90320-P [DOI] [PubMed] [Google Scholar]
- Reinhardt J. A., Wanjiru B. M., Brant A. T., Saelao P., Begun D. J., Jones C. D. (2013). De novo ORFs in Drosophila are important to organismal fitness and evolved rapidly from previously noncoding sequences. PLoS Genet. 9:e1003860. 10.1371/journal.pgen.1003860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J. L., Chang H. Y. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81 145–166. 10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrig H., Schmidt J., Miklashevichs E., Schell J., John M. (2002). Soybean ENOD40 encodes two peptides that bind to sucrose synthase. Proc. Natl. Acad. Sci. U.S.A. 99 1915–1920. 10.1073/pnas.022664799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M., Yandell M. D., Wortman J. R., Gabor Miklos G. L., Nelson C. R., Hariharan I. K., et al. (2000). Comparative genomics of the eukaryotes. Science 287 2204–2215. 10.1126/science.287.5461.2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggles K. V., Krug K., Wang X., Clauser K. R., Wang J., Payne S. H., et al. (2017). Methods, tools and current perspectives in proteogenomics. Mol. Cell. Proteomics 16 959–981. 10.1074/mcp.MR117.000024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Orera J., Messeguer X., Subirana J. A., Alba M. M. (2014). Long non-coding RNAs as a source of new peptides. eLife 3:e03523. 10.7554/eLife.03523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A., Stottmeister C., Glažar P., Jens M., Pino N., Giusti S., et al. (2015). Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 58 870–885. 10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]
- Sanger H. L., Klotz G., Riesner D., Gross H. J., Kleinschmidt A. K. (1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U.S.A. 73 3852–3856. 10.1073/pnas.73.11.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz-Knappe P., Schrader M., Zucht H. D. (2005). The peptidomics concept. Comb. Chem. High Throughput Screen. 8 697–704. 10.2174/138620705774962418 [DOI] [PubMed] [Google Scholar]
- Slavoff S. A., Heo J., Budnik B. A., Hanakahi L. A., Saghatelian A. (2014). A human short open reading frame (sORF)-encoded polypeptide that stimulates DNA end joining. J. Biol. Chem. 289 10950–10957. 10.1074/jbc.C113.533968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starke S., Jost I., Rossbach O., Schneider T., Schreiner S., Hung L. H., et al. (2015). Exon circularization requires canonical splice signals. Cell Rep. 10 103–111. 10.1016/j.celrep.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Stein C. S., Jadiya P., Zhang X., McLendon J. M., Abouassaly G. M., Witmer N. H., et al. (2018). Mitoregulin: a lncRNA-encoded microprotein that supports mitochondrial supercomplexes and respiratory efficiency. Cell Rep. 23 3710–3720.e8. 10.1016/j.celrep.2018.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover C. K., Pham X. Q., Erwin A. L., Mizoguchi S. D., Warrener P., Hickey M. J., et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1: an opportunistic pathogen. Nature 406 959–964. 10.1038/35023079 [DOI] [PubMed] [Google Scholar]
- Taggart A. J., DeSimone A. M., Shih J. S., Filloux M. E., Fairbrother W. G. (2012). Large-scale mapping of branchpoints in human pre-mRNA transcripts in vivo. Nat. Struct. Mol. Biol. 19 719–721. 10.1038/nsmb.2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y., Rinn J., Pandolfi P. P. (2014). The multilayered complexity of ceRNA crosstalk and competition. Nature 505 344–352. 10.1038/nature12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I., Bartel D. P. (2013). lincRNAs: genomics, evolution, and mechanisms. Cell 154 26–46. 10.1016/j.cell.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulveling D., Francastel C., Hubé F. (2011). Identification of potentially new bifunctional RNA based on genome-wide data-mining of alternative splicing events. Biochimie 93 2024–2027. 10.1016/j.biochi.2011.06.019 [DOI] [PubMed] [Google Scholar]
- Valluy J., Bicker S., Aksoy-Aksel A., Lackinger M., Sumer S., Fiore R., et al. (2015). A coding-independent function of an alternative Ube3a transcript during neuronal development. Nat. Neurosci. 18 666–673. 10.1038/nn.3996 [DOI] [PubMed] [Google Scholar]
- van Heesch S., van Iterson M., Jacobi J., Boymans S., Essers P. B., de Bruijn E., et al. (2014). Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 15:R6. 10.1186/gb-2014-15-1-r6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter J. C., Adams M. D., Myers E. W., Li P. W., Mural R. J., Sutton G. G., et al. (2001). The sequence of the human genome. Science 291 1304–1351. 10.1126/science.1058040 [DOI] [PubMed] [Google Scholar]
- Verheggen K., Volders P. J., Mestdagh P., Menschaert G., Van Damme P., Gevaert K., et al. (2017). Noncoding after all: biases in proteomics data do not explain observed absence of lncRNA translation products. J. Proteome Res. 16 2508–2515. 10.1021/acs.jproteome.7b00085 [DOI] [PubMed] [Google Scholar]
- Volders P. J., Helsens K., Wang X., Menten B., Martens L., Gevaert K., et al. (2013). LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 41 D246–D251. 10.1093/nar/gks915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. C., Chang H. Y. (2011). Molecular mechanisms of long noncoding RNAs. Mol. Cell 43 904–914. 10.1016/j.molcel.2011.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. L., Bao Y., Yee M. C., Barrett S. P., Hogan G. J., Olsen M. N., et al. (2014). Circular RNA is expressed across the eukaryotic tree of life. PLoS One 9:e90859. 10.1371/journal.pone.0090859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang Z. (2015). Efficient backsplicing produces translatable circular mRNAs. RNA 21 172–179. 10.1261/rna.048272.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Lease K. A., Walker J. C. (2004). DVL, a novel class of small polypeptides: overexpression alters Arabidopsis development. Plant J. 37 668–677. 10.1111/j.1365-313X.2003.01994.x [DOI] [PubMed] [Google Scholar]
- Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., et al. (2018). Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 110 304–315. 10.1093/jnci/djx166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeasmin F., Yada T., Akimitsu N. (2018). Micropeptides encoded in transcripts previously identified as long noncoding RNAs: a new chapter in transcriptomics and proteomics. Front. Genet. 9:144. 10.3389/fgene.2018.00144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J. H., Abdelmohsen K., Gorospe M. (2014). Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 34 9–14. 10.1016/j.semcdb.2014.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Wang J., Wang X., Zhu J., Liu Q., Shi Z., et al. (2014). Proteogenomic characterization of human colon and rectal cancer. Nature 513 382–387. 10.1038/nature13438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Liu T., Zhang Z., Payne S. H., Zhang B., McDermott J. E., et al. (2016). Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell 166 755–765. 10.1016/j.cell.2016.05.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Huang N., Yang X., Luo J., Yan S., Xiao F., et al. (2018a). A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37 1805–1814. 10.1038/s41388-017-0019-9 [DOI] [PubMed] [Google Scholar]
- Zhang M., Zhao K., Xu X., Yang Y., Yan S., Wei P., et al. (2018b). A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 9:4475. 10.1038/s41467-018-06862-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Vashisht A. A., O’Rourke J., Corbel S. Y., Moran R., Romero A., et al. (2017). The microprotein Minion controls cell fusion and muscle formation. Nat. Commun. 8:15664. 10.1038/ncomms15664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. O., Wang H. B., Zhang Y., Lu X., Chen L. L., Yang L. (2014). Complementary sequence-mediated exon circularization. Cell 159 134–147. 10.1016/j.cell.2014.09.001 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Orre L. M., Johansson H. J., Huss M., Boekel J., Vesterlund M., et al. (2018). Discovery of coding regions in the human genome by integrated proteogenomics analysis workflow. Nat. Commun. 9:903. 10.1038/s41467-018-03311-y [DOI] [PMC free article] [PubMed] [Google Scholar]


