Summary
Global food production is set to keep increasing despite a predicted decrease in total arable land [1]. To achieve higher production, denser planting will be required on increasingly degraded soils. When grown in dense stands, crops elongate and raise their leaves in an effort to reach sunlight, a process termed shade avoidance [2]. Shade is perceived by a reduction in the ratio of red (R) to far-red (FR) light and results in the stabilization of a class of transcription factors known as PHYTOCHROME INTERACTING FACTORS (PIFs) [3, 4]. PIFs activate the expression of auxin biosynthesis genes [4, 5] and enhance auxin sensitivity [6], which promotes cell-wall loosening and drives elongation growth. Despite our molecular understanding of shade-induced growth, little is known about how this developmental program is integrated with other environmental factors. Here, we demonstrate that low levels of NaCl in soil strongly impair the ability of plants to respond to shade. This block is dependent upon abscisic acid (ABA) signaling and the canonical ABA signaling pathway. Low R:FR light enhances brassinosteroid (BR) signaling through BRASSINOSTEROID SIGNALING KINASE 5 (BSK5) and leads to the activation of BRI1 EMS SUPPRESSOR 1 (BES1). ABA inhibits BSK5 upregulation and interferes with GSK3-like kinase inactivation by the BR pathway, thus leading to a suppression of BES1:PIF function. By demonstrating a link between light, ABA-, and BR-signaling pathways, this study provides an important step forward in our understanding of how multiple environmental cues are integrated into plant development.
Keywords: plant photobiology, salt response, phytohormones, abscisic acid, brassinosteroids, salt stress, phytochrome, PIF
Highlights
-
•
Low-level soil salinity inhibits plant shade avoidance
-
•
The effect of salt is dependent upon abscisic acid
-
•
Salt antagonizes the shade-mediated upregulation of brassinosteroid signaling
Intensively farmed crops often experience multiple stresses simultaneously. Here, Hayes et al. show that low-level soil salinity suppresses shade avoidance in plants. Through investigation of the mechanisms underlying this trait, they uncover a regulatory pathway that converges at the level of brassinosteroid signaling.
Results and Discussion
Soil Salinity Inhibits Plant Shade Avoidance
Soil salinity is detrimental to plants and often results in a reduction in stem and root biomass accumulation [7]. Most previous studies have, however, focused on the effects of salt at a range of very high concentrations (at a median concentration of 150 mM NaCl) [8]. We opted for a nuanced approach to investigate how low concentrations of NaCl may affect plant shade avoidance. Growing plants in tissue culture often masks genes involved in NaCl sensitivity [9, 10], and so for phenotypic experiments plants were grown on soil. Arabidopsis thaliana (Arabidopsis) seeds were germinated under white light (Wl) for 3 days, before transfer to new soil that had been pre-treated with NaCl solution. Following this, plants were returned to Wl for a further day to acclimate, before they were shifted to Wl or Wl with supplementary far-red LEDs (+FR). Hypocotyl lengths were measured on day 7 (for a schematic diagram of the treatments see Figure S1A). Water or NaCl solution was applied from below, and the soil was kept saturated to avoid dehydration.
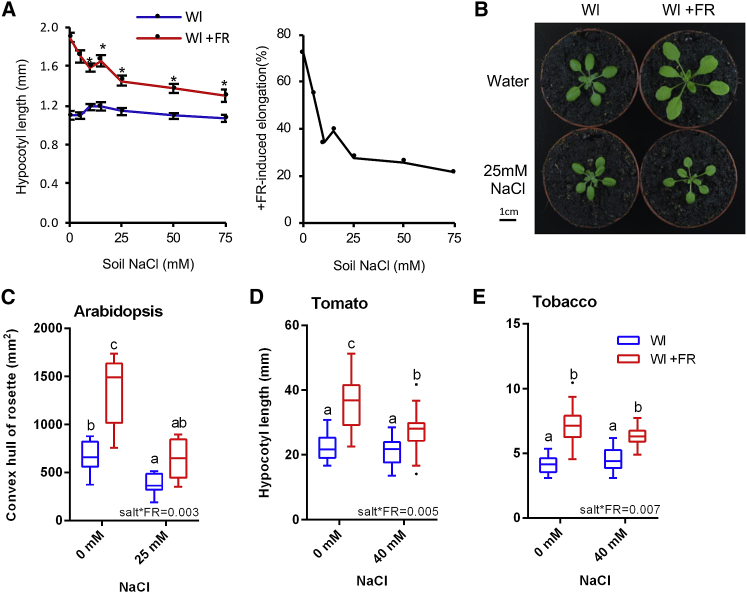
We observed a strong NaCl-mediated reduction in +FR-induced hypocotyl elongation when plants were watered with as low as 10 mM NaCl (Figure 1A). Increasing NaCl concentrations beyond 25–75 mM NaCl provided no further inhibition of +FR-induced hypocotyl elongation, and so 25 mM NaCl was selected for further investigation. NaCl inhibited +FR-induced elongation across the 3 days of +FR treatment (Figure S1B). In adult plants, 25 mM NaCl exerted a strong inhibition of +FR-induced rosette expansion, mostly through an inhibition of lamina elongation (Figures 1B, 1C, S1C, and S1D). Additionally, we found that NaCl inhibited +FR-induced elongation in tomato and tobacco seedlings (Figures 1D and 1E), raising the possibility that this is a general phenomenon in shade-avoiding plants.
Figure 1.
Soil Salinity Inhibits +FR-Induced Elongation
(A) Left: hypocotyl length of 7-day-old Col seedlings germinated in Wl, transferred to NaCl soil of the indicated concentration at day 3, and then shifted to WL ± FR at day 4. Data represent mean (n ≥ 19) ±SE. Asterisks indicate salt-mediated inhibition of hypocotyl elongation compared to 0 mM NaCl control; Student’s t test (p < 0.05). Right: the same data expressed as the percentage of +FR-induced elongation at each salt concentration.
(B) Representative 21-day-old Col plants germinated in Wl and transferred to ±25 mM NaCl at day 3, before shifting to WL ± FR at day 10.
(C) Rosette circumference of plants grown as in (B) (n ≥ 10).
(D) Hypocotyl length of 11-day-old tomato (Solanum lycopersicum var. “Moneymaker”) seedlings germinated in Wl, transferred to ±40 mM NaCl at day 7, and shifted into WL ± FR at day 8 (n ≥ 19).
(E) Hypocotyl length of tobacco (Nicotiana benthamiana) seedlings grown as in (D) (n ≥ 18). Boxplots are visualized by the Tukey method. In all figures, different letters designate significantly different means by 2-way ANOVA + Tukey’s post hoc test (p < 0.05). Interaction p value is shown inset.
See also Figure S1.
Soil Salinity Acts through the ABA Pathway
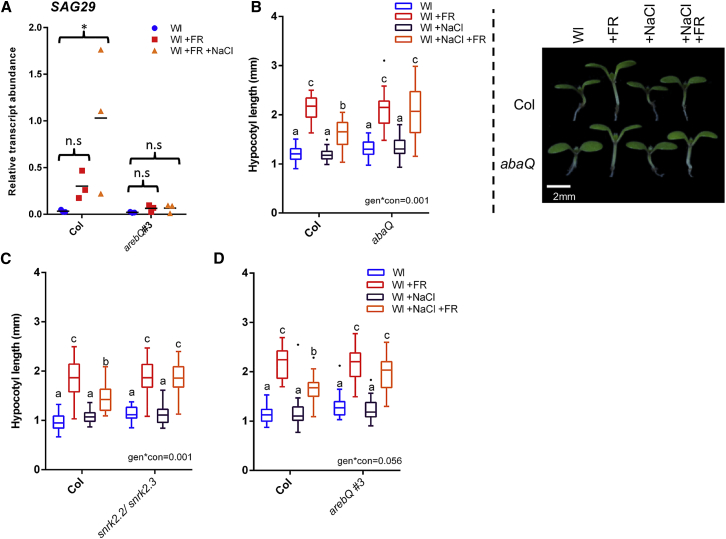
Many plant responses to soil salinity are mediated through the hormone abscisic acid (ABA) [11]. We found that transcripts of SAG29 (an ABA-responsive gene [12]) accumulated to high levels in +NaCl +FR conditions (Figure 2A). This accumulation did not occur in plants lacking four ABA-responsive transcription factors (ABA-responsive element-binding factor 1, 2, 3, and 4 [13], here referred to as arebQ). We therefore decided to assess whether ABA signaling is required for NaCl-mediated inhibition of +FR-induced elongation. In mutants that lacked either four of the ABA receptors (pyr1/pyl1/pyl2/pyl4- here referred to as abaQ [14]) or two ABA-signaling kinases (sucrose non-fermenting 1-related protein kinase 2.2 and 2.3- snrk2.2snrk2.3) [15], NaCl no longer had an effect on +FR-induced hypocotyl elongation (Figures 2B and 2C). Whereas plants lacking a single ABA activated transcription factor, ABA INSENSITIVE 5 (abi5-1), still showed a strong NaCl-mediated inhibition of hypocotyl elongation under +FR light (Figure S2A), plants lacking the AREB quartet (arebQ) elongated to the same extent in the presence and absence of NaCl (Figure 2D). Taken together, these results suggest that the effect of NaCl on +FR-induced elongation is dependent upon enhanced ABA synthesis or signaling.
Figure 2.
Salinity-Mediated Inhibition of +FR-Induced Elongation Requires ABA Signaling
(A) Relative SAG29 transcript abundance in the hypocotyls of Col and arebQ mutant plants grown 3 days in Wl, transferred to ±25 mM NaCl soil at day 3, and then shifted to WL ± FR at day 4. Tissues were harvested at Zeitgeber time (ZT) 4.5 on day 6 (n = 3). ∗Significant difference from Wl control.
(B–D) Hypocotyl lengths of 7-day-old wild-type and (B) abaQ, (C) snrk2.2/snrk2.3, and (D) arebQ mutants germinated in Wl, transferred to ±25 mM NaCl soil at day 3, and then shifted to WL ± FR at day 4 (n ≥ 22).
Right image of (A) depicts representative seedlings grown in these conditions. Boxplots are visualized by the Tukey method. Gene expression studies show individual values, with a horizontal bar representing the mean. Different letters designate significantly different means by 2-way ANOVA + Tukey’s post hoc test (p < 0.05). Interaction p value is shown inset.
See also Figure S2 and STAR Methods.
In addition to these mutant and gene expression analyses, we found that applying increasing concentrations of ABA directly between the cotyledons had a similar effect as applying increasing NaCl concentrations to the soil (Figure 1A versus Figures S2B). Low concentrations of ABA provided a strong break on hypocotyl elongation in +FR light; this inhibition was saturated by 10 μM ABA and remained constant until at least 100 μM ABA. As with NaCl, the inhibition of +FR-induced hypocotyl elongation by exogenous ABA was absent in the abaQ (Figure S2C) and snrk2.2/snrk2.3 mutants (Figure S2D). Despite the clear requirement for ABA signaling, NaCl still inhibited +FR-induced elongation in mutants that have reduced levels of ABA synthesis, such as aba2-1 and aba3-1 (Figures S2E and S2F), and we were unable to detect any increase in ABA levels in whole NaCl-grown seedlings (Figure S2G). It may be that ABA signaling is enhanced only locally, ABA distribution is altered, or that there is direct activation of the ABA signal pathway by an unknown mechanism.
Salt and ABA Impede upon PIF4/PIF5 Action
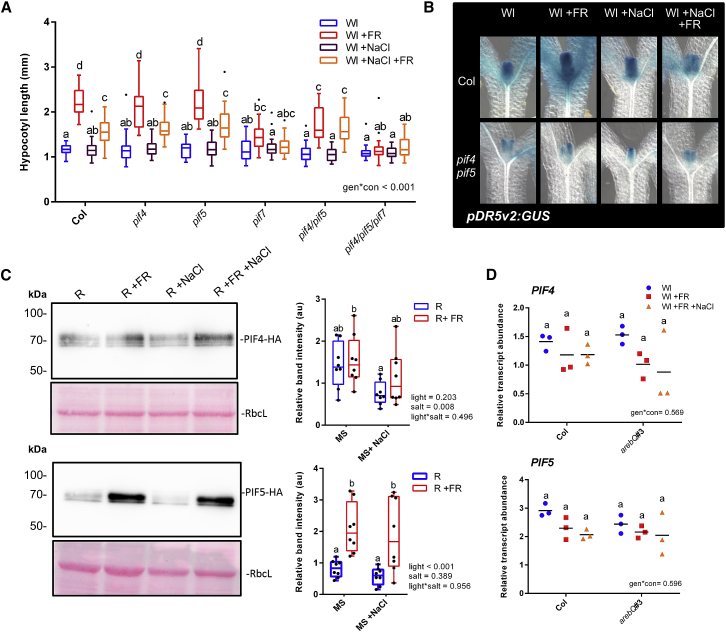
We next investigated the manner in which ABA and NaCl inhibit +FR-induced elongation. The induction of hypocotyl elongation by +FR requires PIF4, PIF5, and PIF7 [3, 4, 16] (Figure 3A). In our conditions, PIF7 was the dominant PIF driving +FR-induced hypocotyl elongation, as the pif7 single mutant showed no significant +FR-induced elongation and there were no additional effects in the pif4/pif5/pif7 triple mutant. Single pif4 or pif5 mutants appeared similar to the wild-type, with strong +FR-induced hypocotyl elongation that was inhibited by NaCl. The pif4/pif5 double mutant did however show reduced +FR-induced elongation, suggesting that these genes act redundantly. Notably, the hypocotyl length of the pif4/pif5 mutant in +FR light was the same as NaCl +FR-treated wild-type plants, and NaCl did not further suppress elongation in this mutant (Figure 3A). In a similar fashion to NaCl, ABA inhibited +FR-induced hypocotyl elongation in the wild-type to the same length as the pif4/pif5 mutant, and ABA had no further effect on elongation in this mutant (Figure S2C). These findings suggest that NaCl and ABA could act to suppress PIF4/PIF5 function.
Figure 3.
Soil Salinity Suppresses PIF Function and +FR-Induced Gene Expression and Auxin Signaling
(A) Hypocotyl length of 7-day-old Col, pif4-101, pif5, and pif7-1 double- and triple-mutant seedlings germinated in Wl, transferred to 25 mM NaCl soil at day 3, and then shifted to WL ± FR at day 4 (n ≥ 23).
(B) Seedlings expressing a pDR5v2:GUS reporter in the Col or pif4-101/pif5 background were germinated in Wl and transferred to ±25 mM NaCl soil on day 3. On day 4 at plants were shifted to WL ± FR and seedlings were fixed on day 5 at ZT 4.5. Representative hypocotyls of GUS stained seedlings are shown (quantification of the top half of the hypocotyls is shown in Figure S3E).
(C) Western blots of 35S:PIF4-HA (top) and 35S:PIF5-HA (bottom) plants germinated on plates in Wl, transferred to 75 mM NaCl plates, and grown for a further 2 days in R light. At ZT 3 on day 5, plants were moved to R ± FR for 1 h, and seedlings were harvested into liquid N2. Western blots probed with anti-HA. Pink lanes show Ponceau staining of rubisco large subunit (RbcL) as a loading control. The right image depicts relative band intensity after normalization to RbcL (n = 8, across 4 separate experiments).
(D) Relative PIF4 (top) and PIF5 (bottom) transcript abundance in the hypocotyls of Col and arebQ mutant plants grown as in (A) that were harvested at ZT 4.5 on day 6 (n = 3).
Boxplots are visualized by the Tukey method (if n ≥ 10) or as max-min with every value shown (if n < 10). Gene expression studies show individual values, with a horizontal bar representing the mean. In all figures, different letters designate significantly different means by 2-way ANOVA + Tukey’s post hoc test (p < 0.05).
See also Figure S3 and STAR Methods.
We found that the expression of two +FR-induced genes (PRE1 and SAUR16) was suppressed by pre-treatment with NaCl. In the pif4/pif5 mutant, however, NaCl did not affect the expression of these genes (Figure S3A). We used plants expressing luciferase (LUC) reporter constructs to investigate salt-mediated inhibition of +FR-induced genes over a longer timescale. When LUC expression was driven by the PIL1 [17] or IAA19 promoter, +FR light resulted in a rapid induction of LUC activity, and NaCl and ABA caused a sustained suppression of this upregulation over several days (Figure S3B). As both the PIL1 and IAA19 promoters contain several G-boxes (Figure S3C) that are established PIF4 and PIF5 direct targets [5, 18], these results are consistent with NaCl and ABA-mediated inhibition of PIF4 and PIF5 activity.
PIFs promote hypocotyl elongation in +FR light through an increase in auxin signaling [2]. To investigate whether NaCl and ABA suppressed auxin signaling, we generated a DR5v2:GUS (β-glucuronidase) auxin reporter line. DR5v2 is a synthetic promoter that was developed as a more sensitive auxin reporter than the widely used DR5 [19]. Three independent DR5v2:GUS lines were tested and confirmed to be highly auxin responsive (Figure S3D). We found that, when grown under supplementary FR light, DR5v2:GUS plants had increased GUS staining at the upper part of the hypocotyl, but that this response was attenuated in the presence of NaCl (Figures 3B and S3E). We then crossed this line into the pif4/pif5 mutant background. In the absence of PIF4 and PIF5, we no longer detected a +FR-mediated increase in GUS staining, and salt had no further effect on this mutant (Figures 3B and S3E). Again, this is consistent with an NaCl-mediated inhibition of PIF4/ PIF5 activity.
In wild-type plants, auxin application promoted hypocotyl elongation in all of our growth conditions, but NaCl still significantly inhibited +FR-induced elongation in the presence of supplementary auxin (Figure S3F), suggesting that NaCl may act to suppress auxin sensitivity or transport [20]. Supplementary auxin also enhanced hypocotyl elongation in the pif4/pif5 mutant in most of the conditions tested but did not rescue elongation in this mutant to the same length as wild-type plants treated with auxin and +FR (Figure S3F). NaCl had no effect on +FR-induced elongation in the pif4/pif5 mutant, even in the presence of supplementary auxin, which is in accordance with the role of PIF4 and PIF5 in the control of auxin signaling [6].
PIFs are often regulated by light at the level of protein stability [3, 21], so we assessed the abundance of constitutively expressed PIF4-HA and PIF5-HA under +FR and +NaCl conditions. While 2-way ANOVAs revealed an effect of NaCl on PIF4 stability, we did not see a statistically significant reduction in PIF4 protein levels in any of the individual light conditions tested (Figure 3C). PIF5 was stabilized by +FR light, but NaCl had no significant effect on this stabilization (Figure 3C). Additionally, we observed no changes in PIF4 or PIF5 expression in NaCl or in the arebQ mutant (Figure 3D).
Brassinosteroid Pathway Is Inhibited by Salt and ABA
We hypothesized that PIF4 and PIF5 action could be inhibited through a change in activity of one of their interaction partners. PIF action is suppressed by competitive heterodimerization with DELLA proteins [22, 23]. It has previously been shown that NaCl stabilizes DELLA proteins [24] and DELLA degradation is required for +FR-induced shade avoidance to occur [25]. Mutants lacking all five DELLA proteins showed longer hypocotyls than the wild-type in all of our conditions (Figure S4A) but still exhibited a strong NaCl-mediated inhibition of +FR-induced elongation. This suggests that NaCl inhibits shade avoidance through another mechanism. A recent study found that the stabilization of DELLAs by NaCl in the root is transient [26], and so it may be that their role is restricted to growth arrest upon initial NaCl exposure.
In NaCl-treated roots, ABA production precedes a reduction of BR signaling [26] and it is known that BR signaling is necessary for +FR-induced hypocotyl elongation to occur [16]. This raises the possibility that during NaCl exposure ABA acts to inhibit BR signals. We tested the effect of brassinazole (BRZ- an inhibitor of BR synthesis) on hypocotyl elongation under +FR light. We found that BRZ, much like both NaCl and ABA, inhibited hypocotyl elongation very readily at low concentrations and that this inhibition remained constant as concentrations increased (Figure S4B). Intriguingly, hypocotyl length at this plateau was equal to that of +FR-treated plants grown on saline soil in the absence of BRZ. Furthermore, simultaneous application of ABA, BRZ, and NaCl had no further effect on the hypocotyl length of +FR-treated plants than NaCl treatment alone (Figure S4C), consistent with these compounds converging on the same pathway. Supporting this, application of epi-brassinolide (BL) rescued +FR-induced hypocotyl elongation in NaCl-exposed plants (Figure S4D). Hypocotyl length was rescued at low concentrations of epi-BL, up to the extent of +FR-treated control plants grown in the absence of NaCl (Figure S4D). We also observed that epi-BL could not rescue hypocotyl elongation in the yucca2589 mutant (Figure S4E), which probably indicates that in our conditions, BR-mediated rescue of hypocotyl elongation is auxin dependent.
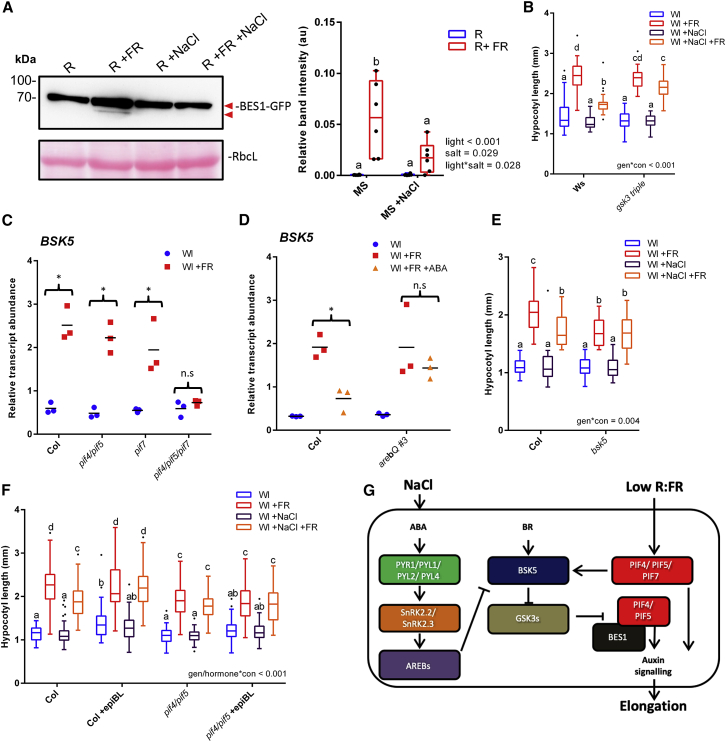
Although it is known that inhibition of BR signaling inhibits shade avoidance, it is currently unclear whether +FR enhances BR signals. We assessed BR activity through looking at BES1 phosphorylation levels in a 35S:BES1-GFP line. We found that short-term +FR treatments rapidly resulted in a faster running form of BES1 (Figure 4A), which has previously been shown to correspond to the active, de-phosphorylated form of the protein [27]. We found that NaCl inhibited the +FR-mediated induction of this faster running band (Figure 4A), suggesting that BES1 activation by +FR light is inhibited by NaCl. Despite this, we found that plants that hyper-accumulate BES1 (bes1-D mutants) actually had shorter hypocotyls under +FR than wild-type plants (Figure S4F). Recently, it has been shown that the balance between BES1 and PIF4 levels dictates whether BES1 acts as an activator or repressor of BR synthesis genes [28]. Over-accumulation of BES1 suppresses BR synthesis genes and, since PIF4 activity is directly suppressed in the absence of BR [29], this may explain the reduced +FR response in the bes1-D line.
Figure 4.
ABA and NaCl Inhibit +FR Elongation through Inhibiting BR Signaling
(A) 35S:BES1-GFP-expressing plants were germinated on plates for 3 days in Wl, transferred to ±75 mM NaCl plates, and then grown in R light for 2 further days. On day 5, plants were shifted to R ± FR at ZT 3 and tissue was harvested at ZT 4. Proteins were identified through western blots with anti-GFP. Right image shows quantified lower band intensity relative to RbcL (n = 6).
(B) Hypocotyl length of 7-day-old Wassilevskija (Ws) and gsk3-triple mutant germinated in Wl, transferred to ±25 mM NaCl soil at day 3, and then shifted to WL ± FR at day 4 (n ≥ 24).
(C) Col and pif4-101/pif5, pif7-1, and pif4-101/pif5/pif7-1 plants were germinated in Wl for 3 days before transfer to new pots. At ZT 2.5 on day 7, plants were shifted to Wl ± FR light. At ZT 4.5, the hypocotyls of approximately 20 seedlings per sample were dissected and RNA was extracted. BSK5 transcripts relative to AT1613320 are shown (n = 3). ∗+FR-induced increase.
(D) Col and arebQ #3 plants were grown as in (C). At ZT 1.5 on day 7, plants were sprayed with 25 μM ABA and exposed to Wl ± FR at ZT 2.5. At ZT 4.5, the hypocotyls of approximately 20 seedlings per sample were dissected and RNA extracted. BSK5 transcripts relative to AT1613320 are shown (n = 3). ∗ABA-mediated decrease.
(E) Hypocotyl length of Col and bsk5 grown as in (B) (n ≥ 22).
(F) Hypocotyl length of 7-day-old Col and pif4-101/pif5 plants grown as in (B), with applications 10 μM epi-BL or ethanol control on days 3–6 (n ≥ 46).
(G) A proposed mechanism for salt-mediated inhibition of low R:FR-induced elongation. Low R:FR light promotes the stability of PIF4, 5 and 7. These PIFs redundantly upregulate the expression of BSK5. BSK5 acts to suppress the activity of GSK3-like kinases, which relieves their suppression of the PIF:BES1 signaling module in a positive feedback loop. NaCl promotes the canonical ABA signal transduction pathway, resulting in the increased activity of AREB transcription factors. AREBs inhibit the upregulation of BSK5 and thereby enhance GSK3-like kinase action. GSK3-like kinases are then free to inhibit the PIF:BES1 signaling module, limiting auxin signaling in the hypocotyl.
Boxplots are visualized by the Tukey method (if n ≥ 10) or as max-min with every value shown (if n < 10). Gene expression studies show individual values, with a horizontal bar representing the mean. Where letters are shown, different letters designate significantly different means by 2-way ANOVA + Tukey’s post hoc test (p < 0.05).
See also Figure S4 and STAR Methods.
BES1 phosphorylation is mediated through BRASSINOSTEROID INSENSITIVE 2 (BIN2- a GSK3-like kinase), which acts as a negative regulator of the BR pathway. Previously, it was shown that ABA enhances the kinase activity of BIN2 toward BES1 [30]. We found that mutants that lack BIN2 and two closely related GSK3 kinases (gsk3 triple) no longer showed inhibition of +FR-induced elongation in response to NaCl, ABA, or BRZ (Figures 4B and S4G).
ABA Inhibits +FR-Induced Expression of BSK5
We reasoned that the inhibition of GSK3 kinase function may be required for full +FR-induced hypocotyl elongation. Upstream of GSK3 kinases in the BR signal cascade are the BRASSINOSTEROID SIGNALING KINASES (BSKs). BSK activation by BR [31] results in the inactivation of GSK3 kinases [31, 32, 33]. Despite high redundancy between the 12 BSK family members [33], mutation of just one (BSK5) alters plant sensitivity to NaCl and ABA [34]. Interestingly, BSK5 transcripts are enhanced in hypocotyls under +FR light [35]. We found that this upregulation was redundantly regulated by PIF4, PIF5, and PIF7 (Figure 4C).
The BSK5 promoter contains a G-box approximately 3.5 kb from its start codon (Figure S4H). Multiple PIFs bind directly to this G-box [18], as does at least one AREB family member, ABF3 (in an ABA-dependent manner) [12]. We found that ABA pre-treatment strongly inhibited the +FR-mediated upregulation of BSK5 transcripts in hypocotyls (Figure 4D) and that ABA-mediated repression of BSK5 transcripts was dependent upon AREB family transcription factors. +FR-induced hypocotyl elongation was dependent upon BSK5 as bsk5 mutants showed only limited +FR-induced elongation (Figure 4E). This elongation was to a similar length as wild-type plants treated with +FR and NaCl, and NaCl had no further effects on +FR-induced elongation in a bsk5 mutant (Figure 4E). These results demonstrate that the +FR-mediated upregulation of BSK5 is required for maximal hypocotyl elongation under +FR light, and they suggest that ABA may at least in part inhibit +FR-induced elongation through a suppression of BSK5 transcription.
Repression of +FR-Induced Elongation Centers on the PIF-BES1 Module
Importantly, the rescue of hypocotyl elongation by epi-BL did not occur in the pif4/pif5 mutant, demonstrating that in this context, BR-induced growth requires PIF4 and/or PIF5 (Figure 4F). Recent reports have demonstrated a close regulatory relationship between PIF and brassinosteroid signaling. Brassinosteroid upregulates each of the shade-induced genes that we tested (PIL1, IAA19, SAUR16, and PRE1) [29, 36, 37]. PIF4 heterodimerizes with BES1 [28] and a BES1 homolog, BRASINAZOLE RESISTANT 1 (BZR1), and inter-dependently they control the expression of thousands of genes [38]. Additionally, BIN2 has a direct inhibitory effect on both PIF4 [29] and PIF3 [39].
From our results, we propose a mechanism (Figure 4G) whereby +FR light promotes the activity of PIF4, PIF5, and PIF7, and these transcription factors then redundantly promote the expression of BSK5. BSK5 suppresses GSK3-like kinases, allowing for the activation of the BES1:PIF4/PIF5 signaling module. In saline soils, activation of the ABA signal transduction pathway results in the enhanced action of AREB/ABF transcription factors that suppress BSK5 transcription, and therefore break the +FR-induced PIF:BES1 feedforward loop. The absence of BSK5 would allow for strong GSK3-like kinase action, leading to a suppression of the BES1:PIF4/PIF5 signal module. We do not rule out other mechanisms by which NaCl may inhibit +FR-induced elongation; indeed, we found that mutants lacking the evening complex component ELF3 had elongated hypocotyls in all of our growth conditions and appeared to lack NaCl-mediated inhibition of +FR-induced hypocotyl elongation (Figure S4H). This implies that NaCl may also suppress PIF4 and PIF5 through the circadian clock.
There may be an adaptive significance to our finding that plants grown in saline soils suppress BES1:PIF signaling when they are presented with shade cues. Indeed, a recent study suggested that overexpression of PIF4 may reduce plant survival in saline conditions [40]. It is possible then that shade avoidance signaling is detrimental to salt survival. It is notable that BR-based chemicals are currently being developed as treatments for salt and drought afflicted crops [41]. It may be that, under concurrent salt and shade conditions, re-activation of the BR signal cascade is damaging to plant health.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| High Affinity Anti-HA-HRP (3F10) | Roche | RRID:AB_390914 |
| Anti-GFP-HRP | Miltenyi Biotec | RRID:AB_2470 |
| Bacterial Strains | ||
| E.coli (DH5α) | N/A | N/A |
| A. tumefaciens (AGL1) | N/A | N/A |
| Chemicals, Peptides and Recombinant Proteins | ||
| Murashige &Skoog (MS) medium | Duchefa Biochemie | M0222.0050 |
| Silwet® L-77 | Phytotech Labs | S7777 |
| Epibrassinolide | Santa-Cruz Biotechnology | sc-211419 |
| Abscisic Acid | Sigma Aldrich | 862169 |
| Brassinazole | Sigma Aldrich | SML1406 |
| Luciferin | Promega | E1601 |
| X-glucuronide | Sigma Aldrich | B5285-10MG |
| SuperSignal West Femto | Thermo Fisher Scientific | 34095 |
| SuperSignal West Pico PLUS | Thermo Fisher Scientific | 34580 |
| FastDigest SacI | Thermo Fisher Scientific | FD1133 |
| FastDigest NotI | Thermo Fisher Scientific | FD0593 |
| FastDigest EcoRI | Thermo Fisher Scientific | FD0274 |
| FastDigest BamHI | Thermo Fisher Scientific | FD0054 |
| FastDigest Green Buffer (10X) | Thermo Fisher Scientific | B72 |
| Critical Commercial Assays | ||
| RNeasy Mini Kit | QIAGEN | 74106 |
| RNase-Free DNase Set (50) | QIAGEN | 79254 |
| QIAprep Spin Miniprep Kit | QIAGEN | 27106 |
| Nucleospin Gel and PCR Clean-up | Macherey-Nagel | 740609 |
| RevertAid First Strand cDNA Synthesis Kit | Thermo Fisher Scientific | K1621 |
| SsoAdvanced Universal SYBR Green Supermix | Bio-Rad | 1725271 |
| Deposited Data | ||
| PIF4/5-HA western blots | https://zenodo.org/record/1343501#.XH_f2ohKg2w | N/A |
| BES1-GFP western blots | https://zenodo.org/record/1480822#.XH_fCohKg2w | N/A |
| GUS staining images | https://zenodo.org/record/2022607#.XH_f74hKg2w | N/A |
| Raw data for figure generation | https://zenodo.org/record/2592526#.XIk36yhKg2w | N/A |
| Experimental Organisms | ||
| Col-0 | N/A | N/A |
| Ws | N/A | N/A |
| Ler | N/A | N/A |
| pif4-101 | 3 | Garlic_114_G06 |
| pif5 (pil6-1) | 43 | SALK_087012 |
| pif7-1 | 44 | SALK_044061 |
| pif4-101/pif5 (aka pil6-1) | 3 | Garlic_114_G06/ SALK_087012 |
| pif4-101/pif5 (aka pil6-1)/ pif7-1 | 45 | Garlic_114_G06/ SALK_087012/ SALK_044061 |
| abaQ (pyr1-1/pyl1-1/pyl2-1/pyl4-1) | 14 | Point/ Salk_054640/ CSHL_GT2864_1/ Sail_517_C08 |
| snrk2.2/snrk2.3 | 15 | GABI_807G04/ Salk_107315 |
| arebQ (areb1/ areb2/ abf3/ abf1-1) | 13 | SALK_002984/ SALK_069523/ SALK_096965/SALK_132819 |
| abi5-1 | 46 | N/A |
| bes1-D | 27 | N/A |
| gsk3-triple (bin2-3bil1-1bil2-1) | 47 | FLAG_593C09/Wisonsin KO /Wisonsin KO |
| bsk5 | NASC (characterized in 34) | SALK_051739C |
| aba2-1 | 48 | N/A |
| aba3-1 | 48 | N/A |
| della global (gai-t6/ rga-t2/ rgl1-1/ rgl2-1/ rgl3-4) | 49 | N/A |
| pPIL1:LUC | 17 | N/A |
| pIAA19:LUC | New line | N/A |
| pDR5V2:GUS | New line | N/A |
| pDR5V2:GUS/ pif4-101/ pif5 (pil6-1) | New line | N/A /Garlic_114_G06/ SALK_087012 |
| p35S:PIF4-HA | Franklin Lab, Bristol, UK | N/A |
| p35S:PIF5-HA | Franklin Lab, Bristol, UK | N/A |
| p35S:BES1-GFP | 28 | N/A |
| Tomato- S.lycopersicum var. “Moneymaker” | Intratuin, NL | EAN# 8717263349600 |
| Tobacco- N. benthamiana | N/A | N/A |
| Recombinant DNA | ||
| pENTR_D-TOPO | Thermo Fisher | K2400-20 |
| LucTrap3 | N/A | GenBank: AY968054.1 |
| pGREENII0179 | N/A | N/A |
| pUC57:DR5v2 | Dolf Weijers (WUR) | N/A |
| Software and Algorithms | ||
| Microsoft Excel 2010 | Microsoft | N/A |
| Microsoft Powerpoint 2010 | Microsoft | N/A |
| Microsoft Word 2010 | Microsoft | N/A |
| SPSS Statistics 24 | IBM | N/A |
| Snapgene | GSL Biotech LLC | N/A |
| ImageJ | NIH | N/A |
| ICY | Quantitative Image Analysis Unit, Institut Pasteur | N/A |
| GIMP2 | The GIMP Development Team | N/A |
| ViiA 7 Software | Thermo Fisher Scientific | N/A |
| Prism 6 | Graphpad | N/A |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ronald Pierik (r.pierik@uu.nl).
Experimental Model and Subject Details
The main experimental organism used in this study was Arabidopsis thaliana. Several mutant Arabidopsis lines were used: pif4-101, pif5 (pil6-1), pif7-1, pif4-101/pif5 (pil6-1), pif4-101/pif5 (pil6-1)/ pif7-1, abaQ (pyr1-1/pyl1-1/pyl2-1/pyl4-1), snrk2.2/snrk2.3, arebQ (areb1/ areb2/ abf3/ abf1-1), bsk5, aba2-1, aba3-1, pPIL1:LUC and p35S:BES1-GFP in the Columbia ecotype; abi5-1 and gsk3-triple (bin2-3/bil1-1/bil2-1) in the Ws ecotype; and della global (gai-t6/ rga-t2/ rgl1-1/ rgl2-1/ rgl3-4), p35S:PIF4-HA and p35S:PIF5-HA in the Ler ecotype. The other species used in this study were tomato (S.lycopersicum var. “Moneymaker”) and tobacco (N. benthamiana). See the Key Resources Table for details.
Plant Propagation
To generate the seed used in this study, plants were grown in 130-140 μmol m-2 s-1 white light with a 16h 8h photoperiod at 21°C. Plants were kept well-watered until silique senescence, after which water was increasingly withheld until the plant was fully senesced. Harvested seeds were stored dry at room temperature for at least 2 weeks before use.
Method Details
Morphological studies
Seeds were sown directly onto wetted soil and stratified at 4°C in darkness. After 3-4 days of stratification, plants were moved to growth chambers with a 16:8 hour photoperiod (PP), 130-140 μmol m-2 s-1 white light at 21°C. After 3 days, plants were transferred to new soil that had been previously wetted with deionised water or the indicated concentration of NaCl and then returned to white light for the indicated period. Wl +FR treatments were provided by supplementary FR LEDs to a R:FR ratio of 0.2.
Development of Transgenic Lines
pDR5v2:GUS- The pGREENII0179:pDR5v2:GUS plasmid was constructed by ligating a pDR5v2 fragment cut from pUC57:DR5v2 [19] with EcoRI and BamHI into pGREENII0179 lacking a promoter. The GUS gene was amplified from a pDR5:GUS Arabidopsis line containing a GUS reporter with primers introducing NotI and SacI restriction sites. The PCR fragment was subsequently cut with NotI and SacI and ligated into pGREENII0179:pDR5v2 to create pGREENII0179:pDR5v2:GUS. This was then transformed into E.coli (strain- DH5α) and sequenced to confirm the correct insertion of GUS. The pDR5v2:GUS Arabidopsis line was made transforming pGREENII0179:pDR5v2:GUS into agrobacterium (strain AGL1) containing the pSOUP plasmid. This was then transformed into Col-0 plants by floral dipping, as in reference [42]. Briefly, agrobacterium were grown in YEBS media to saturation, and Silwet® L-77 was added to 200ul/l. Arabidopsis inflorescence stems were dipped into the solution and seeds harvested after senescence. 21 independent transformants were selected by antibiotic resistance and T2 lines screened for single insertions and GUS staining intensity. T3 homozygous lines were selected by hygromycin resistance and checked for signal strength, distribution and auxin responsiveness.
pIAA19:LUC- The pIAA19:LUC plasmid was constructed by amplifying the IAA19 promoter from Arabidopsis thaliana (Col ecotype), with the primers listed in the key resources table (see online). The promoter fragment was then cloned into pENTR_D-TOPO and mobilized into the LucTrap3 vector. This construct was then transformed into agrobacterium (strain AGL1) and used for transformation of Col-0 plants by floral dip as above.
p35S:PIF4-HA and p35:PIF5-HA- The pCF402 and pCF404 plasmids (described previously in [3]) were introduced into agrobacterium and then transformed into Ler plants by floral dip. Transformed lines were selected based on seed-coat GFP fluorescence. These lines are very similar to existing lines in the Col-0 background [3].
For oligonucleotides used in this study see STAR Methods.
Gene Expression Studies
Seedlings were grown in the indicated conditions, before dissection and flash freezing in liquid nitrogen. RNA was extracted with an RNeasy Mini Kit (QIAGEN) with an on-column treatment with DNase (QIAGEN). cDNA was synthesized using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) with random hexamers. qPCR was performed with SYBR green ready mix (Bio-rad) in a ViiA 7 Real-Time PCR System (Thermo Fisher Scientific). Ct values for the gene of interest are expressed relative to that Ct value found for primers targeted to AT1G13320. For each experiment, 2 technical and a minimum of 3 biological repeats was performed. Primer efficiency tests were performed for each primer pair used.
For oligonucleotides used in this study see STAR Methods.
Luciferase Assay
pPIL1:LUC and pIAA19:LUC seedlings were germinated in 1/2 MS media at 120-140 μmolm-2s-1 white light (16h:8h PP) at 22°C. On cotyledon expansion, seedlings were transferred to microtiter plates containing 170 μl solid 1/2 MS media (6% agar) supplemented with EtOH (mock), 75 mM NaCl (NaCl), or 2 μM ABA, per well. 30 μl of a luciferin solution (8 μl of a 10 mg/ ml luciferin (Promega) stock in DMSO, diluted in 4 mL H2O) were added per well, and plates were sealed with a sealing film (Applied Biosystems). Two holes were then made with a fine needle per well for seedlings transpiration. Plants were moved to red light (30-35 μmol m-2 s-1-16h:8h PP) and luminescence was recorded by a Berthold LB960 station installed in a Percival growth chamber. Luminescence was recorded each hour, with a 2 s read per well. On day 2 at Zt4, R light was supplemented with FR.
ABA Extraction and Quantification
Plants were grown under the same conditions used for morphological studies. At Zt 4 on day 7, 20-50mg of tissue (approximately 50 seedlings) were harvested and flash frozen in liquid nitrogen. Samples were homogenized (Precellys 24, Bertin Technologies, Aix-en-Provence, France) and extracted in ethyl acetate with an internal standard of D6-ABA (C/D/N Isotopes Inc, Canada). Samples were dried on a vacuum concentrator (CentriVap Centrifugal Concentrator, Labconco, Kansas City, MO, USA) at room temperature. Residues were re-suspended in 0.25 mL of 70% methanol (v/v) and samples were analyzed by liquid chromatography-mass spectrometry (LC-MS) on a Varian 320 Triple Quad LC-MS/MS. ABA levels were quantified from the peak area of each sample compared with the internal standard and normalized by fresh weight.
Western Blots
Plants were germinated on 1/2 MS plates in white light (120-140 μmol m-2 s-1 −16:8h PP) for 3 days at 21°C, before being transferred to plates with or without 75mM NaCl. Plates were then grown in R light (30-35 μmol m-2 s-1 −16:8h PP) for a further 2 days. On day 5, at Zt 3, half the plates were moved to R+FR light. Tissues were harvested at Zt 4. 10 seedlings were homogenized and proteins were extracted in 70 μl Cracking Buffer (125mM Tris pH 7.4, 2% SDS, 10% Glycerol, 6M Urea, 5% βME) and 15 μl of the extract was run on a 10% polyacrylamide gel. Blots were probed with anti-HA-HRP (Roche) or anti-GFP-HRP (Miltenyi Biotec) as indicated in the text. Membranes were developed with 50/50 mix of ‘pico’ and ‘femto’ chemiluminescence substrates (Thermo) on a ChemiDoc (Biorad).
GUS Staining
Tissues were harvested an immediately fixed in 90% acetone at −20°C. Tissues were washed twice under a vacuum in GUS wash solution (0.1M Phospho-PI pH 7.0, 10 mM EDTA, 2 mM K3Fe(CN)6) and then stained overnight at 37°C in GUS staining solution (0.1M Phospho-PI pH 7.0, 10 mM EDTA, 1 mM K3Fe(CN)6, 1mM K4Fe(CN)6 ∗ 3H2O, 0.5 mg/ ml X-glucuronide). The GUS reaction was stopped by treatment with 3:1 ethanol: acetic acid mix for 1 hour at 37°C. Tissues were cleared with 70% ethanol over several days and then mounted in 10% chloral hydrate, 30% glycerol. Representative images were taken with a Zeiss Axioskop 2 with Infinity 3 camera attachment and slides were scanned with an Epson v300 scanner for the purposes of GUS quantification.
Quantification and Statistical Analyses
Image quantification
For morphological studies, plants were laid out on 0.8% agar plates and scanned with an Epson v300 scanner. Length measurements were then made in ImageJ. Western blot band intensities were quantified by measuring pixel intensity in ImageJ. Total protein loading was assessed through Rubisco Large Subunit band intensity, as visualized through ponceau stinging. Total protein loading was then used to normalize chemiluminescence blot band intensity. GUS staining quantification was performed in the ImageJ-based program, Icy. Relative GUS levels in the top half of the hypocotyl were defined by the mean intensity in Ch-0 minus the mean intensity in Ch-1.
Data presentation
Data presented as box and whisker diagrams were created in Graphpad Prism, and compiled in Microsoft Powerpoint. Datasets where n ≥ 10 are represented with a Tukey representation. The upper and lower edges of the box represent interquartile range (IQR) and the middle bar represents the median value. values 1.5 x IQR higher than the 75th percentile or 1.5x IQR lower than the 25th percentile are marked as single point outliers. Whiskers are plotted as the maximum and minimum values, or as 1.5x IQR, depending on which is closer to the median. Datasets where n ≤ 10 are presented as box and whisker diagrams, with all points shown. Whiskers represent the maximum and minimum values.
Statistical Analyses
1-way and 2-way ANOVAs were performed in Graphpad Prism or with SPSS statistics. t tests were performed in Microsoft Excel. Significant differences are defined as p > 0.05. In graphs that include letters, each letter represents a statistically significant mean.
Data Availability
The data used in the preparation of this manuscript, including morphological measurements, average Ct values, image analyses, ABA quantifications and photon counts is available at the following site: https://zenodo.org/record/2592526#.XIk36yhKg2w
The images used for western blot quantification are available at the sites listed below:
PIF4-HA and PIF5-HA: https://zenodo.org/record/1343501#.XH_f2ohKg2w
The images used for GUS-staining quantification are available at the site listed below:
DR5v2:GUS in both Col and pif4pif5 mutants: https://zenodo.org/record/2022607#.XH_f74hKg2w
Acknowledgments
We would like to thank Prof. Dolf Weijers (WUR, Wageningen, NL) for providing the pUC57:DR5v2 plasmid, Prof. Christian Fankhauser (UNIL, Lausanne, CH) for the pCF402 and pCF404 plasmids, Prof. Kazuko Yamaguchi-Shinozaki (The University of Tokyo, Tokyo, Japan) for arebQ seeds, Prof. Keara Franklin (The University of Bristol) for the p35S:PIF4-HA and p35S:PIF5-HA seeds ahead of publication, the Plant Ecophysiology Group (Utrecht University, Utrecht, NL) for help with harvesting and gene expression studies, and Dr. Charlotte Gommers (WUR, Wageningen, NL) for critical reading of this manuscript. S.H. was supported through a fellowship from the European Molecular Biology Organisation (EMBO- ATFL 404-2015) and Marie Skłodowska Curie Actions (792624). The research was funded through the Netherlands Organisation for Scientific Research (NWO, awarded to R.P., VIDI-864.12.003), the Spanish Ministry of Economy and Competitiveness (MINECO, awarded to S.P., BIO2017-90056-R), and the UK Biotechnology and Biological Sciences Research Council (BBSRC, grant BB/M008711/1 to support A.S.). ORCIDs for the authors are as follows: 0000-0001-8943-6238 (S.H.), 0000-0001-5412-6029 (C.K.P.), 0000-0001-7809-2812 (K.v.G.), 0000-0003-0299-7900 (A.L.T.) 0000-0002-9137-0888 (A.S.), 0000-0001-6451-5968 (M.d.V.), 0000-0003-2684-5485 (S.P.), 0000-0002-9223-9996 (R.C.S.), 0000-0001-6738-115X (C.T.), and 0000-0002-5320-6817 (R.P.).
Author Contributions
Conceptualization, S.H., C.T., and R.P.; Methodology, S.H., A.L.T., S.P., and R.P.; Formal Analysis, S.H.; Investigation, S.H., C.K.P., K.v.G., A.L.T., E.R., A.S., M.d.V., and S.P.; Resources, S.P., R.C.S., and R.P.; Writing – Original, S.H.; Writing – Review & Editing, S.H., C.K.P., K.v.G., C.T., S.P., R.C.S., and R.P.; Visualization, S.H.; Funding Acquisition, S.H., R.P., and S.P.
Declaration of Interests
The authors declare no competing interests.
Published: May 2, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cub.2019.03.042.
Supplemental Information
References
- 1.Ausubel J.H.J., Wernick I.K.I., Waggoner P.E.P. Peak Farmland and the Prospect for Land Sparing. Popul. Dev. Rev. 2013;38:221–242. [Google Scholar]
- 2.Fraser D.P., Hayes S., Franklin K.A. Photoreceptor crosstalk in shade avoidance. Curr. Opin. Plant Biol. 2016;33:1–7. doi: 10.1016/j.pbi.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- 4.Li L., Ljung K., Breton G., Schmitz R.J., Pruneda-Paz J., Cowing-Zitron C., Cole B.J., Ivans L.J., Pedmale U.V., Jung H.S. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012;26:785–790. doi: 10.1101/gad.187849.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hornitschek P., Kohnen M.V., Lorrain S., Rougemont J., Ljung K., López-Vidriero I., Franco-Zorrilla J.M., Solano R., Trevisan M., Pradervand S. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 2012;71:699–711. doi: 10.1111/j.1365-313X.2012.05033.x. [DOI] [PubMed] [Google Scholar]
- 6.Hersch M., Lorrain S., de Wit M., Trevisan M., Ljung K., Bergmann S., Fankhauser C. Light intensity modulates the regulatory network of the shade avoidance response in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2014;111:6515–6520. doi: 10.1073/pnas.1320355111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Julkowska M.M., Testerink C. Tuning plant signaling and growth to survive salt. Trends Plant Sci. 2015;20:586–594. doi: 10.1016/j.tplants.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Claeys H., Van Landeghem S., Dubois M., Maleux K., Inzé D. What Is Stress? Dose-Response Effects in Commonly Used in Vitro Stress Assays. Plant Physiol. 2014;165:519–527. doi: 10.1104/pp.113.234641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang C., Belfield E.J., Mithani A., Visscher A., Ragoussis J., Mott R., Smith J.A., Harberd N.P. ROS-mediated vascular homeostatic control of root-to-shoot soil Na delivery in Arabidopsis. EMBO J. 2012;31:4359–4370. doi: 10.1038/emboj.2012.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang C., Belfield E.J., Cao Y., Smith J.A., Harberd N.P. An Arabidopsis soil-salinity-tolerance mutation confers ethylene-mediated enhancement of sodium/potassium homeostasis. Plant Cell. 2013;25:3535–3552. doi: 10.1105/tpc.113.115659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu J.K. Abiotic Stress Signaling and Responses in Plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song L., Huang S.C., Wise A., Castanon R., Nery J.R., Chen H., Watanabe M., Thomas J., Bar-Joseph Z., Ecker J.R. A transcription factor hierarchy defines an environmental stress response network. Science. 2016;354:aag1550. doi: 10.1126/science.aag1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshida T., Fujita Y., Maruyama K., Mogami J., Todaka D., Shinozaki K., Yamaguchi-Shinozaki K. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015;38:35–49. doi: 10.1111/pce.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park S.Y., Fung P., Nishimura N., Jensen D.R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T.F. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujii H., Verslues P.E., Zhu J.-K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Wit M., Keuskamp D.H., Bongers F.J., Hornitschek P., Gommers C.M.M., Reinen E., Martínez-Cerón C., Fankhauser C., Pierik R. Integration of Phytochrome and Cryptochrome Signals Determines Plant Growth during Competition for Light. Curr. Biol. 2016;26:3320–3326. doi: 10.1016/j.cub.2016.10.031. [DOI] [PubMed] [Google Scholar]
- 17.Li L., Zhang Q., Pedmale U.V., Nito K., Fu W., Lin L., Hazen S.P., Chory J. PIL1 participates in a negative feedback loop that regulates its own gene expression in response to shade. Mol. Plant. 2014;7:1582–1585. doi: 10.1093/mp/ssu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfeiffer A., Shi H., Tepperman J.M., Zhang Y., Quail P.H. Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol. Plant. 2014;7:1598–1618. doi: 10.1093/mp/ssu087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao C.Y., Smet W., Brunoud G., Yoshida S., Vernoux T., Weijers D. Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods. 2015;12 doi: 10.1038/nmeth.3279. 207–210, 2, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korver R.A., Koevoets I.T., Testerink C. Out of Shape During Stress: A Key Role for Auxin. Trends Plant Sci. 2018;23:783–793. doi: 10.1016/j.tplants.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayes S., Velanis C.N., Jenkins G.I., Franklin K.A. UV-B detected by the UVR8 photoreceptor antagonizes auxin signaling and plant shade avoidance. Proc. Natl. Acad. Sci. USA. 2014;111:11894–11899. doi: 10.1073/pnas.1403052111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng S., Martinez C., Gusmaroli G., Wang Y., Zhou J., Wang F., Chen L., Yu L., Iglesias-Pedraz J.M., Kircher S. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 24.Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., Van Der Straeten D., Peng J., Harberd N.P. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- 25.Djakovic-Petrovic T., de Wit M., Voesenek L.A., Pierik R. DELLA protein function in growth responses to canopy signals. Plant J. 2007;51:117–126. doi: 10.1111/j.1365-313X.2007.03122.x. [DOI] [PubMed] [Google Scholar]
- 26.Geng Y., Wu R., Wee C.W., Xie F., Wei X., Chan P.M., Tham C., Duan L., Dinneny J.R. A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell. 2013;25:2132–2154. doi: 10.1105/tpc.113.112896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 28.Martínez C., Espinosa-Ruíz A., de Lucas M., Bernardo-García S., Franco-Zorrilla J.M., Prat S. PIF4-induced BR synthesis is critical to diurnal and thermomorphogenic growth. EMBO J. 2018;37:99552. doi: 10.15252/embj.201899552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernardo-García S., de Lucas M., Martínez C., Espinosa-Ruiz A., Davière J.M., Prat S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 2014;28:1681–1694. doi: 10.1101/gad.243675.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang S., Cai Z., Wang X. The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc. Natl. Acad. Sci. USA. 2009;106:4543–4548. doi: 10.1073/pnas.0900349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang W., Kim T.W., Oses-Prieto J.A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A.L., Wang Z.Y. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim T.W., Guan S., Sun Y., Deng Z., Tang W., Shang J.-X., Sun Y., Burlingame A.L., Wang Z.Y. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 2009;11:1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sreeramulu S., Mostizky Y., Sunitha S., Shani E., Nahum H., Salomon D., Hayun L.B., Gruetter C., Rauh D., Ori N., Sessa G. BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J. 2013;74:905–919. doi: 10.1111/tpj.12175. [DOI] [PubMed] [Google Scholar]
- 34.Li Z.Y., Xu Z.S., He G.Y., Yang G.X., Chen M., Li L.C., Ma Y.Z. A mutation in Arabidopsis BSK5 encoding a brassinosteroid-signaling kinase protein affects responses to salinity and abscisic acid. Biochem. Biophys. Res. Commun. 2012;426:522–527. doi: 10.1016/j.bbrc.2012.08.118. [DOI] [PubMed] [Google Scholar]
- 35.Kohnen M.V., Schmid-Siegert E., Trevisan M., Petrolati L.A., Sénéchal F., Müller-Moulé P., Maloof J., Xenarios I., Fankhauser C. Neighbor Detection Induces Organ-Specific Transcriptomes, Revealing Patterns Underlying Hypocotyl-Specific Growth. Plant Cell. 2016;28:2889–2904. doi: 10.1105/tpc.16.00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu X., Li L., Zola J., Aluru M., Ye H., Foudree A., Guo H., Anderson S., Aluru S., Liu P. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011;65:634–646. doi: 10.1111/j.1365-313X.2010.04449.x. [DOI] [PubMed] [Google Scholar]
- 37.Oh E., Zhu J.Y.J., Bai M.Y., Arenhart R.A., Sun Y., Wang Z.Y. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife. 2014;3:1–19. doi: 10.7554/eLife.03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh E., Zhu J.Y., Wang Z.Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012;14:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ling J.J., Li J., Zhu D., Deng X.W. Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proc. Natl. Acad. Sci. USA. 2017;114:3539–3544. doi: 10.1073/pnas.1700850114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakuraba Y., Bülbül S., Piao W., Choi G., Paek N.C. Arabidopsis EARLY FLOWERING3 increases salt tolerance by suppressing salt stress response pathways. Plant J. 2017;92:1106–1120. doi: 10.1111/tpj.13747. [DOI] [PubMed] [Google Scholar]
- 41.Kahlaoui B., Misle E., Khaskhoussy K., Jaouadi I., Hachicha M. Brassinosteroids and drought tolerance in plants. In: Ahmad P., editor. Water Stress and Crop Plants: A Sustainable Approach. Wiley; 2016. pp. 600–607. [Google Scholar]
- 42.Davis A.M., Hall A., Millar A.J., Darrah C., Davis S.J. Protocol: Streamlined sub-protocols for floral-dip transformation and selection of transformants in Arabidopsis thaliana. Plant Methods. 2009;5:3. doi: 10.1186/1746-4811-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in the preparation of this manuscript, including morphological measurements, average Ct values, image analyses, ABA quantifications and photon counts is available at the following site: https://zenodo.org/record/2592526#.XIk36yhKg2w
The images used for western blot quantification are available at the sites listed below:
PIF4-HA and PIF5-HA: https://zenodo.org/record/1343501#.XH_f2ohKg2w
The images used for GUS-staining quantification are available at the site listed below:
DR5v2:GUS in both Col and pif4pif5 mutants: https://zenodo.org/record/2022607#.XH_f74hKg2w