Abstract
Proprioceptive feedback from Group Ia/II muscle spindle afferents and Group Ib Golgi tendon afferents is critical for the normal execution of most motor tasks, yet how these distinct proprioceptor subtypes emerge during development remains poorly understood. Using molecular genetic approaches in mice of either sex, we identified 24 transcripts that have not previously been associated with a proprioceptor identity. Combinatorial expression analyses of these markers reveal at least three molecularly distinct proprioceptor subtypes. In addition, we find that 12 of these transcripts are expressed well after proprioceptors innervate their respective sensory receptors, and expression of three of these markers, including the heart development molecule Heg1, is significantly reduced in mice that lack muscle spindles. These data reveal Heg1 as a putative marker for proprioceptive muscle spindle afferents. Moreover, they suggest that the phenotypic specialization of functionally distinct proprioceptor subtypes depends, in part, on extrinsic sensory receptor organ-derived signals.
SIGNIFICANCE STATEMENT Sensory feedback from muscle spindle (MS) and Golgi tendon organ (GTO) sensory end organs is critical for normal motor control, but how distinct MS and GTO afferent sensory neurons emerge during development remains poorly understood. Using (bulk) transcriptome analysis of genetically identified proprioceptors, this work reveals molecular markers for distinct proprioceptor subsets, including some that appear selectively expressed in MS afferents. Detailed analysis of the expression of these transcripts provides evidence that MS/GTO afferent subtype phenotypes may, at least in part, emerge through extrinsic, sensory end organ-derived signals.
Keywords: Golgi tendon organ, muscle spindle, neuronal identity, proprioception, sensory
Introduction
The vertebrate musculoskeletal system is extremely versatile in its ability to generate movement. However, most motor behaviors demand precise interlimb coordination and temporal control of individual muscles to achieve movement accuracy (Akay et al., 2014; Drew and Marigold, 2015). Both these features are informed by a continuous stream of sensory feedback from skeletal muscle, joints, and skin (Proske and Gandevia, 2012; Tuthill and Azim, 2018). Muscle afferent proprioceptive sensory neurons (pSNs), which provide a large component of this feedback, reside in Dorsal Root Ganglia (DRG) and innervate one of two distinct receptor organs in muscle: muscle spindles (MSs) or Golgi tendon organs (GTOs) (Matthews, 1972; Windhorst, 2007). MSs are innervated by Group Ia and Group II afferents and relay the rate and magnitude of changes in muscle length, respectively (Hunt, 1990; Macefield and Knellwolf, 2018). GTOs, in contrast, are innervated by Group Ib afferents and provide information on muscle tension (Houk and Henneman, 1967; Jami, 1992). Yet, while both pSN subclasses convey critical aspects of the status of the musculo-skeletal system, the developmental mechanisms that underlie the acquisition of MS and GTO subtype identities remain unknown.
In contrast to MS/GTO afferent subtype diversification, the events that lead to a generic proprioceptive sensory identity are relatively well understood. Presumptive pSNs progenitors are born during the first wave of DRG sensory neurogenesis at embryonic day (e)9.5–10 and segregate into two lineages: TrkB+Shox2+ and TrkC+Rx3+ neurons (Ma et al., 1999; Kramer et al., 2006; Lallemend and Ernfors, 2012). TrkB+Shox2+ progenitors give rise to rapidly adapting (RA) low threshold mechanoreceptors (LTMRs), such as Meissner and Pacinian afferents (Abdo et al., 2011; Li et al., 2011). The TrkC+Rx3+ lineage instead gives rise to proprioceptive muscle afferents and slowly adapting (SA)-LTMRs, including Merkel cell afferents (Levanon et al., 2002; de Nooij et al., 2013). TrkC+Rx3+ proprioceptors subsequently (∼e14.5) express parvalbumin (PV), marking their commitment to the pSN lineage, and associate with their nascent MS and GTO receptor organs (Tourtellotte and Milbrandt, 1998; Hippenmeyer et al., 2002). It has been shown that the development of MS sensory receptors depends on inductive signals from the innervating sensory axons (Hippenmeyer et al., 2002). This observation, together with findings that individual muscles exhibit a stereotypical pattern and number of MSs and GTOs, has led to the belief that MS/GTO pSN subclass identity is established through intrinsic genetic determinants, before the innervation of presumptive MS and GTO receptor organs (Matthews, 1972; Banks, 2006). However, the molecular correlates of distinct MS/GTO subclass identities have remained surprisingly difficult to identify.
The apparent difficulty to find transcripts that segregate with MS afferents (Groups Ia and II) or GTO afferents (Group Ib) at early developmental stages has cast doubt on the idea that MS/GTO proprioceptor subtype identity is fixed from the outset, and raises the possibility that pSN subtype identity is acquired through extrinsic, possibly target-derived, signals. Recent advances in molecular profiling techniques have yet to shed light on this issue, in part because pSNs are difficult to distinguish from other mechanoreceptors in DRG (e.g., the aforementioned RA-LTMRs and SA-LTMRs) (Lee et al., 2012; Chiu et al., 2014). In addition, pSNs make up <10% of all DRG neurons, severely limiting their coverage in unbiased single-cell DRG transcriptome studies (Usoskin et al., 2015; Li et al., 2016). Thus, molecular profiling of embryonic or adult muscle pSNs has thus far failed to help distinguish between pSN MS/GTO subtypes, let alone provide information on when or how these subtypes emerge during development.
To permit analysis of the proprioceptor transcriptome more selectively, we leveraged various genetic tools to isolate combinations of muscle pSNs, SA-LTMRs, and/or RA-LTMRs. RNA sequencing (RNA-seq) and differential expression analysis of these sensory cohorts identified 316 transcripts that are enriched in pSNs relative to RA-LTMR sensory neurons. Of these, we thus far identified 24 transcripts that are largely confined to proprioceptors with little DRG expression outside of this neuronal subset. In addition, we find that at least 13 of these transcripts localize to subsets of pSNs, and that the combinatorial expression of these transcripts delineates molecular distinct pSN classes. We also find that a large number of these transcripts (12 of 24) is first expressed after pSNs connect with their sensory receptors. Intriguingly, expression of four of these markers, most notably the heart development protein heart of glass (Heg1), is significantly altered in Egr3−/− mice that lack MSs. These data support the notion that MS/GTO afferent subtype acquisition involves a protracted developmental process, and are consistent with the idea that pSN subtype identity is influenced by retrograde signals from their sensory organs.
Materials and Methods
Animal husbandry and mouse strains.
All experiments were performed according to National Institutes of Health guidelines and approved by the Institutional Animal Care and Use Committee of Columbia University. Mouse strains used were Runx3−/− (IMSR, catalog #JAX:008773, RRID:IMSR_JAX:008773) (Taniuchi et al., 2002; Kramer et al., 2006), PV:Cre (IMSR, catalog #JAX:008069, RRID:IMSR_JAX:008069) and Tau-lxp-STOP-lxp:GFP-ires-nLZ (Hippenmeyer et al., 2005), Ai9:tdTomato (Madisen et al., 2010), TrkC:GFP (MMRRC, catalog #000269-MU, RRID:MMRRC_000269-MU) (Gong et al., 2003), Isl2:DTAflx (Yang et al., 2001), Egr3−/− (MGI, catalog #5489936, RRID:MGI:5489936) (Tourtellotte and Milbrandt, 1998), and Egr3:WGA (de Nooij et al., 2013).
Isolation of DRG neuronal subsets through FACS.
DRG from neonates (of either sex) were dissected in ice-cold Hank's balanced salt solution and dissociated by enzymatic digestion (Papain, Collagenase IV, Dispase Type II; Worthington Biochemical) and mechanical trituration, essentially as described previously (Malin et al., 2007). Cell suspensions were passed through 35 μm gauze filters to clear suspension from remaining cellular aggregates. Fluorescently labeled neuronal subsets were isolated through FACS using a Beckman Coulter Altra Hypersort (BD Biosciences). Neurons were sorted at 12–13 psi using a 100 μm nozzle and collected in lysis buffer (Agilent Technologies) and stored at −80°C until processed. For each genotype/neuronal cohort, at least three samples were obtained: for TrkC:GFP/[pSN+SA], sample 1: ∼2100 neurons from four p6 pups; sample 2: ∼1700 neurons from two p2 pups; sample 3: ∼5900 neurons from four p1 pups; for Rx3+/+;PV:tdT/[pSN+RA], sample 1: ∼4900 neurons from four p6 pups; sample 2: ∼10,400 neurons from three p4 pups; sample 3: ∼8400 neurons from three p3 pups; and for Rx3−/−;PV:tdT/[RA], sample 1: ∼260 cells from one p3 pup; sample 2: ∼145 cells from one p0 pup; sample 3: ∼210 neurons from four p0 pups.
RNA isolation, cDNA preparation, and RNA sequencing.
Total RNA was extracted from all neuronal samples (Absolutely RNA nanoprep kit; Agilent Technologies), and RNA integrity was assessed by Bioanalyzer (quality threshold was set at RIN > 8). Amplified cDNA was generated using the Ovation RNA-Seq System V2 kit (NuGen). cDNA sample quality and fragmentation were measured by Bioanalyzer (DNA1000 chip). Library preparation and single-end RNA sequencing were performed at the JP Sulzberger Columbia Genome Center (25–30 million reads/sample). RNA-seq data were mapped by OLego version 1.1.5 (Wu et al., 2013) to the reference genome (mm10), and a comprehensive database of exon junctions was provided for read mapping. (All data have been deposited in NCBI's GEO database, accession #GSE127161.) Only reads unambiguously mapped to the genome or exon junctions (single hits) were used for downstream analysis. Gene expression was quantified using the Quantas pipeline as we described previously (Yan et al., 2015). Hierarchical clustering (Eisen et al., 1998) was performed using the subset of 1091 genes filtered by abundance (reads per kilobase million [RPKM] ≥ 5 in ≥ 3 samples) and variation (SD ≥ 0.6 in log2 scale). Differential expression was evaluated by edgeR (Robinson et al., 2010), as part of the Quantas pipeline. Differentially expressed transcripts were identified based on the following criteria: Benjamini–Hochberg FDR < 0.03, |log2(fold change)| ≥ 2, and RPKM >1 in at least one of the compared conditions (Benjamini and Hochberg, 1995). Transcripts meeting these criteria were validated, to a first degree, using the Allen Brain Atlas (Lein et al., 2007) to assess their abundance of expression in DRG. Transcripts expressed in relatively small subsets of DRG neurons, as well as those not present in the Allen Brain Atlas, were selected for further analysis by ISH analysis.
RNA ISH, RNAscope, and immunohistochemistry.
ISH histochemistry was performed on 15 μm cryostat sections using digoxigenin-labeled cRNA probes (Arber et al., 2000). Hybridizations were performed at 65°C–72°C. cDNA templates for probes were cloned by PCR from DRG or brain (cortex/cerebellum) cDNA samples. Primer sequences used to clone cDNA fragments used for ISH probes were as follows: B3galt2 forward, TCACAGGGCTGCAGAACA, and B3Galt2 reverse, TGCCTGCCTTTTCCTTTG; Cacna1i forward, CCATCTTCCACCACTACTCCTC, and Cacna1i reverse, AGACTGTCCTCCAGTGTGGTCT; Clec2l forward, TCGGAGGAACCTAGGGACTG, and Clec2l reverse, ATATAACCCGTGTGTCGCCC; Cyp2s1 forward, TTGGCATCCGTTTGCCCTAT, and Cyp2s1 reverse, GAAGCAAGTGGTCCTCGTGA; Gprc5b forward, GTGACACGAAGCCAGCCT, and Gprc5b reverse, GGACGATTCCGTGTTTGC; Grm3 forward, TATGGAGCCATCACCCTGGA, and Grm3 reverse, TGCCCAGTGGCCAACTTTTA; Heatr5a forward, TAACTGCTAAGCATCCAGGAGCA, and Heatr5a reverse, CTGTAGTGCCAAGGTTGTGG; Heg1 forward, ACTTCCAAATGTCCCCATACAC, and Heg1 reverse, CCAGCCCAATCTATTAAAGTGC; Inhbb forward, CCCTGACTTGTCCCAGGTTC, and Inhbb reverse, GCCACTTCTGGACACACGTA; Itga2 forward, AGTCTCACAGAGGGGACCAG, and Itga2 reverse, ACACGCAGATCCAAAGGGTT; Mctp2 forward, AGCAGTGACCACAGCCATAA, and Kcnc1 reverse, TTCTGCGGGAGAGGCATAGA; Mtfp1 forward, TGGGACCCAATTCTGTCTGC, and Mtfp1 reverse, GGGTTGTGATTGTCCTGCCT; Ndst4 forward, ATACATCCAAAACTGACCCCAC, and Ndst4 reverse, AAAAGCACTGGCTGGTAGGTAG; Nxph1 forward, TCGCTTGATCGTCAGCCTTG, and Nxph1 reverse, AACAATGGGCCGTCTTTTGG; Pcdh8 forward, ATGTTCGACGTGCTCACCTT, and Pcdh8 reverse, AAGGTTGACATCTGGGCTGG; Plekhg1 forward, ATGCCACACCTGTCTAACTCCT, and Plekhg1 reverse, TTGCAGTGAGCCTTCTACACAT; Pln forward, ACCGAAGCCAAGGTCTCC, and Pln reverse, TGTTGAGGAAATTGGCAGC; Ppt1 forward, AAGATGGCGTCGTCCTGTTC, and Ppt1 reverse, TTGGAGTAAGCACCGGCATT; Pygm forward, GCCTGGGACGTAACAGTGAA, and Pygm reverse, AGAGCGAGTTGGGGTTGATG; Reg3b forward, GAGGTGAAGTTGCCCTATGTCT, and Reg3b reverse, CCCAAACTTATACCAAAAGGAC; Rhbdl3 forward, ATGCTGGAGGCTGTAGGTTG, and Rhbdl3 reverse, AGCCCTTGAGGTCACTCCCT; Tnni1 forward, GGTTAAGGCCTGCAGCAA, and Tnni1 reverse, GTGCCATTTCATCCTGGC; Wls forward, TGTTTCCCTGGGTTACCGTG, and Wls reverse, ATCATGTGCTCGCCACAGAA; and Wnt7a forward, GGGACTATGACCCGGAAAGC, and Wnt7a reverse, ACGTAGCCTAGCTCTCGGAA.
In situ probes for Runx3 and PV were generated from nucleotide sequences encompassing the full coding regions. A T3 polymerase site (5′-AATTAACCCTCACTAAAGGG-3′) was included in all reverse primers to facilitate direct probe generation from amplified PCR products. All PCR products were sequenced for validation.
RNAscope analysis was performed using the ACDbio RNAscope kit for nonfixed tissue (ACD #320851) as described previously (Hoffman et al., 2018). In short, DRG were dissected on ice and fixed for 15 min in 4% PFA. Fixed DRG were washed in PBS and allowed to equilibrate in 30% sucrose for 2 h. before embedding in OCT (Tissue Tek; Sakura Finetek). DRG were stored at −80°C until use. On day of experiment, DRG were sectioned at 20 μm, dried for 1 h at 30°C, washed in PBS, dehydrated in a series of 50%, 70%, and 100% EtOH, and pretreated with protease IV digestion for 30 minutes at room temperature. Hybridization was performed in a humidified oven (ACDbio) at 40°C for 2 h. Following hybridization, tissues were washed and processed for probe amplification and detection using Amp1 (30′ at 40°C), Amp2 (15′ at 40°C), Amp3 (30′at 40°C), and Alt4 (A, B, or C; 15′ at 40°C). RNAscope probes used in experiments included Rx3 (ACD #451271), PV (ACD #421931), Heg1 (ACD #510581-C2), Inhhb (ACD #475271), Pcdh8 (ACD #558101-C3), Plekhg1 (ACD #411501), Nxph1 (ACD #463401), Wnt7a (ACD #401121), b3Galt2 (ACD #491711), Itga2 (ACD #441081), and Grm3 (ACD #317821). Following RNAscope, tissue slides were mounted with Fluoromount-G and analyzed directly, or processed for immunohistochemical analysis using anti-tdTomato antibodies as described below.
Immunohistochemistry was performed on cryostat sections (15–20 μm) or whole-mount muscle tissue as described previously (de Nooij et al., 2013). Primary antibodies used in immunohistochemistry experiments were as follows: Rb anti-Runx3 (Kramer et al., 2006), Rb and Ck anti-Pv (de Nooij et al., 2013), Ck anti-β-galactosidase (Abcam, catalog #ab9361, RRID:AB_307210), Shp anti-GFP (Bio-Rad/AbD Serotec, catalog #4745-1051, RRID:AB_619712), Rb anti-vGluT1 (Demireva et al., 2011), Gt anti-wheat germ agglutinin (WGA) (Vector Laboratories, catalog #AS2024, RRID:AB_2315609), Rb anti-Pln (Sigma-Aldrich, catalog #HPA026900, RRID:AB_1855314), and Gp anti-tdTomato (S.L. Brenner-Morton and T.M.J., unpublished reagent). Fluorophore-conjugated secondary antibodies were obtained from Jackson ImmunoResearch Laboratories. Images were acquired on an Axioskop2 (Carl Zeiss), or on LSM510 Meta or LSM700 confocal microscopes (Carl Zeiss).
Experimental design and statistical analysis.
For DRG neuronal counts (e.g., for Rx3, PV, WGA), analyses were performed on serial cryostat sections (30 μm) of individual ganglia, with each section counted. For some cervical DRG, every other section was counted, and the total number of neurons was obtained by multiplying the original count by 2. Counted cell bodies and nuclei were required to be of near full-size diameter to avoid double counting of cells or nuclei at the edge of sections. Except when stated otherwise, a minimum of three DRG, obtained from three experimental or WT animals, were counted/segmental level/experimental condition. Average counts/DRG and SEM were calculated through Sigmaplot (Systat Software). For neuronal counts on RNA in situ DRG tissue sections, individual sections (15 μm) from cervical, thoracic, or lumbar DRG were analyzed. Neurons positive for a given pSN-enriched transcript were all counted (unless cell diameter was ∼<10% of normal neuronal diameter), and the average number of neurons/section were calculated by dividing the sum of all counted cells by the total number of counted sections. To estimate the proportion of pSNs that express a given pSN-enriched transcript, we also calculated the average number of PV+ neurons/section (used in most experiments), or Rx3+ neurons/section in neighboring tissue sections, and determined the fraction of marker-positive pSNs by dividing the average number of marker cells/section with the average number of PV or Rx3 neurons per section. Analyses of neuronal counts were largely restricted to thoracic or rostral lumbar segmental levels, at which the number of PVon neurons is approximately consistent with the number of pSNs (see Fig. 1). Therefore, the average number of PV+ neurons/section was not adjusted for the number of PVonRx3off neurons because this population is ∼≤5% at these segmental levels. When using Rx3+ neurons as our “pSN-reference level,” we calculated the average number of Rx3+ neurons/section by subtracting 15% of the total Rx3+ population before dividing this number by the total number of sections, to correct for the number of PvoffRx3on neurons (∼15% at thoracic and rostral lumbar levels; see Fig. 1) that are not part of the pSN population. For colocalization studies, counts on RNAscope DRG tissue sections were performed similarly as described above, with the exception that sections were 20 μm and restricted to L3-L5 lumbar DRG. Total number of PV or marker-expressing neurons were counted across multiple tissue sections (at least 6 per probe combination). All marker+ neurons were gated to PV+ neurons (i.e., any non-PV+ neurons expressing the marker were not included). Percentage of PV neurons expressing a certain marker-combination were calculated by dividing the number of marker+ neurons for each section with the number of PV neurons for the same section. Average percentages and SEM were calculated through Sigmaplot (Systat Software).
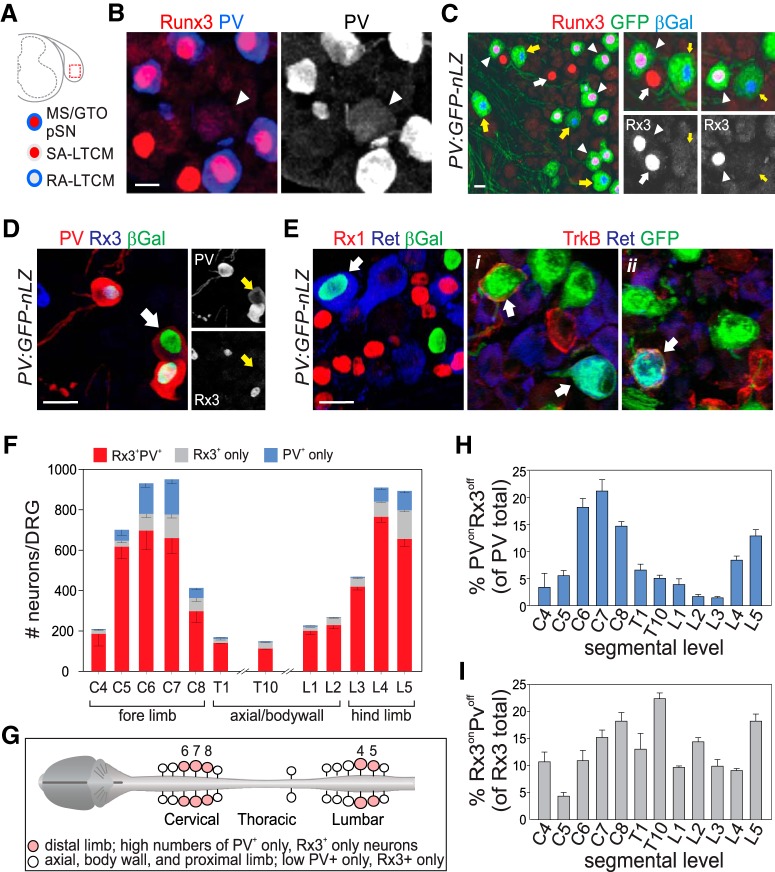
Figure 1.
Distinct somatic mechanoreceptive sensory subclasses delineated through expression of PV and Runx3. A, Schematic representation of mechanoreceptor subtypes marked by the coexpression of PV and Runx3 (all pSNs; red/blue), or the singular expression of PV (blue; RA-LTMR) or Runx3 (red; SA-LTMR). B, Expression of PV and Runx3 in p0 lumbar DRG. PV+Rx3off RA-LTMR neurons (arrowhead) generally express lower levels of PV than PVonRx3on pSNs. C, Genetic labeling of PV+ neurons using a PV:Cre allele and a Tau:lxp-stp-loxp:GFP-ires-nLZ reporter (PV:GFP-nLZ). Immunohistochemistry for Runx3, GFP, and β-galatosidase (βGal) reveals the presence of PVonRx3on (arrowheads), PVonRx3off (yellow arrows), and PVoffRx3on (white arrows) sensory neurons in P10 L5 DRG. D, Expression of PV, Runx3, and βGal in p0 DRG of PV:GFP-nLZ mice. White arrow indicates a βGal+PVon Rx3off neuron. Side images represent identical region but for individual expression of PV and Runx3 (βGal+PVonRx3off neuron marked by yellow arrows). E, Expression of Runx1, Ret, TrkB, and βGal or GFP in p0 DRG of PV:GFP-nLZ mice. βGal colocalizes with Ret in some neurons but never with Runx1, indicating that PV is expressed in a subset of Ret+ LTMRs but not in nociceptive Ret+Runx1+ sensory neurons. Expression of TrkB, Ret, and GFP in PV:GFP-nLZ mice shows that PV+ neurons can coexpress TrkB, Ret, or TrkB and Ret. F, Mean (± SEM) number of PV+Rx3+, PV+Rx3off (PV only), and Rx3+PVoff (Rx3 only) sensory neurons in cervical (C), thoracic (T) or lumbar (L) ganglia. Counts derived from 2–7 DRG obtained from 2–5 animals. C4, n = 2 DRG; C5, n = 4; C6, n = 4; C7, n = 4; C8, n = 3; T1, n = 3; T10, n = 3; L1, n = 3; L2, n = 5; L3, n = 5; L4, n = 3; L5, n = 7. G, Schematic summary of distribution of PV+Rx3+, PV+Rx3off (PV only), and Rx3+PVoff (Rx3 only) sensory neurons along the rostrocaudal extent of the spinal cord. Numbers indicate C6,7,8 and L4,5 ganglia. H, Mean (± SEM) percentage of PV+Rx3off neurons (of all PV+ neurons) in cervical (C), thoracic (T) or lumbar (L) DRG. Calculations based on F. I, Mean (± SEM) percentage of Rx3+PVoff neurons (of all Rx3+ neurons) in cervical (C), thoracic (T) or lumbar (L) DRG. Calculations based on F. Scale bars: B, 10 μm; C–E, 20 μm.
Measurements of cell body size were performed using the ruler tool of Photoshop image analysis software (Adobe). For each cell, measurement of cell body diameter was recorded for the largest measured diameter. Average cell diameter/cell was determined by dividing the sum of all diameters by the number of measured cells per probe and/or genotype. Statistical analysis (Student's t test or Mann–Whitney U test) on all counted neuronal populations or cell diameters was performed using Sigmaplot (Systat Software). Significance was accepted for p < 0.05.
Results
PV and Runx3 delineate distinct classes of low threshold mechanoreceptors in DRG
PV has long been known as a molecular marker for proprioceptive muscle afferents in DRG, and expression of PV protein and genetic labeling of PV+ neurons has been shown to be associated with most, if not all, MS and GTO sensory endings in skeletal muscle (Arber et al., 2000). However, in addition to proprioceptors, PV marks subsets of RA-LTMRs (e.g., Meissner and Pacinian afferents), indicating that expression of PV is not selective for muscle proprioceptors (Fig. 1A–E) (Luo et al., 2009; de Nooij et al., 2013). Similarly, we showed that another proprioceptive marker, the transcription factor Runx3 (Rx3), labels SA-LTMRs, including afferents that associate with Merkel cells in skin (Fig. 1A–C) (de Nooij et al., 2013). In addition to their partial overlap in molecular identity, PVonRx3on muscle proprioceptors, PVonRx3off RA-LTMRs, and Rx3onPVoff SA-LTMRs share several developmental, morphological, and functional features: They all originate from Ngn2+ sensory progenitors, are large-caliber myelinated neurons, have low activation thresholds, and associate with dedicated mechanoreceptive sensory receptor organs (Lallemend and Ernfors, 2012; Abraira and Ginty, 2013). Considering these similarities between proprioceptors, RA-LTMRs, and SA-LTMRs, we reasoned that a differential transcriptome analysis between these neurons could offer an opportunity to identify molecules uniquely associated with a proprioceptor sensory identity.
To explore this possibility, we first assessed the relative proportions of RA-LTMRs (PVonRx3off), SA-LTMRs (Rx3onPvoff), and muscle pSNs (PvonRx3on) in postnatal DRG along the rostrocaudal extent of the spinal cord. Because PV expression levels in (PVonRx3off) RA-LTMRs typically are much lower than in pSNs (Fig. 1B,D), we examined PV expression using PV:Cre; Tau:lxP-stop-lxP:mGFP-ires-nLZ (PV:GFP-nLZ) reporter mice to facilitate the visualization of PV-expressing neurons (Hippenmeyer et al., 2005). In PV:GFP-nLZ mice, expression of GFP and nuclear β-galatosidase (nLZ) is activated following Cre-mediated deletion of a transcriptional stop-cassette (Fig. 1C). In neonate (p8–10) PV:GFP-nLZ mice all nLZ+ neurons coexpress PV protein (Fig. 1D), indicating that the use of this genetic PV reporter is a reliable strategy to assess PV expression. Expression of Rx3 was examined by immunohistological analysis. When assessing the distribution of (PVonRx3off) RA-LTMRs, we observed few PVonRx3off neurons at rostral cervical (C4–5), thoracic (T1, T10), or rostral lumbar (L1–3) levels (mean percentage PVonRx3off of PV total ± SEM for C4, C5: 4.8 ± 1.0%; for T1, T10: 5.8 ± 0.6%; for L1–3: 2.1 ± 0.4%) (Fig. 1F,H). Thus, at these spinal levels, almost all PV neurons coexpress Rx3 and likely correspond to muscle pSNs. In contrast, in caudal cervical and caudal lumbar DRG (containing sensory neurons that innervate distal limbs), we observed many more PVonRX3off neurons (mean percentage PVonRx3off of PV total ± SEM for C7: 21.1 ± 2.1%; for L5: 12.9 ± 1.1%) (Fig. 1F,H). These findings indicate that PVonRx3off sensory neurons are most numerous at caudal limb levels, consistent with the notion that the glabrous skin of fore and hindlimb paws is densely innervated by RA-LTMRs involved in discriminative touch (Johnson, 2001; Abraira and Ginty, 2013). We similarly find that Rx3onPVoff SA-LTMRs are most abundant in caudal cervical and caudal lumbar DRG (mean percentage Rx3onPVoff of all Rx3 ± SEM: 18.2 ± 1.6% for C8; 18.2 ± 1.3% for L5) (Fig. 1F,I). Unlike PVonRx3off RA-LTMR neurons, however, Rx3onPVoff neurons are also prevalent at other spinal levels (compare Fig. 1H,I). The relative abundance of Rx3onPVoff neurons across all ganglia is consistent with the notion that SA-LTMR afferents associate with Merkel cell/guard hair complexes that are present throughout hairy skin (Abraira and Ginty, 2013). Together, these results indicate that, in addition to PVonRx3on muscle proprioceptors, RA- and SA-LTMR neurons that exhibit singular expression of PV or Rx3, respectively, are present at all spinal levels, but are particularly abundant at cervical and lumbar limb levels (Fig. 1G).
Transcriptome analysis of DRG mechanoreceptor subclasses identifies new proprioceptor markers
In view of the prevalence of PVonRx3off and PVoffRx3on sensory neurons in DRG, we sought to conduct a molecular analysis of pSN, RA-LTMR, and SA-LTMR cohorts on the basis of their differential PV and RX3 expression pattern. To perform a transcriptome analysis of neonatal (p0–6) pSNs and RA-LTMRs, we took advantage of a previously generated Runx3 mutant allele, the PV:Cre driver described above, and the Cre-dependent Ai9:tdTomato (tdT) reporter (Taniuchi et al., 2002; Kramer et al., 2006; Madisen et al., 2010), and separately crossed PV:Cre and Ai9:tdT alleles into the Rx3 mutant background. Subsequent intercrosses between these two strains resulted in WT Rx3 PV:Cre;Ai9:tdT animals, and Rx3 mutant PV:Cre;Ai9:tdT animals (hereafter referred to as Rx3+/+;PV:tdT and Rx3−/−;PV:tdT, respectively). In Rx3 mutants, pSNs fail to survive, and all remaining PVon neurons correspond to RA-LTMR sensory neurons (Kramer et al., 2006; de Nooij et al., 2013). We isolated PV:tdT+ sensory neurons from dissociated Rx3+/+ (comprising pSNs and RA-LTMRs) and Rx3−/− DRG (comprising only RA-LTMRs), using FACS (Fig. 2A). In addition, we used TrkC:GFP BAC transgenic mice, in which the GFP reporter is selectively expressed in Rx3onTrkCon neurons but not in Rx3offTrkCon neurons, to isolate a mixed population of pSNs and SA-LTMRs (Fig. 2B) (Lee et al., 2012; de Nooij et al., 2013). We performed global transcriptome analysis by RNA-seq on each of the three isolated mechanoreceptor populations: Rx3+/+;PV:tdT+ (hereafter referred to as [pSN+RA]), Rx3−/−;PV:tdT+ (hereafter referred to as [RA-only]), and TrkC:GFP+ (hereafter referred to as [pSN+SA]). RNA-seq data were mapped to the reference genome (mm10) and reads that unambiguously mapped to the genome or exon junctions were used for downstream analysis (GEO accession #GSE127161; see Materials and Methods) (Wu et al., 2013; Yan et al., 2015).
Figure 2.
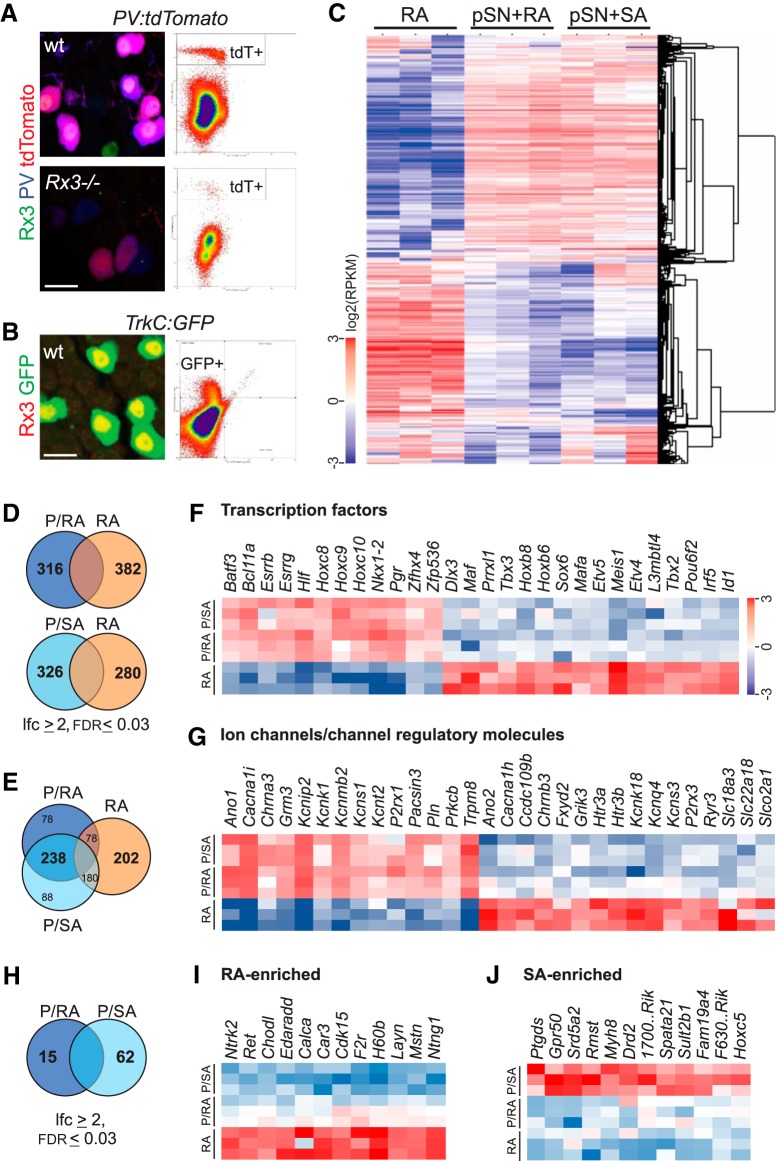
Molecular genetic strategy to identify proprioceptor selective molecular markers. A, Expression of Runx3, PV, and tdT in p0 DRG in Rx3+/+;PV:Cre;Ai9:tdTomato (Rx3+/+;PV:tdTomato) (wt) and Rx3−/−;PV:tdTomato mice (Rx3−/−) and profiles of FACS-isolated tdT+ neurons from animals with identical genotypes. B, Expression of Runx3 and GFP in p0 DRG of TrkC:GFP mice and profile of FACS-isolated GFP+ neurons from mice with similar genotype. C, Relative transcript levels in RA, pSN+RA and pSNA+SA mechanoreceptor populations. The heatmap shows log2 (RPKM) values with the mean expression across samples subtracted. Hierarchical clustering (Eisen et al., 1998) of genes and samples were performed using the subset of 1091 genes filtered by abundance (RPKM ≥ 5 in ≥ 3 samples) and variation (SD ≥ 0.6 in the log2 scale). D, Venn diagrams of upregulated transcripts comparing the pSN+RA (P/RA) and RA only (RA) neuronal cohorts (top), and comparing pSN+SA (P/SA) and RA only cohorts (bottom). E, Venn diagram of upregulated transcripts shared between the pSN+RA (P/RA) and pSN+SA (P/SA) cohorts compared with RA only (RA) neurons. F, Heatmap of transcription factors differentially expressed between pSN+RA (P/RA), pSN+SA (P/SA), and RA only (RA) mechanoreceptor populations (|log2 (fold change)| ≥ 2; FDR ≤ 0.03). Color scale represents log2 (RPKM) values with mean values subtracted. G, Heatmap of ion channels and regulatory channel molecules differentially expressed between pSN+RA (P/RA), pSN+SA (P/SA), and RA only (RA) mechanoreceptor populations (|log2 (fold change)| ≥ 2; FDR ≤ 0.03). Color scale as in F. H, Venn diagram of differentially expressed transcripts between the pSN+RA (P/RA) and pSN+SA (P/SA) cohorts. I, Heatmap of select transcripts differentially upregulated in the RA only (RA) mechanoreceptor population compared with both pSN+RA and pSN+SA populations (|log2 (fold change)| ≥ 2; FDR ≤ 0.03). Color scale as in F. J, Heatmap of select transcripts differentially upregulated in the pSN+SA only (P/SA) mechanoreceptor population compared with the pSN+RA and RA only populations (|log2 (fold change)| ≥ 2; FDR ≤ 0.03). Color scale as in F. 1700..Rik, 1700123O21Rik; F630..Rik, F630111L10Rik. Scale bars: A, B, 10 μm.
To identify transcripts enriched in proprioceptors, we first performed a differential expression analysis between the transcriptomes of the [pSN+RA] and [RA-only] populations (Fig. 2C,D). Differentially expressed transcripts were identified based on the following criteria: FDR < 0.03, |log2(fold change)| ≥ 2, and RPKM ≥ 1 in at least one of the compared conditions. This comparison revealed 316 transcripts that are enriched in the [pSN+RA] population, suggesting that these transcripts are preferentially expressed in proprioceptors compared with RA-LTMRs (Fig. 2D). We find that these pSN-enriched transcripts include several molecules (seven in total) previously known to be restricted to, or enriched in, proprioceptors (e.g., Whrn, Esrrg, Vstm2b, Runx3, Plxd1) (Fig. 2F; and data not shown) (de Nooij et al., 2013, 2015; Pecho-Vrieseling et al., 2009; A. Norovich and T.M.J., personal communication). In addition, the vast majority of these transcripts (238 of 316) were also found to be enriched in the [pSN+SA] neuronal cohort compared with [RA-only] neurons, providing additional validation for our differential expression analysis (Fig. 2D,E). Differentially expressed genes shared between [pSN+RA] and [pSN+SA] compared with [RA-only] include transcription factors, as well as molecules that relate to neural function (e.g., ion channels, channel regulatory molecules, neurotransmitter receptors) (Fig. 2F,G). While the underlying molecular logic of the differential expression for the majority of these transcripts remains to be determined, these data will likely help to provide new insight into the physiological differences between pSNs and RA-LTMRs.
A reverse differential expression analysis between [RA-only] and [pSN+RA] neurons identified 382 transcripts that are enriched in RA-LTMRs (FDR < 0.03, |log2(fold change)| ≥ 2, RPKM ≥ 1) (Fig. 2D). These include known RA-LTMR markers, such as Ntrk2, MafA, Ret, and Kcnq4 as well as transcripts not previously associated with a RA-LTMR identity (Fig. 2F,G,I) (Bourane et al., 2009; Luo et al., 2009; Lecoin et al., 2010; Heidenreich et al., 2011). In addition, a comparison between [pSN+SA] and [pSN+RA] neurons identified 62 transcripts that are enriched in the [pSN+SA] cohort (FDR < 0.03, |log2(fold change)| ≥ 2, RPKM ≥ 1) (Fig. 2H). Some of these transcripts are preferentially expressed in (PVoffRx3on) SA-LTMRs (Fig. 2J), a sensory population for which currently few molecular markers are known.
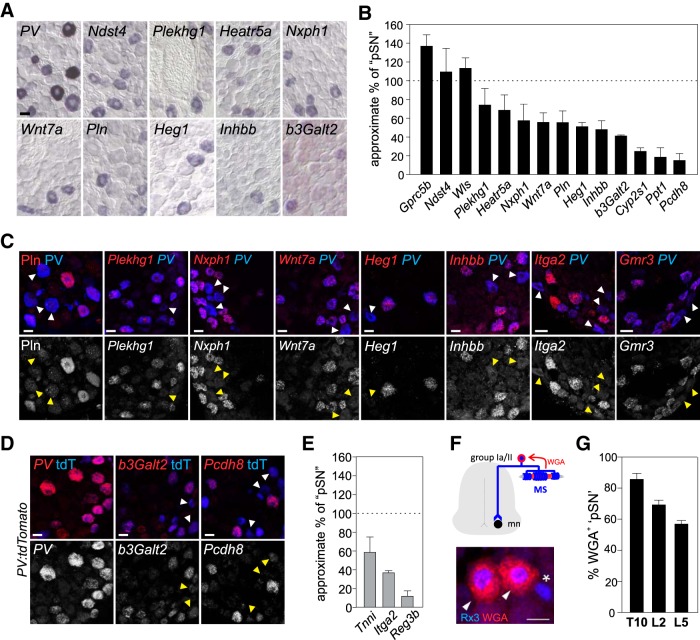
Concentrating on the pSN population, we next attempted to validate the transcripts that show enriched expression in the [pSN+RA] neuronal subset, but which have not previously been observed in proprioceptors (309 of 316). This group includes some transcripts that have been linked with PV+ neurons through other molecular screens but have not yet been confirmed as pSN-selective (Lee et al., 2012; Chiu et al., 2014; de Nooij et al., 2015). To determine the relevance of these transcripts in pSN development, we first assessed their expression pattern in DRG using the Allen Brain Atlas (P4 spinal cord dataset) (Lein et al., 2007). Of the transcripts that were present in the Allen Brain Atlas (220 of 309), many exhibited relative widespread expression in DRG. This suggests that, while these markers are enriched in pSNs compared with RA-LTMRs, they are shared with other DRG sensory subsets. Nevertheless, we also found that ∼30% of our Allen markers (88 of 220) were expressed in DRG in a pattern that resembled the pattern of expression observed for Rx3 or PV. Transcripts for which we observed such pSN-like expression patterns, or for those that were not included in the Allen dataset (89 of 309), were further examined through our own RNA ISH analysis. To determine directly to what extent these transcripts are selective for pSNs, we performed these in situs in WT DRG tissue as well as in DRG tissue obtained from Rx3−/− mice that lack proprioceptors (Fig. 3A,C). Additionally, we examined their expression in DRG of PVCre;Isl2:DTAFlx (PV:DTA) mice, in which pSNs and RA-LTMRs are genetically ablated while SA-LTMRs are preserved (Fig. 3B,C) (Yang et al., 2001; Hippenmeyer et al., 2005; Akay et al., 2014). Use of the PV:DTA mice permitted us to assess the expression in pSNs at later developmental stages (∼p8–10) due to the prolonged survival of these mice compared with Rx3−/− mice (Kramer et al., 2006; Akay et al., 2014). Of ∼90 transcripts thus far tested in these studies, we identified 24 molecular markers which, at the developmental stage examined initially (p0–p8), were either absent or significantly reduced in DRG that lack proprioceptors. This suggests that these genes constitute new proprioceptor-biased molecular markers (Fig. 3). All other identified transcripts that showed enrichment in pSNs relative to RA- and SA-LTMRs were either not expressed, or exhibited more widespread expression in DRG. While these latter transcripts may have relevance for proprioceptor development or function, we did not pursue them for further analysis here.
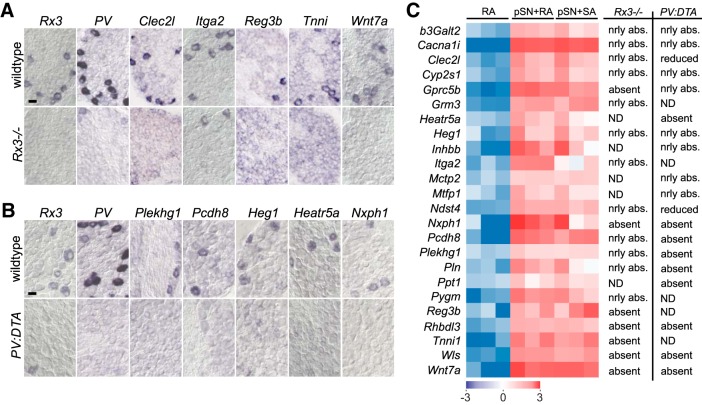
Figure 3.
Identification of proprioceptor-enriched transcripts. A, Expression of pSN-enriched transcripts in p0 DRG of WT (top) and Rx3−/− (bottom) mice. Most markers are absent in DRG of Rx3 mutant mice. For some markers (e.g., Itga2), expression is occasionally still observed in Rx3−/− DRG, indicating that these markers are also expressed in a few other DRG sensory neurons. B, Expression of pSN-enriched transcripts in p8–10 DRG of WT (top) and PV:Cre; Isl2:lxp-STOP-lxp:DTA (PV:DTA; bottom) mice. Many markers are completely absent from PV:DTA DRG, whereas others are still observed, but in fewer neurons. The persistent expression in PV:DTA mice could result from an inefficient Cre-mediated recombination of the floxed-STOP cassette, expression in a subset of PVoffRx3+ SA-LTMRs, or expression in small numbers of other types of sensory neurons (non-pSN or LTMR). C, Heatmap of relative transcript levels in RA, pSN+RA, and pSNA+SA mechanoreceptor populations of our 24 selected transcripts (in alphabetical order) enriched in the [pSN+RA] neuronal cohort compared with the [RA] only cohort (|log2 (fold change)| ≥ 2; FDR ≤ 0.03; RPKM ≥ 1). Columns to the right represent relative loss of expression in DRG of Runx3−/− or PV:DTA mice. Selected transcripts have little or no expression outside the pSN population between p0 and p10. Color scale represents log2(RPKM) values with mean values subtracted. nrly abs., Nearly absent; ND, not determined. Scale bars: A, B, 20 μm.
Together, our differential transcriptome analysis of DRG mechanoreceptor subclasses effectively revealed transcripts that may help define RA- or SA-LTMR neuronal identities and, moreover, identified numerous molecules that show preferential enrichment in proprioceptive muscle afferents.
Proprioceptor markers are first observed after pSNs innervate their receptor targets
MS and GTO afferent pSNs exhibit unique morphological sensory endings and activation properties, and project to separate sets of target neurons in spinal cord, features that are apparent shortly after the neurons are born. As such, MS/GTO pSN subtype identity is thought to be established at an early developmental stage, before the afferents engage with their cognate receptor target. Therefore, to assess our proprioceptor markers in terms of their relative importance with respect to MS/GTO afferent subtype differentiation, we performed a more detailed analysis of their developmental pattern of expression. Specifically, we examined transcript expression at e12.5, when proprioceptor identity is first established, at e14.5, when pSN afferents first innervate their nascent receptor targets, at e17.5 when the pattern of peripheral innervation is largely completed, and at different postnatal stages (p0-p21) during pSN maturation (Hippenmeyer et al., 2002; Kramer et al., 2006; de Nooij et al., 2013). This analysis revealed that our newly identified pSN markers fall into three developmental classes: (1) transcripts that are expressed before target innervation, (2) transcripts that are transiently expressed during the period of target innervation, and (3), transcripts that are first expressed at late embryonic or even postnatal stages (Fig. 4; Table 1). We find that the first class consists of only three transcripts (Plekhg1, Itga2, Gmr3), indicating that a surprisingly small number of our pSN transcripts is present shortly after pSNs are generated (Fig. 4A–D; Table 1). More transcripts can be detected at e14.5, when MS/GTO subtypes first distinguish themselves by virtue of their peripheral target selection, including Cacnai1, Clec2l, Gprc5b, Mctp2, Ndst4, Pcdh8, Wls, Wnt7a, and Reg3b (Table 1). In contrast, for all other pSN-enriched transcripts (12 of 24), expression was first observed well after pSNs innervated their peripheral targets (e.g., Heg1, Inhbb, Heatr5a) (Fig. 4E,F; Table 1). The late onset of expression of this many of our newly identified pSN markers could suggest that aspects of pSN identity are acquired during postnatal maturation. We also noted that a few new pSN markers (Reg3b, Itga2, Gmr3, Tnni, and Pygm) exhibit a transient pattern of expression, with their expression largely extinguished by p8 (Fig. 4C,D; Table 1), indicating that these transcripts may have a temporal role in pSN development. However, for the majority of our new pSN markers (19 of 24), expression is maintained until at least p21. Expression at these later developmental stages, when proprioceptors are considered to be mature, may indicate a role in proprioceptor maintenance or function. Together, the dynamic pattern and late onset of expression of many of our newly identified pSN transcripts indicate that the phenotypic specialization of proprioceptors may be acquired gradually, in a developmental process that extends well beyond the time pSNs first innervate their peripheral receptor targets.
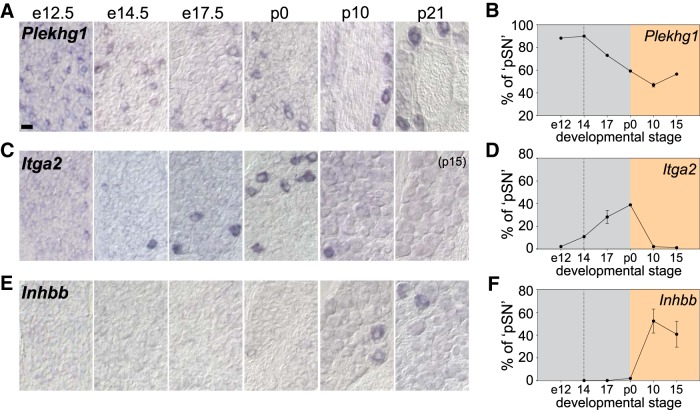
Figure 4.
Expression of newly identified pSN markers across pSN development. A–C, Expression of Plekhg1 (A), Itga2 (B), and Inhbb (C) in embryonic (e12.5, e14.5, e17.5) and postnatal (p0, p10, p21) DRG. D–F, Mean percentage of pSNs that express Plekhg1 (D), Itga2 (E), or Inhbb (F) at different developmental stages. Number of pSN neurons is approximated based on the number of PV+ or Rx3+ neurons in neighboring tissue sections (see Materials and Methods). Plekhg1: e12.5, n = 5 sections, 164 neurons total; e14.5, n = 7, 68 neurons; e17.5, n = 6, 129 neurons; p0, n = 10, 136 neurons; p8–10, n = 12, 59 neurons; p15, n = 20, 137 neurons. Itga2: e12.5, n = 5 sections, 4 neurons total; e14.5, n = 8, 11 neurons; e17.5, n = 20, 129 neurons; p0, n = 9, 75 neurons; p8–10, n = 7, 5 neurons; p15, n = 7, 1 neuron. Inhbb: e12.5, ND; e14.5, n = 4 sections, 0 neurons total; e17.5, n = 4, 0 neurons; p0, n = 7, 3 neurons; p8–10, n = 29, 153 neurons; p15, n = 10, 45 neurons. Gray area represents embryonic developmental stages. Yellow area represents postnatal period. Dotted line indicates developmental stage when pSN axons first engage with their cognate receptor organs. Error bars indicate SEM for data acquired from multiple experiments. Scale bars: A–C, 20 μm.
Table 1.
Expression of pSN-enriched transcripts during developmenta
| Transcript | e14.5 | e17.5 | p0 | p5 | p10 | p15 | p21 | PVss |
|---|---|---|---|---|---|---|---|---|
| Cacna1i | + | + | + | + | ND | + | ND | ND |
| Clec2l | + | + | + | + | ND | + | + | ND |
| Gprc5b | + | + | + | + | ND | + | + | − |
| Mctp2 | + | + | + | + | + | + | ND | ND |
| Ndst4 | + | + | + | + | ND | + | + | − |
| Pcdh8 | + | + | + | + | + | + | + | + |
| Plekhg1 | + | + | + | + | + | + | ND | ± |
| Wls | + | + | + | + | + | + | + | − |
| Wnt7a | + | + | + | + | + | + | + | + |
| Reg3b | + | + | + | ± | ND | − | ND | + |
| Itga2 | + | + | + | + | ± | − | ND | + |
| Gmr3 | + | + | + | + | ± | − | ND | + |
| Tnni | − | + | + | + | ND | − | ND | + |
| Pygm | − | + | + | + | + | − | ND | ND |
| Pln | − | + | + | + | + | + | + | + |
| Ppt1 | − | + | + | + | + | + | + | + |
| Heg1 | − | −/+ | + | + | + | + | + | + |
| b3Galt2 | − | − | + | + | + | + | + | + |
| Rhbdl3 | − | − | + | + | + | + | + | ND |
| Heatr5a | − | − | + | + | + | + | + | + |
| Mtfp1 | − | − | −/+ | + | + | + | ND | ND |
| Inhbb | − | − | − | + | + | + | + | + |
| Cyp2s1 | − | − | − | + | + | + | + | ND |
| Nxph1 | − | − | − | + | + | + | + | + |
aTranscript expression, as measured by RNA ISH, at embryonic and postnatal stages. Transcripts ordered based on their onset and extinction of expression. Analysis based on at least five sections/developmental stage. +, Clearly expressed; −, no expression; ±, just detectable expression; −/+, ND, not determined. Transcript expression observed in subsets of PV+ neurons (PVss) is indicated by +.
Newly identified pSN-enriched transcripts mark proprioceptor subsets
We next examined whether our pSN-enriched molecules label all, or select subsets of pSNs, and determined the proportion of pSN neurons that expresses each transcript. Transcripts that were no longer restricted to pSNs at late developmental stages (Cacna1i, Clec2l), or that were expressed at levels too low (Mctp2, Pygm, Rhbdl3, Mtfp1, Gmr3) to obtain consistent results, were excluded from these analyses. For all other transcripts (17 of 24), we compared the mean number of positive neurons/DRG section with the number of PV+ neurons in neighboring DRG sections. We mostly limited this analysis to thoracic or rostral lumbar DRG, given that at these segmental levels the number of PVon neurons is approximately consistent with the number of pSNs (Fig. 1). On a few occasions, Rx3 was used as the pSN-reference level; in these analyses, the total number of Rx3+ neurons was adjusted by subtracting the average number of PVoffRx3on neurons; see Materials and Methods). In addition, we mostly performed this analysis between p15 and p21. We chose this developmental stage because, in experiments in which we used Cre/Flp-dependent genetic reporters for two of our new markers (Wnt7a and b3Galt2), we observed discrepancies in the number of reporter-labeled neurons and the number of neurons labeled by RNA in situ (data not shown). This indicates that individual transcripts initially may be expressed in a larger neuronal subset than at later developmental stages. Alternatively, transcripts may gradually expand their presence within the pSN population. Thus, by performing our pSN subset expression analysis at p15–p21 (when proprioceptor differentiation is thought to be complete), expression patterns of pSN markers are more likely to be stabilized. For the markers that are no longer expressed at these developmental stages (e.g., Itga2, Tnni, Reg3b), we estimated their relative proportion in p0 DRG.
In performing this analysis, we find that three transcripts, including Gprc5b, Wls, and Ndst4, are observed in equivalent or nearly equivalent numbers as PV+ (or Rx3+) neurons, suggesting that these molecules mark the entire population of proprioceptive muscle afferents (Fig. 5A,B). Interestingly, for most other transcripts (14 of 17), the number of positive DRG neurons is smaller than the number of PV+ neurons (Fig. 5B,E). We estimate that the percentage of pSNs that express these markers ranges between 15% (e.g., Ppt1, Pcdh8, Reg3b) and 75% (e.g., Plekhg1 and Heatr5a) (Fig. 5A,B,E). The observation that these transcripts are expressed in subsets of PV or Rx3 neurons suggests that these molecules mark select proprioceptor subtypes. To validate the subtype-selective expression of these transcripts more directly, we sought to perform colocalization studies with PV. Because immunological reagents for all but one of our markers (Pln) were either unavailable or unreliable, we largely performed these experiments using RNAscope technology (Hoffman et al., 2018). For most of these analyses, we assessed expression of our marker genes in PV neurons defined by PV transcript expression (Fig. 5C). In cases where RNAscope probes were incompatible with our PV probe, we assessed PV expression through use of the genetic PV:tdTomato reporter (Fig. 5D). Consistent with our conventional RNA in situ analysis on proprioceptor mutant tissues, we find that for all 10 markers tested in this coexpression assay, their expression almost exclusively localizes to PV+ neurons. Moreover, in each case, we detected PV+ neurons clearly devoid of marker coexpression, albeit that, for some probes (e.g., Plekhg1), the number of PV+Plekhg1+ neurons appears more abundant than predicted on the basis of our original RNA in situ analysis. We attribute this to the increased sensitivity of RNAscope technology over conventional RNA in situ analysis. Nevertheless, these analyses confirm a pSN subset-selective expression pattern for at least nine of our new pSN markers.
Figure 5.
pSN-enriched transcripts include candidate MS or GTO pSN subtype markers. A, Expression of a sample of pSN-enriched transcripts in p21 WT DRG. B, Mean (± SEM) percentage of pSN neurons expressing an individual pSN-enriched transcript in WT thoracic/rostral lumbar DRG at p15–21. Number of pSN neurons is approximated based on the number of PV+ or Rx3+ neurons in neighboring tissue sections (see Materials and Methods). Quantifications based on DRG sections obtained from at least two experiments and involving a minimum of 5 different animals. Number of sections/total neurons counted: Gprc5b, n = 9 sections, 102 neurons; Ndst4, n = 16, 103 neurons; Wls, n = 13, 134 neurons; Plekhg1, n = 26, 181 neurons; Heatr5a, n = 12, 84 neurons; Nxph1, n = 11, 73 neurons; Wnt7a, n = 26, 175 neurons; Pln, n = 21, 98 neurons; Heg1, n = 25, 127 neurons; Inhbb, n = 18, 95 neurons; b3Galt2, n = 82, 246 neurons; Cyp2s1, n = 14, 43 neurons; Ppt1, n = 18, 54 neurons; Pcdh8, n = 10, 13 neurons. Transcripts not included in analysis are omitted either because of increased expression outside the pSN population (Cacna1i, Clec2l) or because of overall lower expression levels (Mctp2, Pygm, Rhbdl3, Mtfp1, Gmr3). C, D, Colocalization of candidate pSN subset markers with PV protein, PV transcript (C), or genetic reporter expression (PV:tdTomato) (D). With exception of Pln (in C), expression of pSN-enriched transcripts was examined using RNAscope. Probe selection was based on availability. Top, White arrowheads indicate PV neurons devoid of marker expression. Bottom, Yellow arrowheads indicate corresponding neurons. D, Expression of PV is generally in agreement with PV:tdTomato reporter expression, but few PVofftdT+ and PV+tdToff neurons can be observed. E, Mean (± SEM) percentage of pSN neurons expressing Tnni, Itga2, or Reg3b in WT thoracic/rostral lumbar DRG at e18.5-p0. Number of pSN neurons is approximated based on the number of PV+ or Rx3+ neurons in neighboring tissue sections (see Materials and Methods). Quantifications based on DRG sections obtained from at least 2 experiments and involving a minimum of 8 different animals (Tnni, n = 23 sections, 125 neurons; Itga2, n = 19 sections, 138 neurons; Reg3b, n = 24 sections, 60 neurons). F, Genetic strategy to identify MS-innervating pSNs using Egr3:WGA transgenic mice (de Nooij et al., 2013). Expression of WGA in intrafusal muscle fibers labels Rx3+ MS-innervating pSNs (arrowheads) but not presumptive Rx3+ GTO afferents (asterisk). G, Estimated mean (± SEM) percentage of MS and GTO-innervating pSNs on the basis of WGA-labeled pSNs in T10 (n = 3), L2 (n = 6), and L5 (n = 6) DRG, in p8–12 Egr3:WGA mice. Scale bars: A, C, D, 20 μm.
The subset-restricted expression of our pSN transcripts could mean that these markers correspond to a particular muscle-type identity (e.g., flexor, extensor, axial, limb). Alternatively, it could mean that they align with proprioceptor Group Ia/II MS or Group Ib GTO subtype identities. To try to distinguish between these possibilities, we first asked whether any of our candidate pSN subtype markers overlapped with a set of markers identified in a screen for molecules that segregate with proprioceptors innervating distinct hindlimb muscles (Poliak et al., 2016). This inventory revealed just four molecules: Tnni, Wls, Nxph1, and Itga2. While the peripheral biases of these four transcripts have yet to be confirmed experimentally, the notion that only few of our candidate markers have been implicated as muscle-type markers increases the likelihood that some of our pSN subtype markers are indicative of a pSN subtype identity other than muscle-type identity.
We next considered whether our candidate pSN subtype markers could align with either a MS or GTO afferent subtype identity. We previously established that the percentage of MS-innervating proprioceptors at lumbar levels ranges between ∼70% (in L2 DRG) and ∼55% (in L5 DRG) (de Nooij et al., 2013). We derived these numbers from the use of Egr3:WGA transgenic mice, in which expression of WGA is targeted to the intrafusal fibers of MSs and retrogradely labels MS afferent cell bodies in DRG (Fig. 5F) (Yoshihara et al., 1999; de Nooij et al., 2013). An analysis of the number of WGA+ PVonRx3on neurons thus provides us with an estimate of the number of MS-innervating pSNs per ganglia. We now show that the MS afferent bias seen at rostral lumbar levels is also apparent at thoracic segmental levels with 85.8 ± 3.7% SEM of pSNs (PvonRx3on) labeled by WGA in Egr3:WGA transgenic mice (Fig. 5G). These data further emphasize that the distribution of MS and GTO afferents differs along the rostrocaudal axis of the spinal cord, with a higher prevalence of MS-innervating afferents (relative to GTO afferents) at thoracic spinal levels than at L2 and L5 lumbar levels. Given that, in mice, each MS is innervated by a single Group Ia afferent and 1–2 Group II afferents, we predict that the distribution of Group Ia/Group II/Group Ib afferents is ∼34%:51%:15% at thoracic levels, and 28%:42%:30% at rostral lumbar levels. Based on these estimates, we determined that for several of our pSN subtype markers their pattern of expression aligns with a general (i.e., Group Ia and II) MS afferent identity (e.g., Heatr5a), a Group Ia or Group II identity (e.g., Wnt7a, Heg1, Inhbb, b3Galt2), or a GTO afferent identity (e.g., Pcdh8) (Fig. 5A–D). Together, these analyses establish that a substantial number of our newly identified pSN markers define subsets of proprioceptors. Moreover, for many of these pSN subtype markers their pattern of expression is consistent with a MS or GTO afferent subtype identity.
Combinatorial expression of candidate subtype markers reveals distinct pSN subclasses
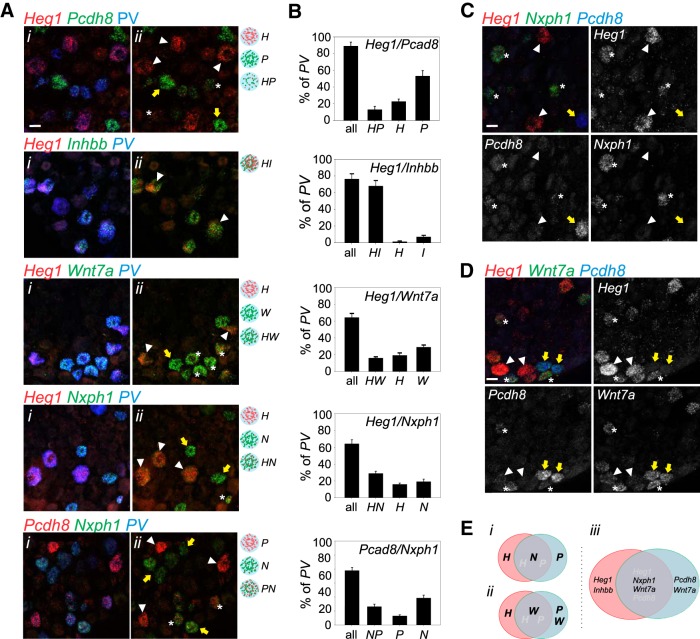
To determine the extent to which our candidate MS/GTO pSN subtype markers capture different or similar proprioceptor subsets, we performed combinatorial RNAscope analysis for some of our markers. We focused on Wnt7a, Inhbb, Heg1, Nxph1, and Pcdh8, markers expressed in smaller PV subsets and with relative robust expression levels. We performed these analyses in p18 lumbar (L3-L5) DRG where we expect sizable numbers of proprioceptors, including GTO afferents. We find that the percentage of PV+ neurons that expresses a given marker when observed with RNAscope is largely in agreement with our analysis based on conventional RNA in situ (Fig. 5B). A notable exception is Pcdh8, for which the percentage of positive neurons appears at odds with our RNA in situ data (Figs. 5B, 6B). Presumably, the increased sensitivity of the RNAscope assay enables the detection of neurons with much lower levels of Pcdh8 (Fig. 6A,B). Assessing different marker combinations, we observed that the pattern of expression of Heg1 and Inhbb almost entirely overlaps, implying that they localize to the same proprioceptor subset. In contrast, all other marker combinations show both overlapping and exclusive expression, suggesting that they delineate multiple pSN populations (Fig. 6A,B). Interestingly, considering that up to 15% of PV+ neurons may represent RA-LTMRs at these (L3–L5) segmental levels (Fig. 1), the combined expression of Heg1 and Pcdh8 appears to capture most proprioceptors (Fig. 6B). This implies that the combinatorial expression of Heg1 and Pcdh8 segregates the pSN population into at least three classes: PV neurons that predominantly express Heg1, PV neurons that predominantly express high levels of Pcdh8, and PV neurons that coexpress low levels of both Heg1 and Pcdh8. This molecular division is supported further by the combinatorial expression of Heg1 and Pcdh8 with either Nxph1 or Wnt7a. While Wnt7a segregates with Heg1low/Pcdh8low and Pcdh8high neurons, Nxph1 predominantly colocalizes with Heg1low/Pcdh8low neurons (Fig. 6C,D). Together, these data reveal that the combinatorial expression of our candidate pSN subtype markers, Heg1, Pcdh8, Nxph1, and Wnt7a, defines as least three molecularly distinct PV neuronal subsets (Fig. 6E).
Figure 6.
Combinatorial expression of pSN-enriched transcripts reveals distinct molecular subclasses. A, Expression analysis (RNAscope) of different combinations of pSN-enriched transcripts, in p18–p21 lumbar DRG, shown in the presence (i) or absence (ii) of expression of PV. Expression of PV is assessed through RNAscope (PV) or through immunological analysis of tdTomato+ (tdT) neurons in PV:tdTomato reporter mice. Schematics indicate the major subtypes defined by each probe combination. B, Quantification of PV+ neurons that (i) express any probe combination ('all'; including singular expression of probe A, singular expression of probe B or coexpression of probe A and B); (ii) coexpress A and B probes; (iii) predominantly express probe A; or (iv) predominantly express probe B. Total number of PV+ neurons and tissue sections assessed for each probe combination: 322/10 for Heg1/Nxph1, 283/13 for Pcdh8/Nxph1, 114/6 for Heg1/Pcdh8, 178/8 for Heg1/Inhbb, and 129/7 for Heg1/Wnt7a. C, Combinatorial expression of Nxph1, Heg1, and Pcdh8 in lumbar DRG at p21. In each panel, white arrowheads indicate neurons predominantly expressing Heg1. Yellow arrows indicate neurons that predominantly express Pcdh8. *Expression of Nxph1 neurons (coexpressing low levels of Heg1 and Pcdh8). D, Combinatorial expression of Wnt7a, Heg1, and Pcdh8 in lumbar DRG at p21. In each panel, white arrowheads indicate neurons predominantly expressing Heg1. Yellow arrows indicate neurons that express Pcdh8 and Wnt7a. *Expression of Wnt7a neurons (coexpressing low levels of Heg1 and Pcdh8). E, Schematic Venn diagram indicating the three main classes of PV+ neurons as revealed by expression of Heg1, Nxph1, Pcdh8 (i), Heg1, Wnt7a, Pcdh8 (ii), and summarized in (iii). H, Heg1; P, Pcdh8; HP, Heg1 and Pcdh8; HI, Heg1 and Inhbb; W, Wnt7a; HW, Heg1 and Wnt7a; N, Nxph1; HN, Heg1 and Nxph1; PN, Pcdh8 and Nxph1. Scale bars: A, C, D, 20 μm.
Altered expression patterns of pSN subtype markers in Egr3−/− mice lacking MSs
The dynamic expression and late onset of many of our pSN markers could indicate that these candidate pSN subtype markers are induced as a consequence of an intrinsic subtype differentiation program, and represent specific features of maturing MS or GTO pSN subtype phenotypes. Alternatively, these observations raise the possibility that aspects of MS/GTO afferent subtype identity are imposed through extrinsic signals. The notion that certain features of MS/GTO pSN subtypes are acquired postnatally and may depend on extrinsic, possibly target-derived, signals has precedent in the expression of Wnt7a in γMNs. Expression of Wnt7a in γMNs was shown to depend on MS innervation, suggesting that spindle-derived instructive signals contribute to the distinction between γ and αMN subtypes (Ashrafi et al., 2012). Could MS- or GTO-derived signals similarly instruct Group Ia/II/Ib afferent subclass identities?
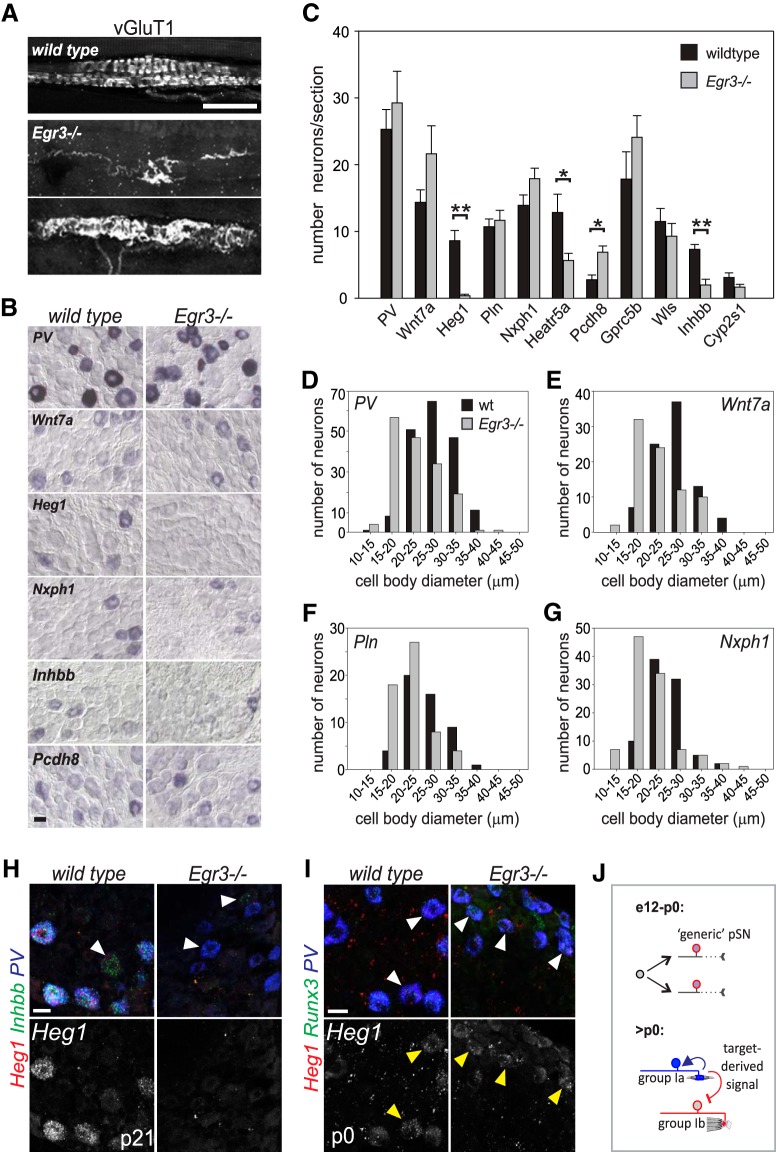
To test this idea, we examined the expression of our candidate pSN subtype markers in DRG of mice that are mostly devoid of normal spindles, using animals that are mutant for the zinc-finger transcription factor Egr3. In WT animals, Egr3 is expressed in a few (non-pSN) DRG neurons, in Schwann cells, and in the intrafusal fibers of the MS (Gao et al., 2007; Oliveira Fernandes and Tourtellotte, 2015). While proprioceptor and Schwann cell development appears unaffected in Egr3 mutants, spindle development is impaired (Tourtellotte and Milbrandt 1998; Oliveira Fernandes and Tourtellotte, 2015). Mutant spindles remain in an immature state; and after an initial contact, many spindle-innervating proprioceptors retract from the intrafusal muscle fibers. The few remaining afferents appear grossly abnormal (Fig. 7A) (Tourtellotte et al., 2001). In contrast, Egr3 is not expressed in GTOs, and GTO-sensory endings are unaffected in Egr3−/− mice (Tourtellotte and Milbrandt, 1998; de Nooij et al., 2013). Thus, to test whether extrinsic signaling factors secreted from the MS intrafusal muscle fibers could influence the phenotype of pSN afferents, we examined the expression of our candidate pSN subtype markers in DRG of p21 Egr3−/− mice. Consistent with previous findings, the number of PV+ neurons was unaffected in p21 Egr3−/− DRG (Fig. 7B,C). However, we detected a reduction in the average pSN cell body size, a consequence of a reduction in the number of larger (>30 μm) cells, and an increase in the number of pSNs in the 20–25 μm size range (Fig. 7D). Mature MSs normally express Neurotrophin 3 (NT3); therefore, the decrease in cell body size in Egr3−/− mice likely results from diminished trophic support. Despite the requirement for MSs in the induction of Wnt7a expression in γMNs, we observed normal numbers of Wnt7a+ neurons in Egr3−/− DRG (Fig. 7B,C). Nevertheless, as for PV, we find that the cell bodies of Wnt7a+ neurons are reduced in size compared with WT (Fig. 7E). This suggests that at least a portion of Wnt7a+ neurons normally supply MSs. Other markers we tested (e.g., Pln and Nxph1) were similarly unchanged in the total number of positive DRG neurons but were found to be predominantly expressed by pSNs of smaller average diameter (Fig. 7B,C,F,G). In contrast to Wnt7a, we found that the transcript level of heart of glass 1 (Heg1), a transmembrane protein involved in cardiovascular development (Kleaveland et al., 2009), is severely diminished if not absent in Egr3−/− DRG (Fig. 7B,C,H). The sparse Heg1+ neurons that remain may reflect the remaining MSs occasionally present in Egr3 mutants (Fig. 7A). Low levels of Heg1 transcript are still observed in Egr3−/− DRG at p0, shortly after the normal onset of Heg1 expression, suggesting that initiation of Heg1 expression may be independent of spindle innervation (Fig. 7I). Nevertheless, the absence of Heg1 expression in Egr3−/− DRG suggests that maintained expression of this candidate pSN subtype marker may be limited to MS-innervating afferents and may depend on spindle innervation. In addition to Heg1, we noted a significant reduction in the number of Heat repeat-containing protein 5a (Heatr5a) and Inhibin β-B (Inhbb) neurons (Fig. 7B,C,G). The partial loss of these markers may indicate that their expression is affected in only one of the two (i.e., Group Ia or II) MS-innervating pSN subtypes and/or reflects their expression in sensory neurons other than pSNs at this stage (e.g., Inhbb) (Fig. 7H). In contrast to Heg1, Heatr5a, and Inhbb, the number of Protocadherin (Pcdh8) neurons appears increased compared with WT DRG (Fig. 7B,C), possibly indicating that spindle contact is required for suppressing alternate pSN subtypes. These data identify Heg1, Inhbb, and Heatr5a as candidate markers for MS afferents, and demonstrate that expression of pSN subtype-selective molecules can be influenced by signals derived from sensory end organs (Fig. 7J).
Figure 7.
Expression of pSN subtype markers depends on the presence of MS sensory end organs. A, Group Ia/II MS sensory endings in hindlimb muscle of WT and Egr3−/− mice as visualized by the expression of vGlut1. While most MS afferents retract from spindles in Egr3 mutants, a few afferents remain. Remaining afferents exhibit severely abnormal morphology. B, Expression of select candidate pSN subtype markers in WT and Egr3−/− limb level DRG at p21. C, Mean (± SEM) number of sensory neurons that express a given candidate pSN subtype marker in WT and Egr3−/− DRG sections. Quantifications performed on p21 cervical DRG and based on at least 6 sections/marker. Similar results were obtained in at least 3 mutant animals. The number of Heg1+, Heatr5a+, and Inhbb+ neurons is significantly reduced in Egr3−/− DRG (Mann–Whitney rank sum test, p < 0.001 for Heg1; t test, p = 0.017 for Heatr5a; t test, p < 0.001 for Inhbb). In contrast, the number of Pcdh8 neurons is increased in Egr3−/− DRG (t test, p = 0.003). D–G, Mean (± SEM) cell body diameter of PV (D), Wnt7a (E), Pln (F), and Nxph1 (G) neurons in WT and Egr3−/− DRG. Similar as for PV, mean cell body diameters of Wnt7a, Pln, and Nxph1 neurons are reduced in Egr3−/− mice, a consequence of a loss in the proportion of larger (25–35 μm) neurons and an increase in the proportion of smaller sized (15–25 μm) neurons (PV: 27.5 ± 0.4 μm for wt, 22.9 ± 0.4 μm for Egr3−/−; Wnt7a: 26.4 ± 0.5 μm for wt, 22.3 ± 0.6 μm for Egr3−/−; Pln: 26.0 ± 0.6 μm for wt, 22.2 ± 0.6 μm for Egr3−/−; Nxph1: 24.5 ± 0.4 μm for wt, 22.9 ± 0.4 μm for Egr3−/−; for all Mann–Whitney rank sum test, p < 0.001). H, RNAscope analysis of Heg1 and Inhbb in PV+ neurons in p21 WT and Egr3−/− lumbar DRG. Heg1 is completely absent, and Inhbb is nearly absent from PV+ neurons. White arrowhead indicates a PV neuron in which low levels of Inhbb expression remain. Yellow arrowheads indicate Inhbb expression in non-PV neurons. I, RNAscope analysis of Heg1 in PV+Rx3+ neurons in p0 WT and Egr3−/− lumbar DRG. J, Schematic depicting a possible mechanism for proprioceptor muscle afferent specification. pSNs are born without subtype identity and may innervate their target muscles in a largely naive state. Once contact is made with nascent MS or GTO receptor targets, a pSN subtype specific identity is imposed or reinforced through MS or GTO-derived signals. Scale bars: A, H, I, 20 μm; B, 50 μm. *p < 0.05, **p < 0.001.
Discussion
We performed transcriptome analysis on proprioceptive muscle afferents and two other classes of mechanoreceptive sensory neurons and identified 24 molecules that are largely specific for muscle proprioceptors. A number of these markers have also been identified as pSN-enriched transcripts in other studies but had not yet been validated as such (Lee et al., 2012; Chiu et al., 2014; Usoskin et al., 2015; Li et al., 2016). We demonstrate here that these transcripts, as well as those not previously associated with pSN identity, constitute new markers for proprioceptive muscle afferents. We also show that 13 of these molecules mark subsets of pSNs, suggesting they could define pSN MS/GTO subtypes. Indeed, at least two of these candidate subtype markers, Heg1 and Inhbb, appear almost exclusively expressed in MS-innervating afferents. The observations that several of these newly identified pSN subtype markers are first expressed after proprioceptors engage with their nascent receptor organs, and that their expression depends on interactions with these organs, support the possibility that aspects of MS and GTO subtype identity are instructed through target-derived signals.
Extrinsic control of pSN subtype differentiation
Proprioceptor subtype differentiation has been thought to follow a developmental trajectory similar to spinal motor neurons, with intrinsic genetic determinants that dictate peripheral target selection at an early stage (Dasen et al., 2005; Stifani, 2014). Yet, embryonic markers that segregate with either MS or GTO afferent phenotypes have yet to be uncovered, raising the possibility that Group Ia/II/Ib subtype identity could be more akin the differentiation of motor neurons into force-generating (α, β) and fusimotor (γ) subtypes (Friese et al., 2009; Shneider et al., 2009). Several of our findings lend support to this idea. First, some of our pSN subtype markers exhibit a dynamic pattern of expression, suggesting that pSN identity may evolve over an extended postnatal period. Second, many of the markers we identified (12 of 24) are first expressed after pSN end organ innervation. Third, expression of some pSN-enriched transcripts is altered in Egr3−/− mice, which lack MS sensory receptor organs. Thus, similar as has been observed for γ motor neurons, MS/GTO afferent subtype acquisition may involve a protracted developmental process.
The observation that proprioceptors acquire their MS/GTO subclass identity over an extensive developmental period favors a larger influence from peripheral signals over intrinsic molecular determinants. Indeed, expression of at least three of our identified pSN subtype markers, Heg1, Inhbb, and Heatr5a, is absent or reduced in mice that lack MSs, suggesting that the developing sensory organs may provide an inductive or maintenance signal for these molecules (Fig. 7J). In contrast, the number of Pcdh8-expressing neurons appears increased, possibly indicating that MS-derived signals repress alternative pSN (possibly GTO afferent) identities. Due to technical limitations, we performed our screen in neonates, ∼6–10 d after pSNs first establish contact with their receptor targets. Therefore, it remains possible that our set of candidate pSN subtype markers represent aspects of pSN subtype differentiation that are a consequence of earlier, transiently acting, transcriptional determinants. Nevertheless, while the notion that environmental cues contribute to sensory neuron differentiation is not new (Patel et al., 2000), to our knowledge these studies are the first to indicate that the development of functionally distinct proprioceptor subtypes may rely on receptor-organ-derived signals. Recent studies indicate that the influence of receptor targets on sensory subtype specification may emerge as a more general developmental strategy in sensory systems (Shrestha et al., 2018; Sun et al., 2018).
A comparative transcriptome analysis of low threshold mechanoreceptors in DRG
In addition to proprioceptors, our transcriptome analysis provides molecular insight into RA-LTMR and SA-LTMR touch, vibration, and pressure receptor neurons (Johnson, 2001). Although molecular markers for individual RA-LTMR subtypes have been described in recent years (Abraira and Ginty, 2013), our studies could help to elucidate the molecules that underlie the functional features of different RA-LTMR subclasses beyond the markers presently known. Perhaps of even more impact is the identification of transcripts enriched in SA-LTMRs. Apart from their association with Merkel cells, selective markers for SA-LTMR afferents in DRG remain lacking. Thus, while these findings require further corroboration, some of the molecules we identified may selectively mark this sensory population.
Our transcriptome analysis between pSNs and cutaneous LTMRs effectively identified new pSN markers, yet the comparison of pSNs with RA/SA-LTMRs also complicated our attempt to uncover molecules that distinguish between MS- and GTO-innervating pSNs. A given pSN afferent subtype may exhibit commonalities with RA/SA-LTMR subtypes but not with other pSN subtypes. For instance, Group Ia MS afferents are unique among pSNs in their exquisite sensitivity to vibration, a property that underlies their dynamic sensitivity and that they share with RA-LTMR afferents supplying Pacinian corpuscles (Sato, 1961; Brown et al., 1967). In addition, Group Ib GTO afferent sensory endings are similar in morphology to SA-LTMR Ruffini endings (Matthews, 1972; Jami, 1992). Consequently, transcripts that underlie the vibration sensitivity of Group Ia pSNs or the morphological specializations of Group Ib pSNs may have remained undetected in our differential transcriptome analysis. Nevertheless, despite this potential caveat, we were able to uncover several transcripts that mark distinct proprioceptor subsets, including two that appear preferentially expressed in MS-innervating proprioceptors (see below).
Identification of molecular markers of MS and GTO afferent proprioceptors
In addition to pan-proprioceptor markers (e.g., Gprc5b, Ndst4, Wls, Clec2l), our analysis uncovered 13 transcripts that appear selective for proprioceptor subsets. The challenge to assign these molecules to specific functional pSN subclasses is not trivial, however. Apart from Group Ia/II/Ib pSN subclasses, pSNs can be distinguished according to their muscle target, which vastly expands the number of possible pSN subtypes. Previous studies have uncovered numerous molecules that exhibit a biased expression pattern in neonate proprioceptors with regionally distinct muscle innervation patterns (e.g., limb or axial, proximal-distal), or that supply functionally different muscles (e.g., flexor, extensor, abductor, adductor) (Poliak et al., 2016). We found that only four of these muscle-type markers overlapped with our candidate pSN subtype markers, thus increasing the likelihood that our subtype-selective molecules represent aspects of MS or GTO subtype identity. Intriguingly, the combinatorial expression analysis with some of our candidate pSN subtype markers reveals that expression of Heg1, Pcdh8, Nxph1, and Wnt7a divides the PV/pSN population into three molecularly distinct subtypes. While we have yet to definitively assign these molecular subtypes to known functional pSN phenotypes, it is tantalizing to speculate they represent MS Group Ia, MS Group II, and GTO Group Ib proprioceptors.
In assigning our pSN subset-selective transcripts to distinct pSN subtypes, use of Egr3 mutant mice proved particularly informative. In WT animals, pSN cell bodies are on a continuum size scale, but Group Ia and Group Ib afferents tend to occupy the “larger” side of the size spectrum, whereas Group II neurons are generally of smaller caliber (Matthews, 1972). This size distribution is altered in Egr3−/− mice, presumably because of their impairment in spindle development and the associated reduction in spindle-derived trophic support for MS afferents (Oliveira Fernandes and Tourtellotte, 2015). Several of our pSN candidate subtype markers (e.g., Wnt7a, Pln, Nxph1) are expressed in neurons that are reduced in size in Egr3−/− DRG, suggesting that at least some of the neurons that express these markers normally innervate MSs. Remarkably, our analysis showed a near complete loss of Heg1 expression, as well as a significant reduction in Inhbb and Heatr5a in Egr3−/− DRG. This suggests that these molecules normally are expressed in all or a subset of MS afferents, and that expression of these transcripts may depend on the presence of the spindle. Consistent with this idea, MSs serve as a signaling source for proprioceptors and γMNs, and express NT3 and GDNF (Shneider et al., 2009; Oliveira Fernandes and Tourtellotte, 2015). Although expression of NT3 is reduced and GDNF is abolished in Egr3 mutant spindles, we do not consider it likely that either of these signaling factors may be responsible for the biased loss/reduced expression of Heg1, Inhbb, or Heatr5a. First, NT3 is expressed in both MS and GTO sensory receptors; and second, pSNs are not known to express a receptor for GDNF. An alternative possibility is that the reduced expression of Heg1, Inhbb, or Heatr5a results from the loss of Egr3 in Schwann cells (Gao et al., 2007). However, unless we invoke distinct classes, it is difficult to envision how Schwann cells would direct the selective expression of Heg, Inhbb, or Heatr5a to a subset of pSNs. Therefore, at present, we favor the idea that an as of yet unknown spindle-derived signaling molecule may help impart MS afferent identity on naive proprioceptors.
What is the relevance of these pSN subtype-selective molecules in MS afferent differentiation? Heg1 encodes a transmembrane receptor important in cardiovascular development but has no known role in neuronal differentiation or function (Kleaveland et al., 2009). In the heart, Heg1 is expressed in endothelial cells and supports vascular development by stabilizing endothelial cell–cell junctions (Kleaveland et al., 2009; de Kreuk et al., 2016). Recent studies indicate that the level of Heg1 expression is controlled by increased blood flow, implicating Heg1 as a mechanosensitive downstream effector in regulating cardiac valve morphology (Donat et al., 2018). Thus, Heg1 similarly may be activated by mechanoreceptive signaling in pSNs, possibly to regulate interactions with intrafusal myofibers. Heg1 mutant mice do not exhibit any overt motor coordination phenotype, however, indicating that Heg1 protein is not essential for MS afferent development or function (Kleaveland et al., 2009; M.L. Kahn, personal communication). Like Heg1, neither Inhbb (a member of the transforming growth factor-β family) nor Heatr5a (a protein of unknown function) has previously been implicated in neuronal differentiation. (Vassalli et al., 1994; Diggle et al., 2014; Bursać et al., 2018). Thus, regardless of their assignment to specific pSN subsets, since many of our pSN-enriched subtype markers have not previously been associated with a pSN identity, they should offer new insight into proprioceptor development, mechanotransduction, or sensory processing.
Footnotes
J.C.d.N. was supported by National Institutes of Health Grant NS090067 and the Thompson Family Foundation Initiative. T.M.J. was supported by National Institutes of Health Grant NS033245 and Project A.L.S. We thank Staceyann Doobar and Ummea Urmi for technical help; Barbara Han, Susan Brenner-Morton, Monica Mendelsohn, and Nataliya Zabello for help with generating antibodies and mouse strains; Jeff Milbrandt (Washington University) for the Egr3 mutant strain; Theanne Griffith and Ellen Lumpkin for sharing their RNAscope protocol; and Amy Norovich, George Mentis, and Eiman Azim for discussions and comments on the manuscript.
The authors declare no competing financial interests.
References
- Abdo H, Li L, Lallemend F, Bachy I, Xu XJ, Rice FL, Ernfors P (2011) Dependence on the transcription factor Shox2 for specification of sensory neurons conveying discriminative touch. Eur J Neurosci 34:1529–1541. 10.1111/j.1460-9568.2011.07883.x [DOI] [PubMed] [Google Scholar]
- Abraira VE, Ginty DD (2013) The sensory neurons of touch. Neuron 79:618–639. 10.1016/j.neuron.2013.07.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akay T, Tourtellotte WG, Arber S, Jessell TM (2014) Degradation of mouse locomotor pattern in the absence of proprioceptor sensory feedback. Proc Natl Acad Sci U S A 111:16877–16882. 10.1073/pnas.1419045111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber S, Ladle DR, Lin JH, Frank E, Jessell TM (2000) ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell 101:485–498. 10.1016/S0092-8674(00)80859-4 [DOI] [PubMed] [Google Scholar]
- Ashrafi S, Lalancette-Hébert M, Friese A, Sigrist M, Arber S, Shneider NA, Kaltschmidt JA (2012) Wnt7A identifies embryonic γ-motor neurons and reveals early postnatal dependence of γ-motor neurons on a muscle spindle-derived signal. J Neurosci 32:8725–8731. 10.1523/JNEUROSCI.1160-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks RW. (2006) An allometric analysis of the number of muscle spindles in mammalian skeletal muscles. J Anat 208:753–768. 10.1111/j.1469-7580.2006.00558.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300. [Google Scholar]
- Bourane S, Garces A, Venteo S, Pattyn A, Hubert T, Fichard A, Puech S, Boukhaddaoui H, Baudet C, Takahashi S, Valmier J, Carroll P (2009) Low-threshold mechanoreceptor subtypes selectively express MafA and are specified by ret signaling. Neuron 64:857–870. 10.1016/j.neuron.2009.12.004 [DOI] [PubMed] [Google Scholar]
- Brown MC, Engberg I, Matthews PB (1967) The relative sensitivity to vibration of muscle receptors of the cat. J Physiol 192:773–800. 10.1113/jphysiol.1967.sp008330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursać S, Jurada D, Volarević S (2018) New insights into HEATR1 functions. Cell Cycle 17:143–144. 10.1080/15384101.2017.1411325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu IM, Barrett LB, Williams EK, Strochlic DE, Lee S, Weyer AD, Lou S, Bryman GS, Roberson DP, Ghasemlou N, Piccoli C, Ahat E, Wang V, Cobos EJ, Stucky CL, Ma Q, Liberles SD, Woolf CJ (2014) Transcriptional profiling at whole population and single cell levels reveals somatosensory neuron molecular diversity. Elife 3:e04660. 10.7554/eLife.04660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasen JS, Tice BC, Brenner-Morton S, Jessell TM (2005) A hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell 123:477–491. 10.1016/j.cell.2005.09.009 [DOI] [PubMed] [Google Scholar]
- de Kreuk BJ, Gingras AR, Knight JD, Liu JJ, Gingras AC, Ginsberg MH (2016) Heart of glass anchors Rasip1 at endothelial cell–cell junctions to support vascular integrity. Elife 5:e11394. 10.7554/eLife.11394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demireva EY, Shapiro LS, Jessell TM, Zampieri N (2011) Motor neuron position and topographic order imposed by β- and γ-catenin activities. Cell 147:641–652. 10.1016/j.cell.2011.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nooij JC, Doobar S, Jessell TM (2013) Etv1 inactivation reveals proprioceptor subclasses that reflect the level of NT3 expression in muscle targets. Neuron 77:1055–1068. 10.1016/j.neuron.2013.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nooij JC, Simon CM, Simon A, Doobar S, Steel KP, Banks RW, Mentis GZ, Bewick GS, Jessell TM (2015) The PDZ-domain protein whirlin facilitates mechanosensory signaling in mammalian proprioceptors. J Neurosci 35:3073–3084. 10.1523/JNEUROSCI.3699-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle CP, Moore DJ, Mali G, zur Lage P, Ait-Lounis A, Schmidts M, Shoemark A, Garcia Munoz A, Halachev MR, Gautier P, Yeyati PL, Bonthron DT, Carr IM, Hayward B, Markham AF, Hope JE, von Kriegsheim A, Mitchison HM, Jackson IJ, Durand B, et al. (2014) HEATR2 plays a conserved role in assembly of the ciliary motile apparatus. PLoS Genet 10:e1004577. 10.1371/journal.pgen.1004577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donat S, Lourenço M, Paolini A, Otten C, Renz M, Abdelilah-Seyfried S (2018) Heg1 and Ccm1/2 proteins control endocardial mechanosensitivity during zebrafish valvulogenesis. Elife 7:e28939. 10.7554/eLife.28939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew T, Marigold DS (2015) Taking the next step: cortical contributions to the control of locomotion. Curr Opin Neurobiol 33:25–33. 10.1016/j.conb.2015.01.011 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95:14863–14868. 10.1073/pnas.95.25.14863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friese A, Kaltschmidt JA, Ladle DR, Sigrist M, Jessell TM, Arber S (2009) Gamma and alpha motor neurons distinguished by expression of transcription factor Err3. Proc Natl Acad Sci U S A 106:13588–13593. 10.1073/pnas.0906809106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Daugherty RL, Tourtellotte WG (2007) Regulation of low affinity neurotrophin receptor (p75(NTR)) by early growth response (Egr) transcriptional regulators. Mol Cell Neurosci 36:501–514. 10.1016/j.mcn.2007.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N (2003) A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425:917–925. 10.1038/nature02033 [DOI] [PubMed] [Google Scholar]
- Heidenreich M, Lechner SG, Vardanyan V, Wetzel C, Cremers CW, De Leenheer EM, Aránguez G, Moreno-Pelayo MÁ, Jentsch TJ, Lewin GR (2011) KCNQ4 K(+) channels tune mechanoreceptors for normal touch sensation in mouse and man. Nat Neurosci 15:138–145. 10.1038/nn.2985 [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Shneider NA, Birchmeier C, Burden SJ, Jessell TM, Arber S (2002) A role for neuregulin1 signaling in muscle spindle differentiation. Neuron 36:1035–1049. 10.1016/S0896-6273(02)01101-7 [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S (2005) A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol 3:e159. 10.1371/journal.pbio.0030159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman BU, Baba Y, Griffith TN, Mosharov EV, Woo SH, Roybal DD, Karsenty G, Patapoutian A, Sulzer D, Lumpkin EA (2018) Merkel cells activate sensory neural pathways through adrenergic synapses. Neuron 100:1401–1413.e6. 10.1016/j.neuron.2018.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houk J, Henneman E (1967) Responses of Golgi tendon organs to active contractions of the soleus muscle of the cat. J Neurophysiol 30:466–481. 10.1152/jn.1967.30.3.466 [DOI] [PubMed] [Google Scholar]
- Hunt CC. (1990) Mammalian muscle spindles: peripheral mechanisms. Physiol Rev 70:643–663. 10.1152/physrev.1990.70.3.643 [DOI] [PubMed] [Google Scholar]
- Jami L. (1992) Golgi tendon organs in mammalian skeletal muscle: functional properties and central actions. Physiol Rev 72:623–666. 10.1152/physrev.1992.72.3.623 [DOI] [PubMed] [Google Scholar]
- Johnson KO. (2001) The roles and functions of cutaneous mechanoreceptors. Curr Opin Neurobiol 11:455–461. 10.1016/S0959-4388(00)00234-8 [DOI] [PubMed] [Google Scholar]
- Kleaveland B, Zheng X, Liu JJ, Blum Y, Tung JJ, Zou Z, Sweeney SM, Chen M, Guo L, Lu MM, Zhou D, Kitajewski J, Affolter M, Ginsberg MH, Kahn ML (2009) Regulation of cardiovascular development and integrity by the heart of glass-cerebral cavernous malformation protein pathway. Nat Med 15:169–176. 10.1038/nm.1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer I, Sigrist M, de Nooij JC, Taniuchi I, Jessell TM, Arber S (2006) A role for Runx transcription factor signaling in dorsal root ganglion sensory neuron diversification. Neuron 49:379–393. 10.1016/j.neuron.2006.01.008 [DOI] [PubMed] [Google Scholar]
- Lallemend F, Ernfors P (2012) Molecular interactions underlying the specification of sensory neurons. Trends Neurosci 35:373–381. 10.1016/j.tins.2012.03.006 [DOI] [PubMed] [Google Scholar]
- Lecoin L, Rocques N, El-Yakoubi W, Ben Achour S, Larcher M, Pouponnot C, Eychène A (2010) MafA transcription factor identifies the early ret-expressing sensory neurons. Dev Neurobiol 70:485–497. 10.1002/dneu.20790 [DOI] [PubMed] [Google Scholar]
- Lee J, Friese A, Mielich M, Sigrist M, Arber S (2012) Scaling proprioceptor gene transcription by retrograde NT3 signaling. PLoS One 7:e45551. 10.1371/journal.pone.0045551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, et al. (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168–176. 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Levanon D, Bettoun D, Harris-Cerruti C, Woolf E, Negreanu V, Eilam R, Bernstein Y, Goldenberg D, Xiao C, Fliegauf M, Kremer E, Otto F, Brenner O, Lev-Tov A, Groner Y (2002) The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J 21:3454–3463. 10.1093/emboj/cdf370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CL, Li KC, Wu D, Chen Y, Luo H, Zhao JR, Wang SS, Sun MM, Lu YJ, Zhong YQ, Hu XY, Hou R, Zhou BB, Bao L, Xiao HS, Zhang X (2016) Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res 26:967. 10.1038/cr.2016.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Rutlin M, Abraira VE, Cassidy C, Kus L, Gong S, Jankowski MP, Luo W, Heintz N, Koerber HR, Woodbury CJ, Ginty DD (2011) The functional organization of cutaneous low-threshold mechanosensory neurons. Cell 147:1615–1627. 10.1016/j.cell.2011.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Enomoto H, Rice FL, Milbrandt J, Ginty DD (2009) Molecular identification of rapidly adapting mechanoreceptors and their developmental dependence on ret signaling. Neuron 64:841–856. 10.1016/j.neuron.2009.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Fode C, Guillemot F, Anderson DJ (1999) Neurogenin1 and neurogenin2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev 13:1717–1728. 10.1101/gad.13.13.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macefield VG, Knellwolf TP (2018) Functional properties of human muscle spindles. J Neurophysiol 120:452–467. 10.1152/jn.00071.2018 [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H (2010) A robust and high-throughput cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malin SA, Davis BM, Molliver DC (2007) Production of dissociated sensory neuron cultures and considerations for their use in studying neuronal function and plasticity. Nat Protoc 2:152–160. 10.1038/nprot.2006.461 [DOI] [PubMed] [Google Scholar]
- Matthews PB. (1972) Mammalian muscle receptors and their central actions. London: Edward Arnold. [Google Scholar]
- Oliveira Fernandes M, Tourtellotte WG (2015) Egr3-dependent muscle spindle stretch receptor intrafusal muscle fiber differentiation and fusimotor innervation homeostasis. J Neurosci 35:5566–5578. 10.1523/JNEUROSCI.0241-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel TD, Jackman A, Rice FL, Kucera J, Snider WD (2000) Development of sensory neurons in the absence of NGF/TrkA signaling in vivo. Neuron 25:345–357. 10.1016/S0896-6273(00)80899-5 [DOI] [PubMed] [Google Scholar]
- Pecho-Vrieseling E, Sigrist M, Yoshida Y, Jessell TM, Arber S (2009) Specificity of sensory-motor connections encoded by Sema3e-Plxnd1 recognition. Nature 459:842–846. 10.1038/nature08000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliak S, Norovich AL, Yamagata M, Sanes JR, Jessell TM (2016) Muscle-type identity of proprioceptors specified by spatially restricted signals from limb mesenchyme. Cell 164:512–525. 10.1016/j.cell.2015.12.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proske U, Gandevia SC (2012) The proprioceptive senses: their roles in signaling body shape, body position and movement, and muscle force. Physiol Rev 92:1651–1697. 10.1152/physrev.00048.2011 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M. (1961) Response of Pacinian corpuscles to sinusoidal vibration. J Physiol 159:391–409. 10.1113/jphysiol.1961.sp006817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shneider NA, Brown MN, Smith CA, Pickel J, Alvarez FJ (2009) Gamma motor neurons express distinct genetic markers at birth and require muscle spindle-derived GDNF for postnatal survival. Neural Dev 4:42. 10.1186/1749-8104-4-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha BR, Chia C, Wu L, Kujawa SG, Liberman MC, Goodrich LV (2018) Sensory neuron diversity in the inner ear is shaped by activity. Cell 174:1229–1246.e17. 10.1016/j.cell.2018.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stifani N. (2014) Motor neurons and the generation of spinal motor neuron diversity. Front Cell Neurosci 8:293. 10.3389/fncel.2014.00293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Babola T, Pregernig G, So KS, Nguyen M, Su SM, Palermo AT, Bergles DE, Burns JC, Müller U (2018) Hair cell mechanotransduction regulates spontaneous activity and spiral ganglion subtype specification in the auditory system. Cell 174:1247–1263.e15. 10.1016/j.cell.2018.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi I, Sunshine MJ, Festenstein R, Littman DR (2002) Evidence for distinct CD4 silencer functions at different stages of thymocyte differentiation. Mol Cell 10:1083–1096. 10.1016/S1097-2765(02)00735-9 [DOI] [PubMed] [Google Scholar]
- Tourtellotte WG, Milbrandt J (1998) Sensory ataxia and muscle spindle agenesis in mice lacking the transcription factor Egr3. Nat Genet 20:87–91. 10.1038/1757 [DOI] [PubMed] [Google Scholar]
- Tourtellotte WG, Keller-Peck C, Milbrandt J, Kucera J (2001) The transcription factor Egr3 modulates sensory axon-myotube interactions during muscle spindle morphogenesis. Dev Biol 232:388–399. 10.1006/dbio.2001.0202 [DOI] [PubMed] [Google Scholar]
- Tuthill JC, Azim E (2018) Proprioception. Curr Biol 28:R194–R203. 10.1016/j.cub.2018.01.064 [DOI] [PubMed] [Google Scholar]
- Usoskin D, Furlan A, Islam S, Abdo H, Lönnerberg P, Lou D, Hjerling-Leffler J, Haeggström J, Kharchenko O, Kharchenko PV, Linnarsson S, Ernfors P (2015) Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 18:145–153. 10.1038/nn.3881 [DOI] [PubMed] [Google Scholar]
- Vassalli A, Matzuk MM, Gardner HA, Lee KF, Jaenisch R (1994) Activin/inhibin beta B subunit gene disruption leads to defects in eyelid development and female reproduction. Genes Dev 8:414–427. 10.1101/gad.8.4.414 [DOI] [PubMed] [Google Scholar]
- Windhorst U. (2007) Muscle proprioceptive feedback and spinal networks. Brain Res Bull 73:155–202. 10.1016/j.brainresbull.2007.03.010 [DOI] [PubMed] [Google Scholar]
- Wu J, Anczuków O, Krainer AR, Zhang MQ, Zhang C (2013) Olego: fast and sensitive mapping of spliced mRNA-seq reads using small seeds. Nucleic Acids Res 41:5149–5163. 10.1093/nar/gkt216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Weyn-Vanhentenryck SM, Wu J, Sloan SA, Zhang Y, Chen K, Wu JQ, Barres BA, Zhang C (2015) Systematic discovery of regulated and conserved alternative exons in the mammalian brain reveals NMD modulating chromatin regulators. Proc Natl Acad Sci U S A 112:3445–3350. 10.1073/pnas.1502849112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Arber S, William C, Li L, Tanabe Y, Jessell TM, Birchmeier C, Burden SJ (2001) Patterning of muscle acetylcholine receptor gene expression in the absence of motor innervation. Neuron 30:399–410. 10.1016/S0896-6273(01)00287-2 [DOI] [PubMed] [Google Scholar]
- Yoshihara Y, Mizuno T, Nakahira M, Kawasaki M, Watanabe Y, Kagamiyama H, Jishage K, Ueda O, Suzuki H, Tabuchi K, Sawamoto K, Okano H, Noda T, Mori K (1999) A genetic approach to visualization of multisynaptic neural pathways using plant lectin transgene. Neuron 22:33–41. 10.1016/S0896-6273(00)80676-5 [DOI] [PubMed] [Google Scholar]