Abstract
Disease relapse in B-cell acute lymphoblastic leukemia (ALL), either due to development of acquired resistance after therapy or because of de novo resistance, remains a therapeutic challenge. In the present study, we have developed a cytarabine (Ara-C)-resistant REH cell line (REH/Ara-C) as a chemoresistance model. REH/Ara-C 1) was not crossresistant to vincristine or methotrexate; 2) showed a similar proliferation rate and cell surface marker expression as parental REH; 3) demonstrated decreased chemotaxis toward bone marrow stromal cells; and 4) expressed higher transcript levels of cytidine deaminase (CDA) and mitoNEET (CISD1) than the parental REH cell line. Based on these findings, we tested NL-1, a mitoNEET inhibitor, which induced a concentration-dependent decrease in cell viability with a comparable IC50 value in REH and REH/Ara-C. Furthermore, NL-1 decreased cell viability in six different ALL cell lines and showed inhibitory activity in a hemosphere assay. NL-1 also impaired the migratory ability of leukemic cells, irrespective of the chemoattractant used, in a chemotaxis assay. More importantly, NL-1 showed specific activity in inducing death in a drug-resistant population of leukemic cells within a coculture model that mimicked the acquired resistance and de novo resistance observed in the bone marrow of relapsed patients. Subsequent studies indicated that NL-1 mediates autophagy, and inhibition of autophagy partially decreased NL-1–induced tumor cell death. Finally, NL-1 showed antileukemic activity in an in vivo mouse ALL model. Taken together, our study demonstrates that mitoNEET has potential as a novel antileukemic drug target in treatment refractory or relapsed ALL.
Introduction
In B-cell acute lymphoblastic leukemia (ALL) patients, there is a positive correlation between drug resistance and poor prognostic clinical outcome, independent of age, gender, and initial leukemic burden (Pieters et al., 1991; Kaspers et al., 1997). Emergence of drug resistance after chemotherapy is a common phenomenon in newly relapsed ALL patients, and unfortunately the drug-resistance profile worsens with subsequent relapse (Klumper et al., 1995; Pogorzala et al., 2015). Additionally, throughout disease progression, leukemic cells are constantly modifying their bone marrow (BM) microenvironment to form a leukemic niche, which affords de novo chemoresistance (Colmone et al., 2008). This de novo resistance leads to survival of leukemic cells after therapy, and the residual surviving cell number is a measure of minimal residual disease. Importantly, minimal residual disease is correlated with a poor prognosis in patients (Stow et al., 2010).
To study drug resistance in leukemic cells, we previously developed an in vitro coculture model consisting of human-derived ALL cells cultured in the presence of either primary human bone marrow stromal cells (BMSCs) or primary human osteoblasts (HOBs) (Moses et al., 2016b). Using this model, we developed a protocol to isolate and characterize a subset of phase dim (PD) leukemic cells buried under the BM-derived adherent layer (Slone et al., 2016b). PD cells share the characteristics typical of relapsed/refractory leukemic cells, in that they are quiescent and resistant to chemotherapy, have impaired microRNA biogenesis, and are relevant to the current study on manifested altered mitochondrial metabolism (Moses et al., 2016a,b; Slone et al., 2016a). This drug-resistant phenotype was context dependent and transient, since removing the PD cells from the coculture restored their sensitivity to chemotherapy treatment. In the present study, we have developed a cytarabine (Ara-C)-resistant REH cell line (REH/Ara-C) and cocultured it with BMSCs or HOBs to mimic both the acquired and de novo drug-resistant phenotype generally found in the BM of relapsed ALL patients.
Central to the rationale of this investigation is work by others in which mitochondrial dysfunction has been shown to be associated with chemoresistance and has been targeted as a strategy for anticancer therapy (Neuzil et al., 2013). Evasion of programmed cell death is one of the major mechanisms of drug resistance and cancer progression (Hanahan and Weinberg, 2011), and targeting mitochondrial function may be a therapeutic option in targeting this population of cells (Fu et al., 2017; Kuntz et al., 2017).
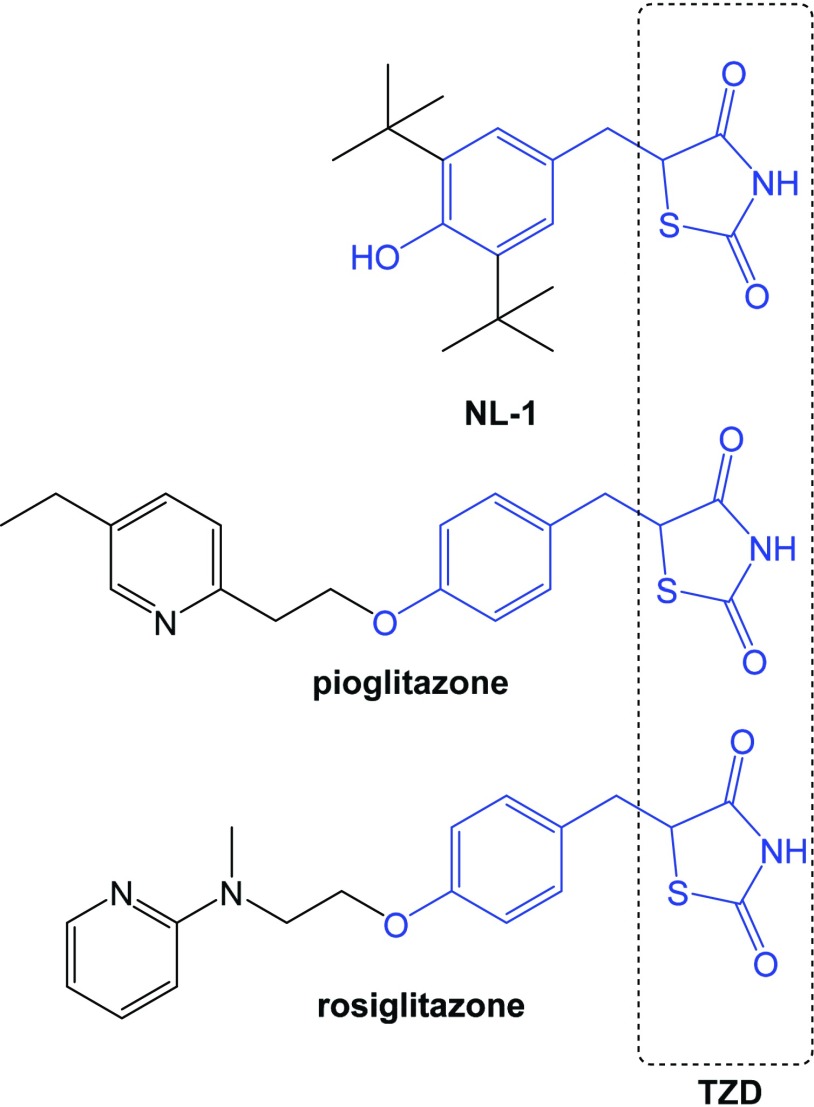
MitoNEET (CISD1) is a mitochondrial outer membrane protein that plays a critical role in maintaining mitochondrial homeostasis and is frequently overexpressed in cancer (Bai et al., 2015). This 2Fe-2S cluster-containing protein was first discovered as a secondary target for the antidiabetic drug pioglitazone (Colca et al., 2004). In the present study, we have used a small molecule ligand of mitoNEET called NL-1, which was derived from pioglitazone (Geldenhuys et al., 2010) and demonstrated its antileukemic activity in both acquired and de novo drug-resistant cells (Fig. 1). NL-1, pioglitazone, and rosiglitazone belong to the glitazone or thiazolidinedione class of compounds. Additionally, we show that NL-1–mediated death in leukemic cells requires the activation of the autophagic pathway. These results led us to propose the use of NL-1 for the treatment of relapsed ALL, leveraging the vulnerability of the mitochondria as a therapeutic strategy.
Fig. 1.
Structure of NL-1. The thiazolidinedione (TZD) warhead scaffold is shown in blue, alongside pioglitazone and rosiglitazone, which are classic TZDs.
Materials and Methods
Cell Cultures.
Nine human B-cell ALL cell lines were purchased from ATCC or DSMZ. SUP-B15 (CRL-1929), TOM1 (ACC 578), and JM1 (CRL-10423) cells were maintained in RPMI 1640 containing 10% FBS, 0.05 mM β-mercaptoethanol, and 1x streptomycin/penicillin. REH (CRL-8286), NALM-1 (CRL-1567), NALM-6 (ACC 128), BV-173 (ACC 20), RS4; 11 (CRL-1873), and SD-1 (ACC 366) were maintained in RPMI 1640 containing 10% FBS and 1x streptomycin/penicillin. Primary CD3+ T cells, CD19+ B cells, peripheral blood mononuclear cells, and bone marrow mononuclear cells were purchased from AllCells (Allcells.com) and maintained in Lymphocyte Growth Medium-3 (Lonza) containing 10% FBS and 1x streptomycin/penicillin. HOBs were purchased from PromoCell (Heidelberg, Germany) and cultured according to the vendor’s recommendations.
A chemotherapy-resistant REH ALL cell line (REH/Ara-C) was developed by exposing REH cells to increasing concentrations of Ara-C starting at 0.2 μM. Following a 1-week period, if cell viability of the Ara-C–treated culture was determined to be greater than 70%, the concentration of Ara-C was increased by 0.1 µM. The resulting REH/Ara-C cell line was maintained in growth media containing 1 μM Ara-C, and all experiments utilizing REH/Ara-C cells were carried out following 48 hours of exposure to 1 µM Ara-C.
Deidentified primary BMSCs were provided by the WVU Cancer Institute Biospecimen Processing Core and the WVU Pathology Laboratory Tissue Bank. The ALL cell lines used in the current study were authenticated by short tandem repeat analysis (University of Arizona Genetics Core, Tucson, AZ) and maintained in 6% CO2 in normoxia at 37°C.
Cell Proliferation Assays.
ALL cell lines were plated at 5 × 104 cells/well in a 96-well plate. Following treatment with chemotherapy (pharmacologic inhibitors or NL-1), viable cells were quantitated using a Cell Counting Kit-8 (Dojindo Molecular Technology Inc.) according to the manufacturer’s instructions.
Cell Surface Expression Analysis.
Cell surface proteins were stained with fluorochrome-conjugated antibodies to CD19 (PerCP), CD34 (PE), CD38 (APC), CD44 (FITC), and CD45 (PE-Cy5) in staining buffer (PBS + 10% FBS) for 1 hour at room temperature. Cells were washed with PBS, fixed with 1% paraformaldehyde, and resuspended in PBS for analysis of mean fluorescence intensity by flow cytometry (BD LSR Fortessa). Fluorochrome-conjugated isotype controls were used for measuring nonspecific binding.
Chemotaxis Assay.
BMSCs or HOBs were plated at 90% confluency in 24-well Transwell plates (Costar). After overnight incubation, untreated ALL cells or pretreated NL-1 (60 μM) cells (1 hour pretreatment) were plated at 1.5 × 105 cells in the top of the Transwell insert. The cells were allowed to migrate toward adherent layers for 4 hours. Migrated leukemic cells were collected, fixed by 2% paraformaldehyde, and counted using flow cytometry. SDF-1 (100 ng/ml) served as the positive control for the chemoattractant and media alone served as the negative control.
Real-Time Quantitative Reverse Transcriptase Polymerase Chain Reaction.
REH and REH/Ara-C cells were plated at 1 × 106 cells/ml and cultured overnight, and then the cells were pelleted and processed for RNA extraction using an RNeasy kit (Qiagen) according to the manufacturer’s instruction. RNA (50 ng) was then amplified by real-time polymerase chain reaction using the Power SYBR Green RNA-to-CT 1-Step kit (ThermoFisher Scientific) with primers specific for cytidine deaminase (CDA), deoxycytidine kinase, ribonuclease reductase M1, and ACTB (VHPS-110) purchased from RealTimePrimers.com, and primers for SCL19A1 (FWD 5′-GAA CCC GTT TGA GCT CGG TA-3′and REV 5′-GCT GCG ATC CAT TCA ACT CG-3′) and CISD1 (FWD 5′-CCT CCT GGT GTC AGC AAG CT-3′ and REV 5′-CCC GAG AGT CAC TGG TTC ACA-3′) purchased from Integrated DNA Technologies. The gene expression in REH and REH/Ara-C was normalized to ACTB and the fold difference in gene expression between the two cells lines was subsequently analyzed.
Hemosphere Assay.
SD1 leukemic cells were plated at 1 × 105 cells/well in a 96-well plate and incubated for 4 days to allow sphere formation. Following confirmation of spheroids in culture by light microscopy, the cells were treated with 60 μM NL-1 for 72 hours. At the end of the treatment, the resultant spheroids were counted via microscopy and the images were captured (DM IL LED microscope/camera; Leica). Cells were collected at the end of the study and counted with Trypan Blue (0.4% v/v) to determine the number of live cells.
Coculture Study.
Coculture conditions have been previously described (Slone et al., 2016b). Briefly, 1 × 106 REH or REH/Ara-C cells were seeded on either BMSC or HOB adherent layers and maintained in 5% O2. The coculture was fed every 4 days, and on the ninth day the coculture was treated with 60 μM NL-1 for 72 hours. Following exposure, ALL cells that were in suspension were collected [suspended (S) cells]. The ALL cells that were buried under the adherent layer were trypsinized and separated from BMSCs or HOBs by size exclusion using Sephadex G-10 columns (GE Healthcare). These buried ALL cells were designated as PD cells and have been previously described to share relevant characteristics with cells that contribute to treatment-resistant minimal residual disease (Moses et al., 2016a; Slone et al., 2016a).
Autophagy Detection Assay.
REH cells were plated at 1 × 106 cells/ml and then treated with 5 μM chloroquine for 1 hour. Following chloroquine treatment, the cells were exposed to 60 μM NL-1 for 6 hours. At the end of the treatment, the autophagic flux was measured using CYTO-ID Autophagy Detection Kit 2.0 (Enzo Life Sciences) according the manufacturer’s instructions. Briefly, treated cells were washed with 1x assay buffer and resuspended in CYTO-ID green stain solution for 30 minutes at 37°C. Following incubation, the cells were thoroughly washed and cell density was adjusted to 5 × 104 cells/ml. Then, 100 μl of this cell suspension was plated in triplicate in a 96-well plate and the fluorescence intensity was measured with Biotek Plate Reader Synergy HT using a FITC filter (excitation 480 nm, emission 530 nm).
Cell Death Assay.
Primary immune cells were plated at 1 × 106 cells/ml in 24-well plates. The cells were treated with 60 μM NL-1 for 24 hours and then stained using a LIVE/DEAD Fixable Dead Cell Stain Kit (ThermoFisher Scientific) following the manufacturer’s instructions. The number of dead cells was then analyzed using flow cytometry.
In Vivo Study.
All animal procedures were reviewed and approved by the WVU Institutional Animal Care and Use Committee. Ten 6–8 month old female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice were injected intravenously with 1 × 106 luciferase-expressing TOM1 ALL cells. Two days postinjection, the mice were injected with luciferin and the tumor burden was analyzed using an IVIS SpectrumCT (PerkinElmer). After confirmation of tumor engraftment, the mice were randomized into two groups of five mice each and treated with 1 mg/kg (i.p.) NL-1 or vehicle control for five consecutive days. Tumor burden was monitored weekly using IVIS imaging. The study was terminated when the control mice showed signs of morbidity due to high tumor burden.
Statistical Analysis.
In vitro experiments were performed in triplicate in at least three independent experiments, unless otherwise stated. For studies consisting of more than two treatment groups, statistical significance between groups was determined using one-way ANOVA followed by a post hoc Tukey’s test. For the chemotaxis assay, two-way ANOVA followed by a post hoc Sidak’s multiple comparisons test was used to determine statistical significance. For the in vivo study, an unpaired one-tailed t test was performed to determine the statistical significance between the NL-1–treated and control vehicle–treated mice. Data are shown as mean ± S.E.M., and P < 0.05 was considered statistically significant.
Results
Development of a Chemotherapy-Resistant ALL Cell Line.
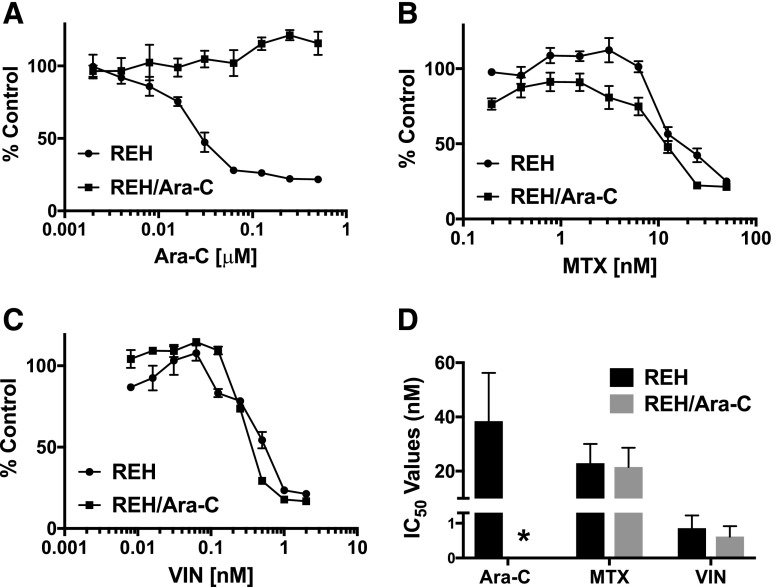
The emergence of chemotherapy-resistant tumors presents a major clinical challenge in the long-term treatment of ALL patients. To generate a model to address chemotherapy-resistant disease, the REH ALL cell line was treated with increasing doses of Ara-C over the course of several months to generate a cell line that is resistant to this drug (REH/Ara-C). A dose-response curve using Ara-C was performed to compare the REH/Ara-C cells to the parental REH cells. As expected, there was a dose-dependent decrease in the number of viable REH cells, with an IC50 value of 19.5 nM; however, Ara-C had no effect on the number of viable REH/Ara-C cells, even at doses 10 times higher than the IC50 value for the parental line (Fig. 2A). The chemotherapy resistance in the REH/Ara-C cell line was specific to that treatment, since those cells remained sensitive to other chemotherapy reagents including methotrexate (Fig. 2B) and vincristine (Fig. 2C), showing IC50 values comparable to the parental cell line, REH (Fig. 2D).
Fig. 2.
REH/Ara-C cell line is resistant to Ara-C. REH and REH/Ara-C were plated at 5 × 104 cells per well in a 96-well plate and treated with indicated doses of (A) Ara-C, (B) methotrexate (MTX), or (C) vincristine (VIN). The number of viable cells was analyzed following 72 hours of treatment. (D) The concentration-dependent curve was used to calculate the IC50 value using CompuSyn software. All experiments were performed in triplicate at least three different times and the data are represented as mean ± S.E.M. *Ara-C treatment of the resistant REH/Ara-C cells did not induce cell death.
REH/Ara-C Cell Line Shows Increased CISD1 Levels.
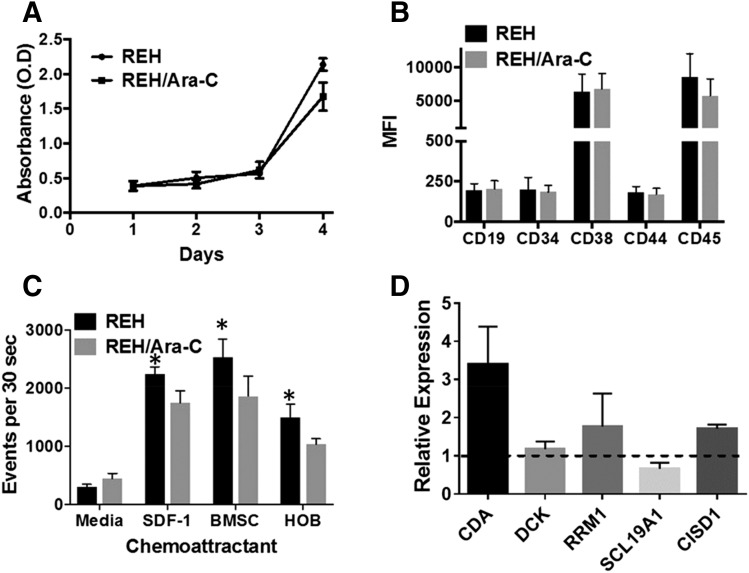
Further experiments were performed using the REH and REH/Ara-C cells to assess the functional or expression differences that may contribute to the differential sensitivity to chemotherapy. REH and REH/Ara-C had similar cell proliferation rates; the modest difference noted at day 4 was not statistically significant (Fig. 3A). Analysis of cell surface receptors demonstrated that CD19, CD34, CD38, CD44, and CD45 were present at the same levels in both REH and REH/Ara-C cell lines (Fig. 3B). Interestingly, the chemotactic ability of the REH/Ara-C cells to migrate toward BMSCs, HOBs, and SDF-1 was diminished when compared with the REH parental cell line. However, no difference was observed between the two cell lines when media were used as the chemoattractant (Fig. 3C). Gene expression analysis showed a 3-fold increase in the level of CDA transcripts (CDA) and a 1.7-fold increase in mitoNEET (CISD1) transcripts (Fig. 3D); however, deoxycytidine kinase (DCK), ribonuclease reductase M1 polypeptide (RRM1), and solute carrier family 19 (folate transporter) member 1 (SCL19A1) transcripts were comparable to the parental cell line.
Fig. 3.
Characterization of the REH/Ara-C cell line. (A) REH and REH/Ara-C were plated at 5 × 104 cells per well in a 96-well plate and cell viability was analyzed every 24 hours for 4 days. (B) REH and REH/Ara-C cells were stained with flurochrome-conjugated antibodies targeting the different cell surface receptors. The intensity of the staining was acquired and analyzed using flow cytometry. (C) REH or REH/Ara-C cells were plated on the insert and allowed to migrate toward the bottom well containing no chemoattractant (media), 100 ng/ml SDF-1, BMSCs, or HOBs. The number of migrated cells was counted using flow cytometry after 4 hours. (D) REH and REH/Ara-C cells were plated at 1 × 106 cells/ml for 24 hours and then processed for RNA and subjected to real-time reverse transcriptase polymerase chain reaction protocol as described in Materials and Methods. The ΔΔCT values were used to compare the fold change in expression of the indicated genes in REH/Ara-C cells compared with REH cells. All experiments were performed in triplicate at least three different times and the data are represented as mean ± S.E.M. *P < 0.05, statistical significance compared with REH cell numbers that migrated toward the BMSCs at the end of 4 hours.
NL-1 Shows Anticancer Activity in ALL Cell Lines.
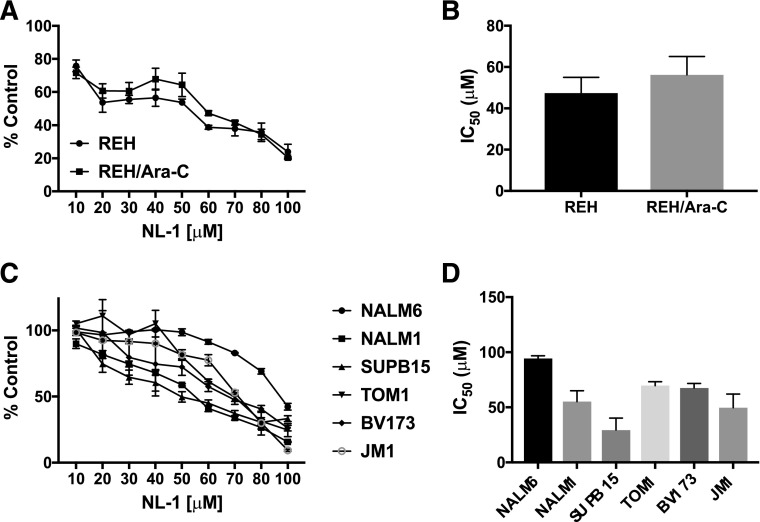
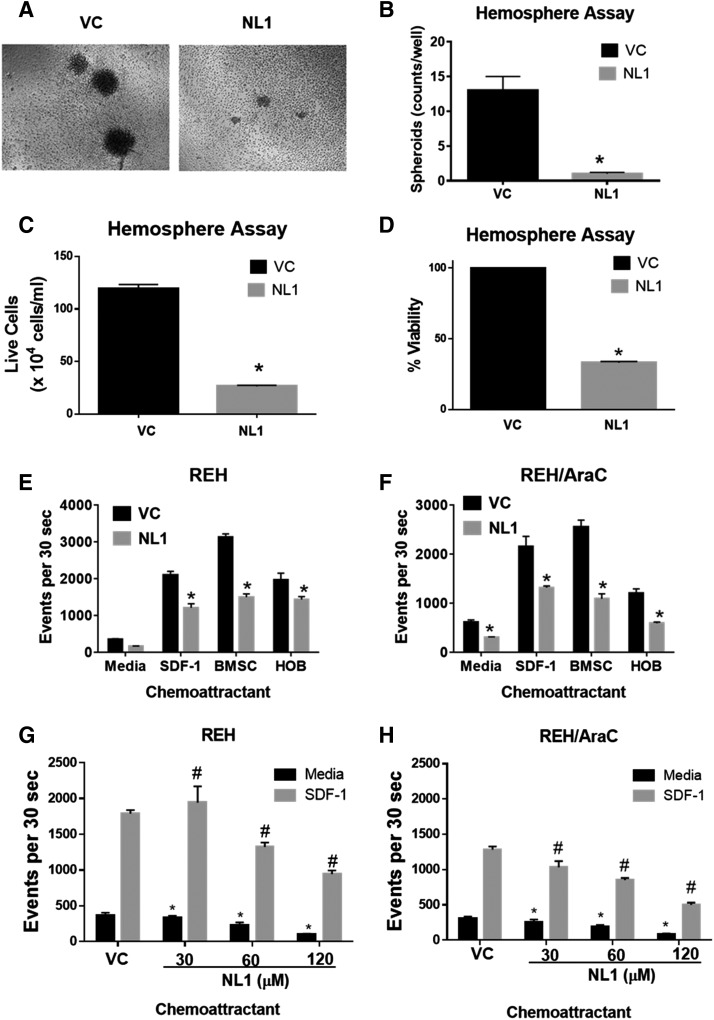
Based on the increased expression of mitoNEET in the drug-resistant cell line, we tested the activity of a mitoNEET ligand, NL-1. Treatment with NL-1 reduced the number of viable cells in both REH and REH/Ara-C cell lines in a concentration-dependent manner (Fig. 4A). The IC50 value of NL-1 was comparable in REH (47.35 ± 7.7 μM) and REH/Ara-C (56.26 ± 8.8 μM) cells (Fig. 4B). Furthermore, NL-1 successfully decreased the number of viable cells in a concentration-dependent manner in six additional ALL cell lines (Fig. 4C). SUPB15 was the most sensitive to NL-1 with an IC50 value of 29.48 ± 10.66 μM, and NALM6 demonstrated the most resistance to NL-1 with an IC50 value of 94.26 ± 2.60 μM (Fig. 4D). In contrast, TOM1, BV173, NALM1, and JM1 all had similar IC50 values of around 60 μM for NL-1 (Fig. 4D). Furthermore, we used SD1 ALL cells to form hemospheres for evaluation of NL-1 in a model of tumor stem cell–like behavior. In contrast to the REH cells, SD1 cells will form hemospheres suited for this experimental design to identify compounds, which will target this stem cell population. SD1 cells formed 10–15 spheroids/well. After the spheroids formed, they were treated with NL-1. The NL-1 treatment consistently reduced the size and number of preformed SD1 spheroids (Fig. 5, A and B). Additionally, we found that both the number of live cells (Fig. 5C) and the percentage of viability (Fig. 5D) of the cells was reduced when treated with NL-1.
Fig. 4.
NL-1 decreases cell viability in ALL cell lines. ALL cell lines, (A) REH, REH/Ara-C and (C) NALM6, NALM1, SUPB15, TOM1, BV173, and JM1, were plated at 5 × 104 cells/well in a 96-well cell culture plate and treated with indicated concentrations of NL-1. The number of live cells was measured after 72 hours of treatment (B and D). IC50 values were calculated from the concentration-dependent curves of each cell line using the CompuSyn software. The data are represented as mean ± S.E.M. of a study performed in triplicate and are representative of experiments that were carried out at least three independent times.
Fig. 5.
NL-1 activity in hemosphere and chemotaxis assays. (A) SD1 cells were plated at 1 × 105 cells/well and allowed to form spheroids by culturing them for 4 days. Spheroid formation was confirmed by observation using light microscopy and then treated with 60 μM of NL-1 or PBS [vehicle control (VC)] for 72 hours. At the end of treatment, the resulting hemosphere images were captured (A) and counted (B). (C) Number of live SD-1 cells and percent viability (D) after treatment of NL-1. REH (E and G) or REH/Ara-C (F and H) cells were pretreated for 1 hour with either PBS (VC) or 60 μM of NL-1. (E–H) After treatment the cells were plated on the insert and allowed to migrate toward the bottom well containing no chemoattractant (media), 100 ng/ml of SDF-1, BMSCs, or HOBs. The number of migrated cells was counted using flow cytometry after 4 hours. The data are represented as mean ± S.E.M. and are representative of experiments that were carried out at least three independent times. *P < 0.05, when compared with VC-treated group migrating toward media. #P < 0.05 when compared with the VC-treated group migrating toward SDF-1.
NL-1 Impairs Chemotaxis in ALL Cells.
Soluble factor gradients driving chemotaxis play an important role in leukemic niche development (Möhle et al., 2000; Colmone et al., 2008). Our initial studies showed that parental REH and drug-resistant REH/Ara-C cells both had comparable chemotaxis profiles, with REH/Ara-C showing only a modest decrease in its migratory capacity. NL-1 pretreatment inhibited the chemotactic ability of both REH (Fig. 5E) and REH/Ara-C (Fig. 5F) cells to migrate toward multiple chemoattractants. This inhibitory NL-1 activity was most pronounced in REH and REH/Ara-C cells migrating toward the BMSCs, although there were also significant differences in all of the conditions. The cells treated with NL-1 showed a dose-dependent decrease in chemotaxis in both the REH (Fig. 5G) and REH/Ara-C (Fig. 5H) cells.
NL-1 Induces Death in the Drug-Resistant Phase Dim ALL Cell Population.
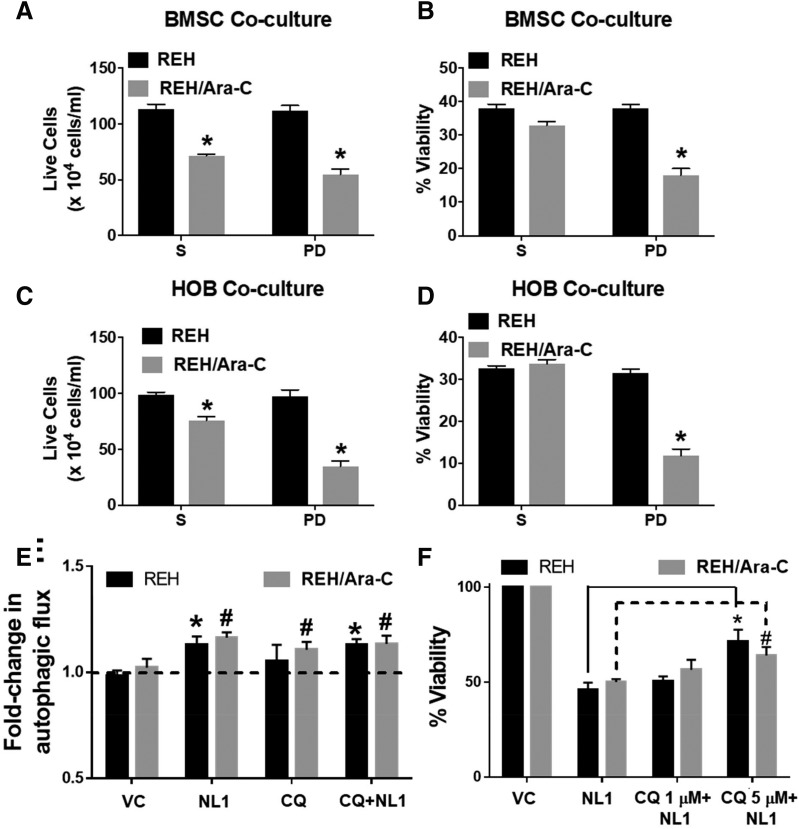
To model the bone marrow microenvironment, which can provide a site of sanctuary for leukemia, our laboratory has previously developed a coculture model where ALL cells are grown with either primary human BMSCs or HOBs. The leukemia cells form three populations in the coculture: those floating in suspension (the S cells), those loosely adhered to the top of the stromal cell layer, and the cells that bury underneath the stromal cell layer (phase dim cells; PD). The PD leukemic cells consistently show increased resistance to a variety of chemotherapies. To determine whether NL-1 could affect cells that are protected by both BM microenvironment cues and acquired drug resistance, NL-1 was tested in cocultures including either REH or REH/Ara-C cells. Treatment of BMSC and HOB cocultures with NL-1 decreased the number of live cells recovered from the S and PD cell populations (Fig. 6, A and C) and decreased viability of the PD population (Fig. 6, B and D), where the number of live cells were determined with Trypan Blue live staining. In these conditions, the S cells showed a small but significant decrease in the number of live cells in the REH/Ara-C suspension cells, but the viability was not decreased.
Fig. 6.
NL-1 induces cell death in coculture through an autophagy-dependent mechanism. REH and REH/Ara-C cells were grown in a coculture with either BMSCs or HOBs in a 24-well plate for 12 days. On day 9 in coculture, the cells were treated with 60 μM NL-1, chloroquine (CQ), or vehicle/DMSO. At the end of treatment, the total live cells in suspension (S) and the leukemic cells buried under the adherent layer (PD) were counted using the trypan blue dye exclusion method (A and C). The total cells and the live cell population were used to calculate the percentage of viability (B and D). *P < 0.05, when compared with REH cells. (E) REH and REH/Ara-C cells were plated in a 96-well plate and selected wells were pretreated for 1 hour with CQ. Following CQ treatment, the cells were treated with 60 μM NL-1. After 6 hours of treatment, the autophagic flux was measured. *P < 0.05, when compared with REH control, #P < 0.05, when compared with REH-Ara-C control group. (F) REH and REH/Ara-C cells were plated at 5 × 104 cells per well in a 96-well plate and treated with indicated concentrations of CQ. Following 1 hour of CQ treatment the cells were exposed to 60 μM NL-1. After 72 hours of NL-1 treatment the cell viability was analyzed as described in Materials and Methods. *P < 0.05, when compared with NL-1 REH–treated group. #P < 0.05, when compared with NL-1 REH/Ara-C–treated group. All data are presented as mean ± S.E.M. and are representative of a study performed in triplicate and conducted three independent times.
NL-1–Mediated Induction of Autophagy.
MitoNEET has been shown to play a role in mitophagy during autophagy (Lazarou et al., 2013). To determine if NL-1 modulates autophagic flux in ALL cells, we used a fluorescent dye that specifically labels autophagosomes. Treatment with NL-1 increased the fluorescence intensity compared with untreated cells, indicating an increase in autophagy (Fig. 6E). The specificity of the assay was confirmed by pretreating the cells with chloroquine, an inhibitor of autophagy. Treating ALL cells with chloroquine before the addition of NL-1 slightly reduced the increase in fluorescence (Fig. 6E). To investigate if autophagy is critical in NL-1–mediated induction of death in leukemic cells, we pretreated cells with increasing doses of chloroquine followed by NL-1 and then analyzed cell viability. Treatment with NL-1 by itself caused death in REH cells (Fig. 6F). Pretreatment with 5 μM chloroquine to inhibit autophagy reduced the amount of NL-1–mediated cell death significantly in both the REH and REH/Ara-C cells (Fig. 6F). We repeated this pharmacological inhibitory study in TOM1 and observed a similar reduction in NL-1–mediated death (Supplemental Material).
NL-1 Has Antileukemic Activity In Vivo.
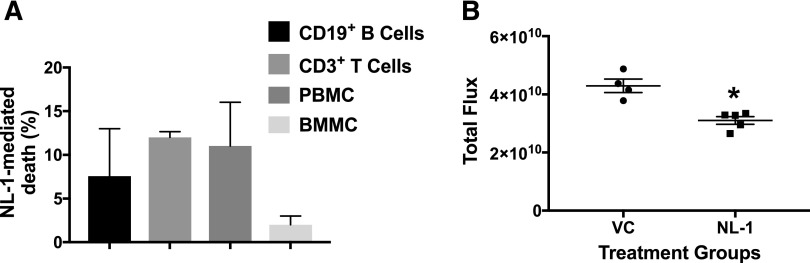
To determine whether NL-1 would negatively affect the viability of other hematopoietic cells, we treated primary immune cells with NL-1 and found that NL-1 did not substantially decrease cellular viability in those cells (Fig. 7A). NL-1 caused the highest increase in death in CD3 T cells (12.00% ± 0.66%), followed by peripheral blood mononuclear cells (11.00% ± 5.00%), then CD19 B cells (7.57% ± 5.44%), and finally bone marrow mononuclear cells (2.00% ± 1.00%). To determine whether NL-1 would affect leukemic cell growth in vivo, we evaluated NL-1 in a pilot study with a single dose of NL-1, where we injected TOM1 ALL cells expressing luciferase into NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice. After confirming leukemic cell engraftment by IVIS luminescent imaging, the mice were treated with 1 mg/kg NL-1 every day for 5 days. The dose was based on our previous pharmacokinetic study (Pedada et al., 2014). Out of the 10 mice, one control mouse did not show tumor progression after engraftment and was not included in the study. On day 14 postinjection, the NL-1–treated group showed significantly lower tumor burden than the control vehicle–treated group (Fig. 7B).
Fig. 7.
NL-1 does not affect primary immune cells but demonstrated antileukemic activity in a pilot in vivo study. (A) Normal primary B cells, T cells, peripheral blood mononuclear cells (PBMC) and bone marrow mononuclear cells (BMMC) were plated at 1 × 106 cells/ml in a 24-well plate and then treated with 60 μM NL-1 for 24 hours. The cells were then processed for flow cytometry using a LIVE/DEAD stain following the manufacturer’s instructions. (B) NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ female mice were injected with 1 × 106 luciferase expressing TOM1 cells. Two days after engraftment the mice were treated with 10 mg/kg NL-1 for 5 days. The tumor burden was analyzed by IVIS imaging at the end of 14 days. All data are represented as mean ± S.E.M. *P < 0.05, when compared with vehicle-treated control.
Discussion
In the present study, we have developed an Ara-C–resistant leukemic cell line (REH/Ara-C) to model the development of chemotherapy resistance observed in patients with ALL. REH/Ara-C cells overexpress CDA and CISD1 (mitoNEET protein) transcripts. This overexpression did not result in crossresistance to methotrexate or vincristine but was associated with altered migratory capacity toward BMSCs, HOBs, and SDF-1. Targeting the mitochondrial protein mitoNEET (CISD1) with NL-1 decreased the viability in several ALL cell lines with comparable IC50 vaues. Moreover, NL-1 efficiently disrupted preformed spheroids in a hemosphere assay and was able to inhibit the chemotaxis of both parental REH and REH/Ara-C cell lines. More importantly, NL-1 was able to specifically decrease the viability of REH/Ara-C cells in a coculture model that models the combination of acquired and de novo drug resistance found in relapsed patients. The NL-1–mediated cell death was attributed to its ability, in part, to induce autophagy since pretreatment with the autophagy inhibitor chloroquine was able to reduce NL-1–mediated cell death. Finally, NL-1 was nontoxic to normal immune cells and showed significant antileukemic activity in a pilot study using a mouse model of ALL.
B-cell ALL relapsed patients are treated with a combination of chemotherapy and hematopoietic stem cell transplantation (Eckert et al., 2013). The antitumor effect of Ara-C varies widely in the clinic and is related to genetic variation in the key genes involved in the uptake, activation, and metabolism of Ara-C within the tumor cells (Lamba, 2009). SCL19A1 is a folate transporter that is responsible for the uptake of Ara-C and has been shown to be downregulated in Ara-C–resistant cells (Gati et al., 1997). Once inside the cells, Ara-C is activated by deoxycytidylate deaminase (DCK) and its expression has been reported to be decreased in relapsed patients (Kakihara et al., 1998). Ara-C is inactivated by cytidine deaminase (CDA), which has been shown to be upregulated in drug-resistant cells (Schröder et al., 1998). Finally, ribonucleotide reductase M1 (RRM1) catalyzes and maintains the intracellular pools of dCTP, which directly competes with activated Ara-C for incorporation within the DNA (Liliemark and Plunkett, 1986). In our study, CDA was upregulated in the REH/Ara-C cell line and may be the cause of resistance to Ara-C. Since the upregulated CDA has little role to play in modulation of vincristine and methotrexate activity, not surprisingly, these drugs showed similar sensitivity in REH/Ara-C compared with REH cells. This lack of crossresistance has been previously demonstrated in T-lymphoblastic leukemia cells that were resistant to Ara-C (Cai et al., 2008). Since there are no clinical therapies available to mitigate the overexpression of CDA, we sought to identify a druggable target for treatment of relapsed ALL patients. Our previous studies have shown that the drug resistance phenotype is accompanied by mitochondrial dysfunction (Moses et al., 2016b). In light of this, we looked at the mitochondrial protein mitoNEET expression in REH/Ara-C cells and found it to be upregulated in the drug-resistant cells compared with parental REH cells.
MitoNEET has been previously described as being overexpressed in cancer cells of epithelial origins (Sohn et al., 2013). Specifically, constitutive overexpression of mitoNEET increases tumor size in breast cancer, and conversely knocking down the expression of mitoNEET decreased proliferation and tumor development in human breast cancer cells (Sohn et al., 2013; Tamir et al., 2015). Furthermore, a bioinformatics study of gene expression analysis has revealed that mitoNEET expression is elevated in several other solid tumors (Stelzer et al., 2011). MitoNEET is an iron-sulfur 2Fe-2S cluster protein located on the outer mitochondrial membrane, which acts as a redox sensor and regulates mitochondrial bioenergetics (Mittler et al., 2019). However, the role of mitoNEET as a potential drug target in ALL had not yet been investigated. In our studies, we have found that mitoNEET, along with CDA, is overexpressed in our acquired drug-resistant cell line, REH/Ara-C. Overexpression in REH/Ara-C cells was not accompanied by changes in cell proliferation or cell surface receptor expression when compared with the parental cell line, REH.
Impaired apoptosis is one of the most crucial phenotypes associated with resistance to different classes of drugs in ALL (Holleman et al., 2003). Since mitochondria modulate the molecular players involved in apoptosis, targeting the same is an attractive strategy for therapy in cancer (Indran et al., 2011). Toward this goal, we have used a selective mitoNEET ligand, called NL-1, a derivative of the antidiabetic drug pioglitazone (Geldenhuys et al., 2010, 2011). The use of NL-1 as an anticancer agent has not been reported, and our studies indicated that NL-1 has significant antileukemic activity. Our data show that NL-1 was effective in decreasing the cell viability of different ALL cell lines and had similar IC50 values in the REH and resistant REH/Ara-C cell lines. We had previously demonstrated the enrichment of ALL cells with overexpression of stem cell markers like SOX2 in hemospheres (Nair et al., 2018). We evaluated the effect of NL-1 in the SD1 ALL cells since these have a propensity to form hemospheres: NL-1 effectively disrupted preformed spheroids in a hemosphere assay, and the cells had significantly decreased viability (Zhang et al., 2015). We found it interesting that NL-1 was able to target and induce death in the PD cells of the REH/Ara-C cells in coculture with either BMSCs or HOBs. This observation is very notable since the coculture study mimics the acquired and de novo drug-resistant phenotype associated with relapse in ALL patients (Mudry et al., 2000; Fortney et al., 2001; Moses et al., 2016b).
Previous studies have reported that suppression of mitoNEET is accompanied by mitochondrial dysfunction and accumulation of autophagosomes (Sohn et al., 2013). Conversely, overexpression of mitoNEET confers autophagic resistance and promotes tumor growth (Salem et al., 2012). Interestingly, autophagy plays a key role in ALL survival and leukemogenic transformation in the bone tumor microenvironment (Altman et al., 2011). Several drugs used in ALL therapy use activation of autophagy as a mechanism for inducing death in the leukemic cells. For example, glucocorticoids activate the autophagic machinery to induce death in primary ALL cells (Laane et al., 2009). Furthermore, idarubicin mediated cytotoxic autophagy in ALL cells that could be partially reversed by pretreating cells with pharmacologic inhibitors of autophagy (Ristic et al., 2014). Similarly, everolimus induced autophagy-mediated death in ALL primary cell lines and in NOD/SCID mice xenografted with ALL cell lines, which could be partially reversed by genetic ablation of beclin-1, a key regulator of autophagy (Crazzolara et al., 2009; Neri et al., 2014). In the present study, NL-1 treatment in ALL cells led to increased autophagy, which could be reduced by pretreating cells with an autophagy inhibitor, chloroquine (Kimura et al., 2013). More importantly, inhibition of autophagy by the use of chloroquine also significantly reduced NL-1–mediated cell death in ALL. These studies suggest that the decrease in proliferation and increased cell death of the PD cells may in part be due to involvement of the autophagic pathways. Future studies will focus on elucidating the key mechanisms important for cell death seen with NL-1 treatment.
In addition to survival, autophagy also plays a key role in chemotaxis. Specifically, repression of autophagy facilitates membrane trafficking and compartmentalization of critical proteins to aid in the directional chemotactic migration of cells (Coly et al., 2017). Chemotaxis plays a very important role in the niche development within the BM of leukemia patients (Gómez et al., 2015). Previous studies using chemokine inhibitors like AMD3100 reduced tumor burden and increased leukemic death when used in combination with chemotherapy (Fei et al., 2010). Our group has demonstrated the utility of inhibiting chemotaxis as a mechanism for inducing death in drug-resistant ALL cells (Nair et al., 2018). In the present study, NL-1 was sufficient to inhibit ALL chemotaxis, irrespective of the chemoattractant used in the study. The combination of the autophagy-induced death and chemotaxis might partially explain the in vivo antileukemic activity observed in our study. However, future studies to include evaluation of a stable cell line with impaired autophagy will be required to definitively validate this correlative observation made during our study.
In summary, we have demonstrated that targeting of mitoNEET by using the first-in-class compound NL-1 is a clinically viable strategy to consider in order to overcome the drug resistance observed in relapsed/refractory ALL patients. This corroborates findings published by other groups, which indicate that targeting mitochondrial respiration in ALL is a tractable therapeutic strategy (Fu et al., 2017; Kuntz et al., 2017). In addition to its antileukemic activity, NL-1 also showed a nontoxic profile when primary immune cells were exposed to it. NL-1 efficiently reduced viability in several ALL cell lines harboring different molecular drivers of leukemogenesis. Induction of death of leukemic cells in a coculture model of acquired and de novo drug resistance indicates the promise of targeting mitoNEET in combination with the standard of care in the treatment of ALL.
Abbreviations
- ALL
acute lymphoblastic leukemia
- Ara-C
cytarabine
- BM
bone marrow
- BMSC
bone marrow stromal cell
- CDA
cytidine deaminase
- HOB
human osteoblast
- PD
phase dim
- S
suspended
Authorship Contributions
Participated in research design: Geldenhuys, Nair, Piktel, Martin, Gibson.
Conducted experiments: Nair, Piktel.
Performed data analysis: Geldenhuys, Nair, Piktel.
Wrote or contributed to the writing of the manuscript: Nair, Piktel, Martin, Gibson.
Footnotes
This work was supported by the National Institutes of Health National Institute of General Medicine [Grants U54GM104942, P20GM121322, P20GM103488, P20GM103434, S10OD016165, P20GM109098, and P20RR016440] and the Alexander B. Osborn Hematopoietic Malignancy and Transplantation Endowed Professorship.
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Altman BJ, Jacobs SR, Mason EF, Michalek RD, MacIntyre AN, Coloff JL, Ilkayeva O, Jia W, He YW, Rathmell JC. (2011) Autophagy is essential to suppress cell stress and to allow BCR-Abl-mediated leukemogenesis. Oncogene 30:1855–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F, Morcos F, Sohn YS, Darash-Yahana M, Rezende CO, Lipper CH, Paddock ML, Song L, Luo Y, Holt SH, et al. (2015) The Fe-S cluster-containing NEET proteins mitoNEET and NAF-1 as chemotherapeutic targets in breast cancer. Proc Natl Acad Sci USA 112:3698–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Damaraju VL, Groulx N, Mowles D, Peng Y, Robins MJ, Cass CE, Gros P. (2008) Two distinct molecular mechanisms underlying cytarabine resistance in human leukemic cells. Cancer Res 68:2349–2357. [DOI] [PubMed] [Google Scholar]
- Colca JR, McDonald WG, Waldon DJ, Leone JW, Lull JM, Bannow CA, Lund ET, Mathews WR. (2004) Identification of a novel mitochondrial protein (“mitoNEET”) cross-linked specifically by a thiazolidinedione photoprobe. Am J Physiol Endocrinol Metab 286:E252–E260. [DOI] [PubMed] [Google Scholar]
- Colmone A, Amorim M, Pontier AL, Wang S, Jablonski E, Sipkins DA. (2008) Leukemic cells create bone marrow niches that disrupt the behavior of normal hematopoietic progenitor cells. Science 322:1861–1865. [DOI] [PubMed] [Google Scholar]
- Coly PM, Gandolfo P, Castel H, Morin F. (2017) The autophagy machinery: a new player in chemotactic cell migration. Front Neurosci 11:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crazzolara R, Bradstock KF, Bendall LJ. (2009) RAD001 (Everolimus) induces autophagy in acute lymphoblastic leukemia. Autophagy 5:727–728. [DOI] [PubMed] [Google Scholar]
- Eckert C, Henze G, Seeger K, Hagedorn N, Mann G, Panzer-Grümayer R, Peters C, Klingebiel T, Borkhardt A, Schrappe M, et al. (2013) Use of allogeneic hematopoietic stem-cell transplantation based on minimal residual disease response improves outcomes for children with relapsed acute lymphoblastic leukemia in the intermediate-risk group. J Clin Oncol 31:2736–2742. [DOI] [PubMed] [Google Scholar]
- Fei F, Stoddart S, Müschen M, Kim YM, Groffen J, Heisterkamp N. (2010) Development of resistance to dasatinib in Bcr/Abl-positive acute lymphoblastic leukemia. Leukemia 24:813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortney JE, Zhao W, Wenger SL, Gibson LF. (2001) Bone marrow stromal cells regulate caspase 3 activity in leukemic cells during chemotherapy. Leuk Res 25:901–907. [DOI] [PubMed] [Google Scholar]
- Fu X, Liu W, Huang Q, Wang Y, Li H, Xiong Y. (2017) Targeting mitochondrial respiration selectively sensitizes pediatric acute lymphoblastic leukemia cell lines and patient samples to standard chemotherapy. Am J Cancer Res 7:2395–2405. [PMC free article] [PubMed] [Google Scholar]
- Gati WP, Paterson AR, Larratt LM, Turner AR, Belch AR. (1997) Sensitivity of acute leukemia cells to cytarabine is a correlate of cellular es nucleoside transporter site content measured by flow cytometry with SAENTA-fluorescein. Blood 90:346–353. [PubMed] [Google Scholar]
- Geldenhuys WJ, Funk MO, Awale PS, Lin L, Carroll RT. (2011) A novel binding assay identifies high affinity ligands to the rosiglitazone binding site of mitoNEET. Bioorg Med Chem Lett 21:5498–5501. [DOI] [PubMed] [Google Scholar]
- Geldenhuys WJ, Funk MO, Barnes KF, Carroll RT. (2010) Structure-based design of a thiazolidinedione which targets the mitochondrial protein mitoNEET. Bioorg Med Chem Lett 20:819–823. [DOI] [PubMed] [Google Scholar]
- Gómez AM, Martínez C, González M, Luque A, Melen GJ, Martínez J, Hortelano S, Lassaletta Á, Madero L, Ramírez M. (2015) Chemokines and relapses in childhood acute lymphoblastic leukemia: a role in migration and in resistance to antileukemic drugs. Blood Cells Mol Dis 55:220–227. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. (2011) Hallmarks of cancer: the next generation. Cell 144:646–674. [DOI] [PubMed] [Google Scholar]
- Holleman A, den Boer ML, Kazemier KM, Janka-Schaub GE, Pieters R. (2003) Resistance to different classes of drugs is associated with impaired apoptosis in childhood acute lymphoblastic leukemia. Blood 102:4541–4546. [DOI] [PubMed] [Google Scholar]
- Indran IR, Tufo G, Pervaiz S, Brenner C. (2011) Recent advances in apoptosis, mitochondria and drug resistance in cancer cells. Biochim Biophys Acta 1807:735–745. [DOI] [PubMed] [Google Scholar]
- Kakihara T, Fukuda T, Tanaka A, Emura I, Kishi K, Asami K, Uchiyama M. (1998) Expression of deoxycytidine kinase (dCK) gene in leukemic cells in childhood: decreased expression of dCK gene in relapsed leukemia. Leuk Lymphoma 31:405–409. [DOI] [PubMed] [Google Scholar]
- Kaspers GJ, Veerman AJ, Pieters R, Van Zantwijk CH, Smets LA, Van Wering ER, Van Der Does-Van Den Berg A. (1997) In vitro cellular drug resistance and prognosis in newly diagnosed childhood acute lymphoblastic leukemia. Blood 90:2723–2729. [PubMed] [Google Scholar]
- Kimura T, Takabatake Y, Takahashi A, Isaka Y. (2013) Chloroquine in cancer therapy: a double-edged sword of autophagy. Cancer Res 73:3–7. [DOI] [PubMed] [Google Scholar]
- Klumper E, Pieters R, Veerman AJ, Huismans DR, Loonen AH, Hählen K, Kaspers GJ, van Wering ER, Hartmann R, Henze G. (1995) In vitro cellular drug resistance in children with relapsed/refractory acute lymphoblastic leukemia. Blood 86:3861–3868. [PubMed] [Google Scholar]
- Kuntz EM, Baquero P, Michie AM, Dunn K, Tardito S, Holyoake TL, Helgason GV, Gottlieb E. (2017) Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nat Med 23:1234–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laane E, Tamm KP, Buentke E, Ito K, Kharaziha P, Oscarsson J, Corcoran M, Björklund AC, Hultenby K, Lundin J, et al. (2009) Cell death induced by dexamethasone in lymphoid leukemia is mediated through initiation of autophagy [published correction appears in Cell Death Differ (2009) 16:1071]. Cell Death Differ 16:1018–1029. [DOI] [PubMed] [Google Scholar]
- Lamba JK. (2009) Genetic factors influencing cytarabine therapy. Pharmacogenomics 10:1657–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Narendra DP, Jin SM, Tekle E, Banerjee S, Youle RJ. (2013) PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J Cell Biol 200:163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liliemark JO, Plunkett W. (1986) Regulation of 1-β-D-arabinofuranosylcytosine 5′-triphosphate accumulation in human leukemia cells by deoxycytidine 5′-triphosphate. Cancer Res 46:1079–1083. [PubMed] [Google Scholar]
- Mittler R, Darash-Yahana M, Sohn YS, Bai F, Song L, Cabantchik IZ, Jennings PA, Onuchic JN, Nechushtai R. (2019) NEET proteins: a new link between iron metabolism, reactive oxygen species, and cancer. Antioxid Redox Signal 30:1083–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möhle R, Schittenhelm M, Failenschmid C, Bautz F, Kratz-Albers K, Serve H, Brugger W, Kanz L. (2000) Functional response of leukaemic blasts to stromal cell-derived factor-1 correlates with preferential expression of the chemokine receptor CXCR4 in acute myelomonocytic and lymphoblastic leukaemia. Br J Haematol 110:563–572. [DOI] [PubMed] [Google Scholar]
- Moses BS, Evans R, Slone WL, Piktel D, Martinez I, Craig MD, Gibson LF. (2016a) Bone marrow microenvironment niche regulates mir-221/222 in acute lymphoblastic leukemia. Mol Cancer Res 14:909–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses BS, Slone WL, Thomas P, Evans R, Piktel D, Angel PM, Walsh CM, Cantrell PS, Rellick SL, Martin KH, et al. (2016b) Bone marrow microenvironment modulation of acute lymphoblastic leukemia phenotype. Exp Hematol 44:50–59.e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudry RE, Fortney JE, York T, Hall BM, Gibson LF. (2000) Stromal cells regulate survival of B-lineage leukemic cells during chemotherapy. Blood 96:1926–1932. [PubMed] [Google Scholar]
- Nair RR, Piktel D, Geldenhuys WJ, Gibson LF. (2018) Combination of cabazitaxel and plicamycin induces cell death in drug resistant B-cell acute lymphoblastic leukemia. Leuk Res 72:59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri LM, Cani A, Martelli AM, Simioni C, Junghanss C, Tabellini G, Ricci F, Tazzari PL, Pagliaro P, McCubrey JA, et al. (2014) Targeting the PI3K/Akt/mTOR signaling pathway in B-precursor acute lymphoblastic leukemia and its therapeutic potential. Leukemia 28:739–748. [DOI] [PubMed] [Google Scholar]
- Neuzil J, Dong LF, Rohlena J, Truksa J, Ralph SJ. (2013) Classification of mitocans, anti-cancer drugs acting on mitochondria. Mitochondrion 13:199–208. [DOI] [PubMed] [Google Scholar]
- Pedada KK, Zhou X, Jogiraju H, Carroll RT, Geldenhuys WJ, Lin L, Anderson DJ. (2014) A quantitative LC-MS/MS method for determination of thiazolidinedione mitoNEET ligand NL-1 in mouse serum suitable for pharmacokinetic studies. J Chromatogr B Analyt Technol Biomed Life Sci 945–946:141–146. [DOI] [PubMed] [Google Scholar]
- Pieters R, Huismans DR, Loonen AH, Hählen K, van der Does-van den Berg A, van Wering ER, Veerman AJ. (1991) Relation of cellular drug resistance to long-term clinical outcome in childhood acute lymphoblastic leukaemia. Lancet 338:399–403. [DOI] [PubMed] [Google Scholar]
- Pogorzala M, Kubicka M, Rafinska B, Wysocki M, Styczynski J. (2015) Drug-resistance profile in multiple-relapsed childhood acute lymphoblastic leukemia. Anticancer Res 35:5667–5670. [PubMed] [Google Scholar]
- Ristic B, Bosnjak M, Arsikin K, Mircic A, Suzin-Zivkovic V, Bogdanovic A, Perovic V, Martinovic T, Kravic-Stevovic T, Bumbasirevic V, et al. (2014) Idarubicin induces mTOR-dependent cytotoxic autophagy in leukemic cells. Exp Cell Res 326:90–102. [DOI] [PubMed] [Google Scholar]
- Salem AF, Whitaker-Menezes D, Howell A, Sotgia F, Lisanti MP. (2012) Mitochondrial biogenesis in epithelial cancer cells promotes breast cancer tumor growth and confers autophagy resistance. Cell Cycle 11:4174–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder JK, Seidelmann M, Kirch HC, Seeber S, Schütte J. (1998) Assessment of resistance induction to cytosine arabinoside following transfer and overexpression of the deoxycytidylate deaminase gene in vitro. Leuk Res 22:619–624. [DOI] [PubMed] [Google Scholar]
- Slone WL, Moses BS, Evans R, Piktel D, Martin KH, Petros W, Craig M, Gibson LF. (2016a) Modeling chemotherapy resistant leukemia in vitro. J Vis Exp 108:e53645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slone WL, Moses BS, Hare I, Evans R, Piktel D, Gibson LF. (2016b) BCL6 modulation of acute lymphoblastic leukemia response to chemotherapy. Oncotarget 7:23439–23453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn YS, Tamir S, Song L, Michaeli D, Matouk I, Conlan AR, Harir Y, Holt SH, Shulaev V, Paddock ML, et al. (2013) NAF-1 and mitoNEET are central to human breast cancer proliferation by maintaining mitochondrial homeostasis and promoting tumor growth. Proc Natl Acad Sci USA 110:14676–14681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer G, Dalah I, Stein TI, Satanower Y, Rosen N, Nativ N, Oz-Levi D, Olender T, Belinky F, Bahir I, et al. (2011) In-silico human genomics with GeneCards. Hum Genomics 5:709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow P, Key L, Chen X, Pan Q, Neale GA, Coustan-Smith E, Mullighan CG, Zhou Y, Pui CH, Campana D. (2010) Clinical significance of low levels of minimal residual disease at the end of remission induction therapy in childhood acute lymphoblastic leukemia. Blood 115:4657–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamir S, Paddock ML, Darash-Yahana-Baram M, Holt SH, Sohn YS, Agranat L, Michaeli D, Stofleth JT, Lipper CH, Morcos F, et al. (2015) Structure-function analysis of NEET proteins uncovers their role as key regulators of iron and ROS homeostasis in health and disease. Biochim Biophys Acta 1853:1294–1315. [DOI] [PubMed] [Google Scholar]
- Zhang X, de Milito A, Olofsson MH, Gullbo J, D’Arcy P, Linder S. (2015) Targeting mitochondrial function to treat quiescent tumor cells in solid tumors. Int J Mol Sci 16:27313–27326. [DOI] [PMC free article] [PubMed] [Google Scholar]