Abstract
Molecular docking analysis of twenty two phytoconstituents from Hibiscus rosa-sinensis, against seven targets of obesity like pancreatic lipase, fat and obesity protein (FTO protein), cannabinoid receptor, hormones as ghrelin, leptin and protein as SCH1 and MCH1 is detailed in this data article. Chemical structures of phytoconstituents were downloaded from PubChem and protein structures were retrieved from RCSB protein databank. Docking was performed using FlexX software Lead IT version 2.3.2; Bio Solved IT. Visualization and analysis was done by Schrodinger maestro software. The docking score and interactions with important amino acids were analyzed and compared with marketed drug, orlistat. The findings suggest exploitation of best ligands experimentally to develop novel anti-obesity agent.
Specifications table
| Subject area | Chemistry |
| More specific subject area | Computational chemistry |
| Type of data | Table, figure |
| How data was acquired | Ligand based molecular docking using FlexX and Maestro software |
| Data format | Raw and analyzed |
| Experimental factors | Phytoconstituents structures downloaded from PubChem were subjected to Avogadro software for energy minimization. |
| Experimental features | Minimized ligands structures were docked with seven selected protein structure using FlexX software. |
| Data source location | Department of bioinformatics, Dr. D. Y. Patil Biotechnology & Bioinformatics Institute Tathawade, Pune |
| Data accessibility | Data is only with this article |
| Related research article | K. H. Min, J. Yoo, H. Park, Computer-Aided Identification of Ligands for GPCR Anti-Obesity Targets, Curr Top Med Chem. 9 (2009) 539–553 [1]. |
Value of the data
|
1. Data
This dataset contains docking analysis of phytoconstituents of Hibiscus rosa-sinensis on different targets of obesity. Different secondary metabolites present in Hibiscus rosa-sinensis were selected. Chemical structures of selected phytoconstituents were taken from database and were subjected to energy minimization. Seven receptor structures were selected as potential targets of obesity [1], [2], [3], [4], [5], [6], [7], [8]. Protein structures available in database were downloaded from RCSB protein databank. Table 1 gives details of the selected receptors. Two receptors model were prepared using I-TASSER server online. Table 2 summarizes FASTA sequence of Ghrelin and MCH1 receptor subjected to model preparation. Phytoconstituents were docked on the above targets to understand binding interactions. Table 3, Table 4, Table 5, Table 6, Table 7, Table 8, Table 9 summarizes the dock score, bond distance and interacting amino acid residue of all phytoconstituents on seven different targets. Fig. 1-14 gives docked images of phytoconstituents with lowest dock score and standard drug orlistat with seven receptor proteins.
Table 1.
Table summarizing details of targets selected.
| Target | PDB ID | Description | Resolution | R value free | R value work |
|---|---|---|---|---|---|
| Pancreatic Lipase | 1LPB | The 2.46 Å resolution structure of the pancreatic lipase colipase complex inhibited by a c11 alkyl phosphonate | 2.46 Å | 0.285 | 0.183 |
| Fat And Obesity Protein | 3LFM | Crystal structure of the fat mass and obesity associated (FTO) protein reveals basis for its substrate specificity | 2.5 Å | 0.285 | 0.239 |
| Cannabinoid Receptor | 5TGZ | Crystal structure analysis of w35f/h207w mutant of human clic1 | 2.3 Å | 0.306 | 0.240 |
| Leptin | 1AX8 | Human obesity protein, leptin | 2.4 Å | 0.283 | 0.185 |
| SCH1 Protein | 4XWX | Crystal structure of the PTB domain of SHC | 1.87 Å | 0.191 | 0.168 |
Table 2.
Uniprot ID and FASTA sequence of ghrelin and MCH1 receptor.
| Target | UniProt ID | FASTA sequence |
|---|---|---|
| Ghrelin receptor | Q9UBU3 | MPSPGTVCSLLLLGMLWLDLAMAGSSFLSPEHQRVQQRKESKKPPAKLQPRALAGWLRPEDGGQAEGAEDELEVRFNAPFDVGIKLSGVQYQQHSQALGKFLQDILWEEAKEAPADK |
| MCH 1 receptor | Q99705 | MSVGAMKKGVGRAVGLGGGSGCQATEEDPLPNCGACAPGQGGRRWRLPQPAWVEGSSARLWEQATGTGWMDLEASLLPTGPNASNTSDGPDNLTSAGSPPRTGSISYINIIMPSVFGTICLLGIIGNSTVIFAVVKKSKLHWCNNVPDIFIINLSVVDLLFLLGMPFMIHQLMGNGVWHFGETMCTLITAMDANSQFTSTYILTAMAIDRYLATVHPISSTKFRKPSVATLVICLLWALSFISITPVWLYARLIPFPGGAVGCGIRLPNPDTDLYWFTLYQFFLAFALPFVVITAAYVRILQRMTSSVAPASQRSIRLRTKRVTRTAIAICLVFFVCWAPYYVLQLTQLSISRPTLTFVYLYNAAISLGYANSCLNPFVYIVLCETFRKRLVLSVKPAAQGQLRAVSNAQTADEERTESKGT |
Table 3.
Summary of docking analysis with pancreatic lipase (PDB ID 1LPB).
| Sr. No | Posename | Score | Interacting Residues | Bond Type | Bond Distance |
|---|---|---|---|---|---|
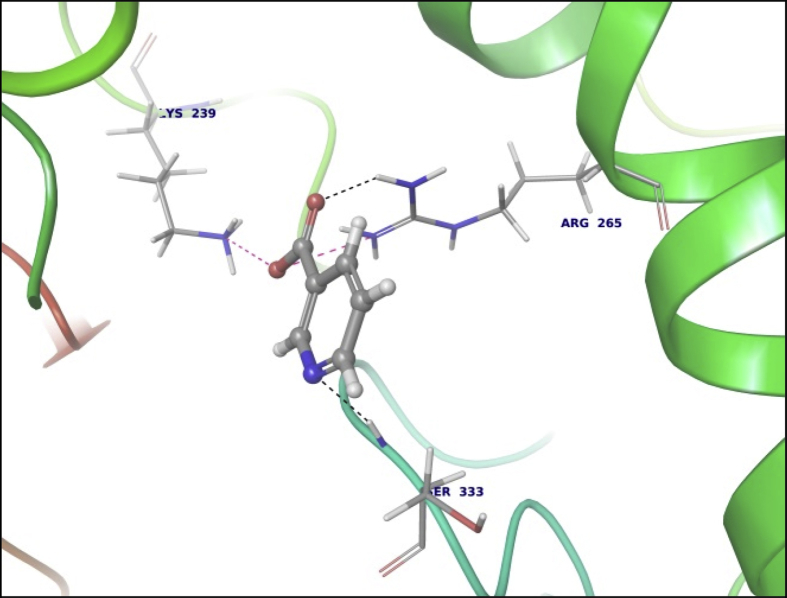
| 1 | Niacin | −27.2868 | SER 333 | HB | 2.01 |
| ARG 265 | Pi-Pi Stacking | 5.21 | |||
| HB | 1.79 | ||||
| Salt bridge | 3.08 | ||||
| LYS 239 | Salt bridge | 2.73 | |||
| 2 | Quercetin 3, 7 diglucoside | −21.223 | LYS 239 | HB | 1.93 |
| ASP 247 | HB | 1.93 | |||
| ASP 257 | HB | 1.71 | |||
| TRY 267 | HB | 1.98 | |||
| THR 271 | HB | 2.70 | |||
| LYS 268 | HB | 1.48 | |||
| Pi cation | 5.10 | ||||
| ASP 249 | HB | 2.12 | |||
| 3 | Ascorbic acid | −20.6315 | SER 333 | HB | 2.32 |
| ASP 247 | HB | 2.06 | |||
| ARG 265 | HB | 2.18 | |||
| ASP 257 | HB | 2.17 | |||
| ASP 249 | HB | 1.79 | |||
| 4 | Quercetin 3, 3′ diglucoside | −20.3198 | SER 333 | 2HB | 2.12,2.31 |
| ASP 247 | HB | 2.34 | |||
| ASP 331 | HB | 1.84 | |||
| ARG 265 | HB | 2.20 | |||
| ASP 257 | HB | 2.66 | |||
| TYR 267 | HB | 2.2.3 | |||
| ASN 88 | HB | 2.52 | |||
| 5 | Quercetin 3,4′ diglucoside | −18.4448 | ASP 331 | 2HB | 2.18, 2,07 |
| ARG 265 | HB | 1.90 | |||
| SER 333 | HB | 2.29 | |||
| PHE 335 | Pi Pi stacking | 5.46 | |||
| ASN 88 | HB | 2.61 | |||
| 6 | 8 nonynoic acid | −17.7764 | LYS 239 | HB | 2.04 |
| Salt Bridge | 3.91 | ||||
| ARG 265 | HB | 1.93 | |||
| 7 | 9 Decynoic acid | −17.4676 | ARG 265 | 2HB | 2.11, 1.85 |
| LYS 239 | HB | 1.94 | |||
| Salt bridge | 4.67 | ||||
| 8 | Cyanidine 3, 5 diglucoside | −15.6327 | ARG 265 | PI Pi stacking | 4.92 |
| ASP 247 | HB | 1.91 | |||
| ASP 257 | HB | 1.89 | |||
| ASP 249 | HB | 2.11 | |||
| Salt Bridge | 4.60 | ||||
| GLU 253 | HB | 2,29 | |||
| SER 333 | HB | 2.39 | |||
| LYS 268 | HB | 2.29 | |||
| ASP 272 | HB | 2.08 | |||
| 9 | Riboflavin | −15.3182 | ASP 249 | HB | 1.36 |
| SER 333 | HB | 1.60 | |||
| GLU 253 | HB | 2.16 | |||
| LYS 268 | HB | 1.71 | |||
| 10 | Thiamine | −14.7694 | ASP 249 | HB | 1.97 |
| Salt Bridge | 4.85 | ||||
| ARG 265 | Pi Pi stacking | 4.94 | |||
| LYS 268 | Pi cation | 2.86 | |||
| ASP 247 | HB | 2.07 | |||
| 11 | Beta rosasterol | −9.4736 | LYS 239 | HB | 2.16 |
| ARG 265 | HB | 2.16 | |||
| 12 | Cyanidin 3-sophoroside-5-glucoside | −8.2017 | ASP 249 | 3HB | 1.46, 2.02, 1.91 |
| ASP 272 | HB | 2.02 | |||
| GLU 253 | 2HB | 1.82, 1.81 | |||
| 13 | Methyl non-8-ynoate | −7.2264 | LYS 239 | HB | 2.05 |
| ARG 265 | HB | 1.94 | |||
| 14 | Methyl Dec-9-ynoate | −5.9149 | LYS 239 | HB | 2.05 |
| ARG 265 | HB | 1.94 | |||
| 15 | Methyl (E)-11-methoxy-9-oxononadec-10-enoate | −4.9341 | SER 333 | HB | 2.14 |
| ARG 265 | HB | 2.15 | |||
| TRY 267 | HB | 2.14 | |||
| LYS 268 | HB | 2.12 | |||
| 16 | Methyl malvalate | −3.6439 | LYS 239 | HB | 2.05 |
| ARG 265 | HB | 1.94 | |||
| 17 | Methyl 8-oxooctadec-9-ynoate | −2.8512 | SER 333 | HB | 2.10 |
| LYS 239 | HB | 1.89 | |||
| ARG 265 | HB | 2.02 | |||
| 18 | Methyl Sterculate | −1.1816 | ARG 265 | HB | 1.98 |
| LYS 239 | HB | 2.06 | |||
| 19 | Campesterol | 1.5909 | GLU 253 | HB | 2.28 |
| 20 | Stigmasterol | 2.651 | ASP 249 | HB | 1.90 |
| GLU 253 | HB | 2.12 | |||
| 21 | Beta sitosterol | 3.2084 | No interaction | ||
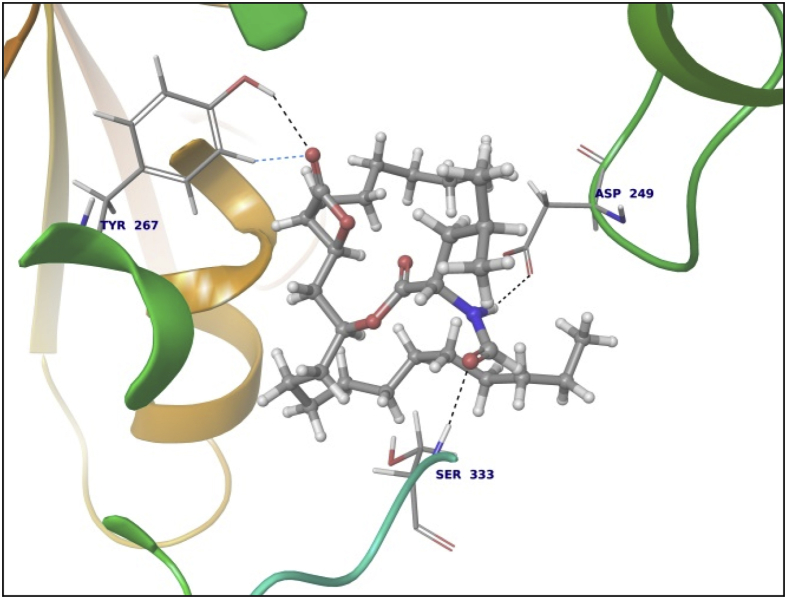
| 22 | Orlistat | 0.1075 | ASP 249 | HB | 1.68 |
| SER 333 | HB | 1.94 | |||
| TYR 267 | HB | 2.23 | |||
| Ar HB | 1.91 | ||||
Orlistat, as only standard drug used in market is used as standard reference for docking studies. Hence the docking result of orlistat in all tables is bold for ease of comparison.
Table 4.
Summary of docking analysis with fat and obesity protein (PDB ID 3LFM).
| Sr. No. | Ligand | Score | Interacting Residues | Bond Type | Bond Distance |
|---|---|---|---|---|---|
| 1 | Riboflavin | −27.3248 | ARG 96 | HB | 1.62 |
| SER 229 | HB | 2.07 | |||
| GLU 234 | HB | 2.01 | |||
| Ar HB | 2.35 | ||||
| 2 | Niacin | −21.5279 | ARG 322 | HB | 1.93 |
| GLU 234 | HB | 1.84 | |||
| ARG 96 | Pi-Pi Stacking | 4.26 | |||
| 3 | Thiamine | −19.313 | TRY 108 | Pi-Pi Stacking | 4.78 |
| HIP 231 | Pi-Pi Stacking | 3.68 | |||
| Pi-Pi Stacking | 5.45 | ||||
| Pi Cation | 4.42 | ||||
| Pi Cation | 3.78 | ||||
| SER 229 | HB | 1.38 | |||
| TYR 106 | HB | 2.00 | |||
| 4 | Ascorbic acid | −16.8546 | ASP 233 | HB | 1.99 |
| ARG 322 | HB | 2.45 | |||
| ARG 96 | HB | 1.99 | |||
| GLU 234 | HB | 1.90 | |||
| 5 | Cyanidine 3, 5 diglucoside | −14.6454 | ARG 322 | Pi Cation | 5.23 |
| HB | 1.52 | ||||
| TRY 106 | HB | 2.08 | |||
| HB | 1.81 | ||||
| HIP 232 | HB | 1.82 | |||
| GLU 234 | HB | 2.25 | |||
| HIP 231 | Pi-Pi Stacking | 4.81 | |||
| Pi-Pi Stacking | 5.43 | ||||
| VAL 94 | HB | 1.53 | |||
| 6 | Quercetin 3,4′ diglucoside | −12.747 | VAL 94 | HB | 2.35 |
| GLU 234 | HB | 2.00 | |||
| HIP 232 | HB | 1.57 | |||
| HB | 1.91 | ||||
| GLN 306 | HB | 2.18 | |||
| HIP 231 | Pi Cation | 6.38 | |||
| 7 | 8 nonynoic acid | −12.149 | ASN 205 | HB | 1.96 |
| ARG 322 | HB | 2.05 | |||
| Salt bridge | 3.78 | ||||
| ARG 96 | HB | 1.78 | |||
| 8 | 9 Decynoic acid | −11.8069 | ARG 322 | HB | 1.97 |
| GLU 234 | HB | 1.97 | |||
| 9 | Quercetin 3,3′ diglucoside | −11.2637 | TYR 108 | Pi-Pi Stacking | 4.55 |
| ARG 96 | HB | 2.66 | |||
| VAL 94 | HB | 1.79 | |||
| ALA 227 | HB | 2.24 | |||
| GLU 234 | HB | 1.62 | |||
| 10 | Quercetin 3,7 diglucoside | −7.7494 | GLU 234 | HB | 2.04 |
| HB | 2.16 | ||||
| TYR 108 | Pi-Pi Stacking | 4.94 | |||
| TYR 106 | HB | 2.29 | |||
| ARG 322 | HB | 1.71 | |||
| HIP 231 | Pi-Pi Stacking | 3.98 | |||
| HIP 232 | HB | 2.06 | |||
| 11 | Methyl 8-oxooctadec-9-ynoate | −6.5642 | HIP 232 | HB | 1.93 |
| ARG 96 | HB | 1.52 | |||
| 12 | Methyl Dec-9-ynoate | −4.8041 | ARG 96 | HB | 2.15 |
| 13 | Methyl non-8-ynoate | −4.4543 | ARG 96 | HB | 2.15 |
| 14 | (9) Methyl (E)-11-methoxy-9-oxononadec-10-enoate | −2.4721 | ARG 96 | HB | 2.15 |
| 15 | Beta rosasterol | −1.029 | VAL 94 | HB | 1.78 |
| 16 | Methyl Sterculate | 0.5157 | ARG 96 | HB | 1.84 |
| 17 | Methyl malvalate | 0.7329 | ARG 96 | HB | 1.88 |
| 18 | Beta sitosterol | 1.2521 | ALA 227 | HB | 2.2 |
| 19 | Campesterol | 1.447 | ALA 227 | HB | 2.21 |
| 20 | Orlistat | −7.2466 | ARG 322 | HB | 2.20 |
| GLU 234 | HB | 2.04 | |||
| HB | 1.66 | ||||
| HIP 232 | HB | 2.17 |
Orlistat, as only standard drug used in market is used as standard reference for docking studies. Hence the docking result of orlistat in all tables is bold for ease of comparison.
Table 5.
Summary of docking analysis with cannabinoid receptor (PDB ID 3TGZ).
| Sr. No. | Ligand | Score | Interacting Residues | Bond Type | Bond Distance |
|---|---|---|---|---|---|
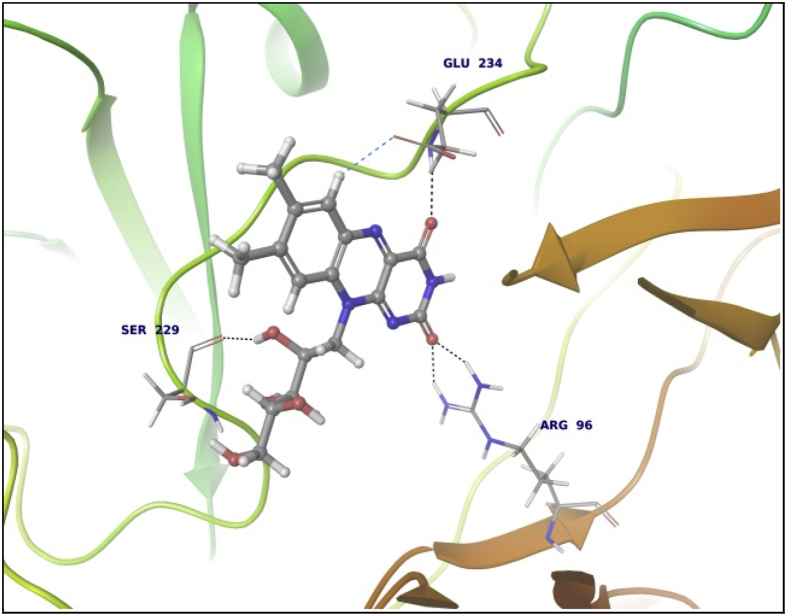
| 1 | Niacin | −14.7132 | MET 103 | HB | 1.84 |
| ASP 104 | HB | 2.07 | |||
| 2 | Thiamine | −13.5476 | PHE 102 | Pi-Pi Stacking | 4.92 |
| SER 383 | HB | 1.81 | |||
| SER 123 | HB | 1.69 | |||
| 3 | Ascorbic acid | −11.9942 | ASP 163 | HB | 2.30 |
| TRP 356 | HB | 1.70 | |||
| CYS 386 | HB | 1.86 | |||
| SER 199 | HB | 2.44 | |||
| ALA 162 | HB | 2.12 | |||
| 4 | Riboflavin | −9.4202 | PHE 170 | Pi-Pi Stacking | 5.43 |
| MET 103 | HB | 1.96 | |||
| SER 383 | HB | 2.06 | |||
| 5 | 8 nonynoic acid | −4.3902 | ASP 104 | HB | 1.84 |
| 6 | 9 Decynoic acid | −3.8828 | ASP 104 | HB | 1.95 |
| MET 103 | HB | 1.83 | |||
| 7 | Methyl 8-oxooctadec-9-ynoate | −3.0906 | ASN 389 | HB | 2.66 |
| TRP 356 | HB | 1.86 | |||
| 8 | Methyl non-8-ynoate | −2.5398 | TRP 356 | HB | 1.95 |
| 9 | Methyl Dec-9-ynoate | −2.4934 | TRP 356 | HB | 1.95 |
| 10 | (9) Methyl (E)-11-methoxy-9-oxononadec-10-enoate | −2.1673 | TRP 356 | HB | 1.95 |
| 11 | Quercetin 3,3′ diglucoside | −1.505 | SER 383 | HB | 2.61 |
| TRP 356 | HB | 2.51 | |||
| SER 390 | HB | 1.50 | |||
| 12 | Methyl malvalate | −0.5677 | No Interaction | ||
| 13 | Methyl Sterculate | −0.2554 | ASN 389 | HB | 2.50 |
| TRP 356 | HB | 1.85 | |||
| 14 | Quercetin 3,4′ diglucoside | −0.201 | PHE 174 | Pi-Pi Stacking | 5.44 |
| ASP 104 | HB | 2.14 | |||
| 15 | Campesterol | 3.5794 | No Interaction | ||
| 16 | Beta rosasterol | 6.6198 | |||
| 17 | Beta sitosterol | 6.6198 | |||
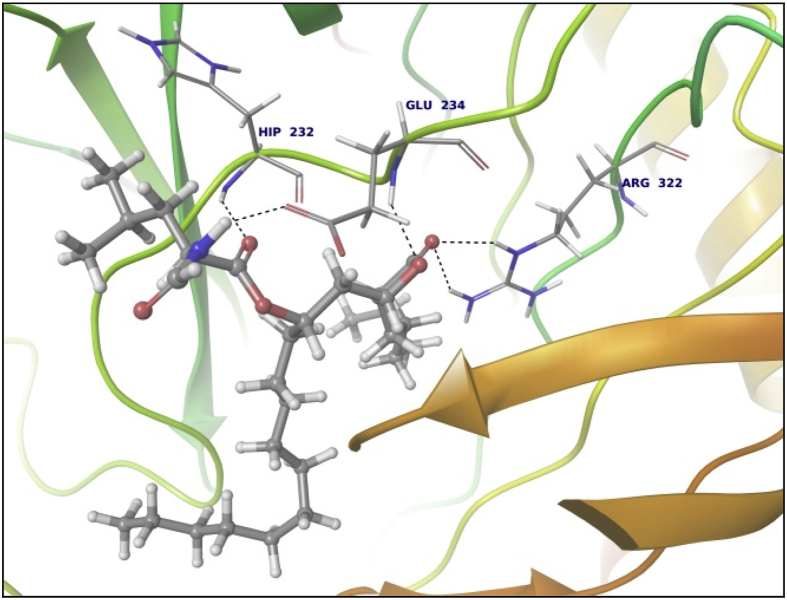
| 18 | Orlistat | −1.7877 | MET 103 | HB | 1.82 |
| ASP 104 | HB | 2.09 | |||
| SER 383 | HB | 1.65 | |||
Orlistat, as only standard drug used in market is used as standard reference for docking studies. Hence the docking result of orlistat in all tables is bold for ease of comparison.
Table 6.
Summary of docking analysis with leptin (PDB ID 1AX8).
| Sr. No. | Ligand | Dock Score | Interacting residues | Bond Type | Bond distance |
|---|---|---|---|---|---|
| 1 | Riboflavin | −18.4869 | GLN 134 | HB | 2.22 |
| HB | 1.91 | ||||
| GLN 130 | HB | 2.12 | |||
| HB | 1.72 | ||||
| ASP 40 | HB | 2.08 | |||
| HB | 1.58 | ||||
| Ar HB | 2.21 | ||||
| ILE 21 | HB | 1.80 | |||
| 2 | Cyanidine 3, 5 diglucoside | −13.4683 | ASP 40 | HB | 1.49 |
| HB | 2.40 | ||||
| HB | 1.75 | ||||
| HB | 1.68 | ||||
| GLN 130 | HB | 1.87 | |||
| HB | 1.83 | ||||
| GLN 134 | HB | 2.41 | |||
| ILE 21 | HB | 1.86 | |||
| HB | 1.53 | ||||
| 3 | Thiamine | −11.3807 | GLN 134 | HB | 2.21 |
| ASP 40 | HB | 2.15 | |||
| ILE 42 | HB | 1.84 | |||
| 4 | Ascorbic acid | −11.1364 | GLY 44 | HB | 1.94 |
| GLN 134 | HB | 1.98 | |||
| HB | 2.20 | ||||
| 5 | Quercetin 3,4′ diglucoside | −10.9657 | GLY 44 | HB | 2.57 |
| HB | 2.27 | ||||
| ASP 135 | HB | 2.15 | |||
| GLN 130 | HB | 1.90 | |||
| ASP 40 | HB | 2.05 | |||
| LEU 39 | HB | 1.84 | |||
| HB | 1.92 | ||||
| 6 | Quercetin 3,3′ diglucoside | −10.3108 | ASP 40 | HB | 2.29 |
| SER 127 | HB | 1.60 | |||
| 7 | Quercetin 3,7 diglucoside | −10.2723 | PHE 41 | Pi-Pi Stacking | 5.04 |
| GLN 130 | HB | 1.97 | |||
| ASP 40 | HB | 2.07 | |||
| GLY 131 | HB | 1.56 | |||
| GLY 44 | HB | 1.64 | |||
| ASP 135 | HB | 1.56 | |||
| 8 | Niacin | −9.3776 | ASP 40 | HB | 1.84 |
| 9 | Beta rosasterol | −6.3064 | GLY 44 | HB | 1.82 |
| 10 | Cyanidin 3-sophoroside-5-glucoside | −5.2426 | GLN 134 | HB | 1.84 |
| ASP 135 | HB | 2.07 | |||
| HB | 2.54 | ||||
| LEU 39 | HB | 1.84 | |||
| GLN 130 | HB | 1.99 | |||
| PHE 41 | HB | 1.84 | |||
| HB | 1.91 | ||||
| 11 | Campesterol | −3.5982 | No interaction | ||
| 12 | Stigmasterol | −2.8915 | ASP 135 | HB | 2.22 |
| GLY 44 | HB | 2.40 | |||
| 13 | 8 nonynoic acid | 0.1127 | OHE 41 | HB | 2.01 |
| 14 | Beta sitosterol | 0.4685 | ASP 135 | HB | 1.97 |
| GLY 44 | HB | 2.48 | |||
| 15 | 9 Decynoic acid | 1.1976 | ASP 40 | HB | 1.88 |
| PHE 41 | HB | 1.86 | |||
| 16 | Methyl non-8-ynoate | 2.0473 | PHE 41 | HB | 1.89 |
| 17 | Methyl Dec-9-ynoate | 2.8153 | PHE 41 | HB | 1.95 |
| 18 | Methyl 8-oxooctadec-9-ynoate | 5.4298 | PHE 41 | HB | 1.87 |
| 19 | (9) Methyl (E)-11-methoxy-9-oxononadec-10-enoate | 6.5759 | PHE 41 | HB | 1.83 |
| 20 | Methyl Sterculate | 6.9274 | PHE 41 | HB | 1.87 |
| 21 | Methyl malvalate | 8.0895 | PHE 41 | HB | 1.87 |
| 22 | Orlistat | 8.3009 | ASP 40 | HB | 1.71 |
| GLU 134 | HB | 1.80 | |||
| GLY 44 | HB | 1.99 | |||
Orlistat, as only standard drug used in market is used as standard reference for docking studies. Hence the docking result of orlistat in all tables is bold for ease of comparison.
Table 7.
Summary of docking analysis with SCH1 protein (PDB ID 4XWX).
| Sr. No. | Ligand | Dock score | Interacting residues | Bond type | Bond angle |
|---|---|---|---|---|---|
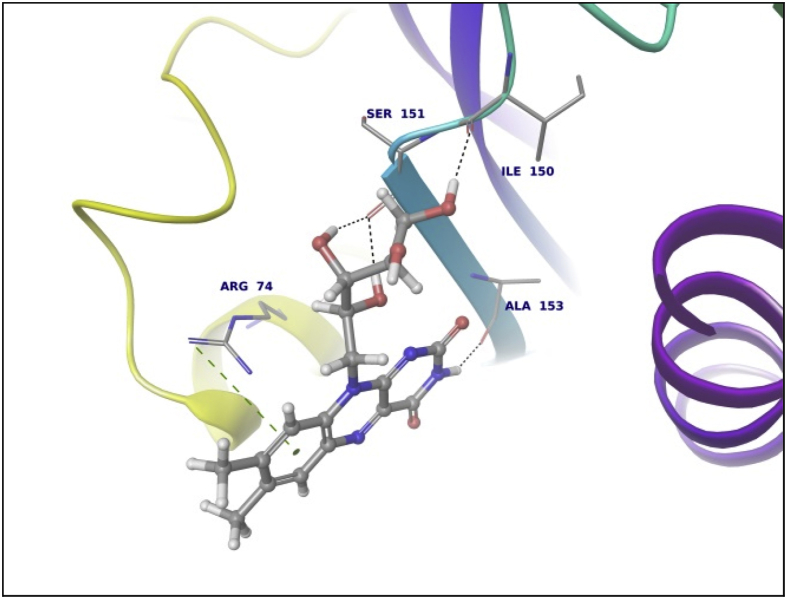
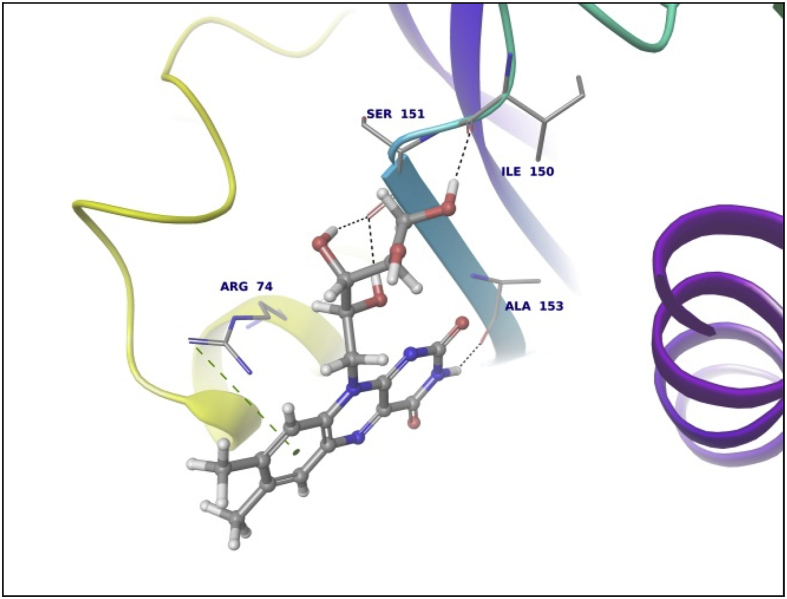
| 1 | Riboflavin | −13.553 | ARG 74 | Pi cation | 5.25 |
| Pi Pi stacking | 4.72 | ||||
| ILE 150 | HB | 1.68 | |||
| ALA 153 | HB | 1.84 | |||
| SER 151 | HB | 1.95 | |||
| HB | 2.19 | ||||
| 2 | Niacin | −11.0861 | PHE 198 | Pi-Pi Stacking | 4.93 |
| 3 | Ascorbic acid | −8.3129 | ALA 153 | HB | 2.20 |
| HB | 1.90 | ||||
| SER 151 | HB | 1.58 | |||
| HB | 1.88 | ||||
| 4 | Thiamine | −8.2065 | ALA 153 | HB | 1.53 |
| PHE 198 | Pi-Pi Stacking | 4.09 | |||
| ILE 150 | HB | 1.73 | |||
| 5 | Quercetin 3,3′ diglucoside | −6.2583 | GLY 195 | HB | 1.94 |
| ALA 153 | HB | 1.58 | |||
| ILE 191 | HB | 2.10 | |||
| HB | 2.49 | ||||
| PHE 198 | Pi – Pi Stacking | 5.06 | |||
| SER 151 | HB | 2.24 | |||
| ILE 150 | HB | 1.61 | |||
| HB | 1.75 | ||||
| 6 | Cyanidine 3, 5′ diglucoside | −4.9771 | GLU 199 | Salt Bridge | 2.92 |
| PHE 198 | Pi Pi Stacking | 4.73 | |||
| ALA 153 | HB | 2.15 | |||
| HB | 2.17 | ||||
| SER 151 | HB | 1.84 | |||
| WATER | HB | 2.43 | |||
| ILE 150 | HB | 1.81 | |||
| HB | 1.77 | ||||
| 7 | Campesterol | −4.5453 | ALA 153 | HB | 1.92 |
| 8 | Beta sitosterol | −1.7076 | ILE 191 | HB | 1.95 |
| 9 | 9 Decynoic acid | −1.6636 | ARG 74 | Salt Bridge | 4.96 |
| 10 | 8 nonynoic acid | −1.5286 | ARG 74 | Salt Bridge | 4.96 |
| 11 | Stigmasterol | −1.0801 | No interaction | ||
| 12 | Quercetin 3,4′ diglucoside | 0.3325 | GLY 155 | HB | 2.43 |
| WATER | HB | 2.17 | |||
| ALA 153 | HB | 1.88 | |||
| HB | 2.07 | ||||
| HB | 2.33 | ||||
| SER 151 | HB | 2.02 | |||
| PHE 198 | Pi Pi Stacking | 5.32 | |||
| GLY 195 | HB | 2.26 | |||
| 13 | Beta rosasterol | 0.3474 | No interaction | ||
| 14 | Methyl non-8-ynoate | 0.477 | |||
| 15 | Methyl Dec-9-ynoate | 1.0452 | |||
| 16 | Quercetin 3,7 diglucoside | 2.2611 | WATER | HB | 1.14 |
| HB | 0.61 | ||||
| HB | 2.50 | ||||
| ILE 150 | HB | 1.76 | |||
| HB | 1.66 | ||||
| SER 151 | HB | 1.76 | |||
| ARG 74 | Pi Cation | 3.84 | |||
| 17 | Methyl 8-oxooctadec-9-ynoate | 3.4243 | No Interaction | ||
| 18 | Methyl malvalate | 5.8575 | |||
| 19 | Methyl Sterculate | 6.8808 | |||
| 20 | (9) Methyl (E)-11-methoxy-9-oxononadec-10-enoate | 7.7443 | |||
| 21 | Cyanidin 3-sophoroside-5-glucoside | 8.5222 | |||
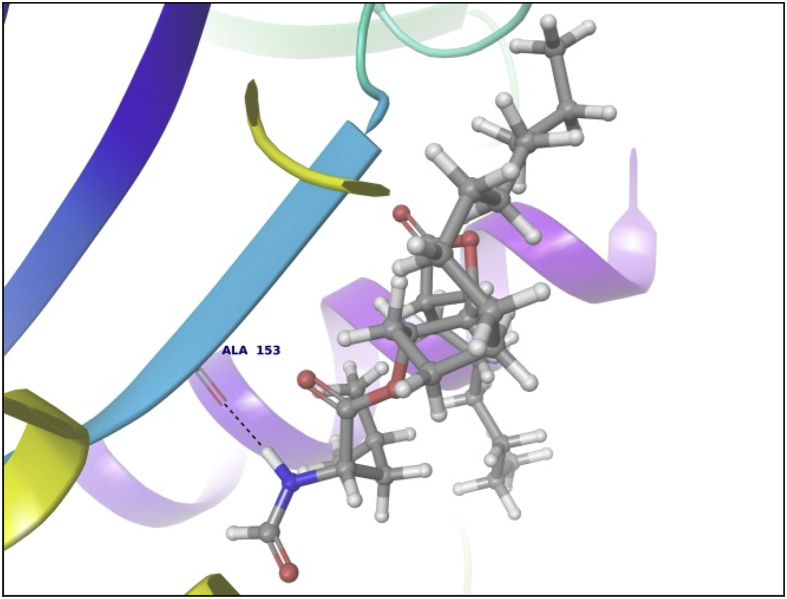
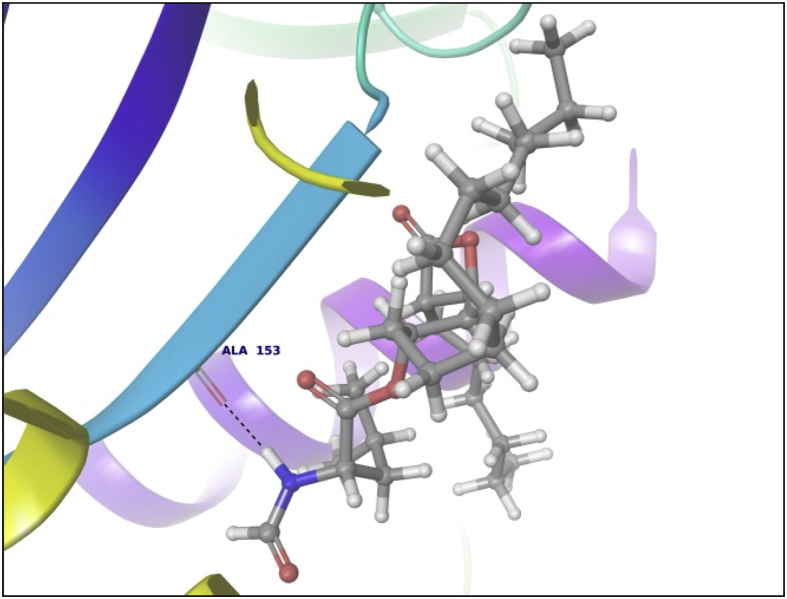
| 22 | Orlistat | 10.3508 | ALA 153 | HB | 1.86 |
Orlistat, as only standard drug used in market is used as standard reference for docking studies. Hence the docking result of orlistat in all tables is bold for ease of comparison.
Table 8.
Summary of docking analysis with ghrelin.
| Sr. No. | Ligand | Dock score | Interacting residues | Bond type | Bond angle |
|---|---|---|---|---|---|
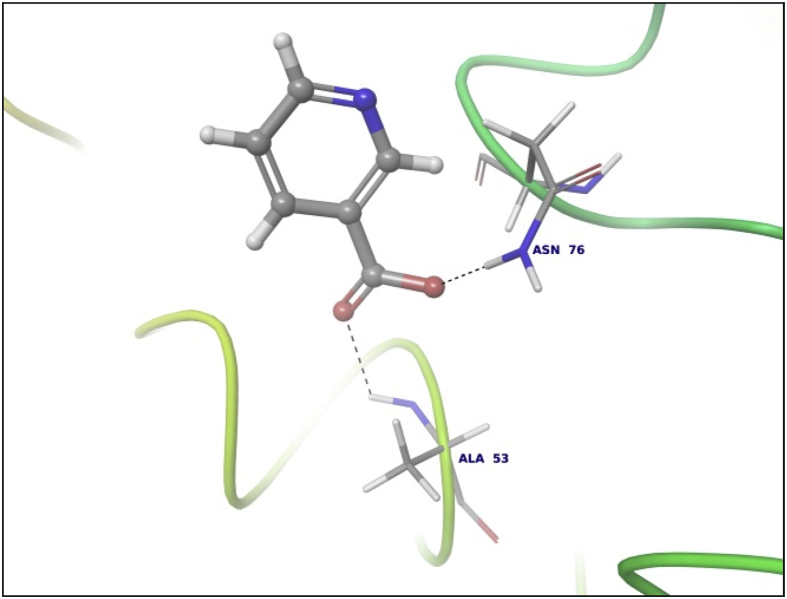
| 1 | Niacin | −11.1374 | ALA 53 | HB | 2.36 |
| ASN 76 | HB | 1.85 | |||
| 2 | Ascorbic acid | −7.2393 | PRO 49 | HB | 2.15 |
| HB | 1.86 | ||||
| GLN 36 | HB | 1.77 | |||
| ALA 77 | HB | 2.18 | |||
| 3 | Riboflavin | −7.0131 | ALA 77 | HB | 2.40 |
| GLN 36 | HB | 1.68 | |||
| HB | 1.59 | ||||
| ASN 76 | HB | 1.62 | |||
| 4 | Thiamine | −4.7344 | GLN 36 | HB | 1.77 |
| ALA 77 | HB | 2.13 | |||
| HIE 32 | Pi-Pi Stacking | 5.33 | |||
| 5 | 8 nonynoic acid | 1.9189 | ASN 76 | HB | 1.84 |
| 6 | 9 Decynoic acid | 2.7981 | ALA 77 | HB | 2.13 |
| 7 | Methyl non-8-ynoate | 2.9037 | No interaction | ||
| 8 | Methyl Dec-9-ynoate | 4.0035 | |||
| 9 | Campesterol | 6.9115 | |||
| 10 | Methyl 8-oxooctadec-9-ynoate | 8.7284 | GLU 36 | HB | 2.20 |
| ASN 76 | HB | 1.92 | |||
| 11 | Methyl malvalate | 11.8293 | ALA 77 | HB | 2.18 |
| 12 | Methyl Sterculate | 12.0917 | No interaction | ||
| 13 | Beta sitosterol | 12.2015 | |||
| 14 | (9) Methyl (E)-11-methoxy-9-oxononadec-10-enoate | 13.2915 | |||
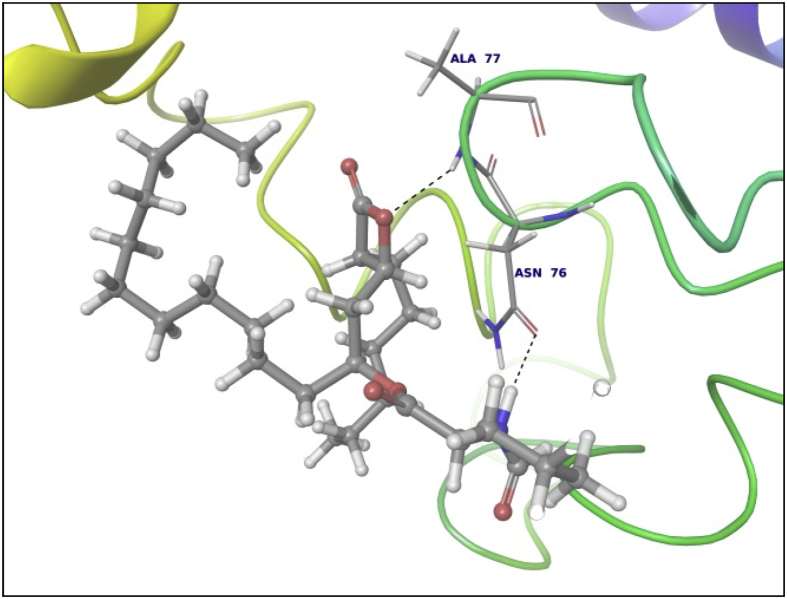
| 15 | Orlistat | 15.8166 | ASN 76 | HB | 1.65 |
| ALA 77 | HB | 2.20 | |||
Orlistat, as only standard drug used in market is used as standard reference for docking studies. Hence the docking result of orlistat in all tables is bold for ease of comparison.
Table 9.
Summary of docking analysis with MCH1.
| Sr. No. | Ligand | Dock score | Interacting residues | Bond type | Bond angle |
|---|---|---|---|---|---|
| 1 | Quercetin 3,3′ diglucoside | −13.7266 | ASP 91 | HB | 1.96 |
| HB | 1.74 | ||||
| GLY 80 | HB | 1.76 | |||
| GLY 18 | HB | 2.00 | |||
| SER 57 | HB | 1.73 | |||
| 2 | Riboflavin | −12.9742 | GLY 18 | HB | 2.19 |
| HB | 2.10 | ||||
| SER 87 | HB | 2.36 | |||
| SER 57 | HB | 1.55 | |||
| 3 | Thiamine | −9.527 | LEU 16 | HB | 1.82 |
| HB | 1.90 | ||||
| GLU 54 | Salt Bridge | 4.99 | |||
| HB | 1.90 | ||||
| 4 | Quercetin 3,7 diglucoside | −8.7967 | VAL 3 | HB | 2.02 |
| LEU 76 | HB | 1.63 | |||
| ACE 0 | HB | 2.10 | |||
| GLU 80 | HB | 2.24 | |||
| ASP 91 | HB | 2.35 | |||
| 5 | Cyanidine 3, 5′ diglucoside | −8.3388 | LEU 76 | HB | 1.68 |
| ACE 0 | HB | 1.63 | |||
| HB | 1.51 | ||||
| VAL 3 | HB | 2.31 | |||
| ASP 91 | HB | 1.55 | |||
| HB | 1.58 | ||||
| SER 87 | HB | 1.76 | |||
| GLY 18 | HB | 2.31 | |||
| 6 | Ascorbic acid | −7.7733 | SER 57 | HB | 2.19 |
| HB | 1.80 | ||||
| GLU 54 | HB | 1.76 | |||
| HB | 1.65 | ||||
| 7 | Cyanidin 3-sophoroside-5-glucoside | −5.7144 | LEU 76 | HB | 1.70 |
| ASP 91 | HB | 1.62 | |||
| HB | 1.65 | ||||
| GLY 18 | HB | 1.73 | |||
| ACE 0 | HB | 1.92 | |||
| HB | 1.72 | ||||
| 8 | Quercetin 3,4′ diglucoside | −5.236 | GLY 15 | HB | 2.09 |
| VAL 14 | HB | 2.35 | |||
| SER 57 | HB | 1.74 | |||
| SER 87 | HB | 2.20 | |||
| ASP 91 | HB | 1.56 | |||
| HB | 1.42 | ||||
| 9 | Niacin | −5.127 | VAL 3 | HB | 1.84 |
| 10 | Campesterol | −0.843 | GLU 54 | HB | 2.16 |
| 11 | Stigmasterol | −0.787 | GLU 54 | HB | 1.93 |
| 12 | Beta sitosterol | 1.849 | GLU 54 | HB | 2.00 |
| 13 | Beta rosasterol | 2.848 | No interaction | ||
| 14 | 8 nonynoic acid | 3.749 | VAL 3 | HB | 1.84 |
| 15 | 9 Decynoic acid | 4.120 | VAL 3 | HB | 1.84 |
| 16 | Methyl Dec-9-ynoate | 5.030 | VAL 3 | HB | 1.89 |
| 17 | Methyl non-8-ynoate | 5.193 | VAL 3 | HB | 1.89 |
| 18 | (9) Methyl (E)-11-methoxy-9-oxononadec-10-enoate | 8.373 | TRP 61 | HB | 1.75 |
| 19 | Methyl 8-oxooctadec-9-ynoate | 8.384 | SER 2 | HB | 2.03 |
| 20 | VAL 3 | HB | 1.89 | ||
| 21 | Methyl malvalate | 11.003 | VAL 3 | HB | 1.89 |
| 22 | Methyl Sterculate | 11.447 | VAL 3 | HB | 1.89 |
| 23 | Orlistat | 11.712 | Glu 54 | HB | 2.09 |
Orlistat, as only standard drug used in market is used as standard reference for docking studies. Hence the docking result of orlistat in all tables is bold for ease of comparison.
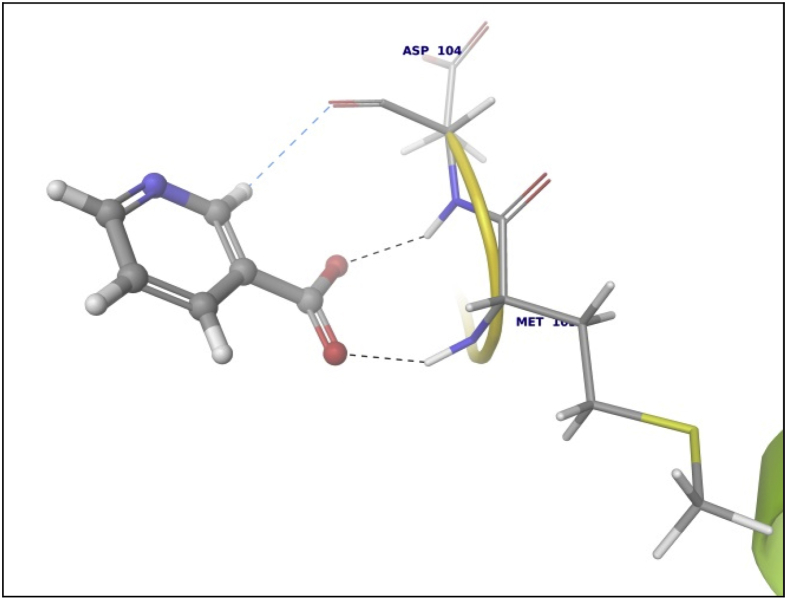
Fig. 1.
1LPB interaction with Niacin.
2. Experimental design, materials, and methods
2.1. Ligand preparation
Twenty two phytoconstituents present in Hibiscus rosa-sinensis were selected. Structures of all phytoconstituents were downloaded from PubChem database. Orlistat (PubChem CID 3034010) only available synthetic drug was used as reference standard.
2.2. Energy minimization
All structures were subjected to energy minimization using Avogadro software where universal force field (UFF) and first order steepest descent algorithm were used. This gave energetically stable conformations for the structures. Avogadro is free open source molecular builder software used for molecular modeling. It calculates the lowest energy conformation from the bond lengths and bond angles with smallest steric energy. Energy minimization helps in attaining structure conformation with lower delta G values which is considered close to biological system.
2.3. Retrieval of protein structure and preparation
Seven targets which play important role in maintaining energy balance of body and thus address obesity were selected. Protein structures of ligands were downloaded from the RCSB Protein Data Bank, database for 3D structures of large biological molecules, including proteins and nucleic acids. Downloaded protein structures were prepared X ray crystal structure of PDB ID 1LPB, 3LFM, 3TGZ, 1AX8, 4XWX for pancreatic lipase [2], FTO protein [3], cannabinoid receptor [4], hormones leptin [5] and protein SCH1 [6] respectively were selected. Data summarized in Table 1.
X- Ray crystal structure for Ghrelin [7] and MCH1 [8] receptor is not available in PDB databank so model protein structure was created using I-TASSER server online. FASTA sequence was taken from Uniprot ID of protein and submitted for model preparation. Table 2 summarizes FASTA sequence of Ghrelin and MCH1. Model was evaluated for C-score, TM score and RMSD. Model with C-score between −5 and 2, TM score greater than 0.5 were selected. Finalized model were validated on PROSA, Saves v5.0, Ramachandran plot and ProQ and then were used as receptors.
2.4. Molecular docking studies
Molecular docking techniques dock small molecules into the protein binding site. In order to understand how these ligands bind to the enzyme, docking analysis were performed using FlexX software. The receptor ligand interactions were done using Maestro software. Interacting amino acid residue, bond type and bond distance were noted.
Data summarized in Table 3, Table 4, Table 5, Table 6, Table 7, Table 8, Table 9 and Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7, Fig. 8, Fig. 9, Fig. 10, Fig. 11, Fig. 12, Fig. 13, Fig. 14.
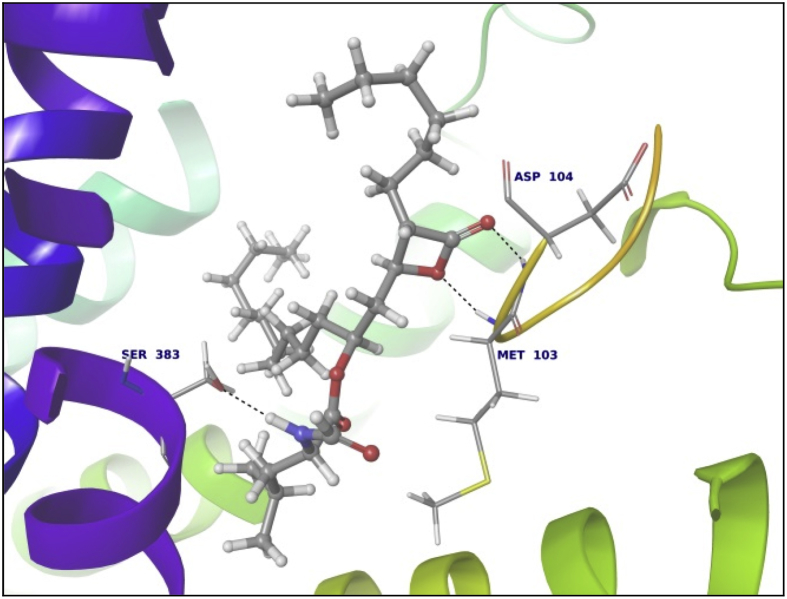
Fig. 2.
1LPB interaction with Orlistat.
Fig. 3.
3LFM interaction with Riboflavin.
Fig. 4.
3LFM interaction with Orlistat.
Fig. 5.
3TGZ interaction with Niacin.
Fig. 6.
3TGZ interaction with Orlistat.
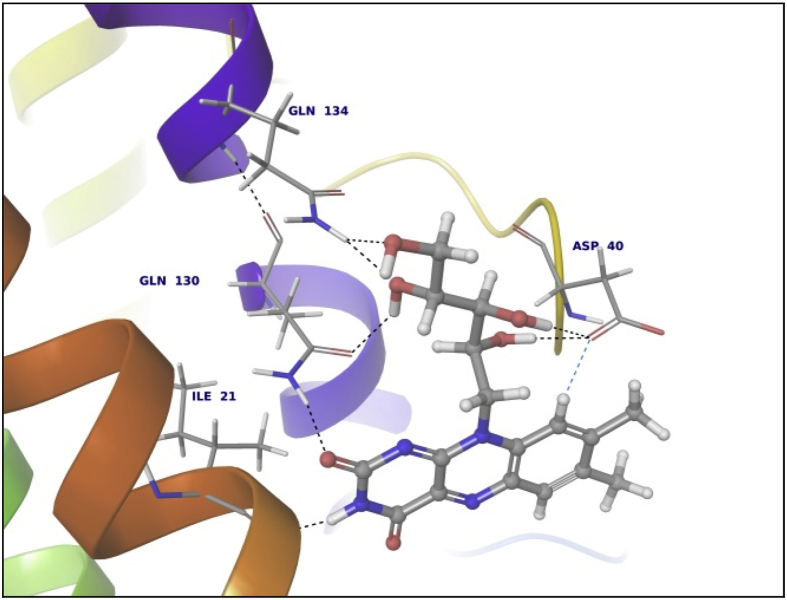
Fig. 7.
1AX8 interaction with Riboflavin.
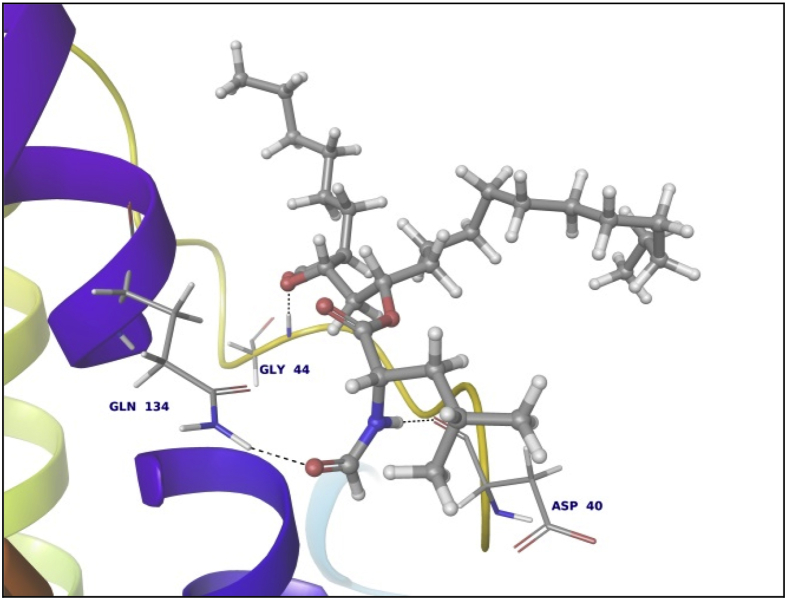
Fig. 8.
1AX8 interaction with Orlistat.
Fig. 9.
4XWX interaction with Riboflavin.
Fig. 10.
4XWX interaction with Orlistat.
Fig. 11.
Ghrelin interaction with Niacin.
Fig. 12.
Ghrelin interaction with Orlistat.
Fig. 13.
MCH1 interaction with Riboflavin.
Fig. 14.
MCH1 interaction with Orlistat.
Acknowledgments
The work was supported by Dr. D. Y. Patil Biotechnology & Bioinformatics Institute, Tathawade, Pune.
Footnotes
Transparency document associated with this article can be found in the online version at https://doi.org/10.1016/j.dib.2019.103994.
Transparency document
The following is the transparency document related to this article:
References
- 1.Min K.H., Yoo J., Park H. Computer-aided identification of ligands for GPCR anti-obesity targets. Curr. Top. Med. Chem. 2009;9:539–553. doi: 10.2174/156802609788897871. [DOI] [PubMed] [Google Scholar]
- 2.Sankar V., Engels S.M. Synthesis, biological evaluation, molecular docking and in silico ADME studies of phenacyl esters of N-phthaloyl amino acids as pancreatic lipase inhibitors. FJPS. 2018;4:276–283. [Google Scholar]
- 3.Fawcett K.A., Barroso I. The genetics of obesity: FTO leads the way. Trends Genet. 2010;26:266–274. doi: 10.1016/j.tig.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luigi B., Giacomo M., Valentina V., Renato P., Uberto P. Cannabinoid receptors as therapeutic targets for obesity and metabolic diseases. Curr. Opin. Pharmacol. 2006;6:586–591. doi: 10.1016/j.coph.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Tutone M., Lauria A., Almerico A.M. Leptin and the ob-receptor as anti-obesity target: recent in silico advances in the comprehension of the protein-protein interaction and rational drug design of antiobesity lead compounds. Curr. Pharmaceut. Des. 2014;20:001–010. doi: 10.2174/13816128113196660743. [DOI] [PubMed] [Google Scholar]
- 6.Tomilov A., Bettaieb A., Kim K., Sahdeo S., Tomilova N., Lam A., Hagopian K., Connell M., Fong J., Rowland D., Griffey S., Ramsey J., Haj F., Cortopassi G. SHC depletion stimulates brown fat activity in vivo and in vitro. Aging Cell. 2014;13:1049–1058. doi: 10.1111/acel.12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cummings D.E., Shannon M.H. Roles for ghrelin in the regulation of appetite and body weight. Arch. Surg. 2003;138:389–396. doi: 10.1001/archsurg.138.4.389. [DOI] [PubMed] [Google Scholar]
- 8.Sakurai T., Ogawa K., Ishihara Y., Kasai S., Nakayama M. The MCH1 receptor, an anti-obesity target, is allosterically inhibited by 8-methylquinoline derivatives possessing subnanomolar binding and long residence times. Br. J. Pharmacol. 2014;171:1287–1298. doi: 10.1111/bph.12529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.