Abstract
Homing peptides to the spinal cord were identified and isolated using phage display technology. In vivo biopanning was performed by intravenous systemic injection of a phage library to screen specific peptides targeting the spinal cord of mice. Analyses of the sequences of targeted phages yielded two candidate peptides targeting the spinal cord: SP1 (C-LHQSPHI-C) and SP2 (C-PTNNPRS-C). These peptides were synthesized and intravenously injected into mice to evaluate their tissue specificity and potential as gene delivery carriers. The complexes between SP1 or SP2 peptides and the plasmid vector expressing the reporter gene could induce gene transduction in the spinal cord through systemic injection without gene expression in the brain, liver, and kidney. In addition, intravenous injection of the complex between SP1 and the vectors induced interleukin-4 expression in the spinal cord, resulting in effective suppression of lipopolysaccharide-induced hyperalgesia. Therefore, intravenously administered spinal cord homing peptides complexed with a plasmid vector provided tissue-specific treatment featuring gene delivery to the CNS through systemic circulation. This novel method of gene delivery is feasible and has great potential for clinical application.
Keywords: homing peptides, gene delivery, spinal cord, systemic circulation, targeting
Graphical Abstract
Introduction
Homing peptides to many kinds of tissues have been identified, and cell- or tissue-specific delivery of drugs and genes has been anticipated when they are coupled together.1, 2 These peptides are typically very short, consisting of 3–12 amino acids.1, 3 However, these peptides may have great potential value, given their avid affinity and specificity against target ligands, such as the arginine-glycine-aspartic acid (RGD) motif against αv integrins.4, 5 Insertion of homing peptides into virus vectors to produce tissue-specific targeting virus vectors for gene therapy have improved the transduction efficacy by over 100 times.6, 7 These homing peptides were identified by screening with in vivo or in vitro phage display technology using M13 filamentous phages as the platform for the phage library, which displayed short random peptides in minor coat protein (pIII).8 Cell- or tissue-specific binding peptides have been isolated by several repeats of a procedure called biopanning with phage.2
Previously, we identified the successful targeting of homing peptides to the neurons in the dorsal root ganglion (DRG) in mice.9 Three kinds of DRG homing peptides were used, which recognized different sizes of neurons.9 The peptides were inserted into helper-dependent adenovirus vectors (termed gutless adenovirus vectors) and developed for clinical use. The result was a novel technology of DRG-targeted tissue-specific gene therapy.6 In another study, homing peptides to microglia and astrocytes were also identified and applied for the delivery of small interfering RNA (siRNA) to treat neuropathic pain in a mouse model.10 Although homing peptides have great potential applicability and could become powerful tools for drug and gene delivery, these experiments showed successful results of gene therapy only by the intrathecal route of administration.6, 10 Intrathecal injection after lumbar puncture is routinely performed clinically. However, a less-invasive route, such as intravenous injection, is desirable for patients, especially if repeated injection is required for therapy.
In this study, phage display technology was applied to identify the specific peptide motif that recognized the spinal cord through the systemic circulation in mice. These peptides were combined with plasmid vectors for a gene delivery trial. In particular, we performed gene therapy for inflammation-induced allodynia by the delivery of interleukin (IL)-4 expression vector with homing peptides, since IL-4 has been reported to be effective for allodynia.11, 12, 13 The spinal cord is a potential target for the treatment of motor neuron diseases, spinal injury, spastic paraplegia, multiple sclerosis, sensory ataxia, and neuropathic pain.14, 15, 16, 17, 18, 19
In this study, the demonstrated delivery of vectors containing homing peptides to the spinal cord through the systemic circulation verified the potential value of homing peptides for disease treatment in combination with therapeutic genes.
Results
Screening of Specific Phage Homing to the Spinal Cord by Phage Display
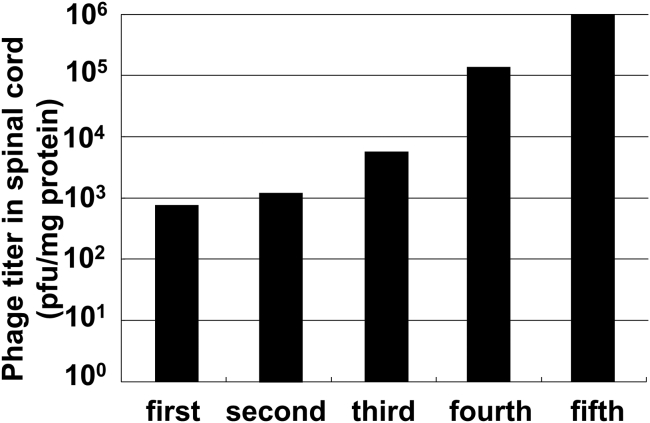
In the first biopanning of in vivo phage display, phages were collected at an absolute titer of 103 plaque-forming units (PFU)/mg spinal cord protein (Figure 1). The biopanning was repeated five times, and the titers of the total recovered phages gradually increased with each repeat, finally reaching 106 PFU/mg spinal cord protein (Figure 1). It is likely that the phages with high affinity to the spinal cord were concentrated by the in vivo phage display, because the titers gradually increased and the final titer was quite high. DNA sequence analysis of each phage displaying avid affinity to the spinal cord was performed after the third, fourth, and fifth rounds of biopanning.
Figure 1.
Titer of Total Recovered Phage in the Spinal Cord
Bars show the numbers of recovered phages per milligram of protein weight of the spinal cord in each biopanning round. In each round, the phages contained in the spinal cords of 3–5 mice were combined after the injection of 1011 plaque-forming units (PFU) of phage into each mouse.
The DNA sequences coding the SP1 (C-LHQSPHI-C) peptide were observed four times among 44 phage plaques in the third round (9.1%), eight times among 49 phage plaques in the fourth round (16.3%), and 31 times among 47 phage plaques in the fifth round (66.0%). The SP1 population gradually increased from the third to the fifth rounds of biopanning, and the final frequency was the highest of the phages examined (Table 1; Tables S1 and S2). The second most frequent peptide sequence was SP2 (C-PTNNPRS-C), which was observed 12 times among 47 phage plaques in the fifth round (25.5%). As with the SP1 peptide, the frequency of the SP2 peptide gradually increased (Table 1; Tables S1 and S2). The other three peptides (SP3–SP5) were observed among 47 phage plaques in the fifth round, but only once or twice (Table 1). SP1 and SP2 were considered candidate homing peptides specific for the spinal cord.
Table 1.
Amino Acid Sequences and Phage Plaque Frequencies of Peptides that Specifically Home to Spinal Cord in the Fifth Round of Biopanning
| Homing Peptide | Amino Acid Sequence | Frequency (/Total) |
|---|---|---|
| SP1 | C-LHQSPHI-C | 31/47 |
| SP2 | C-PTNNPRS-C | 12/47 |
| SP3 | C-NMRTLMQ-C | 2/47 |
| SP4 | C-DMHQGKT-C | 1/47 |
| SP5 | C-SPDKNRS-C | 1/47 |
High Affinity and Specificity of SP1 and SP2 Peptides to the Spinal Cord
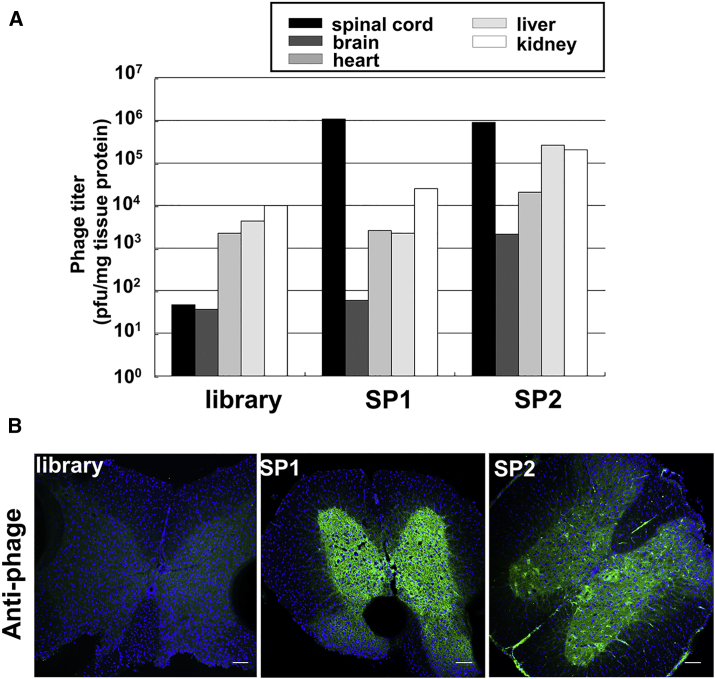
To demonstrate the spinal cord specificity of SP1 and SP2, each phage expressing SP1 or SP2 was individually amplified to 10–11 PFU and injected into the tail veins of C57BL/6 mice. Subsequently, the spinal cord, brain, and several organs were isolated, and the titer of the inclusive phages was measured and compared with the phage library (Figure 2A). In the SP1 group, a large number of phages accumulated in the spinal cord. The phage numbers were approximately 100–1,000 times higher compared to that in the brain and other organs with abundant blood flow. In addition, the accumulation of the injected SP2 phages in the spinal cord was 10–500 times greater than that in the brain and other organs (Figure 2A). In addition, an affinity of over 104-fold to the spinal cord was observed in both the SP1 and SP2 groups against the library injection group. These phages mainly accumulated in the gray matter of the spinal cord (Figure 2B). Immunohistochemistry showed that the distribution of phage antibodies was similar between SP1 and SP2 phages (Figure 2B). These results suggest that the SP1 and SP2 peptides have high affinity and specificity to the spinal cord.
Figure 2.
Comparison of Recovered Phage Titers with Other Organs and Localization of Phages in the Spinal Cord
(A) Phage titers in multiple organs after intravenous injection of phages. Bars show the numbers of recovered phages per milligram of protein weight of each organ after the injection of library, SP1, or SP2 phage of 1011 PFU. (B) Immunohistochemistry with anti-phage antibodies in spinal cord sections from C57BL/6 mice after the injection of 1011 library, SP1, or SP2 phage. Scale bars, 100 μm.
Characterization of Spinal Cord and Cell Targeting with SP1 and SP2 Peptides
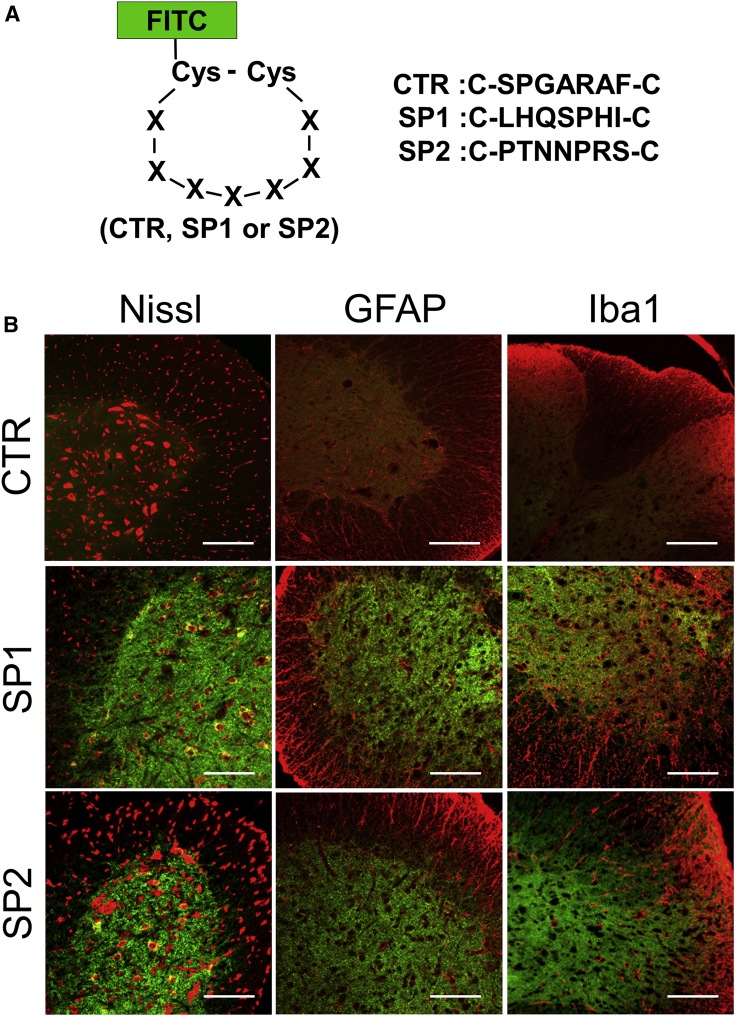
The SP1 and SP2 peptides were synthesized and applied to mice and cultured cells to clarify as to what type of cells each peptide had a high affinity (Figures 3 and 4). Control (CTR [C-SPGARAF-C]), SP1, and SP2 peptides were labeled with fluorescein isothiocyanate (FITC) for the localization analysis (Figure 3A). The FITC-labeled peptides were separately injected into the mice through the tail vein. After systemic circulation of the peptides, the spinal cord was isolated and the localization was analyzed with a marker stain (Figure 3B). FITC signaling from both SP1 and SP2 peptides was mainly observed in gray matter areas of the spinal cord, which mainly overlapped with the Nissl stain as a neuronal marker (Figure 3B).
Figure 3.
In Vivo Distribution of SP1 and SP2 Peptides in the Spinal Cord
(A) Scheme of the control (CTR), SP1, or SP2 peptide structure labeled with FITC and their amino acid sequences. Cys, cysteine; X, amino acid. (B) CTR, SP1, and SP2 peptide distribution in spinal cords after the injection of 3 μg CTR, SP1, or SP2 peptide. Peptides are shown in green. Red color indicates staining with Nissl or immunohistochemistry with anti-GFAP or anti-Iba1 antibodies. Scale bars, 100 μm.
Figure 4.
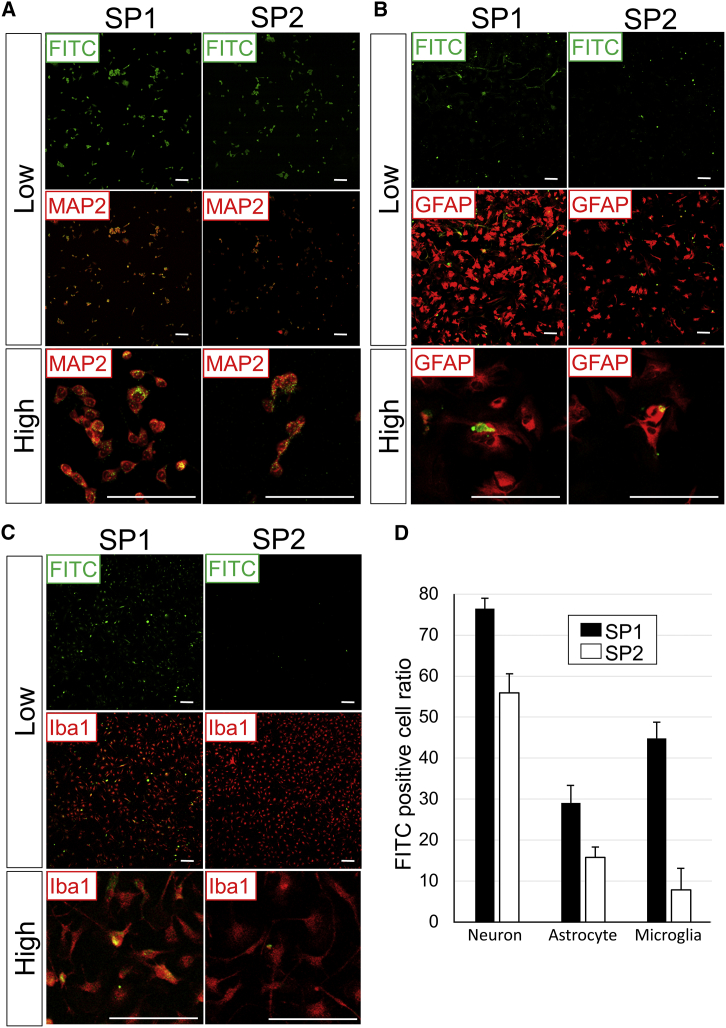
Characterization of the Cell Type Targeted with SP1 and SP2 Peptides
1 μg SP1 or SP2 peptide labeled with FITC was separately incubated with NSC-34 cells, astrocytes, or microglia. (A) SP1 and SP2 peptide distribution (green) and MAP2 immunocytochemistry (red) in NSC-34 cells. (B) SP1 and SP2 peptide distribution (green) and GFAP immunocytochemistry (red) in the primary culture of astrocytes. (C) SP1 and SP2 peptide distribution (green) and Iba1 immunocytochemistry (red) in the primary culture of microglia. (D) Percentage of the cell population labeled with spinal cord homing peptides (SP1 and SP2). Low, low magnification; high, high magnification. Scale bars, 100 μm (A–C). Bars show means ± SD in (D).
In the in vitro studies, the SP1 and SP2 peptides penetrated NSC-34 motor neuron cells at a moderate or high frequency (Figures 4A and 4D). These two peptides were also incubated with primary cultured astrocytes or microglia (Figures 4B and 4C). The SP1 peptide localized in both cell types at frequencies of 29% and 45%, respectively (Figures 4B–4D). In contrast, the SP2 peptide was observed only at low frequencies (16% and 8%, respectively) in the primary culture of astrocytes and microglia (Figures 4B–4D).
Production of Complexes between Peptides and Vectors for Gene Delivery and Their Transduction Efficacy in Cultured Cells
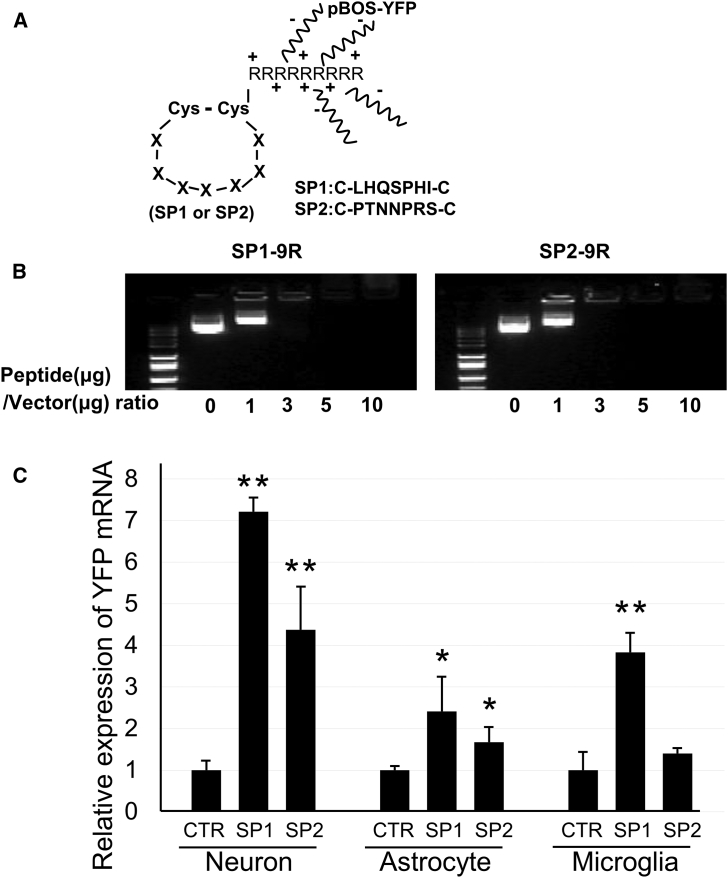
Next, the SP1 and SP2 peptides were applied for tissue-specific gene delivery. Both peptides were synthesized and combined with nine arginine residues (R) at the C-terminal end to harbor positive electric charges (SP1-9R [SP1(C-LHQSPHI-C) + GGG + 9R] and SP2-9R [SP2(C-PTNNPRS-C) + GGG + 9R]) (Figure 5A). To confirm that these peptides were potentially useful for gene delivery, gene delivery of yellow fluorescein protein (YFP)-expressing plasmids with SP1 or SP2 peptides was assessed. Either SP1-9R or SP2-9R was mixed with pBOS-YFP plasmids. Bonding occurred due to electrostatic interaction, because of the negative electric charges of the plasmids related to their constituent nucleic acids (Figure 5A). Mixtures between peptides and plasmids were prepared using several molecular weight ratios to determine the appropriate molecular weight ratio for gene therapy (Figure 5B).
Figure 5.
Design and Stability of Complexes between Peptides and Plasmid Vectors
(A) Scheme for peptide structure and binding of peptides with vectors. Cys, cysteine; R, arginine; X, amino acid; pBOS-YFP, YFP expression vectors driven by the BOS promoter. (B) Electrophoresis of complexes between peptides (SP1-9R or SP2-9R) and vectors at several patterns of molecular weight ratio. 0, 1, 3, 5, and 10 indicate the peptide:plasmid vector ratios of 0:1, 1:1, 3:1, 5:1, and 10:1. (C) Expressions of YFP mRNA in neuron (n = 6/group), astrocyte (n = 6/group), and microglia (n = 6/group) were analyzed by qRT-PCR after transduction with BOS-YFP vector control (CTR), SP1 + BOS-YFP (SP1), or SP2 + BOS-YFP (SP2). YFP mRNA expressions were standardized by β-actin mRNA expression, and their relative expression ratios in SP1 and SP2 groups were calculated against each CTR group. Bars show means ± SD in (C). *p < 0.05 and **p < 0.01 versus the CTR group.
In the SP1-9R group, the binding plasmids (pBOS-YFP) with SP1-9R peptides were loaded at peptide:plasmid vector ratios of 0:1 (0), 1:1 (1), 3:1 (3), 5:1 (5), and 10:1 (10). Of these, broad weak bands were evident at the 3:1 ratio in agarose gels after electrophoresis (Figure 5B, lane 3 at left). Non-binding plasmids with SP1-9R peptides showed bright and sharp bands (Figure 5B, lanes 0 and 1 at left). Similarly, the binding plasmids with SP2-9R peptides were loaded at peptide:plasmid vector ratios of 0:1 (0), 1:1 (1), 3:1 (3), 5:1 (5), and 10:1 (10). The 3:1 ratio produced broad, very weak bands in the SP2-9R + plasmid group (Figure 5B, lane 3 at right). Therefore, the 3:1 molecular weight ratio was defined as the appropriate mixture condition of the two kinds of peptide-plasmid complexes for in vivo studies.
Next, the complexes were applied to NSC-34 neuronal cells, primary cultures of astrocytes, and microglia (Figure 5C). A high level of YFP mRNA expression was observed in neuronal cells for both the SP1 and SP2 groups. In addition, these groups showed significantly higher expression of the YFP gene than the CTR group in astrocytes. In microglia, only the SP1 group produced a significantly higher result than the CTR group. In all three cultured cells, the transduction efficacy of the YFP gene showed higher in the SP1 group than in the SP2 group (Figure 5C).
Gene Delivery of Plasmid Vectors with Homing Peptides to the Spinal Cord in Mice
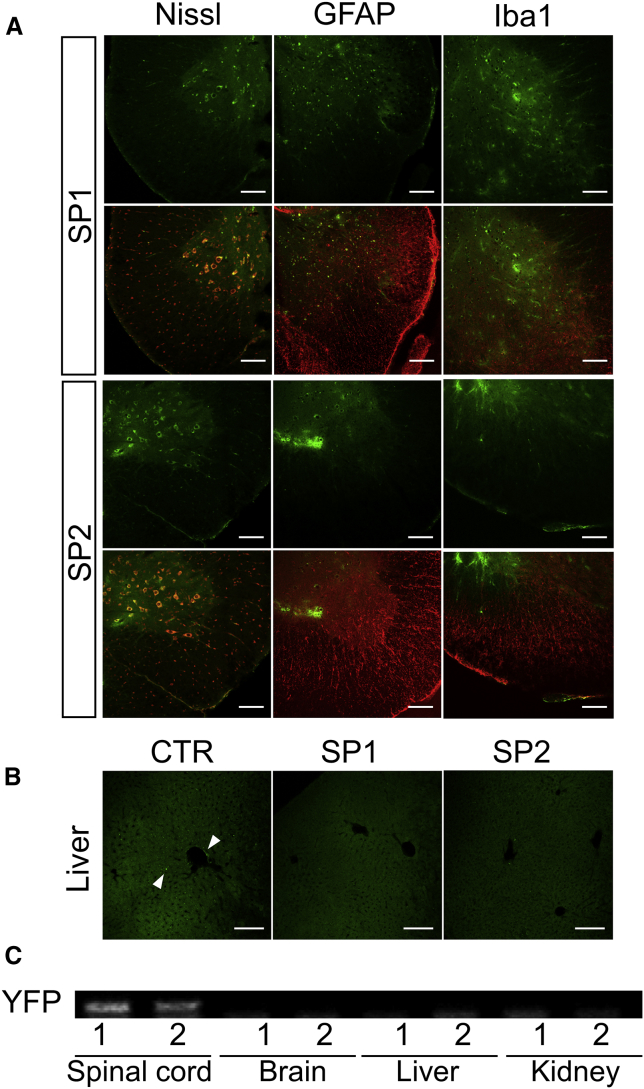
Gene delivery of pBOS-YFP vectors with SP1 or SP2 peptides was performed in wild-type C57BL/6 mice. Sections were prepared from mouse spinal cords 3 days after injection of the peptide-plasmid complexes through the tail vein. Histological analysis of spinal cord sections was performed by Nissl stain, glial fibrillary acidic protein (GFAP), or ionized calcium binding adaptor molecule 1 (Iba1) immunostaining (Figure 6A). YFP signals were mainly observed in white matter lesions of the spinal cord, and they were merged with the Nissl stain in both the SP1 and SP2 groups (Figure 6A). However, the YFP signal was not observed in the vector-only group (Figure S1). Liver sections were observed as a representative control of organs with abundant blood flow (Figure 6B). In liver sections, only YFP-positive spots were observed mainly in the CTR group and rarely in the SP1 and SP2 groups (Figure 6B). In addition, mRNA expression of the YFP gene was analyzed by RT-PCR with mRNA from the spinal cord and other organs (Figure 6C). Expression of mRNA of the YFP gene was detected in the tissues of the spinal cord in the SP1 and SP2 groups (Figure 6C). These results suggested that the SP1 and SP2 peptides have great potential to deliver genes transvenously and to target plasmid vectors specifically to the spinal cord.
Figure 6.
Transduction Efficiency of Gene Delivery with Homing Peptides to the Spinal Cord
(A) Histological analysis of YFP signal (green) with a marker stain by Nissl, GFAP, or Iba1 (red) in spinal cord tissues 3 days after the injection of SP1 + pBOS-YFP (SP1) or SP2 + pBOS-YFP (SP2). Scale bars, 100 μm. (B) Histological analysis of YFP signal (green) in liver sections 3 days after the injection of BOS-YFP only (CTR), SP1, or SP2. Arrowheads show the positive signal of YFP. Scale bars, 100 μm. (C) RT-PCR of the YFP gene in the spinal cord, brain, liver, and kidney from C57BL/6 mice at 3 days after the injection of SP1 + pBOS-YFP (1) or SP2 + pBOS-YFP (2).
Gene Therapy for Inflammation-Induced Mechanical Allodynia by IL-4-Expressing Vectors with SP1 Homing Peptides to the Spinal Cord in Mice
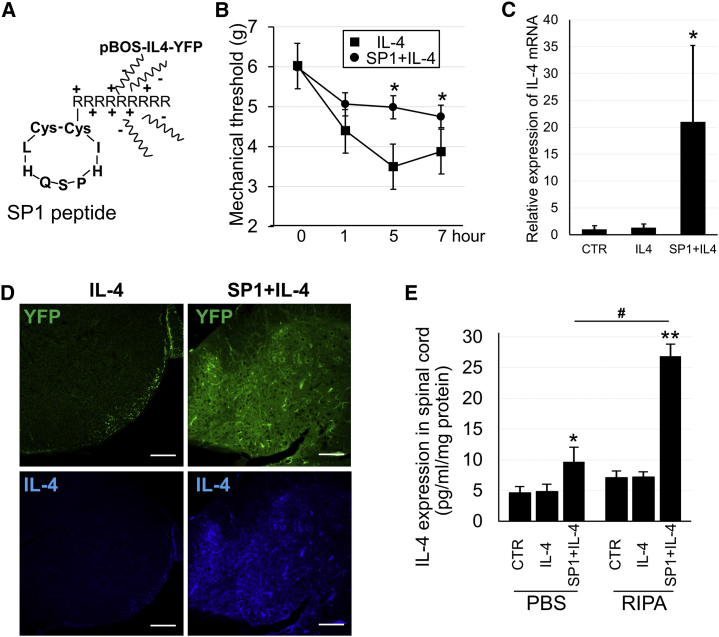
Gene therapy was performed in a mouse model of lipopolysaccharide (LPS)-induced mechanical allodynia by the overexpression of IL-4 using the SP1 peptide + pBOS-IL-4-YFP vector complex (Figure 7A). The SP1 peptide + pBOS-IL-4-YFP vector complex was intravenously injected into mice 2 days before the induction of mechanical allodynia. Upon intrathecal injection of LPS, a reduction in the mechanical threshold against allodynia was observed with a planter test at 5 and 7 h, and the effects of gene therapy were evaluated by the mechanical threshold level 1–6 h after LPS injection (Figure 7B).
Figure 7.
Gene Therapy for Neuropathic Pain by IL-4 Expression with SP1 Homing Peptides to the Spinal Cord
(A) Scheme of SP1 peptide structure and binding of peptides with vectors. (B) Mechanical threshold of the hind paw before and after LPS injection with gene therapy in pBOS-IL4-YFP (IL-4, n = 7) and SP1 + pBOS-IL-4-YFP (SP1 + IL-4, n = 8) groups. (C) mRNA expression of IL-4 in the spinal cord after gene therapy by pBOS-YFP (CTR, n = 5), pBOS-IL4-YFP (IL-4, n = 5), or SP1 + pBOS-IL-4-YFP (SP1 + IL-4, n = 5). IL-4 mRNA expressions were standardized by GAPDH mRNA expression, and their relative expression ratios were calculated against the CTR group. (D) Immunohistochemistry of IL-4 in the spinal cord after gene therapy. Top shows YFP (green) and bottom shows IL-4 staining (blue). Scale bars, 100 μm. (E) IL-4 protein expression by ELISA analysis in spinal cord homogenized in PBS or RIPA buffer after the injection of pBOS-YFP (CTR, n = 5), pBOS-IL-4-YFP (IL-4, n = 5), or SP1 + pBOS-IL-4-YFP (SP1 + IL-4, n = 5). IL-4 protein contents were standardized by spinal cord protein weight. *p < 0.05 and **p < 0.01 versus others; #p < 0.01 in (E).
At 5 and 7 h after LPS injection, the mechanical threshold in the SP1 + IL-4 vector group was significantly increased compared to the threshold in the IL-4 vector-only group (Figure 7B). Expression of IL-4 mRNA in the spinal cord after behavioral tests was upregulated in the SP1 + IL-4 vector group (Figure 7C). In addition, immunohistochemistry experiments were performed using spinal cord sections from neuropathic pain model mice after gene therapy with the SP1 + IL-4 vector; IL-4 immunostaining was much stronger than that in the IL-4 vector-only group (Figure 7D).
Furthermore, to investigate whether IL-4 was secreted or not, IL-4 ELISA was performed in spinal cord homogenized in either PBS or radioimmunoprecipitation (RIPA) buffer (Figure 7E). This was important to determine since secreted IL-4 provides therapeutic benefits. In both PBS- and RIPA-treated spinal cords, IL-4 protein expression was significantly increased in the spinal cord of the SP1 + IL-4 group compared to the CTR and IL-4 groups (Figure 7E). Remarkably, the elevation of IL-4 protein level in the SP1 + IL-4 group compared to CTR and IL-4 groups in PBS-treated spinal cord indirectly indicated the increased secretion of IL-4 protein, because cells do not lyse in PBS, although the expression level was less than in RIPA-treated spinal cord (Figure 7E).
Both mRNA and protein levels of IL-4 were effectively expressed by the SP1-IL-4 vector complexes (Figure 7C–7E), which could lead to a reduction in neuropathic pain in the disease mouse model. These findings highlight the selective targeting ability of SP1 peptides to deliver IL-4 vectors to the spinal cord, and they indicate the potential value in the development of novel gene therapy modalities.
Discussion
Development of an effective gene therapy to the spinal cord requires the identification of homing peptides that can access the CNS through the blood-brain barrier. Therefore, in vivo biopanning of a phage library through systemic circulation was used in this study to screen peptides that target the spinal cord. We identified two homing peptides, SP1 and SP2, which were able to target the spinal cord through blood vessels. In addition, SP1 and SP2 peptides displayed a marked potential to deliver plasmid vectors to the spinal cord, which were transferred into several kinds of cells, and the delivered reporter gene was expressed in the cells. Moreover, reporter gene expression was not seen in the brain, liver, and kidneys as representative organs with abundant blood flow. This suggests the high tissue specificity of these peptides.
In molecular therapy with gene or drug delivery, some side effects are always present, especially in non-target tissues. Merits and demerits are to be considered at the time of therapy. After treatment with anti-cancer drugs, many types of side effects occur systemically because the drugs were delivered to cancer tissues and normally functioning tissues.20, 21 The administration of drugs will be restricted if side effects are not minimized. Recently, molecular target drugs or virus vectors capable of overexpressing therapeutic genes were widely developed for clinical usage.22, 23 However, these therapies also suffer from the same safety issues due to side effects. A drug delivery system (DDS) capable of effective molecular therapy with minimum side effects is needed.24
For the past decade, we have focused on homing peptides and developed novel technologies. Developing a tissue-specific DDS could realize the ideal treatment and allow the administration of potently therapeutic drugs free of undesirable side effects. We previously identified homing peptides to DRG tissues.9 A DRG peptide was inserted into helper-dependent adenovirus vectors, which delivered therapeutic genes to the DRG with high transduction efficiency.6 However, the authors reported that inflammation was induced by the virus vectors in the nervous system.
To improve this DDS, we delivered a gene from naked plasmid DNA with homing peptides. This reflected the observation that virus proteins can become antigens for immune responses in vivo.6 In the spinal cord, the SP1 homing peptide displayed a certain therapeutic effect by the delivery of plasmid DNA without virus vectors. This advantage will accelerate achievement of the safer clinical application of gene therapy.
Non-viral vectors have been explored as another strategy for plasmid vector delivery.25, 26 Cationic liposomes and polymers are classically the most representative materials. They are easily complexed with nucleic acids through electrical interactions.27 In vitro experiments revealed the high transduction efficiency of these cationic carriers. However, they are potentially toxic to cells and are unstable in vivo.28 Considering the safety aspects, biocompatible materials have been recently developed as nucleotide carriers.29, 30, 31 Neutral lipids and lipid nanoparticles are very safe, very stable in vivo, and are very likely to interact with siRNA.25, 32 Lipid nanoparticles are expected to be developed as carriers for plasmid DNA, but they do not have tropism for every tissue. In contrast, homing peptides are tissue specific and very efficiently transduce to the target organ. In addition, these homing peptides bind strongly with plasmid DNA via nine arginine residues. The use of homing peptides will be a powerful strategy for the delivery of plasmid DNA and for gene therapy in combination with lipid nanoparticles.
The route of administration of therapeutic agents is one of the most important points for effective gene therapy. Gene therapy is widely performed by subcutaneous, intramuscular, intraperitoneal, or intravenous injection or by local injection (e.g., joint injection, organ injection, or brain injection).33 In the CNS, the blood-brain barrier protects nervous tissues from external factors.34 Therefore, permeability through this barrier needs to be considered for the development of a DDS to neuronal tissues.35 In previous reports, we demonstrated DRG-targeting gene therapy or microglia-specific gene delivery in the spinal cord by intrathecal injection.6, 10 Lumbar puncture and intrathecal injection are usually performed clinically. Side effects include headache and intracranial hypotension.36 A non-invasive approach is most suitable for patients, and intravenous injection is safer than intrathecal injection. Presently, via a circulation approach dependent on the homing peptides SP1 and SP2, plasmid vector was delivered to the spinal cord and taken up by cells, and the transgene was expressed in the spinal cord. These findings are very encouraging and represent an advance in the delivery of plasmid DNA.
Materials and Methods
Animals
C57BL/6 mice were purchased from Jackson Laboratory (Bar Harbor, ME, USA). All animals were housed and provided with water and mouse chow ad libitum, and they were maintained under a 12-h light and dark cycle. All animal experimental protocols were approved by the Institutional Animal Care and Usage Committee (IACUC) at Shiga University of Medical Science, and they were performed according to the guidelines of the IACUC at Shiga University of Medical Science.
Preparation of Cultured Cells
NSC-34 cells were purchased from CELLutions Biosystems (Burlington, ON, Canada), and they were prepared as the representative neuronal cells. NSC-34 cells were maintained with 10% fetal calf serum in DMEM/F12 medium (Thermo Fisher Scientific, Waltham, MA, USA). Primary cultures of astrocytes and microglia were isolated from the brain tissue of C57BL/6 pups at days 1–3 after birth. Astrocytes were isolated by the passage of whole cells at the initial 5–7 days. After a 14-day incubation of mixed cultured cells, microglia began to grow on the astrocyte cell sheet and were isolated to another culture dish. The isolated microglia were then used for experiments as previously described.37
In Vivo Phage Display and Screening of Spinal Cord Homing Peptides
The CX7C phage library purchased from New England Biolabs (Ipswich, MA, USA) was used for in vivo phage display for the screening of spinal cord homing peptides. The protocols described by Christianson et al.38 were employed with minor modifications. The phage library was injected into C57BL/6 mice (n = 3–5 in each round of biopanning) through the tail vein at a titer of 1011 PFU in 100 μL Tris-buffered saline (TBS; 50 mM Tris-HCl [pH 7.5]). At 5 min after injections, whole spinal cords were isolated after transcardiac removal of blood. The weight of each spinal cord was measured, and each was homogenized in DMEM (Thermo Fisher Scientific) containing protease inhibitor (Sigma-Aldrich, St. Louis, MO, USA). Aliquots from 3–5 mice were combined and infected into Escherichia coli ER2738 (New England Biolabs). The number of phages included in the spinal cord was titrated as PFU. After amplification to 1011 PFU, the phages were again injected into other C57BL/6 mice. This biopanning procedure was repeated five times to select phages that specifically bound to the spinal cord. After the third to fifth rounds of biopanning, each phage genome was isolated from 44–49 phage plaques and analyzed using DNA sequence-coding pIII protein.
Analysis of Spinal Cord Binding Affinity of a Specific Phage
To analyze the binding affinity of a phage to the spinal cord, including candidate homing peptides, the two phages displaying the greatest affinity (including SP1 and SP2) were individually amplified to 1011 PFU and systemically injected through the tail veins of mice. After 5 min, the spinal cord, brain, heart, liver, and kidney were isolated from each mouse after transcardiac removal of blood. Each organ was examined to determine the phage titer.
For the histological analysis of specific phages targeting the spinal cord, the two aforementioned phages (including SP1 and SP2) were individually amplified to 1011 PFU and were systemically injected through the tail veins of mice. After 5 min, transcardiac perfusion and fixation with 4% paraformaldehyde (PFA) were performed. Sections of the spinal cord from each mouse were incubated with an anti-phage antibody (Sigma-Aldrich), and the sections were incubated with a goat anti-rabbit Alexa Fluor 488 antibody (Molecular Probes, Eugene, OR, USA). These sections were mounted with Vectashield mounting medium with DNA staining using DAPI (Vector Laboratories, Burlingame, CA, USA).
Peptides, Their Applications to Mice and Cultured Cells, and Histological Analysis
SP1 and SP2 peptides were synthesized and labeled with FITC (Invitrogen, Carlsbad, CA, USA). CTR peptide labeled with FITC, which targeted the DRG, was prepared as a negative control.6, 9 The peptides (3 μg/100 μL in PBS) were administered to mice through the tail veins. After 5 min, transcardiac perfusion and fixation with 4% PFA were performed. Some sections of the spinal cord from each mouse were treated with Nissl stain and the others were incubated with either anti-GFAP (Promega, Madison, WI, USA) or anti-Iba1 antibody (Wako Pure Chemical Industries, Osaka, Japan). Next, the sections were incubated with a goat anti-rabbit Alexa Fluor 555 antibody (Molecular Probes).
The peptides were applied to NSC-34 cells as representative neuronal cells and a primary culture of astrocytes and microglia at a concentration of 1 μg/mL in 200 μL culture medium in wells of 48-well plates. After 24 h of incubation with the peptides, the cells were fixed with 4% PFA, and immunocytochemistry assays were performed with each primary antibody. The primary antibodies were anti-microtubule-associated protein (MAP)2 antibody (Merck Millipore, Darmstadt, Germany) for neurons, anti-GFAP antibody for astrocytes, and anti-Iba1 for microglia. This was followed by species-matched Alexa Fluor-labeled secondary antibody (Molecular Probes). All cells were observed using a model C1si confocal laser microscope (Nikon, Tokyo, Japan) equipped with EZC1 3.90 software (Nikon).
Complex between Peptides and Plasmid Vectors for Gene Delivery
The SP1-9R and SP2-9R peptides were synthesized by Invitrogen. The pBOS-YFP vector was constructed to insert the YFP sequence into pLPBL-BOS plasmid vectors that contain the elongation factor-1 (EF-1) promoter, internal ribosome entry site (IRES) sequence, and rabbit β-globin polyadenylation signal on the pLPBL-1 backbone (a gift from Dr. Lawrence Chan and Dr. Kazuhiro Oka, Baylor College of Medicine, Houston, TX, USA). The complexes between peptides and vectors were formulated by electrostatic interaction, and the appropriate molecular ratios of peptide:vector were decided by visualization of those oligonucleotides with ethidium bromide under UV excitation after agarose gel electrophoresis. A final peptide:vector molecular weight ratio of 3:1 was used for the gene transduction study.
Gene Delivery of Vectors to Cultured Cells and Spinal Cord in C57BL/6 Mice
NSC-34 neuronal cells and primary cultures of astrocytes and microglia were prepared in 12-well plates 1 day prior to the experiments. Either SP1 (3 μg) + pBOS-YFP (1 μg), SP2 (3 μg) + pBOS-YFP (1 μg), or pBOS-YFP (1 μg) alone was applied to the cells in 1 mL complete medium. After 48 h, transduction effects were evaluated by measuring YFP mRNA expression by qRT-PCR (YFP, forward primer 5′-TCATGGCCGACAAGCAGA-3′ and reverse primer 5′-TCAGGTAGTGGTTGTCGGGCA-3′; and β-actin, forward primer 5′-CGTGCGTGACATCAAAGAGAA-3′ and reverse primer 5′-TGGATGCCACAGGATTCCAT-3′) of each treatment group as well as the control vector group (SP1 + pBOS-YFP, SP2 + pBOS-YFP, and pBOS-YFP alone). These in vitro gene delivery experiments were performed in triplicate.
Next, gene delivery experiments were performed on C57BL/6 adult mice. The peptide-vector complexes (30 and 10 ng/μL) in 100 μL PBS buffer were injected into the C57BL/6 mice through the tail veins. Then, 3 days later these mice were used for the analysis of gene delivery efficacy by the SP1 or SP2 peptide.
For histological analysis of gene delivery with the peptides, spinal cord and liver sections were removed after perfusion fixation with 4% PFA. Some spinal cord sections were treated with Nissl stain (Neuro Trace 435/455; Molecular Probes) and the others were incubated with anti-GFAP antibody or anti-Iba1 as a primary antibody overnight at 4°C. After washing to remove unbound antibody, sections were incubated with goat anti-rabbit Alexa Fluor 555 secondary antibody (Molecular Probes). Liver sections from the SP1, SP2, or control group were prepared as the negative control. All sections were observed by confocal laser microscopy using the aforementioned microscope and software.
RT-PCR Analysis of Delivery Gene
After the gene delivery experiments were performed on C57BL/6 adult mice with SP1-pBOS-YFP or SP2-pBOS-YFP, the spinal cord, brain, liver, and kidneys were obtained at day 3. Total mRNA was purified from each organ, and RT-PCR was performed for the YFP gene with forward and reverse primers as described above. Following agarose gel electrophoresis of the PCR products, the oligonucleotide bands were visualized by ethidium bromide under UV excitation.
Gene Therapy by pBOS-IL-4-YFP Vector with SP1 Peptide for Inflammation-Induced Mechanical Allodynia
The IL-4 cDNA was cloned from C57BL/6 mouse bone marrow into pCR2.1-TOPO (Invitrogen) by RT-PCR using the following primers: 5′ upstream, 5′-ATGGGTCTCAACCCCCAGCTA-3′; and 3′ downstream, 5′-CTACGAGTAATCCATTTGCA-3′. IL-4 cDNA with the Kozak sequence (CCACC) was subcloned into the pBOS-YFP vector, and the pBOS-IL-4-YFP vector was constructed.
Gene therapy experiments were performed using a mouse model of inflammatory mechanical allodynia induced by LPS injection into the intrathecal space. Empty vector control (pBOS-YFP), IL-4 vector control (pBOS-IL-4-YFP), and the peptide-vector complexes (SP1-pBOS-IL-4-YFP) were prepared as a 100 μL PBS solution containing the plasmid at a concentration of 50 ng/μL and the peptide at a concentration of zero or 150 ng/μL. The vectors or complexes were injected once into each mouse through the tail vein 2 days before the induction of allodynia by LPS. Planter tests were performed before and after LPS injection to evaluate the mechanical threshold on the ipsilateral side.
At 7 h after LPS injection, a subset of mice was used to analyze the efficacy of gene delivery by SP1 peptides. Total mRNA was purified from L1–L5 spinal cord tissue, and qRT-PCR was performed to measure transcript levels of the IL-4 gene (forward primer 5′-TCAACCCCCAGCTAGTTGTC-3′ and reverse primer 5′-TGTTCTTCGTTGCTGTTAGG-3′). The IL-4 expression level was standardized by the expression level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA (forward primer 5′-ATGACCACAGTCCATGCCATC-3′ and reverse primer 5′-GAGCTTCCCGTTCAGCTCTG-3′).
For histological analysis of gene delivery with the SP1 peptide, spinal cords were sectioned after perfusion fixation with 4% PFA at the 7-h time point after LPS injection. The sections were incubated with anti-IL-4 primary antibodies overnight at 4°C, washed, and incubated with species-matched secondary antibodies tagged with Alexa Fluor 633 (Molecular Probes). All sections were visualized using confocal laser microscopy, as described above using the aforementioned software. The protein expression of IL-4 in each spinal cord was measured using a mouse IL-4 ELISA kit (R&D Systems, Minneapolis, MN, USA) after homogenization in PBS or RIPA buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, and 1.0% NP-40) containing protease inhibitor (Sigma-Aldrich) to evaluate the secreted or intracellular IL-4.
Statistical Analysis
Data are expressed as mean ± SD. In vitro experiments were performed in triplicate in at least three independent experiments. For the in vivo gene therapy experiments, one-way ANOVA and Tukey’s tests were used. A p value < 0.05 was considered significant.
Author Contributions
T.T. conceived and performed the experiments, wrote the manuscript, and secured funding. N.O., Y.N., and M.K. performed the experiments. T.S. provided reagents and secured funding. J.O., H.M., and H.K. provided expertise and feedback.
Conflicts of Interest
T.S. is employed by Daiichi Sankyo Co., Ltd. The other authors declare no competing interests.
Acknowledgments
We thank T. Yamamoto and F. Kimura for technical support. This work was supported by MEXT KAKENHI grants JP23790988 and JP18K07498 and by research funding from the Daiichi Sankyo TaNeDS Funding Program.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2019.04.008.
Supplemental Information
References
- 1.Kolonin M., Pasqualini R., Arap W. Molecular addresses in blood vessels as targets for therapy. Curr. Opin. Chem. Biol. 2001;5:308–313. doi: 10.1016/s1367-5931(00)00207-6. [DOI] [PubMed] [Google Scholar]; Kolonin, M., Pasqualini, R., and Arap, W. (2001). Molecular addresses in blood vessels as targets for therapy. Curr. Opin. Chem. Biol. 5, 308-313. [DOI] [PubMed]
- 2.Giordano R.J., Cardó-Vila M., Lahdenranta J., Pasqualini R., Arap W. Biopanning and rapid analysis of selective interactive ligands. Nat. Med. 2001;7:1249–1253. doi: 10.1038/nm1101-1249. [DOI] [PubMed] [Google Scholar]; Giordano, R.J., Cardo-Vila, M., Lahdenranta, J., Pasqualini, R., and Arap, W. (2001). Biopanning and rapid analysis of selective interactive ligands. Nat. Med. 7, 1249-1253. [DOI] [PubMed]
- 3.Arap W., Kolonin M.G., Trepel M., Lahdenranta J., Cardó-Vila M., Giordano R.J., Mintz P.J., Ardelt P.U., Yao V.J., Vidal C.I. Steps toward mapping the human vasculature by phage display. Nat. Med. 2002;8:121–127. doi: 10.1038/nm0202-121. [DOI] [PubMed] [Google Scholar]; Arap, W., Kolonin, M.G., Trepel, M., Lahdenranta, J., Cardo-Vila, M., Giordano, R.J., Mintz, P.J., Ardelt, P.U., Yao, V.J., Vidal, C.I., et al. (2002). Steps toward mapping the human vasculature by phage display. Nat. Med. 8, 121-127. [DOI] [PubMed]
- 4.Pasqualini R., Koivunen E., Ruoslahti E. α v integrins as receptors for tumor targeting by circulating ligands. Nat. Biotechnol. 1997;15:542–546. doi: 10.1038/nbt0697-542. [DOI] [PubMed] [Google Scholar]; Pasqualini, R., Koivunen, E., and Ruoslahti, E. (1997). α v integrins as receptors for tumor targeting by circulating ligands. Nat. Biotechnol. 15, 542-546. [DOI] [PubMed]
- 5.Arap W., Pasqualini R., Ruoslahti E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science. 1998;279:377–380. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]; Arap, W., Pasqualini, R., and Ruoslahti, E. (1998). Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science 279, 377-380. [DOI] [PubMed]
- 6.Terashima T., Oka K., Kritz A.B., Kojima H., Baker A.H., Chan L. DRG-targeted helper-dependent adenoviruses mediate selective gene delivery for therapeutic rescue of sensory neuronopathies in mice. J. Clin. Invest. 2009;119:2100–2112. doi: 10.1172/JCI39038. [DOI] [PMC free article] [PubMed] [Google Scholar]; Terashima, T., Oka, K., Kritz, A.B., Kojima, H., Baker, A.H., and Chan, L. (2009). DRG-targeted helper-dependent adenoviruses mediate selective gene delivery for therapeutic rescue of sensory neuronopathies in mice. J. Clin. Invest. 119, 2100-2112. [DOI] [PMC free article] [PubMed]
- 7.White S.J., Nicklin S.A., Büning H., Brosnan M.J., Leike K., Papadakis E.D., Hallek M., Baker A.H. Targeted gene delivery to vascular tissue in vivo by tropism-modified adeno-associated virus vectors. Circulation. 2004;109:513–519. doi: 10.1161/01.CIR.0000109697.68832.5D. [DOI] [PubMed] [Google Scholar]; White, S.J., Nicklin, S.A., Buning, H., Brosnan, M.J., Leike, K., Papadakis, E.D., Hallek, M., and Baker, A.H. (2004). Targeted gene delivery to vascular tissue in vivo by tropism-modified adeno-associated virus vectors. Circulation 109, 513-519. [DOI] [PubMed]
- 8.Koivunen E., Arap W., Rajotte D., Lahdenranta J., Pasqualini R. Identification of receptor ligands with phage display peptide libraries. J. Nucl. Med. 1999;40:883–888. [PubMed] [Google Scholar]; Koivunen, E., Arap, W., Rajotte, D., Lahdenranta, J., and Pasqualini, R. (1999). Identification of receptor ligands with phage display peptide libraries. J. Nucl. Med. 40, 883-888. [PubMed]
- 9.Oi J., Terashima T., Kojima H., Fujimiya M., Maeda K., Arai R., Chan L., Yasuda H., Kashiwagi A., Kimura H. Isolation of specific peptides that home to dorsal root ganglion neurons in mice. Neurosci. Lett. 2008;434:266–272. doi: 10.1016/j.neulet.2008.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]; Oi, J., Terashima, T., Kojima, H., Fujimiya, M., Maeda, K., Arai, R., Chan, L., Yasuda, H., Kashiwagi, A., and Kimura, H. (2008). Isolation of specific peptides that home to dorsal root ganglion neurons in mice. Neurosci. Lett. 434, 266-272. [DOI] [PMC free article] [PubMed]
- 10.Terashima T., Ogawa N., Nakae Y., Sato T., Katagi M., Okano J., Maegawa H., Kojima H. Gene Therapy for neuropathic pain through siRNA-IRF5 gene delivery with homing peptides to microglia. Mol. Ther. Nucleic Acids. 2018;11:203–215. doi: 10.1016/j.omtn.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Terashima, T., Ogawa, N., Nakae, Y., Sato, T., Katagi, M., Okano, J., Maegawa, H., and Kojima, H. (2018). Gene Therapy for neuropathic pain through siRNA-IRF5 gene delivery with homing peptides to microglia. Mol. Ther. Nucleic Acids 11, 203-215. [DOI] [PMC free article] [PubMed]
- 11.Cunha F.Q., Poole S., Lorenzetti B.B., Veiga F.H., Ferreira S.H. Cytokine-mediated inflammatory hyperalgesia limited by interleukin-4. Br. J. Pharmacol. 1999;126:45–50. doi: 10.1038/sj.bjp.0702266. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cunha, F.Q., Poole, S., Lorenzetti, B.B., Veiga, F.H., and Ferreira, S.H. (1999). Cytokine-mediated inflammatory hyperalgesia limited by interleukin-4. Br. J. Pharmacol. 126, 45-50. [DOI] [PMC free article] [PubMed]
- 12.Vale M.L., Marques J.B., Moreira C.A., Rocha F.A., Ferreira S.H., Poole S., Cunha F.Q., Ribeiro R.A. Antinociceptive effects of interleukin-4, -10, and -13 on the writhing response in mice and zymosan-induced knee joint incapacitation in rats. J. Pharmacol. Exp. Ther. 2003;304:102–108. doi: 10.1124/jpet.102.038703. [DOI] [PubMed] [Google Scholar]; Vale, M.L., Marques, J.B., Moreira, C.A., Rocha, F.A., Ferreira, S.H., Poole, S., Cunha, F.Q., and Ribeiro, R.A. (2003). Antinociceptive effects of interleukin-4, -10, and -13 on the writhing response in mice and zymosan-induced knee joint incapacitation in rats. J. Pharmacol. Exp. Ther. 304, 102-108. [DOI] [PubMed]
- 13.Hao S., Mata M., Glorioso J.C., Fink D.J. HSV-mediated expression of interleukin-4 in dorsal root ganglion neurons reduces neuropathic pain. Mol. Pain. 2006;2:6. doi: 10.1186/1744-8069-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hao, S., Mata, M., Glorioso, J.C., and Fink, D.J. (2006). HSV-mediated expression of interleukin-4 in dorsal root ganglion neurons reduces neuropathic pain. Mol. Pain 2, 6. [DOI] [PMC free article] [PubMed]
- 14.Mitchell J.D., Borasio G.D. Amyotrophic lateral sclerosis. Lancet. 2007;369:2031–2041. doi: 10.1016/S0140-6736(07)60944-1. [DOI] [PubMed] [Google Scholar]; Mitchell, J.D., and Borasio, G.D. (2007). Amyotrophic lateral sclerosis. Lancet 369, 2031-2041. [DOI] [PubMed]
- 15.Hausmann O.N. Post-traumatic inflammation following spinal cord injury. Spinal Cord. 2003;41:369–378. doi: 10.1038/sj.sc.3101483. [DOI] [PubMed] [Google Scholar]; Hausmann, O.N. (2003). Post-traumatic inflammation following spinal cord injury. Spinal Cord 41, 369-378. [DOI] [PubMed]
- 16.Dietz V. Spasticity-spastic movement disorder. Spinal Cord. 2008;46:588. doi: 10.1038/sc.2008.45. [DOI] [PubMed] [Google Scholar]; Dietz, V. (2008). Spasticity-spastic movement disorder. Spinal Cord 46, 588. [DOI] [PubMed]
- 17.Nociti V., Cianfoni A., Mirabella M., Caggiula M., Frisullo G., Patanella A.K., Sancricca C., Angelucci F., Tonali P.A., Batocchi A.P. Clinical characteristics, course and prognosis of spinal multiple sclerosis. Spinal Cord. 2005;43:731–734. doi: 10.1038/sj.sc.3101798. [DOI] [PubMed] [Google Scholar]; Nociti, V., Cianfoni, A., Mirabella, M., Caggiula, M., Frisullo, G., Patanella, A.K., Sancricca, C., Angelucci, F., Tonali, P.A., and Batocchi, A.P. (2005). Clinical characteristics, course and prognosis of spinal multiple sclerosis. Spinal Cord 43, 731-734. [DOI] [PubMed]
- 18.Gwathmey K.G. Sensory neuronopathies. Muscle Nerve. 2016;53:8–19. doi: 10.1002/mus.24943. [DOI] [PubMed] [Google Scholar]; Gwathmey, K.G. (2016). Sensory neuronopathies. Muscle Nerve 53, 8-19. [DOI] [PubMed]
- 19.Pandolfo M. Friedreich ataxia: the clinical picture. J. Neurol. 2009;256(Suppl 1):3–8. doi: 10.1007/s00415-009-1002-3. [DOI] [PubMed] [Google Scholar]; Pandolfo, M. (2009). Friedreich ataxia: the clinical picture. J. Neurol. 256 (Suppl 1), 3-8. [DOI] [PubMed]
- 20.Roodhart J.M., Langenberg M.H., Witteveen E., Voest E.E. The molecular basis of class side effects due to treatment with inhibitors of the VEGF/VEGFR pathway. Curr. Clin. Pharmacol. 2008;3:132–143. doi: 10.2174/157488408784293705. [DOI] [PubMed] [Google Scholar]; Roodhart, J.M., Langenberg, M.H., Witteveen, E., and Voest, E.E. (2008). The molecular basis of class side effects due to treatment with inhibitors of the VEGF/VEGFR pathway. Curr. Clin. Pharmacol. 3, 132-143. [DOI] [PubMed]
- 21.Beusterien K., Grinspan J., Kuchuk I., Mazzarello S., Dent S., Gertler S., Bouganim N., Vandermeer L., Clemons M. Use of conjoint analysis to assess breast cancer patient preferences for chemotherapy side effects. Oncologist. 2014;19:127–134. doi: 10.1634/theoncologist.2013-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]; Beusterien, K., Grinspan, J., Kuchuk, I., Mazzarello, S., Dent, S., Gertler, S., Bouganim, N., Vandermeer, L., and Clemons, M. (2014). Use of conjoint analysis to assess breast cancer patient preferences for chemotherapy side effects. Oncologist 19, 127-134. [DOI] [PMC free article] [PubMed]
- 22.Rivière I., Sadelain M. Chimeric antigen receptors: A cell and gene therapy perspective. Mol. Ther. 2017;25:1117–1124. doi: 10.1016/j.ymthe.2017.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]; Riviere, I., and Sadelain, M. (2017). Chimeric antigen receptors: A cell and gene therapy perspective. Mol. Ther. 25, 1117-1124. [DOI] [PMC free article] [PubMed]
- 23.Borel F., Kay M.A., Mueller C. Recombinant AAV as a platform for translating the therapeutic potential of RNA interference. Mol. Ther. 2014;22:692–701. doi: 10.1038/mt.2013.285. [DOI] [PMC free article] [PubMed] [Google Scholar]; Borel, F., Kay, M.A., and Mueller, C. (2014). Recombinant AAV as a platform for translating the therapeutic potential of RNA interference. Mol. Ther. 22, 692-701. [DOI] [PMC free article] [PubMed]
- 24.Zhou J., Rossi J.J. Cell-type-specific, aptamer-functionalized agents for targeted disease therapy. Mol. Ther. Nucleic Acids. 2014;3:e169. doi: 10.1038/mtna.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhou, J., and Rossi, J.J. (2014). Cell-type-specific, aptamer-functionalized agents for targeted disease therapy. Mol. Ther. Nucleic Acids 3, e169. [DOI] [PMC free article] [PubMed]
- 25.Cullis P.R., Hope M.J. Lipid nanoparticle systems for enabling gene therapies. Mol. Ther. 2017;25:1467–1475. doi: 10.1016/j.ymthe.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cullis, P.R., and Hope, M.J. (2017). Lipid nanoparticle systems for enabling gene therapies. Mol. Ther. 25, 1467-1475. [DOI] [PMC free article] [PubMed]
- 26.Chakraborty C., Sharma A.R., Sharma G., Doss C.G.P., Lee S.S. Therapeutic miRNA and siRNA: Moving from bench to clinic as next generation medicine. Mol. Ther. Nucleic Acids. 2017;8:132–143. doi: 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chakraborty, C., Sharma, A.R., Sharma, G., Doss, C.G.P., and Lee, S.S. (2017). Therapeutic miRNA and siRNA: Moving from bench to clinic as next generation medicine. Mol. Ther. Nucleic Acids 8, 132-143. [DOI] [PMC free article] [PubMed]
- 27.Kanasty R., Dorkin J.R., Vegas A., Anderson D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]; Kanasty, R., Dorkin, J.R., Vegas, A., and Anderson, D. (2013). Delivery materials for siRNA therapeutics. Nat. Mater. 12, 967-977. [DOI] [PubMed]
- 28.Lv H., Zhang S., Wang B., Cui S., Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]; Lv, H., Zhang, S., Wang, B., Cui, S., and Yan, J. (2006). Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release 114, 100-109. [DOI] [PubMed]
- 29.Gilleron J., Querbes W., Zeigerer A., Borodovsky A., Marsico G., Schubert U., Manygoats K., Seifert S., Andree C., Stöter M. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]; Gilleron, J., Querbes, W., Zeigerer, A., Borodovsky, A., Marsico, G., Schubert, U., Manygoats, K., Seifert, S., Andree, C., Stoter, M., et al. (2013). Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 31, 638-646. [DOI] [PubMed]
- 30.Sahay G., Querbes W., Alabi C., Eltoukhy A., Sarkar S., Zurenko C., Karagiannis E., Love K., Chen D., Zoncu R. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 2013;31:653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sahay, G., Querbes, W., Alabi, C., Eltoukhy, A., Sarkar, S., Zurenko, C., Karagiannis, E., Love, K., Chen, D., Zoncu, R., et al. (2013). Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 31, 653-658. [DOI] [PMC free article] [PubMed]
- 31.Wittrup A., Ai A., Liu X., Hamar P., Trifonova R., Charisse K., Manoharan M., Kirchhausen T., Lieberman J. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. 2015;33:870–876. doi: 10.1038/nbt.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wittrup, A., Ai, A., Liu, X., Hamar, P., Trifonova, R., Charisse, K., Manoharan, M., Kirchhausen, T., and Lieberman, J. (2015). Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. 33, 870-876. [DOI] [PMC free article] [PubMed]
- 32.Landen C.N., Jr., Chavez-Reyes A., Bucana C., Schmandt R., Deavers M.T., Lopez-Berestein G., Sood A.K. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65:6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]; Landen, C.N., Jr., Chavez-Reyes, A., Bucana, C., Schmandt, R., Deavers, M.T., Lopez-Berestein, G., and Sood, A.K. (2005). Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 65, 6910-6918. [DOI] [PubMed]
- 33.Kumar S.R., Markusic D.M., Biswas M., High K.A., Herzog R.W. Clinical development of gene therapy: results and lessons from recent successes. Mol. Ther. Methods Clin. Dev. 2016;3:16034. doi: 10.1038/mtm.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kumar, S.R., Markusic, D.M., Biswas, M., High, K.A., and Herzog, R.W. (2016). Clinical development of gene therapy: results and lessons from recent successes. Mol. Ther. Methods Clin. Dev. 3, 16034. [DOI] [PMC free article] [PubMed]
- 34.Abbott N.J., Patabendige A.A.K., Dolman D.E.M., Yusof S.R., Begley D.J. Structure and function of the blood-brain barrier. Neurobiol. Dis. 2010;37:13–25. doi: 10.1016/j.nbd.2009.07.030. [DOI] [PubMed] [Google Scholar]; Abbott, N.J., Patabendige, A.A.K., Dolman, D.E.M., Yusof, S.R., and Begley, D.J. (2010). Structure and function of the blood-brain barrier. Neurobiol. Dis. 37, 13-25. [DOI] [PubMed]
- 35.Méndez-Gómez H.R., Galera-Prat A., Meyers C., Chen W., Singh J., Carrión-Vázquez M., Muzyczka N. Transcytosis in the blood-cerebrospinal fluid barrier of the mouse brain with an engineered receptor/ligand system. Mol. Ther. Methods Clin. Dev. 2015;2:15037. doi: 10.1038/mtm.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mendez-Gomez, H.R., Galera-Prat, A., Meyers, C., Chen, W., Singh, J., Carrion-Vazquez, M., and Muzyczka, N. (2015). Transcytosis in the blood-cerebrospinal fluid barrier of the mouse brain with an engineered receptor/ligand system. Mol. Ther. Methods Clin. Dev. 2, 15037. [DOI] [PMC free article] [PubMed]
- 36.Pattichis A.A., Slee M. CSF hypotension: A review of its manifestations, investigation and management. J. Clin. Neurosci. 2016;34:39–43. doi: 10.1016/j.jocn.2016.07.002. [DOI] [PubMed] [Google Scholar]; Pattichis, A.A., and Slee, M. (2016). CSF hypotension: A review of its manifestations, investigation and management. J. Clin. Neurosci. 34, 39-43. [DOI] [PubMed]
- 37.Terashima T., Nakae Y., Katagi M., Okano J., Suzuki Y., Kojima H. Stem cell factor induces polarization of microglia to the neuroprotective phenotype in vitro. Heliyon. 2018;4:e00837. doi: 10.1016/j.heliyon.2018.e00837. [DOI] [PMC free article] [PubMed] [Google Scholar]; Terashima, T., Nakae, Y., Katagi, M., Okano, J., Suzuki, Y., and Kojima, H. (2018). Stem cell factor induces polarization of microglia to the neuroprotective phenotype in vitro. Heliyon 4, e00837. [DOI] [PMC free article] [PubMed]
- 38.Christianson D.R., Ozawa M.G., Pasqualini R., Arap W. Techniques to decipher molecular diversity by phage display. Methods Mol. Biol. 2007;357:385–406. doi: 10.1385/1-59745-214-9:385. [DOI] [PubMed] [Google Scholar]; Christianson, D.R., Ozawa, M.G., Pasqualini, R., and Arap, W. (2007). Techniques to decipher molecular diversity by phage display. Methods Mol. Biol. 357, 385-406. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.