Abstract
The peripheral taste and olfactory systems in mammals are separate and independent sensory systems. In the current model of chemosensation, gustatory, and olfactory receptors are genetically divergent families expressed in anatomically distinct locations that project to disparate downstream targets. Although information from the 2 sensory systems merges to form the perception of flavor, the first cross talk is thought to occur centrally, in the insular cortex. Recent studies have shown that gustatory and olfactory receptors are expressed throughout the body and serve as chemical sensors in multiple tissues. Olfactory receptor cDNA has been detected in the tongue, yet the presence of physiologically functional olfactory receptors in taste cells has not yet been demonstrated. Here we report that olfactory receptors are functionally expressed in taste papillae. We found expression of olfactory receptors in the taste papillae of green fluorescent protein-expressing transgenic mice and, using immunocytochemistry and real-time quantitative polymerase chain reaction experiments, the presence of olfactory signal transduction molecules and olfactory receptors in cultured human fungiform taste papilla (HBO) cells. Both HBO cells and mouse taste papilla cells responded to odorants. Knockdown of adenylyl cyclase mRNA by specific small inhibitory RNA and pharmacological block of adenylyl cyclase eliminated these responses, leading us to hypothesize that the gustatory system may receive olfactory information in the periphery. These results provide the first direct evidence of the presence of functional olfactory receptors in mammalian taste cells. Our results also demonstrate that the initial integration of gustatory and olfactory information may occur as early as the taste receptor cells.
Keywords: adenylyl cyclase, gustatory, HBO cells, odor, olfactory receptors, taste cells
Introduction
The act of eating simultaneously stimulates the gustatory, olfactory, and somatosensory systems. Although these sensory systems use different receptors to transduce sensations, pass information through anatomically distinct systems, and project to disparate downstream targets, they also combine to form the unified percept of flavor. The combination of these independent sensory inputs from each modality is thought to occur in the anterior insula (Small 2012). The cross talk between these senses has implications for food science, as volatiles can alter sweet perception independent of sugar concentration (Bartoshuk and Klee 2013), and nonvolatiles can serve as flavor enhancers by increasing the perception of the volatile components of food (Green et al. 2012).
Recently, both olfactory and gustatory receptors have been shown to be expressed in a wide variety of tissues. The G-protein-coupled sweet, bitter, and umami gustatory receptors and other gustatory signaling transduction proteins are expressed in such tissues as the digestive system (Margolskee et al. 2007; Fujita 1991), respiratory system (Shah et al. 2009; Tizzano et al. 2011), brain (Ren et al. 2009), testicles (Max et al. 2001; Kiuchi et al. 2006; Gong et al. 2016), pancreas (Taniguchi 2004; Nakagawa et al. 2009), urinary bladder (Elliott et al. 2011), liver (Taniguchi 2004), and spermatozoa (Meyer et al. 2012). Similarly, olfactory receptors are expressed in such tissues as sperm (Parmentier et al. 1992), the nervous system (Weber et al. 2002; Conzelmann et al. 2000; Otaki et al. 2004), placenta (Itakura et al. 2006), gut (Braun et al. 2007), kidney (Pluznick et al. 2009), leucocytes (Malki et al. 2015), tongue (Gaudin et al. 2001; Durzyński et al. 2005), and erythroid cells (Feingold et al. 1999). Several studies have found olfactory receptors in cDNA libraries from whole tongue, leading to the hypothesis that the integration of gustatory and olfactory information may occur as early as the taste receptor cells (Gaudin et al. 2001; Durzyński et al. 2005). In Drosophila, odorants can be detected by olfactory receptors in taste neurons expressing sweet- or bitter-sensing receptors (Hiroi et al. 2008). The olfactory and taste receptors both belong to the family of 7-transmembrane-domain proteins, which have overlapping similarities in sequence and expression pattern (Kim and Carlson 2002).
Here we examined whether olfactory receptors are expressed in taste receptor cells and whether they respond to odors. We found that both cultured human fungiform taste papilla (HBO) cells and mouse taste papilla cells respond to odors. In addition, pharmacologically blocking the olfactory transduction component adenylyl cyclase III and knocking down adenylyl cyclase III expression by small inhibitory RNA (siRNA) eliminated these responses. These results provide the first direct evidence that functional olfactory receptors are present in cultured taste cells obtained from mice and humans.
Materials and methods
Transgenic mice expressing Olfr151 (M71) or Olfr73 (mOR-EG) receptor promoter
The olfactory receptor properties of taste cells were investigated by examining the taste papillae of previously generated M71 receptor transgenic mice (Bozza et al. 2002) and EG receptor transgenic mice (Oka et al. 2006). Briefly, the mOR-EG-GFP line was generated using a transgene consisting of 3.0 kb upstream of the mOR-EG transcription start site followed by mOR-EG-IRES-gapEGFP (Oka et al. 2006). The GFP-labeled M71 mouse lines were generated by inserting IRES-tauGFP after the mouse M71 coding sequences (Bozza et al. 2002) and stated that the mouse M71 targeting vectors were derived from genomic fragments isolated from a mouse (129/Sv) and that the mice are in a mixed (129 × C57BL/6J) background. The IRES sequence allows coexpression of an olfactory receptor with a reporter protein. Acetophenone is a known ligand of Olfr151 (M71), and eugenol is a known ligand of Olfr73 (mOR-EG).
Establishment and maintenance of mouse taste papillae culture
We collected taste tissues from mOR-EG-GFP transgenic mice, mOR-M71-GFP transgenic mice, and 2 different wild-type strains (129/Sv and C57BL/6) (>4 months old; both sexes). Mice were killed prior to tongue removal for the isolation of taste tissues (circumvallate and foliate papillae). The care and killing of mice in this study were in accordance with National Institute of Health (NIH) Guidelines for the Care and Use of Laboratory Animals. All of the experimental procedures involving mice in this study were approved by the Institutional Animal Care and Use Committee at Monell Chemical Senses Center (IACUC ACC#1142). Circumvallate and foliate taste papillae tissue were obtained from more than 3 (wild-type [129/Sv, C57BL/6], M71, and EG receptor) transgenic mice were isolated according to previously published procedures (Ozdener et al. 2006). Briefly, after removal mouse tongue was immediately placed into an isolation solution, followed by enzymatic digestion with a mixture of enzymes (pronase and elastase, 1 mg/mL each; Sigma). Mixed-taste papilla tissue was then gently minced with a surgical razor, seeded on collagen type 1–coated coverslips, and incubated at 36 °C in a humidified environment containing 5% CO2.
GFP fluorescence and immunohistochemistry
Tongue circumvallate and foliate taste papilla tissues obtained from wild-type (129/Sv, C57BL/6), M71, and EG receptor transgenic mice were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS, pH 7.2) for 1 h at room temperature and then cryoprotected by sequential 24-h immersions in 10%, 20%, and 30% sucrose in PBS. Sagittal sections of each papilla were cut at 10 μm on a Microm HM 500 OM cryostat. Tissue sections were thaw-mounted onto Superfrost Plus slides and either directly observed for green fluorescent protein (GFP) expression or stained (described later) to localize coexpression of GFP and taste-related molecules. For immunostaining, mouse taste papilla tissues were blocked with antibody dilution solution (3% bovine serum albumin, 0.5% Triton X-100, 3% serum [from animal secondary antibody generated] in PBS) for 1 h at room temperature. Then, phospholipase C-β2 (Santa Cruz), GFP (Cell Signaling), and M71 antibodies (guinea-pig, gifted by Dr Gilad Barnea of Brown University; 1:500) were added and incubated overnight at 4 °C. This step was followed by incubation with appropriate secondary antibodies diluted in blocking buffer for 30 min at room temperature. After washing with PBS and water, slides were mounted using VectaShield with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories). Immunoreactive cells were counted in at least 3 fields at ×20 and ×40 magnification, and images were acquired with a Leica TCS SP2 spectral confocal microscope. The excitation wavelengths used were 488 nm for GFP and 633 nm for Alexa Fluor 633, with emissions detected at appropriate wavelengths. Leica Scanware software was used to acquire confocal images by scanning unidirectionally at a 1024 × 1024 pixel format with 3-line and 2-frame averaging. A computer-controlled digital zoom was used to increase magnification to a maximum of ×2.3 with the ×20 and ×40 objective. Images were arranged and minimally adjusted for contrast and brightness using LCS software (Leica Microsystems) and Adobe Photoshop Elements 2.0 (Adobe Systems).
Human fungiform taste papillae (HBO) cell culture
Human fungiform papilla cells (HBO cells, named after Hasan Basri Ozdener) were previously cultured and maintained according to protocols published elsewhere (Ozdener et al. 2011; Ozdener et al. 2012; Ozdener and Rawson 2013). Briefly, human fungiform taste papillae were digested and then seeded on collagen type 1–coated coverslips and incubated at 36 °C in a humidified environment containing 5% CO2. Cells were maintained and passaged in Iscove’s modified Dulbecco’s medium containing 10% fetal bovine serum, a 1:5 ratio of MCDB 153 medium, and a triple cocktail of antibiotics (100 U/mL penicillin, 100 μg/mL streptomycin, and 0.5 μg/mL fungizone). HBO cells have been maintained in culture for more than 1 year without loss of viability and with retention of the molecular and biochemical properties of acutely isolated taste cells (Subramaniam et al. 2016; Qian et al. 2017).
Immunocytochemistry of human fungiform taste cells
HBO cells between passages 3 and 6 (~4–9 months after the initial establishment) were seeded on coverslips and then fixed with 4% paraformaldehyde for 10 min at room temperature. After blocking, HBO cells were exposed to either goat anti-olfactory marker protein (1:500, Wako Laboratory Chemicals), mouse monoclonal anti-adenylyl cyclase labeled with Alexa 488 (1:25; C-5: Santa Cruz sc-377243 AF488), or mouse monoclonal Gα s/olf antibody labeled with Alexa 488 (1:10; A-5: Santa Cruz sc-55545). Antibodies were used for overnight incubation at 4 °C and then washed 3 times with 1× PBS for 10 min, followed by Millipore water 3 × 20 min, and mounted with VectaShield with DAPI (Vector Laboratories). HBO cells subjected to OMP antibodies were incubated with Alexa 488-conjugated donkey anti-goat (1:500, Invitrogen) secondary antibodies for 30 min at room temperature. After washing with PBS (3 × 10 min) and Millipore water (3 × 20 min), cells were mounted with VectaShield with DAPI. Images were acquired with a Leica TCS SP2 spectral confocal microscope as described earlier.
Quantitative reverse transcription-PCR
Gene expression services were provided by the University of Pennsylvania Molecular Profiling Facility, including RNA isolation with the RNeasy Micro Kit with DNase treatment (Qiagen) and quality control of the total RNA samples by Agilent Bioanalyzer and Nanodrop spectrophotometry. Expression was quantified in quadruplicate by quantitative reverse transcription–polymerase chain reaction (qRT-PCR) using TaqMan Assays (Applied Biosystems by Life Technologies). The reverse transcription reaction was carried out with High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) in 100 μL containing 2.0 μg RNA in 30.0 μL nuclease-free water, 4.0 μL of 25 × (100 mM) dNTPs, 5 μL Multiscribe Reverse Transcriptase (50 U/μL), 10.0 μL of 10 × reverse transcription buffer, and 10.0 μL of 10 × random primer. For synthesis of cDNA, the reaction mixtures were incubated at 25 °C for 10 min, 37 °C for 120 min, and 85 °C for 5 min and then held at 4 °C. Then, 4.5 μL cDNA diluted 1:5 was amplified using 5.0 μL TaqMan 2× Fast Universal PCR Master Mix with No AmpErase UNG (Applied Biosystems), 0.5 μL of assay in a final volume of 10.0 μL. TaqMan probes for GAPDH (glyceraldehyde-3-phosphate dehydrogenase Hs02758991_g1), GNG13 (Hs00213042_m1), ADCY3 (Hs01086502_m1), OR7D4 (Hs02339188_s1), OR5P3 (Hs01029920_s1), OR51E1 (Hs02339849_s1), OR2W1 (Hs01121978_s1), OR5A1 (Hs04980963_s1), T1R3 (Hs01387770_g1), and T2R38 (Hs00604294_s1) were ordered from Applied Biosystems. Quantitative PCR was run on a QuantStudio 12K Flex Real-Time PCR system (Applied Biosystems), and the reaction mixtures were incubated at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The cycle threshold (CT) values were calculated with QuantStudio Software version 1.2.4 (Applied Biosystems). Mean values of CT triplets were calculated (single values differing ≥1 CT were excluded), and ΔCT values were determined using mean CT of GAPDH as reference (ΔCT = CT target − CT reference). Finally, 2 − ΔCT values were calculated based on established methods (Schmittgen and Livak 2001).
Single-cell calcium imaging recording from cultured taste cells
Cultured human fungiform and mouse taste papilla cells were seeded on 15-mm coverslips. The cultured taste cells grown on coverslips were then loaded with the calcium-sensitive dye Fura-2 by incubating the cells in Ringer’s solution (80 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 1 mM Na-pyruvate, and 20 mM HEPES-Na, pH 7.2, with osmolarity adjusted to 300–310 mOsm with 5 M NaCl) supplemented with 1 mM Fura-2 AM (Molecular Probes) and 10 mg/mL Pluronic F127 (Molecular Probes) for 30–60 min at 36 °C. Each coverslip was placed into a p4 chamber system with the delivery and waste pipes attached to the chamber. This allowed for a stable and constant flow of liquid in and out of the chamber and continuously bathed the cells with the Ringer’s solution.
The cells were exposed to various stimuli by switching the superfusion to stimulus solutions, which allowed for a complete change of bath solutions in the chamber within 20 s. Stimuli were dissolved in Ringer’s solution, and pH and osmolality were readjusted if needed. Odorants (eugenol, hexanoic acid, coumarin, acetophenone, heptanal, and lyral) were of the highest purity available (98% pure) and were purchased from Sigma (St. Louis). Individual odorant stocks were made up at 1 M in dimethyl sulfoxide (DMSO) and diluted to a final working concentration in Ringer’s solution. For dose–response analyses, eugenol, a known ligand of Olfr73, was diluted from a stock solution (1 M) to working concentrations (0.01, 0.1, 1, 10, 100, 200 µM). The adenylyl cyclase inhibitor SQ22536 (9-(tetrahydro-2-furanyl)-9H-purin-6-amine) was prepared as 100 mM stocks in DMSO and diluted to 1 µM with Ringer’s solution before each experiment. Bitter mixture (5 mM salicin, 2 mM denatonium, 5 mM phenylthiocarbamide) dissolved in Ringer’s solution was used as bitter stimulus to induce a large number of bitter receptors. Stimuli were bath applied for 60 s using a peristaltic-pump-controlled perfusion system. Each stimulus application was followed by an approximately 2-min washout period with Ringer’s solution.
Calcium imaging recordings were performed using standard imaging techniques. Illumination was via an LSR SpectraMASTER monochromator coupled to the microscope. Cells were illuminated with light emitted by a 75-W xenon lamp alternately filtered with narrow-bandpass filters at 340 and 380 nm. The light emitted from Fura-2 AM in the cells under ×200 microscopic magnification was filtered at 510 nm and passed through an image intensifier coupled with a cooled CCD camera (Olympix, Perkin Elmer Life Sciences). Exposure times were minimized, and the light was shuttered between acquisitions to minimize photobleaching. Cells remained viable in the recording setup for over 2 h without visible effects of dye bleaching. Ratio data for the cells were subsequently analyzed in Excel to determine which cells had responded with a significant change in intracellular calcium. After calcium imaging, coverslips were fixed with 4% paraformaldehyde for 10 min and washed 3 × with PBS. Cells were then observed for GFP-specific signal and colocalized with the odorant-responsive cells.
Knockdown of ADCY3 (adenylyl cyclase III) mRNA by specific siRNA
The RNA interference analysis was performed by transfecting HBO cells with small interfering RNA (siRNA) (FlexiTube siRNA Premix, Qiagen). Three days before transfection, 2000 cells per well were seeded in 12-well plates. HBO cells were transfected with 25 nmol/L ADCY3-specific siRNA (SI00058849) or AllStars scrambled siRNA (SI03650318) as negative control. Scrambled siRNA has no homology to any known mammalian gene, and a variety of cell-based assays have shown no nonspecific effect on gene expression and phenotype. At 3–5 days post-transfection, we performed single-cell calcium imaging of ADCY3-siRNA-treated and scrambled siRNA-treated cells, along with untransfected control cells. The results are representative of at least three independent experiments using a total of 164 cells.
Results
Olfactory receptors are expressed in taste cells
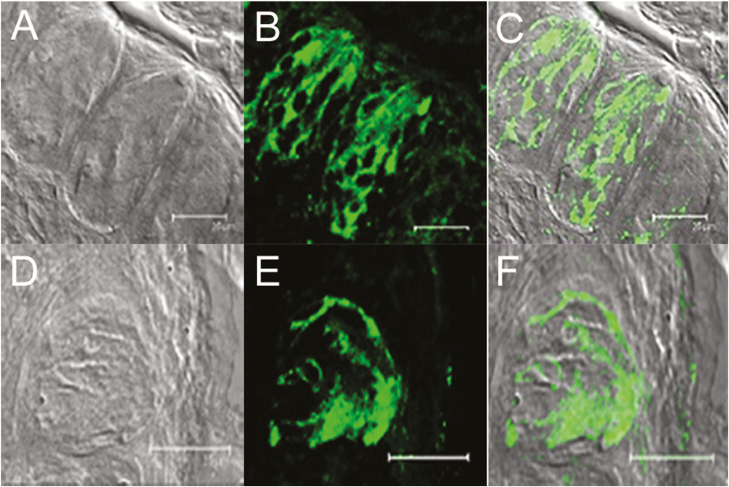
To determine if olfactory receptor proteins are present in taste cells, we examined 2 genetically modified strains of mice coexpressing GFP with either M71 or mOR-EG olfactory receptor. Confocal microscopy revealed that circumvallate taste papillae from transgenic mice expressed GFP (Figure 1A–C, mOR-M71-GFP; Figure 1D–F, OR-EG-GFP), indicating the presence of M71 (Figure 1B) and mOR-EG (Figure 1E) in taste papilla cells. GFP expression was specifically localized in the taste papillae of mOR-M71-GFP (Figure 1C) and mOR-EG-GFP (Figure 1F) transgenic mice and was not observed in the surrounding cells (compare with transmission images of corresponding fields in Figure 1A, D).
Figure 1.
Expression of olfactory receptors in circumvallate taste papillae of mOR-M71-GFP (top row) and mOR-EG-GFP (bottom row) transgenic mice. (A) and (D): DIC brightfield images of the taste papillae from transgenic mice expressing GFP under M71 (A) and eugenol (EG; D) receptor promoter. (B) and (E): GFP fluorescence (green) expression of each transgenic mouse. (C) and (F): Overlays of the images from (A) and (B) and from (D) and (E) demonstrate colocalization of GFP expression with taste cells in the circumvallate taste papillae. Scale bars = 25 µm.
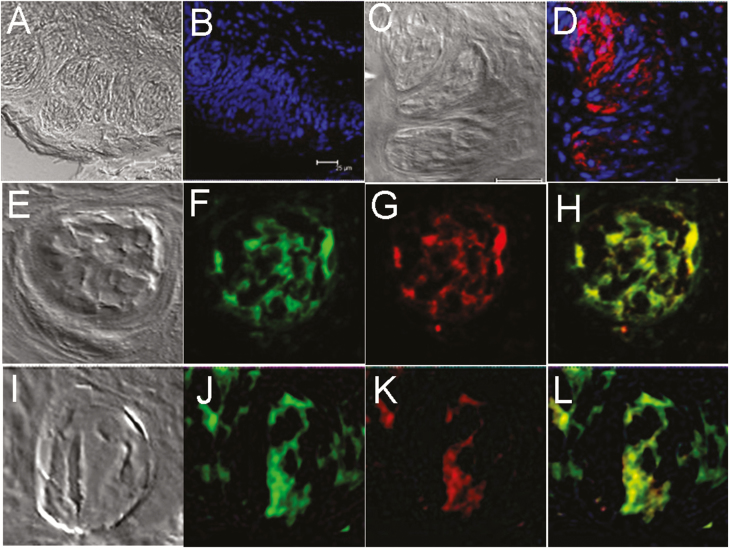
In control experiments, neither the endogenous GFP signal (Figure 2A) nor immunoreactivity to GFP antibodies (Figure 2B) was observed in wild-type mice (C57BL/6), indicating that GFP expression was specific to the taste papillae of transgenic mice. The expression of M71 in taste tissue was also seen in wild-type mice using an antibody to the M71 receptor (Figure 2C, D). In addition, mOR-EG-GFP transgenic mice were stained with anti-GFP antibodies to demonstrate endogenous GFP expression, which colocalized with the immunostaining of GFP antibodies in the fungiform taste papillae (Figure 2E–H). Endogenous GFP expression in taste papillae of mOR-M71-GFP transgenic mouse colocalized with immunoreactivity to anti-M71 antibodies (Figure 2J–L), indicating that GFP expression is driven by M71 olfactory receptors in taste cells.
Figure 2.
Expression of GFP and M71 olfactory receptors in transgenic mice taste tissue is not an artifact or genetic alteration. The expression of GFP and M71 olfactory receptors in transgenic and wild-type (C57BL/6) mice was visualized using GFP and M71 antibodies. (A) and (C): Transmission images of corresponding fields from taste papillae of wild-type mouse. (B): No immunoreactivity (absence of red) to GFP antibody in circumvallate taste tissue of wild-type (C57BL/6) mouse. (D): Immunoreactivity to M71 olfactory receptor antibodies (red) in the taste papillae of wild-type mouse. (E–H): Immunostaining of mOR-EG-GFP mouse taste papilla tissue with GFP antibody (G, red) shows exact colocalization with endogenous GFP expression (F, green; H, merged). (I–L): Taste papilla tissues from mOR-M71-GFP mice demonstrate expression of GFP (green, J) and M71 (red, K) is colocalized (L, merged). E and I are transmission images of corresponding fields in their respective rows. Blue, nuclei staining with DAPI. Scale bars = 25 µm.
To confirm the identity of the cells expressing olfactory receptors, taste tissues from both transgenic mice were stained for the type II taste cell marker phospholipase C-β2 (PLC-β2) (Figure 3E, F). Most PLC-β2–immunoreactive cells in taste papillae from both mOR-M71-GFP and mOR-EG-GFP mice demonstrated overlap with GFP expression (Figure 3C, D), indicating coexpression of type II cell marker PLC-β2 and olfactory receptors in taste cells (Figure 3G, H). Further studies will be required to characterize the relation between the coexpression of taste-cell-specific markers and olfactory-specific markers.
Figure 3.
GFP driven by an olfactory receptor promoter colocalized with phospholipase C-β2 (PLC-β2), a type II taste cell marker, in mouse taste tissue. Taste papillae from mOR-M71-GFP (top row) and mOR-EG-GFP (bottom row) transgenic mice express M71 and eugenol (EG) receptors, respectively. (A) and (B): Transmission images of corresponding fields in (C–H). (C) and (D): GFP expression (green) is specifically localized in taste papillae but is not observed in surrounding cells. (E) and (F): Taste tissues show immunoreactivity to the type II taste cell marker PLC-β2 (red). (G) and (H): GFP expression is colocalized with PLC-β2 expression in taste papillae. Note the excellent overlap of GFP and PLC-β2 expressions (yellow-orange color in merged views) in a subset of taste cells. Images were acquired with a confocal laser scanning microscope and were arranged and minimally adjusted for contrast and brightness. Scale bar = 25 µm.
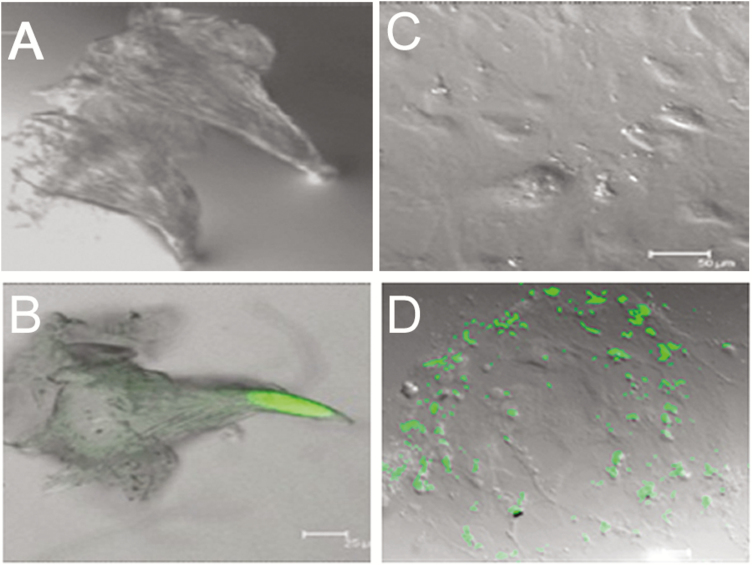
To elaborate the specificity of GFP expression, we did not observe autofluorescence in either freshly isolated wild-type mouse taste cells (Figure 4A) or cultured mouse taste cells obtained from wild-type mice (Figure 4C) using confocal microscopy, indicating the absence of artifactual GFP expression. In contrast, we saw GFP expression in both freshly isolated and cultured taste cells from mOR-M71-GFP mice (Figure 4B, D).
Figure 4.
GFP expression in freshly isolated and cultured mammalian taste papillae. GFP expression is not artifact but under the control of an olfactory receptor promoter (green). (A) and (C): No autofluorescence observed in either freshly isolated (A) or cultured (days 3–5; C) wild-type mouse taste cells. (B) and (D): Both freshly isolated (B) and cultured (D) taste cells from mOR-M71-GFP mice show GFP expression. Scale bars: B and D = 25 µm; C = 50 µm.
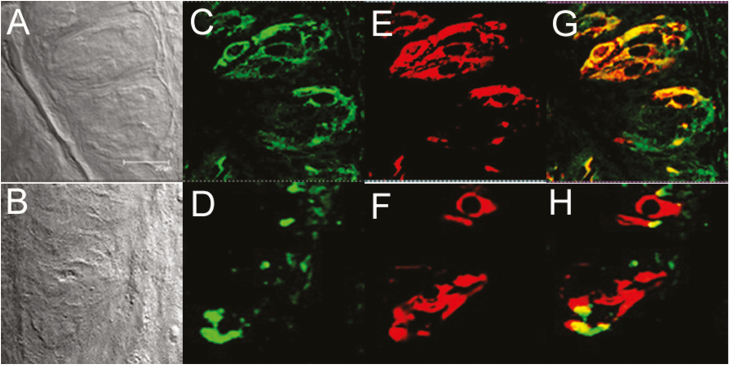
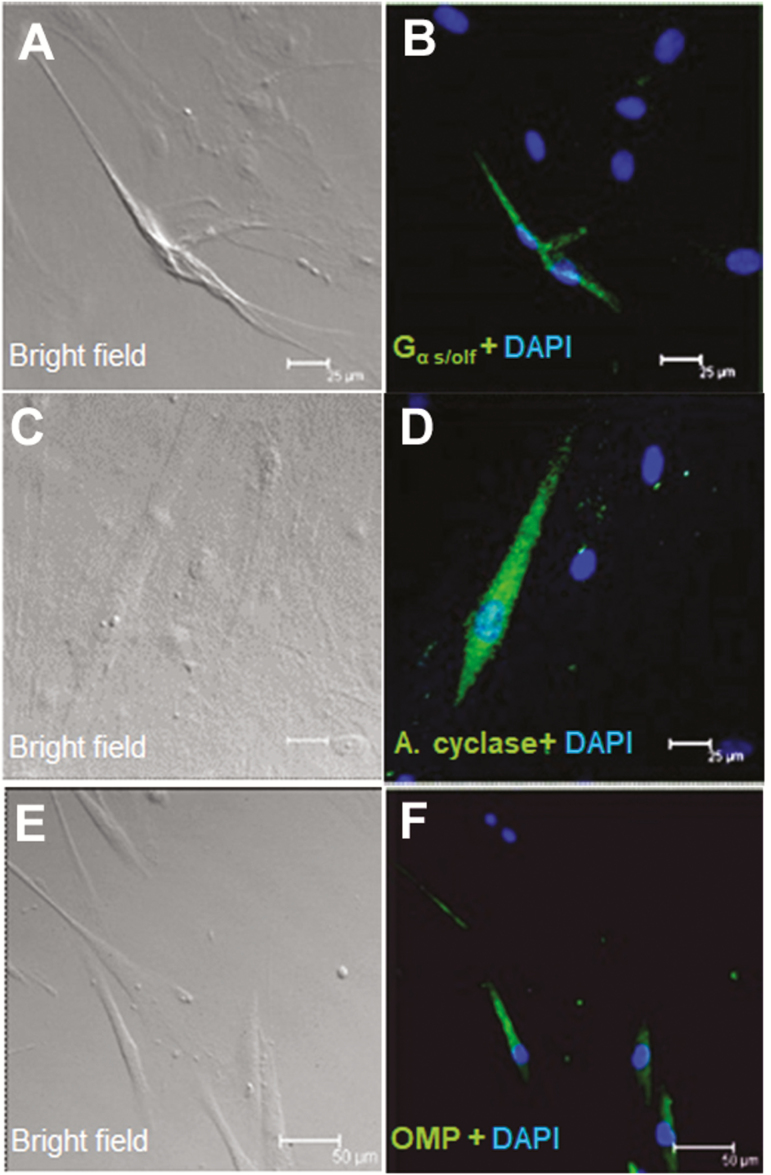
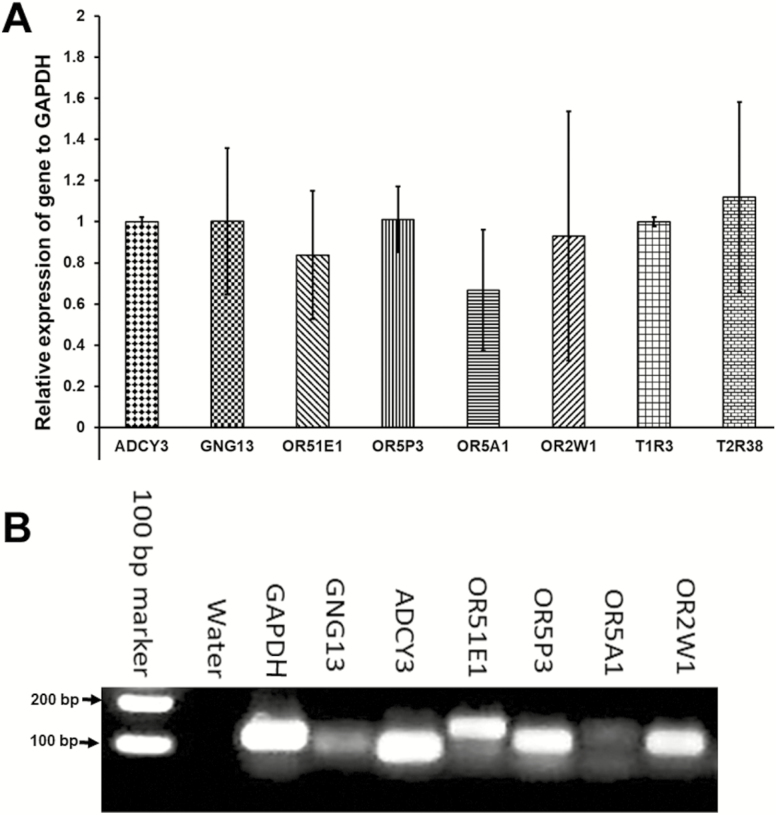
We then examined whether olfactory receptors and signaling molecules are expressed in cultured human taste cells (HBO cells). Using antibodies for Gα s/olf (Figure 5B), adenylyl cyclase (Figure 5D), and olfactory marker protein (OMP; Figure 5F), we found that HBO cells expressed all three olfactory signal transduction molecules, suggesting that the canonical olfactory signal transduction pathway may be intact and may mediate odorant-induced intracellular calcium changes. In addition, we performed quantitative polymerase chain reaction experiments to determine the expression of mRNA for selected olfactory signal molecules: ADCY3 (adenylyl cyclase III), Gng13 (guanine nucleotide binding protein [G protein], gamma 13), OR2W1, OR5A1, OR5P3, and OR51E1 with well-established significance olfactory responses. The amplification of transcript started after cycle 20, depending on target molecules, indicating different expression levels of these mRNAs in HBO cells. The relative expression of mRNA for signal transduction molecules ADCY3 and Gng13, olfactory receptors, and taste receptors in HBO cells, normalized to the housekeeping gene GAPDH. Expression of mRNAs in HBO cells found albeit at different levels, for instance Ggn13 expression. Difference between visualized mRNA and quantification may be due to technical issues (Figure 6 A, B). As seen However, there are more than 400 olfactory receptor genes, so further studies are needed to fully characterize expression of olfactory signaling proteins and olfactory receptors in HBO cells. As control markers, we also demonstrated the expression of mRNAs for sweet- and bitter-specific receptors T1R3 and T2R38 in HBO cells (Figure 6A).
Figure 5.
Immunocytochemical detection of olfactory signal transduction proteins in HBO cells. Brightfield (A, C, and E) and immunofluorescence (B, D, and F) images demonstrate immunoreactivity (green) of Gα s/olf (A and B; n = 15 of 72) in the cytoplasm and membrane and of adenylyl cyclase (C and D; n = 23 of 217) and OMP (E and F n = 13 of 92) in the cytoplasm in a subgroup of HBO cells. Nuclei are stained with DAPI (blue). Scale bars: (A–D) = 25 µm; (E) and (F) = 50 µm.
Figure 6.
mRNA of olfactory receptors and signal molecules is expressed in HBO cells. (A): Quantitative polymerase chain reaction (qPCR) experiments using the Taqman gene expression assay determined the expression of mRNA for ADCY3 (adenylyl cyclase III), Gng13 (guanine nucleotide binding protein (G protein), gamma 13), OR51E1, OR5P3, OR5A1, and OR2W1 olfactory molecules, and for T1R3 (sweet) and T2R38 (bitter) taste receptors, in HBO cells. The values for the relative mRNA expression were normalized based on the housekeeping gene GAPDH. (B): The mRNAs of Gng13, ADCY3, OR51E1, OR5P3, OR5A1, and OR2W1 are expressed in HBO cells at different levels with higher standard deviation.
Mammalian taste cells respond to odorants
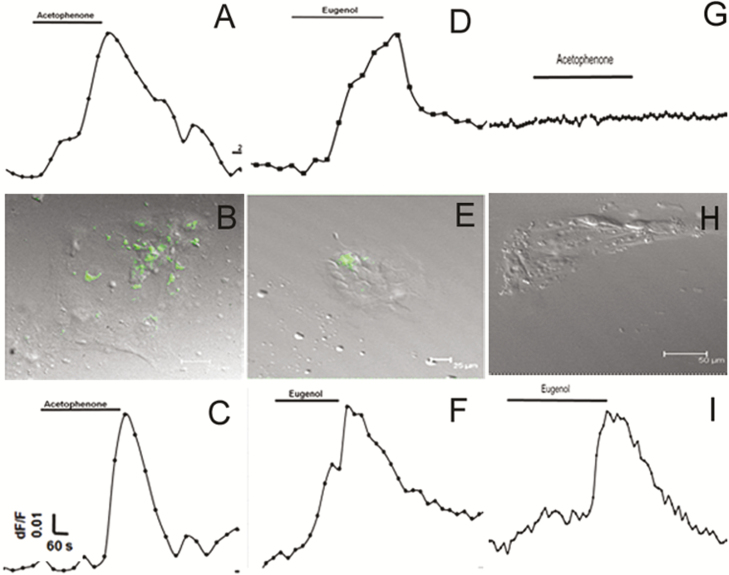
To examine the function of olfactory receptors in mouse and human taste cells, we measured changes in intracellular calcium levels in response to odorants. Figure 7 illustrates representative responses to odorants in individual cultured mouse taste cells. Cultured taste papilla cells from mOR-M71-GFP, mOR-EG-GFP, and wild-type mice (129/Sv and C57BL/6) responded to odorants at a concentration of 100 µM, indicating the presence of odorant-specific receptors in cultured mammalian taste cells. The mOR-M71-GFP and mOR-EG-GFP mouse taste cell cultures exhibited responses to acetophenone, a specific ligand for M71 (Figure 7A), and eugenol, a specific ligand for mOR-EG (Figure 7D). After calcium imaging, taste cells were fixed with 4% paraformaldehyde, and the location of responsive cells was confirmed using confocal microscopy. We found that these odor-responsive cells also expressed GFP (Figure 7B, E). Approximately 10% of taste cells in culture were GFP positive, although some of GFP-negative cells also responded to odorant exposure, indicating the presence of other odorant receptors that may also be targets for the odorants tested. The wild-type mice (129/Sv) also demonstrated responses to the odorants acetophenone and eugenol, indicating that functional odorant receptors are present in the taste cells of wild-type mice (Figure 7C, F). Majority of cells responsive to single odor, however, approximately 10% of cells found responsive to both odorant eugenol and acetophenone. Additional control experiments were performed using cultured taste cells obtained from C57BL/6 wild-type mice, where Olfr151 is a pseudogene (Fleischmann et al. 2008; Jiang et al. 2015). Taste cells obtained from C57BL/6 mice were responsive to eugenol but not to acetophenone (Figure 7G–I).
Figure 7.
Taste cells obtained from transgenic and wild-type mice are responsive to odorants. (A and B) and (D and E): GFP-positive cultured taste cells obtained from mOR-M71-GFP (A and B; n = 124) and mOR-EG-GFP (D and E; n = 118) transgenic mice were responsive to acetophenone (n = 14 of 124) and eugenol (n = 16 of 118), respectively. (A) and (D) are representative recordings from cultured taste cells fixed after calcium imaging; (B) and (E) demonstrate that odorant-responsive cells also express GFP. (C) and (F): Cultured taste cells obtained from a wild-type (129/Sv) mouse responded to odors (n = 6 of 62), indicating odor response is not due to genetic alteration of taste cells. (G–I): Cultured taste cells from wild-type (C57BL/6, n = 57) mice, which have pseudogene for the acetophenone receptor, did not respond to acetophenone but demonstrated response to eugenol (n = 3 of 57).
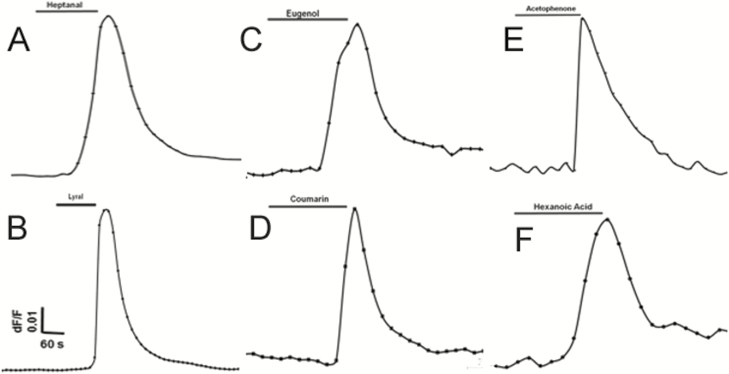
Next, we tested cultured human fungiform taste papilla (HBO) cells. The cells were loaded with the calcium-sensitive dye Fura-2 AM and exposed to individual odor stimuli: eugenol, hexanoic acid, coumarin, acetophenone, heptanal, and lyral. HBO cells showed increased intracellular calcium concentrations (Figure 8), demonstrating response to odorants, thus confirming the presence of functional odorant receptors. Majority of HBO cells were found responsive to single odor, however, approximately 5% of HBO cells (n = 235) were found responsive to two or more odorants. However, further detailed experiments are required to determine the relation between odorant induced response profile.
Figure 8.
Structurally diverse odorants evoke Ca2+ signals in HBO cells. Olfactory properties of HBO cells were examined using single-cell calcium imaging. HBO cells (n = 235) exposed to six different odorants (100 μM; A–F) demonstrated transient Ca2+ signals. Approximately 5% of HBO cells (n = 235) were found responsive to two or more odorants. Representative calcium imaging traces from odorant-stimulated HBO cells show changes in cytosolic intracellular Ca2+ levels as the F340/F380 fluorescence ratio. Each odorant was tested in at least 3 different independent experiments.
We also examined the breadth of tuning between odor-responsive taste cells and taste cells responsive to bitter stimuli. We established taste cell cultures from mOR-M71 mice and from mOR-EG mice and exposed them to the specific ligands acetophenone (M71 ligand) and eugenol (mOR-EG ligand), respectively, followed by a mixed bitter stimulus. All acetophenone-responding cells also responded to the mixed bitter stimulus (Figure 9A). Although most eugenol-responsive cells responded to bitter, a portion did not (Figure 9B). We then tested human taste (HBO) cells; all eugenol-responsive HBO cells also responded to bitter stimuli (Figure 9C), demonstrating that olfactory receptors can colocate with taste receptors. Figure 9D is a representative recording from single cell which responsive to both eugenol and bitter mixture. However, further detailed experiments are needed to examine the relation between odor and different taste modalities, such as sweet, umami, and fat. HBO cells demonstrated an increase in intracellular calcium concentration and the number of responsive cells in a concentration-dependent manner in response to eugenol (Figure 10), yielding an EC50 of 10 μM, similar to previous reports using olfactory receptor expression systems (Mainland et al. 2015; Oka et al. 2004).
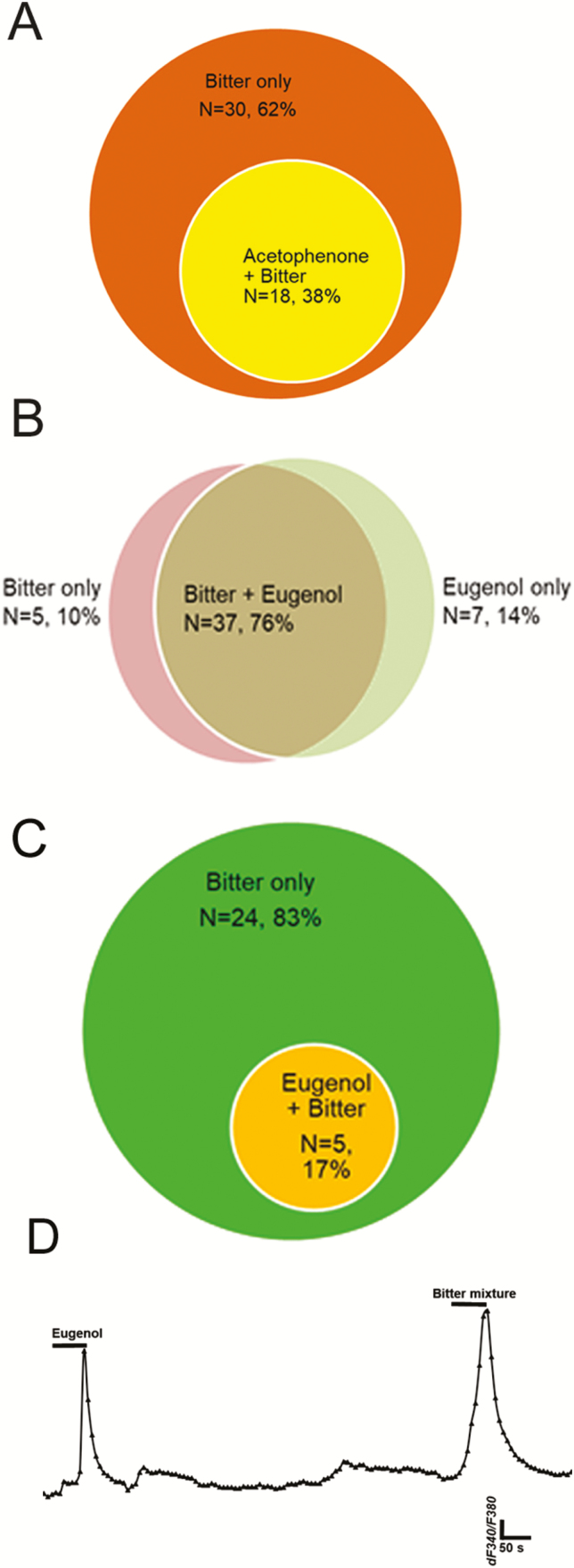
Figure 9.
Breadth of tuning of bitter-responsive taste cells to odorants acetophenone and eugenol. Venn diagrams show numbers of taste cells sensitive to bitter taste stimuli and to olfactory stimuli. (A): 38% (n = 18 of 48) of taste cells responding to the mixed bitter stimulus also responded acetophenone in M71 transgenic mice. (B): 76% of taste cells (n = 37 of 52) responded to both mixed bitter stimulus and eugenol in mEG transgenic mice; 10% and 14% of total responding cells responded to mixed bitter stimulus only or eugenol only, respectively. (C) and (D): In HBO cells, 83% of responding cells (n = 24 of 29) responded only to mixed bitter stimulus; 17% of cells (n = 5 of 29) responded to both eugenol and mixed bitter stimulus, indicating possible peripheral interaction with certain types of odor and taste receptors. (D): A representative recording from single cell which responsive to both eugenol and bitter mixture.
Figure 10.
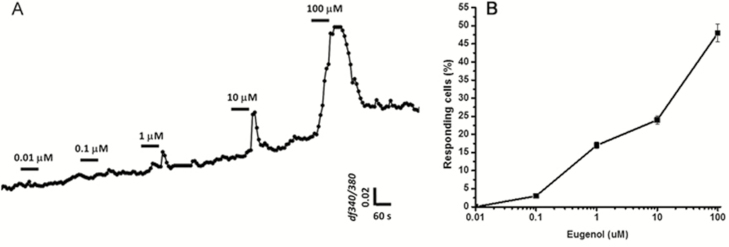
Concentration-dependent responses to eugenol measured in HBO cells. (A): Increasing eugenol concentrations resulted in concentration-dependent increases in the peak amplitude of intracellular calcium concentration changes. HBO cells (n = 9) were stimulated with eugenol at concentrations of 0.01, 0.1, 1, 10, and 100 μM. Data are shown as fluorescence intensity changes (ΔF340/F380) of 9 different cells combined. (B): Dose–response curve for eugenol. Data are shown as the percentage of responding cells compared with the total number of HBO cells (n = 5 experiments) in the same camera field, indicating that increasing concentrations of eugenol induced increasing numbers of responsive cells.
The role of adenylyl cyclase in odor-induced responses in taste cells
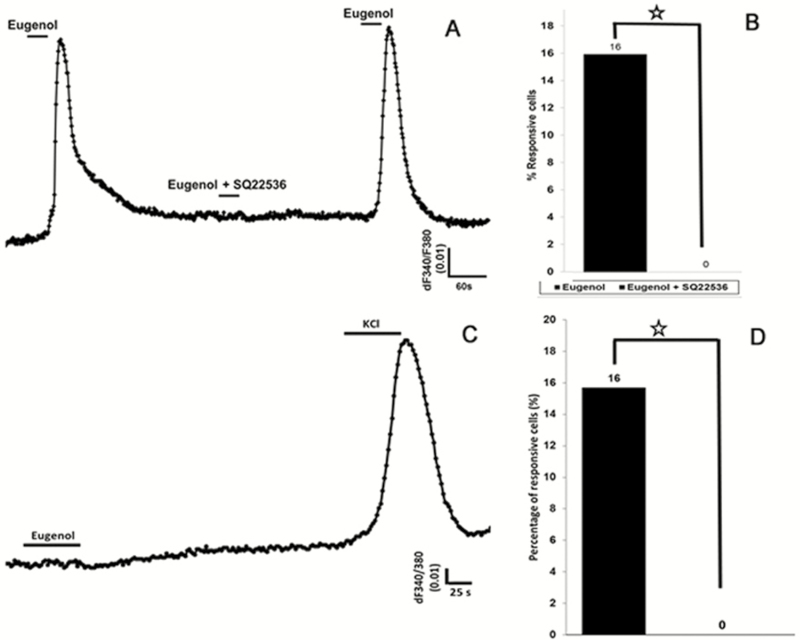
In mammals, the enzyme adenylyl cyclase, a major signaling molecule for olfactory transduction, also may play a role in the sweet signaling transduction pathway in taste cells (Schiffman et al. 1994; Firestein 2001). To test if adenylyl cyclase was indispensable for odor responses in taste cells, we used the potent adenylyl cyclase blocker SQ22536 (9-(tetrahydro-2-furanyl)-9H-purin-6-amine) (Guellaen et al. 1977). The application of SQ22536 resulted in reversible and complete inhibition of eugenol-induced responses in HBO cells (Figure 11A) that was statistically significant (P < 0.05; Figure 11B). We also transfected HBO cells with siRNA against adenylyl cyclase, which also eliminated the response to eugenol (Figure 11C, D; P > 0.05). The siRNA-transfected cells maintained a robust response to potassium chloride, confirming that the cells were still capable of responding (Schild and Restrepo 1998) (Figure 11C). HBO cells transfected with scrambled siRNA showed no significant differences in eugenol-induced responses compared with non-transfected cells (data not shown). Thus, a key component of olfactory signal transduction is present and functional in human taste cells.
Figure 11.
Eugenol-induced intracellular calcium changes in HBO cells are modulated by the enzyme adenylyl cyclase. (A) and (B): In the presence of the adenylyl cyclase III inhibitor SQ22536, the eugenol-induced response is eliminated. (A): Representative calcium recording from single cell showing responses to eugenol (100 µM) before and after treatment with SQ22536. After removing SQ22536, the eugenol-induced response recovered. (B): SQ22536 significantly inhibited the response to eugenol. Percentage of eugenol-responsive cells to total cells was examined with and without SQ22536 treatment. (C) and (D): HBO cells were transfected with small inhibitory RNA (siRNA) against adenylyl cyclase, which resulted in complete elimination of eugenol-induced responses. However, transfected HBO cells were responsive to high-concentration potassium chloride (50 mM), indicating the presence of voltage-gated potassium channels. *P < 0.05, t-test.
Discussion
Olfactory receptors are localized and expressed in the olfactory epithelium; in addition, expression of olfactory receptor mRNA and/or proteins has been demonstrated in a variety of other mammalian tissues (Ferrer et al. 2016; Veitinger and Hatt 2017), including tongue (Gaudin et al. 2001; Durzyński et al. 2005). Here we demonstrate that olfactory receptors are expressed specifically in taste cells on the tongue and are functional, using two transgenic mice, each co-expressing GFP with a different olfactory receptor, as well as cultured human fungiform taste (HBO) cells. To our knowledge, this is the first demonstration of functional olfactory receptor expression in mammalian taste cells.
In the transgenic mice, GFP expression colocalized with the taste-cell-specific marker PLC-β2, and in HBO cells, immunostaining and RT-PCR demonstrated that taste cells express the olfactory signaling molecules adenylyl cyclase, OMP, and Gα s/olf. Single-taste-cell calcium imaging with both transgenic mice lines revealed that GFP-positive cells are responsive to each receptor’s specific ligand, indicating that these olfactory receptors are functional. Calcium imaging experiments also showed that HBO taste cells functionally responded to odorants, and pharmacological and mRNA blockade of the olfactory-cell-specific enzyme adenylyl cyclase reversibly inhibited odorant responses in these taste cells. This report is the first direct evidence, to our knowledge, that physiologically active odorant receptors are expressed in mammalian taste cells, suggesting that olfactory receptors may play a role in the taste system.
Studies with transgenic mice have demonstrated that olfactory receptors detect odor in a combinatory manner: each odorant can be recognized by more than one receptor, and many olfactory receptors recognize multiple odorant molecules (Fleischmann et al. 2008; Godfrey et al. 2004; Oka et al. 2006). We observed here that taste cells obtained from both transgenic mouse models responded to specific odorants, as described earlier (Fleischmann et al. 2008). However, we also observed transgenic mice were also responsive to other odorants, which may be due to mixed genetic background of M71 mice (Bozza et al. 2002). In addition, studies have indicated various interactions of different chemical sensory receptors. An earlier study demonstrated the role of an olfactory binding protein (OBP49a) in inhibition of sweet taste by bitter compounds (Jeong et al. 2013), and the co-expression of TRPV1 (capsaicin-specific receptor) with sweet receptors or bitter receptors in rodent taste receptor cells has been demonstrated (Moon et al. 2010). Furthermore, capsaicin-altered sucrose taste preference has been demonstrated in TRPV1–/– mice, possibly through nonspecific TRPV1-independent mechanisms (Costa et al. 2005). The modulation of sweet and bitter taste with odorants and taste properties of odor molecules has been reported earlier demonstrating odor-taste interactions could take place by peripheral events at the receptor level (Suess et al. 2016; Lim et al. 2014; Barba et al. 2018) (Byram and Frank 1988). However, mechanisms of this interactions need to be examined in details at molecular level such odor – sweet, odor – bitter. Given our present results of coexpression of olfactory and taste receptors in taste cells, the role and function of odorant effects on specific taste receptors should be explored in future studies.
The enzyme adenylyl cyclase is a component of the canonical olfactory signal transduction cascade. We demonstrated by both pharmacological block and knockdown of adenylyl cyclase expression that it is indispensable for odor-induced taste cell responses. In contrast, blocking adenylyl cyclase mRNA by specific siRNA did not affect the activity of voltage-dependent calcium-activated potassium channels in human taste cells, demonstrating that these cells were still able to respond to some stimuli. It has been shown that type II taste cells may respond to multiple taste modalities (glutamate, bitter), as well as potassium (Hacker et al. 2008; Caicedo et al. 2000; Lewandowski et al. 2016). It also been shown that olfactory cells respond to potassium with an increase in calcium, as was also found in response to odorants (Gomez et al. 2005; Schild and Restrepo 1998). The presence and role of potassium conductance in olfactory receptor signaling cascade are well established (Gomez et al. 2000; Yamada and Nakatani 2001). Olfactory receptor neurons have a basal potassium conductance, which keeps the cell hyperpolarized. The presence of adenylyl cyclase in taste cells, specifically in type II cells, which have a role in sweet- and bitter-induced responses, has been reported, although gustducin was found to modulate basal cAMP levels (Clapp et al. 2008; Clapp et al. 2001; Abaffy et al. 2003). Future detailed studies should reveal the role of adenylyl cyclase in taste cell function.
Olfactory receptors expressed in taste cells may detect specific food-borne flavor compounds and alter taste responses peripherally. The olfactory and gustatory systems are known to interact, with olfactory stimuli enhancing specific gustatory qualities (Dalton et al. 2000) and gustatory stimuli lowering olfactory thresholds (Pfeiffer et al. 2005). This interaction is thought to first occur in the insula (Olson et al. 1998), but peripheral expression of olfactory receptors in taste receptor cells would provide an alternate pathway for cross talk. That said, a number of odor-taste interactions are ablated by blocking retronasal airflow (Tieman et al. 2012), suggesting that olfactory receptors expressed in taste cells are not the key drivers for at least a subset of odor-taste interactions. Alternatively, olfactory receptor function may be unrelated to volatile detection, for example, playing a role in of taste cell development.
Olfactory and taste receptors are seven-transmembrane heterotrimeric G protein-coupled receptors, which play a key role in detecting volatile odorants and in detecting taste molecules, respectively. Microarray and RNA-sequencing analyses demonstrate expression of olfactory receptor profiles in nonolfactory tissues (Feldmesser et al. 2006; Flegel et al. 2013). However, the function of olfactory receptors in nonolfactory tissues remains largely elusive. The difficulties of identifying the expression and physiological functions of olfactory receptors in nonolfactory tissues may be due to the low expression levels of olfactory proteins. In addition, there are few specific antibodies to olfactory receptors. The generation of an antibody library and the establishment of a chemical library for agonists and antagonists to specific olfactory receptors are critical for study of olfactory receptor expression outside of the olfactory epithelium.
The current screening platform for olfactory receptors is based on a heterologous expression system, which requires several accessory proteins that enhance the rate of intracellular trafficking for functional expression of olfactory receptors on cellular membranes (Bozza et al. 2002). Recombinant olfactory receptors expressed in heterologous cells are trapped in the endoplasmic reticulum and rarely translocate to the plasma membrane, where the receptors gain access to odorants (McClintock and Sammeta 2003). The coexpression of cell-type-specific chaperone (accessory) proteins may be necessary to facilitate olfactory receptor trafficking and enable effective and functional expression of olfactory receptors on the cellular membrane (Saito et al. 2004). The current heterologous system has identified ligands for only ~50 of the more than 400 olfactory receptors, suggesting that a crucial component may be missing. Alternatively, human taste cells, which we have shown natively express odorant receptors, with responses to odorants similar to those obtained in olfactory cells, may allow researchers to identify ligands for orphan olfactory receptors.
If taste cells express functional olfactory receptors, this raises the intriguing possibility that taste and olfaction might overlap more than previously thought. As seen in insect cells, odor may have modulatory effects in mammalian taste cells. Human perception of food flavors involves the integration of taste and olfactory sensations, and it has been found that odor’s influence on taste perception depends on both odor and taste molecules (Francis et al. 2004; Byram and Frank 1988). Taste and olfactory perception are important for food acceptance and rejection both individually and in combination. However, the olfactory system, in particular, is critical to detect food volatiles before tasting, to discriminate between safe and dangerous foods. Therefore, volatile compounds (i.e., the aroma of food) may have significant contributions to perceived taste. Both ortho- and retronasal olfaction are particularly crucial to detect the flavor of food (Tieman et al. 2012). The olfactory receptors expressed in taste cells may detect specific food-borne flavor compounds and react to food uptake by postprandially responding to the same food-borne chemicals that the chemosensory systems—olfaction and taste—encountered preprandially.
Genetic variation in olfactory receptors can influence eating behaviors as well as adiposity (Choquette et al. 2012). The presence of functional olfactory receptors on taste cells may point to new directions in the manipulation of food flavor to reduce the risk of extreme obesity by controlling overeating in genetically predisposed subjects (Zhe et al. 2017). Further studies are needed to examine whether the odors detected by taste cells acquire a new hedonic value, inducing appetitive or aversive behaviors. Thus, the functional role and interaction of olfactory and taste receptors in taste cells should be further examined in future experiments.
Funding
Monell Center’s Histology and Cellular Localization Core was supported, in part, by National Institutes of Health-NIDCD core [P30DC011735].
Conflict of interest
A patent portfolio covering methods for culturing mammalian taste cells and identifying modulators of olfactory receptors has been filed by Monell Chemical Senses Center, and M.H.O. is named as one of the inventors. M.H.O. received research support and royalties. The other authors declare no competing interests. In accordance with Chemical Senses policy and my ethical obligation as a researcher, I am reporting that I am an inventor on the patent portfolio which covers “culturing mammalian taste cells and identifying modulators of olfactory receptors” and received royalties and research support but not having other financial transaction. I have disclosed those interests fully to Chemical Senses.
Acknowledgments
M.H.O. thanks Dr. Johannes Reisert and Dr. Kazushige Touhara for providing genetically engineered mice used in this study. The authors also thank Dr Gilad Barnea of Brown University for kindly providing M71 antibodies. Histology and imaging experiments were performed at the Monell Center’s Histology and Cellular Localization Core. The authors acknowledge Drs Joel Mainland and Casey Trimmer for editorial assistance and suggestions. Authors thank to Dr Alvaro Garcia-Blanco for technical support during part of this project.
References
- Abaffy T, Trubey KR, Chaudhari N. 2003. Adenylyl cyclase expression and modulation of cAMP in rat taste cells. Am J Physiol Cell Physiol. 284:C1420–C1428. [DOI] [PubMed] [Google Scholar]
- Barba C, Beno N, Guichard E, Thomas-Danguin T. 2018. Selecting odorant compounds to enhance sweet flavor perception by gas chromatography/olfactometry-associated taste (GC/O-AT). Food Chem. 257:172–181. [DOI] [PubMed] [Google Scholar]
- Bartoshuk LM, Klee HJ. 2013. Better fruits and vegetables through sensory analysis. Curr Biol. 23:R374–R378. [DOI] [PubMed] [Google Scholar]
- Bozza T, Feinstein P, Zheng C, Mombaerts P. 2002. Odorant receptor expression defines functional units in the mouse olfactory system. J Neurosci. 22:3033–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T, Voland P, Kunz L, Prinz C, Gratzl M. 2007. Enterochromaffin cells of the human gut: sensors for spices and odorants. Gastroenterology. 132:1890–1901. [DOI] [PubMed] [Google Scholar]
- Byram J, Frank RA. 1988. Taste–smell interactions are tastant and odorant dependent. Chem Senses. 13:445–455. [Google Scholar]
- Caicedo A, Jafri MS, Roper SD. 2000. In situ Ca2+ imaging reveals neurotransmitter receptors for glutamate in taste receptor cells. J Neurosci. 20:7978–7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquette AC, Bouchard L, Drapeau V, Lemieux S, Tremblay A, Bouchard C, Vohl MC, Pérusse L. 2012. Association between olfactory receptor genes, eating behavior traits and adiposity: results from the Quebec Family Study. Physiol Behav. 105:772–776. [DOI] [PubMed] [Google Scholar]
- Clapp TR, Stone LM, Margolskee RF, Kinnamon SC. 2001. Immunocytochemical evidence for co-expression of Type III IP3 receptor with signaling components of bitter taste transduction. BMC Neurosci. 2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapp TR, Trubey KR, Vandenbeuch A, Stone LM, Margolskee RF, Chaudhari N, Kinnamon SC. 2008. Tonic activity of Galpha-gustducin regulates taste cell responsivity. FEBS Lett. 582:3783–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzelmann S, Levai O, Bode B, Eisel U, Raming K, Breer H, Strotmann J. 2000. A novel brain receptor is expressed in a distinct population of olfactory sensory neurons. Eur J Neurosci. 12:3926–3934. [DOI] [PubMed] [Google Scholar]
- Costa RM, Liu L, Nicolelis MAL, Simon SA. 2005. Gustatory effects of capsaicin that are independent of TRPV1 receptors. Chem Senses. 30:i198–i200. [DOI] [PubMed] [Google Scholar]
- Dalton P, Doolittle N, Nagata H, Breslin PA. 2000. The merging of the senses: integration of subthreshold taste and smell. Nat Neurosci. 3:431–432. [DOI] [PubMed] [Google Scholar]
- Durzyński L, Gaudin JC, Myga M, Szydłowski J, Goździcka-Józefiak A, Haertlé T. 2005. Olfactory-like receptor cDNAs are present in human lingual cDNA libraries. Biochem Biophys Res Commun. 333:264–272. [DOI] [PubMed] [Google Scholar]
- Elliott RA, Kapoor S, Tincello DG. 2011. Expression and distribution of the sweet taste receptor isoforms T1R2 and T1R3 in human and rat bladders. J Urol. 186:2455–2462. [DOI] [PubMed] [Google Scholar]
- Feingold EA, Penny LA, Nienhuis AW, Forget BG. 1999. An olfactory receptor gene is located in the extended human beta-globin gene cluster and is expressed in erythroid cells. Genomics. 61:15–23. [DOI] [PubMed] [Google Scholar]
- Feldmesser E, Olender T, Khen M, Yanai I, Ophir R, Lancet D. 2006. Widespread ectopic expression of olfactory receptor genes. BMC Genomics. 7:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer I, Garcia-Esparcia P, Carmona M, Carro E, Aronica E, Kovacs GG, Grison A, Gustincich S. 2016. Olfactory receptors in non-chemosensory organs: the nervous system in health and disease. Front Aging Neurosci. 8:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein S. 2001. How the olfactory system makes sense of scents. Nature. 413:211–218. [DOI] [PubMed] [Google Scholar]
- Flegel C, Manteniotis S, Osthold S, Hatt H, Gisselmann G. 2013. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS One. 8:e55368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann A, Shykind BM, Sosulski DL, Franks KM, Glinka ME, Mei DF, Sun Y, Kirkland J, Mendelsohn M, Albers MW, et al. . 2008. Mice with a “monoclonal nose”: perturbations in an olfactory map impair odor discrimination. Neuron. 60:1068–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis J, Prescott J, Johnstone V. 2004. Odor–taste interactions: effects of attentional strategies during exposure. Chem Senses. 29:331–340. [DOI] [PubMed] [Google Scholar]
- Fujita T. 1991. Taste cells in the gut and on the tongue. Their common, paraneuronal features. Physiol Behav. 49:883–885. [DOI] [PubMed] [Google Scholar]
- Gaudin JC, Breuils L, Haertlé T. 2001. New GPCRs from a human lingual cDNA library. Chem Senses. 26:1157–1166. [DOI] [PubMed] [Google Scholar]
- Godfrey PA, Malnic B, Buck LB. 2004. The mouse olfactory receptor gene family. Proc Natl Acad Sci U S A. 101:2156–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez G, Lischka FW, Haskins ME, Rawson NE. 2005. Evidence for multiple calcium response mechanisms in mammalian olfactory receptor neurons. Chem Senses. 30:317–326. [DOI] [PubMed] [Google Scholar]
- Gomez G, Rawson NE, Hahn CG, Michaels R, Restrepo D. 2000. Characteristics of odorant elicited calcium changes in cultured human olfactory neurons. J Neurosci Res. 62:737–749. [DOI] [PubMed] [Google Scholar]
- Gong T, Wei Q, Mao D, Shi F. 2016. Expression patterns of taste receptor type 1 subunit 3 and α-gustducin in the mouse testis during development. Acta Histochem. 118:20–30. [DOI] [PubMed] [Google Scholar]
- Green BG, Nachtigal D, Hammond S, Lim J. 2012. Enhancement of retronasal odors by taste. Chem Senses. 37:77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guellaen G, Mahu JL, Mavier P, Berthelot P, Hanoune J. 1977. RMI 12330 A, an inhibitor of adenylate cyclase in rat liver. Biochim Biophys Acta. 484:465–475. [DOI] [PubMed] [Google Scholar]
- Hacker K, Laskowski A, Feng L, Restrepo D, Medler K. 2008. Evidence for two populations of bitter responsive taste cells in mice. J Neurophysiol. 99:1503–1514. [DOI] [PubMed] [Google Scholar]
- Hiroi M, Tanimura T, Marion-Poll F. 2008. Hedonic taste in Drosophila revealed by olfactory receptors expressed in taste neurons. PLoS One. 3:e2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura S, Ohno K, Ueki T, Sato K, Kanayama N. 2006. Expression of Golf in the rat placenta: Possible implication in olfactory receptor transduction. Placenta. 27:103–108. [DOI] [PubMed] [Google Scholar]
- Jeong YT, Shim J, Oh SR, Yoon HI, Kim CH, Moon SJ, Montell C. 2013. An odorant-binding protein required for suppression of sweet taste by bitter chemicals. Neuron. 79:725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Gong NN, Hu XS, Ni MJ, Pasi R, Matsunami H. 2015. Molecular profiling of activated olfactory neurons identifies odorant receptors for odors in vivo. Nat Neurosci. 18:1446–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Carlson JR. 2002. Gene discovery by e-genetics: Drosophila odor and taste receptors. J Cell Sci. 115:1107–1112. [DOI] [PubMed] [Google Scholar]
- Kiuchi S, Yamada T, Kiyokawa N, Saito T, Fujimoto J, Yasue H. 2006. Genomic structure of swine taste receptor family 1 member 3, TAS1R3, and its expression in tissues. Cytogenet Genome Res. 115:51–61. [DOI] [PubMed] [Google Scholar]
- Lewandowski BC, Sukumaran SK, Margolskee RF, Bachmanov AA. 2016. Amiloride-insensitive salt taste is mediated by two populations of Type III taste cells with distinct transduction mechanisms. J Neurosci. 36:1942–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Fujimaru T, Linscott TD. 2014. The role of congruency in taste–odor interactions. Food Qual Prefer. 34:5–13 [Google Scholar]
- Mainland JD, Li YR, Zhou T, Liu WL, Matsunami H. 2015. Human olfactory receptor responses to odorants. Sci Data. 2:150002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malki A, Fiedler J, Fricke K, Ballweg I, Pfaffl MW, Krautwurst D. 2015. Class I odorant receptors, TAS1R and TAS2R taste receptors, are markers for subpopulations of circulating leukocytes. J Leukoc Biol. 97:533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolskee RF, Dyer J, Kokrashvili Z, Salmon KS, Ilegems E, Daly K, Maillet EL, Ninomiya Y, Mosinger B, Shirazi-Beechey SP. 2007. T1R3 and gustducin in gut sense sugars to regulate expression of Na+-glucose cotransporter 1. Proc Natl Acad Sci U S A. 104:15075–15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Max M, Shanker YG, Huang L, Rong M, Liu Z, Campagne F, Weinstein H, Damak S, Margolskee RF. 2001. Tas1r3, encoding a new candidate taste receptor, is allelic to the sweet responsiveness locus Sac. Nat Genet. 28:58–63. [DOI] [PubMed] [Google Scholar]
- McClintock TS, Sammeta N. 2003. Trafficking prerogatives of olfactory receptors. Neuroreport. 14:1547–1552. [DOI] [PubMed] [Google Scholar]
- Meyer D, Voigt A, Widmayer P, Borth H, Huebner S, Breit A, Marschall S, de Angelis MH, Boehm U, Meyerhof W, et al. . 2012. Expression of Tas1 taste receptors in mammalian spermatozoa: functional role of Tas1r1 in regulating basal Ca²⁺ and cAMP concentrations in spermatozoa. PLoS One. 7:e32354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon YW, Lee JH, Yoo SB, Jahng JW. 2010. Capsaicin receptors are colocalized with sweet/bitter receptors in the taste sensing cells of circumvallate papillae. Genes Nutr. 5:251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa Y, Nagasawa M, Yamada S, Hara A, Mogami H, Nikolaev VO, Lohse MJ, Shigemura N, Ninomiya Y, Kojima I. 2009. Sweet taste receptor expressed in pancreatic beta-cells activates the calcium and cyclic AMP signaling systems and stimulates insulin secretion. PLoS One. 4:e5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y, Katada S, Omura M, Suwa M, Yoshihara Y, Touhara K. 2006. Odorant receptor map in the mouse olfactory bulb: in vivo sensitivity and specificity of receptor-defined glomeruli. Neuron. 52:857–869. [DOI] [PubMed] [Google Scholar]
- Oka Y, Omura M, Kataoka H, Touhara K. 2004. Olfactory receptor antagonism between odorants. EMBO J. 23:120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson TH, Riedl MS, Vulchanova L, Ortiz-Gonzalez XR, Elde R. 1998. An acid sensing ion channel (ASIC) localizes to small primary afferent neurons in rats. Neuroreport. 9:1109–1113. [DOI] [PubMed] [Google Scholar]
- Otaki JM, Yamamoto H, Firestein S. 2004. Odorant receptor expression in the mouse cerebral cortex. J Neurobiol. 58:315–327. [DOI] [PubMed] [Google Scholar]
- Ozdener H, Spielman AI, Rawson NE. 2012. Isolation and culture of human fungiform taste papillae cells. e3730. [DOI] [PMC free article] [PubMed]
- Ozdener H, Yee KK, Cao J, Brand JG, Teeter JH, Rawson NE. 2006. Characterization and long-term maintenance of rat taste cells in culture. Chem Senses. 31:279–290. [DOI] [PubMed] [Google Scholar]
- Ozdener MH, Brand JG, Spielman AI, Lischka FW, Teeter JH, Breslin PA, Rawson NE. 2011. Characterization of human fungiform papillae cells in culture. Chem Senses. 36:601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdener MH, Rawson NE. 2013. Primary culture of mammalian taste epithelium. In: Randell SH, Fulcher ML, editors. Epithelial cell culture protocols. 2nd ed. Totowa (NJ): Humana Press; p. 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmentier M, Libert F, Schurmans S, Schiffmann S, Lefort A, Eggerickx D, Ledent C, Mollereau C, Gérard C, Perret J. 1992. Expression of members of the putative olfactory receptor gene family in mammalian germ cells. Nature. 355:453–455. [DOI] [PubMed] [Google Scholar]
- Pfeiffer JC, Hollowood TA, Hort J, Taylor AJ. 2005. Temporal synchrony and integration of sub-threshold taste and smell signals. Chem Senses. 30:539–545. [DOI] [PubMed] [Google Scholar]
- Pluznick JL, Zou DJ, Zhang X, Yan Q, Rodriguez-Gil DJ, Eisner C, Wells E, Greer CA, Wang T, Firestein S, et al. . 2009. Functional expression of the olfactory signaling system in the kidney. Proc Natl Acad Sci U S A. 106:2059–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Mummalaneni S, Phan TT, Heck GL, DeSimone JA, West D, Mahavadi S, Hojati D, Murthy KS, Rhyu MR, et al. . 2017. Cyclic-AMP regulates postnatal development of neural and behavioral responses to NaCl in rats. PLoS One. 12:e0171335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Zhou L, Terwilliger R, Newton SS, de Araujo IE. 2009. Sweet taste signaling functions as a hypothalamic glucose sensor. Front Integr Neurosci. 3:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. 2004. RTP family members induce functional expression of mammalian odorant receptors. Cell. 119:679–691. [DOI] [PubMed] [Google Scholar]
- Schiffman SS, Gatlin LA, Suggs MS, Heiman SA, Stagner WC, Erickson RP. 1994. Modulators of the adenylate cyclase system can alter electrophysiological taste responses in gerbil. Pharmacol Biochem Behav. 48:983–990. [DOI] [PubMed] [Google Scholar]
- Schild D, Restrepo D. 1998. Transduction mechanisms in vertebrate olfactory receptor cells. Physiol Rev. 78:429–466. [DOI] [PubMed] [Google Scholar]
- Schmittgen K, Livak T. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔ Ct method. Methods. 25:402–408. [DOI] [PubMed] [Google Scholar]
- Shah AS, Ben-Shahar Y, Moninger TO, Kline JN, Welsh MJ. 2009. Motile cilia of human airway epithelia are chemosensory. Science. 325:1131–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small DM. 2012. Flavor is in the brain. Physiol Behav. 107:540–552. [DOI] [PubMed] [Google Scholar]
- Subramaniam S, Ozdener MH, Abdoul-Azize S, Saito K, Malik B, Maquart G, Hashimoto T, Marambaud P, Aribi M, Tordoff MG, et al. . 2016. ERK1/2 activation in human taste bud cells regulates fatty acid signaling and gustatory perception of fat in mice and humans. FASEB J. 30:3489–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suess B, Brockhoff A, Meyerhof W, Hofmann T. 2016. The Odorant (R)-citronellal attenuates caffeine bitterness by inhibiting the bitter receptors TAS2R43 and TAS2R46. J Agric Food Chem. 66:2301–2311. [DOI] [PubMed] [Google Scholar]
- Taniguchi K. 2004. Expression of the sweet receptor protein, T1R3, in the human liver and pancreas. J Vet Med Sci. 66:1311–1314. [DOI] [PubMed] [Google Scholar]
- Tieman D, Bliss P, McIntyre LM, Blandon-Ubeda A, Bies D, Odabasi AZ, Rodríguez GR, van der Knaap E, Taylor MG, Goulet C, et al. . 2012. The chemical interactions underlying tomato flavor preferences. Curr Biol. 22:1035–1039. [DOI] [PubMed] [Google Scholar]
- Tizzano M, Cristofoletti M, Sbarbati A, Finger TE. 2011. Expression of taste receptors in solitary chemosensory cells of rodent airways. BMC Pulm Med. 11:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitinger S, Hatt H. 2017. Ectopic expression of mammalian olfactory receptors. In: Buettner A, editor. Springer handbook of odor. Cham: Springer International Publishing; p. 83–84. [Google Scholar]
- Weber M, Pehl U, Breer H, Strotmann J. 2002. Olfactory receptor expressed in ganglia of the autonomic nervous system. J Neurosci Res. 68:176–184. [DOI] [PubMed] [Google Scholar]
- Yamada H, Nakatani K. 2001. Odorant-induced hyperpolarization and suppression of cAMP-activated current in newt olfactory receptor neurons. Chem Senses. 26:25–34. [DOI] [PubMed] [Google Scholar]
- Zhe C, Hong Z, Nian F, Linxi C. 2017. The diversified function and potential therapy of ectopic olfactory receptors in non-olfactory tissues. J Cell Physiol. 233:2104–2115. [DOI] [PubMed] [Google Scholar]