Abstract
The novel bioactive actinobacterial strain GSBNT10 obtained from a Saharan soil, was taxonomically characterized using a polyphasic approach. 16S rRNA gene sequence analysis supported the classification of the isolate within the genus Streptomyces indicating it as a novel species. The major metabolite responsible of the bioactivity was purified and structurally characterized as actinomycin D (act-D) by mass spectrometric and nuclear magnetic resonance analyses Plackett-Burman design (PBD) and response surface methodology (RSM) were applied in order to optimize the medium formulation for the production of this bioactive metabolite. By PBD experiments, NaNO3, K2HPO4 and initial pH value were selected as significant variables affecting the metabolite production. Central Composite Design (CCD) showed that adjustment of the fermentative medium at pH 8.25, K2HPO4 at 0.2 gL-1 and NaNO3 at 3.76 gL-1 were the values suiting the production of act-D. Moreover, the results obtained by the statistical approach were confirmed by act-D detection using the HPLC equipped with a diode array detector and coupled online with electrospray-mass spectrometry (ESIMS) technique. act-D production was highly stimulated, obtaining a good yield (656.46 mgL-1) which corresponds to a 58.56% increase compared with the non-optimized conditions and data from LC-ESIMS technique efficiently confirmed the forecast from RSM.
Keywords: Microbiology, Actinomycin D, Central composite design, Optimization, Plackett-Burman design, Saharan saline soil, Streptomyces sp., Electrospray mass spectrometry
1. Introduction
The chromopeptide lactone actinomycin D (act-D, also known as dactinomycin) is structurally composed of a phenoxazone chromophore containing a quinonimine portion, responsible of the red color and its intercalative ability, and two cyclic pentapeptide lactone rings (Williams and Katz, 1977). Since the first isolation of actinomycin D in 1940 from Actinomyces antibioticus, further actinomycins have been isolated from mutants of the original strain and other species of Streptomyces and Micromonospora (Avendaño; Menéndez, 2008).
Although its limited clinical use as antibiotic, due to its toxic effects, act-D remains a commonly used anticancer drug administrated intravenously and used in pediatric tumors and embryonal rhabdomyosarcoma (Eun, 1996), currently employed for the treatment of several human neoplasias and highly aggressive malignancies (Souza et al., 2002; Lohani et al., 2016) such as pancreatic (Kleeff et al., 2000) and Wilm's tumor, and in combined chemotherapies for the treatment of risk cancers as embryonal tumor with multilayerd rosettes brain tumor (ETMR) (Schmidt et al., 2017). It acts as a transcription inhibitor by intercalating into DNA between adjacent guanine–cytosine base pairs inhibiting primarily cellular transcription (Singh et al., 2010). Due to its application as therapeutic agent for the treatment of some types of cancers, a great interest in increasing the yield of act-D was addressed towards different fermentation conditions and various microorganisms (Kurosawa et al., 2006; Dalili and Chau, 1988; Praveen et al., 2008a, 2008b; Hamza et al., 2013; Wei et al., 2017).
The production of act-D obtained so far by most strains is low and an optimization strategy has been generally undertaken, focusing great interest. It is true in the light of the highly complex structure of this molecule, so that fermentation process remains the method of choice for its output and of similar metabolites. This production method gives practical advantages compared to organic synthesis in obtaining the enantiopure form of metabolites which, as act-D, are structurally rich in stereogenic centers.
There is a continuous interest to exploit statistical methodologies in different biotechnological processes, especially in the production of valuable products in an optimized yield (Goupy, 1999). The ability of microorganisms to produce bioactive compounds is greatly affected by different conditions of nutrients and/or cultivation (Krassilnikov, 1960), therefore medium optimization is still a crucial point to be investigated with the aim to achieve the maximum product concentration. It is above all a crucial aspect in the large scale production of bioactive metabolites, to reduce the overall costs and time, and to achieve an industrial process efficiency (Singh et al., 2017).
Recently, many attempts have been made to optimize them using statistical experiment designs such as Taguchi method (Mahalaxmi et al., 2009), factorial fractional design (FFD) (Fontes et al., 2012), central composite design (CCD) (Srinivasulu et al., 2006; Djinni et al., 2018) and Box-Bunken design (BBD) (Vijayabharathi et al., 2012; Kim et al., 2014). The process through statistical approach such as response surface methodology (RSM) is an approach resulting economic, efficient and accurate (Ahsan et al., 2017). Known for the less number of experimental runs and for regarding the interaction among of the process variables involved, it allows to maximize the microbial secondary metabolites yield and to estimate the relevance of the strain for its use in an industrial scale production.
The present work reports on i) the isolation and characterization of the novel actinobacteria strain Streptomyces sp. GSBNT10, ii) the purification and structural elucidation of the therapeutic agent act-D and iii) the optimized production of this main metabolite, as established by HPLC-MS analysis of crude extracts obtained by non-optimized and statistically selected optimized culture conditions.
2. Materials and methods
2.1. Isolation of actinobacterial strains and screening for their antimicrobial activity
The actinobacterial strain studied herein was isolated from a Saharan saline soil samples collected at Beni-Abbes locality in Bechar (South Ouest of Algeria) in 2011. Ten soil samples were collected, kept at 4 °C and transported to the laboratory for processing. Decimal dilutions were made up 10−8 and spread on selective agar media in triplicates Petri plates then incubated at 28 °C for four weeks. Three selective culture media were used: Gausse, SCA and Chitin agar media, all of them were supplemented with K2Cr2O7 (50 μgmL−1) as antifungal agent.
The isolated actinobacteria were initially screened for their antagonistic potency against a standard Gram positive and negative test bacteria at 107 CFUmL−1: E. coli ATCC 25922, S. aureus ATCC 25923, MRSA ATCC 43300 and P. aeruginosa ATCC 27853, using plug agar essay. The inhibition zones were noted by measuring the diameter (in mm) of the clearing zones around the plugs. The most potent strain harboring an important antibacterial potential was selected for further study.
2.2. Characterization and identification of the selected bioactive strain
Cultural features of the selected strain were determined on various international Streptomyces projects (ISP) media (ISP1-ISP7). The strain's morphology was examined under optic (Leica DM 300) and phase contrast microscopes at various magnification (Leica EZ4D) as well as under Scanning Electron Microscopy (SEM, JOEL-JSM-700IF, Field Effect Gun) (Kumar et al., 2011). Physiological properties of the selected isolate were investigated based on the lytic activity towards several organic compounds as described by Gordon et al. (1974) for starch, casein, tyrosine and gelatin hydrolysis. The ability of the strain in utilizing various carbohydrates as sole carbon sources was studied using ISP9 medium supplemented with 1% (w/v) of the carbon source. Moreover, NaCl tolerance was determined on Gausse medium as well as the growth response to different temperatures and pH.
The chemotaxonomic analysis of the selected isolate was carried out using standard procedures. The determination of diaminopimelic acid isomers and the whole cell sugar composition were performed according to Staneck and Roberts (1974) and through Evaporative Light Scattering Detector (ELSD)-HPLC technique using Lechevalier and Lechevalier (1970) procedure for the sample and standards preparation where the concentration was 1 mgmL−1 in acetonitrile/water (70:30).
The analysis was performed by an ELSD-HPLC Agilent 1100 series using reversed phase C18 (Phenomenex Torrance, CA, USA; HELIC, Luna 5 μm, 250 × 4.60mm) column in isocratic mode (acetonitrile/water 90:10, flow:1 mLmin-1, at 45 °C).
After extraction of lipids using chloroform/methanol/water (5: 4.5: 4 v/v) according to Pasciak et al. (2003). Total lipid extracts were analyzed by LC-MS technique, using a reversed phase RP-18 column and gradient mobile phase (solvent A: 100% methanol, solvent B: methanol/water 70:30 with 28 mM aqueous ammonium acetate).
2.3. Taxonomy and phylogenetic analysis of the selected isolate
The 16S rRNA gene was amplified using 16S rRNA universal primers S127F (5′AGAGTTTGATCCTGGCTCAG-3′) and S21525R (5′-AAGGAGGTGATCCAAGCC-3′) (Gürtler and Stanisich, 1996). The PCR product was purified and sequenced using Applied Biosystems 3730XL DNA Analyzer. The almost-complete 16S rRNA gene sequence (1491 nt) of strain Streptomyces sp. GSBNT10 was determined and deposited under accession number MF399050.1 in the GenBank database. EzBioCloud's Identify Service (http://www.ezbiocloud.net/identify) (Yoon et al., 2017) was performed to determine the percentage of similarity with the closest bacteria. Multiple alignment with sequences from closely related species was performed using the program CLUSTAL W (with default parameters) in MEGA version 6 (Tamura et al., 2013). Phylogenetic tree was inferred using the neighbour-joining algorithm (Saitou and Nei, 1987) with the model of Jukes and Cantor (1969). Topology of the resultant tree was evaluated by bootstrap analyses (Felsenstein, 1985) based on 1000 resamplings.
2.4. Strain fermentation and act-D extraction
2.4.1. Spore suspension and culture conditions
Spore suspension of GSBNT10 was prepared in 10 mL distilled water from cultures grown on Czapeck medium at 28 °C for 7 days. The suspension was then agitated on a vortex for about one minute to break up the spore chains. The number of spores mL−1 was adjusted to 1 by measuring the optical density (O.D.) of the spore suspension at 600nm. At O.D. = 1 the number of spores in the suspension is about 107 spores mL−1 (Kieser et al., 2000).
2.4.2. Selection of basal medium
Five culture media (SCA, ISP2, GYEA, Gausse and Czapeck) were used in comparative studies to select the optimal nutrient medium. The strain was cultivated at 107 CFUmL−1 for 7 days at 28 °C. Antimicrobial activity was performed by agar plugs method. The medium in which the strain exhibited the highest bioactivity expressed in terms of inhibition zone was used as the basic medium for the next statistical optimization.
2.4.3. Extraction, purification and identification of act-D
GSBNT10 strain was cultivated on 100 Petri plates containing 40 mL of Czapeck medium at 28 °C. After 6 days of incubation, the seed culture (mycelia and agar) were mixed together then extracted twice with ethyl acetate then subjected to sonication for 1h and maceration overnight. The combined organic phases were filtered, evaporated in vacuo, and the residue was dried to afford an orange crude extract (414 mg). It was subjected to TLC using dichloromethane–methanol (9:1) as mobile phase, then to silica gel column chromatography using a gradient of dichloromethane –methanol yielding two 20 mL fractions. The bioactivity guided purification done for both the fractions was realized based on the well method using Mueller Hinton medium testing 100μL of crude extract.
HPLC analysis was performed on an Agilent 1100 series chromatography system (Agilent Technologies, USA) with a photodiode array detector to purify the active fractions.
Nuclear Magnetic Resonance (NMR) experiments were achieved for the molecular structure elucidation. NMR spectra were recorded by an Advance 400 Bruker spectrometer; 1H at 400 MHz and 13C at 100 MHz in CD3OD (δH 3.31 ppm, δC 49.00ppm, relative to tetramethylsilane). Structural assignements are from Heteronuclear Single Quantum Correlation (HSQC) and Heteronuclear Multiple Bond Correlation (HMBC) experiments. Mass Spectrometry (MS) analysis was carried out to establish molecular mass of each compound. ESI-MS data and tandem fragmentation spectra (MS/MS) were recorded by using a Bruker Esquire LC ion trap mass spectrometer, equipped with an ESI ion source in positive ion mode, through injection of the sample into the source from a methanol solution. MS conditions: source temp. 300 °C, nebulizing gas N2, 4 L min-1, cone voltage 32 V, scan range m/z = 100–1400.
2.5. Statistical optimization of act-D production
2.5.1. Quantitative evaluation of act-D
act-D amount was estimated using UV-Vis spectroscopic method. The concentration expressed in mg/L was determined for each of the prepared crude extract according to Beer-Lambert equation measuring the absorbance at 440 nm. Experiments were conducted in duplicates and the values were combined to give overall production level.
2.5.2. Screening of medium components using Plackett Burman design
In order to determine the most influencing factors for act-D production, Czapeck medium composition was analyzed using Plackett Burman design (PBD). Twelve experiments were conducted to evaluate the effect of six factors (Starch, KCl, NaNO3, K2HPO4, MgSO4·7H2O, pH) at two levels (Table 1) on the metabolite production. The experiments were conducted in duplicates and the average values of act-D concentration (mgL−1) were taken as the response (Table 2). The analysis of variance for the data and the model coefficients were computed using Minitab 16.0 (Minitab Inc., Pensylvania, USA) software.
Table 1.
High (+) and low (-) values in Plackett Burman Design for screening independent variables in act-D production.
| Factor | Variables | Factors levels of (gL−1) |
|
|---|---|---|---|
| -1 | +1 | ||
| Starch | X1 | 5 | 15 |
| KCl | X2 | 0,3 | 0.7 |
| NaNO3 | X3 | 1 | 5 |
| K2HPO4 | X4 | 0.6 | 1.4 |
| MgSO4, 7H2O | X5 | 0.3 | 0.7 |
| pH | X6 | 5.2 | 9.2 |
Table 2.
Plackett Burman experimental design matrix for screening medium components of act-D production.
| Trial | Coded variables |
Actinomycin D (mgL−1) | |||||
|---|---|---|---|---|---|---|---|
| x1 | x2 | x3 | x4 | x5 | x6 | ||
| 1 | - | + | - | - | - | + | 191.59 ± 1.41 |
| 2 | - | + | + | + | - | + | 278.85 ± 14.14 |
| 3 | - | - | - | + | + | + | 290.26 ± 5.64 |
| 4 | + | + | + | - | + | + | 153.28 ± 9.89 |
| 5 | - | + | + | - | + | - | 206.87 ± 9.88 |
| 6 | + | - | + | - | - | - | 180.31 ± 11.31 |
| 7 | + | - | - | - | + | + | 163.68 ± 8.48 |
| 8 | - | - | + | + | + | - | 168.88 ± 7.07 |
| 9 | + | - | + | + | - | + | 122.92 ± 2.82 |
| 10 | - | - | - | - | - | - | 479.74 ± 9.89 |
| 11 | + | + | - | + | - | - | 154.82 ± 4.24 |
| 12 | + | + | - | + | + | - | 103.17 ± 8.48 |
2.5.3. Optimization of significant factors by RSM and central composite design (CCD)
The significant variables identified by PBD experiments were optimized using CC design at different levels (Table 3), while the other non-significant variables were fixed at their initial medium level.
Table 3.
Coded and natural values of selected factors at different levels.
| Factors | Levels |
||||
|---|---|---|---|---|---|
| -1.68 | -1 | 0 | +1 | +1.68 | |
| NaNO3 | 1 | 2 | 3 | 4 | 5 |
| K2HPO4 | 0.2 | 0.6 | 1 | 1.4 | 1.8 |
| pH | 3.2 | 5.2 | 7.2 | 9.2 | 11.2 |
Regression analysis determined the factors that had significant (95 % level (P∖0.05)) effect on the metabolite yield. These factors were evaluated in further optimization experiments. CCD was applied for the elucidation of the variables interactions. The design was composed of 23 factorial (1–8 trials), stars points (9–14 trials) and three replicates at center domain condition were added to provide estimation of the model curvature and allow the estimation of the experimental error (Table 4).
Table 4.
Composite design matrix and results of act-D production.
| Run N° | Natural values |
Coded values |
Y (mgL−1) | (mgL−1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| NaNO3 (gL−1) | K2HPO4 (gL−1) | pH | x0 | x3 | x4 | x6 | |||
| 1 | 2 | 0.6 | 5.2 | 1 | -1 | -1 | -1 | 290.31 | 220.69 |
| 2 | 4 | 0.6 | 5.2 | 1 | +1 | -1 | -1 | 337.40 | 313.50 |
| 3 | 2 | 1.4 | 5.2 | 1 | -1 | +1 | -1 | 169.42 | 93.67 |
| 4 | 4 | 1.4 | 5.2 | 1 | +1 | +1 | -1 | 177.89 | 186.47 |
| 5 | 2 | 0.6 | 9.2 | 1 | -1 | -1 | +1 | 401 | 392.12 |
| 6 | 4 | 0.6 | 9.2 | 1 | +1 | -1 | +1 | 635.86 | 484.93 |
| 7 | 2 | 1.4 | 9.2 | 1 | -1 | +1 | +1 | 408.91 | 265.09 |
| 8 | 4 | 1.4 | 9.2 | 1 | +1 | +1 | +1 | 452,60 | 357.90 |
| 9 | 1 | 1 | 7.2 | 1 | -1.68 | 0 | 0 | 75.20 | 157.38 |
| 10 | 5 | 1 | 7.2 | 1 | +1.68 | 0 | 0 | 253.56 | 313.29 |
| 11 | 3 | 0.2 | 7.2 | 1 | 0 | -1.68 | 0 | 548.73 | 548.17 |
| 12 | 3 | 1.8 | 7.2 | 1 | 0 | +1.68 | 0 | 304.51 | 334.76 |
| 13 | 3 | 1 | 3.2 | 1 | 0 | 0 | -1.68 | 74.07 | 74.12 |
| 14 | 3 | 1 | 11.2 | 1 | 0 | 0 | +1.68 | 220.26 | 362.12 |
| 15 | 3 | 1 | 7.2 | 1 | 0 | 0 | 0 | 493.03 | 441.47 |
| 16 | 3 | 1 | 7.2 | 1 | 0 | 0 | 0 | 420.45 | 441.47 |
| 17 | 3 | 1 | 7.2 | 1 | 0 | 0 | 0 | 436.13 | 441.47 |
The regression analysis was performed to estimate the response function as a second order polynomial equation using the least-square method as shown below.
where is the predicted act-D concentration (response), b0 is the intercept term, bj, buj and bjj are the linear, interaction and quadratic terms, respectively. xj, xu represented the independent factors (medium component) in coded values.
2.6. Validation of the model and experimental detection of act-D by LC-MS analysis
The statistical model as well as the optimized parameters were experimentally validated by culturing the strain GSBNT10 under optimized conditions at 28 °C for 6 days. Bioactive metabolites were extracted using ethyl acetate and its concentration determined as reported above. Experiments were carried out in duplicates and the crude extracts were combined to give overall production level.
In order to compare the metabolite profile of the strain GSBNT10, crude extracts of non-optimized, optimized and one selected culture medium from the experimental design (Table 4) were analyzed by DAD-HPLC online coupled to ESIMS in reversed phase conditions, (Agilent Eclipse XDB-C18 column, methanol-water (75:25), flow rate 1 mLmin-1 at 440 nm).
The three crude extracts were prepared by mixing 0.4 mL of ethyl acetate crude extract with 0.5mL of methanol. The quantitative analysis of act-D based on the relative areas of corresponding peaks in the extracted-ion chromatogram.
3. Results and discussion
3.1. Isolation and screening of actinobacteria
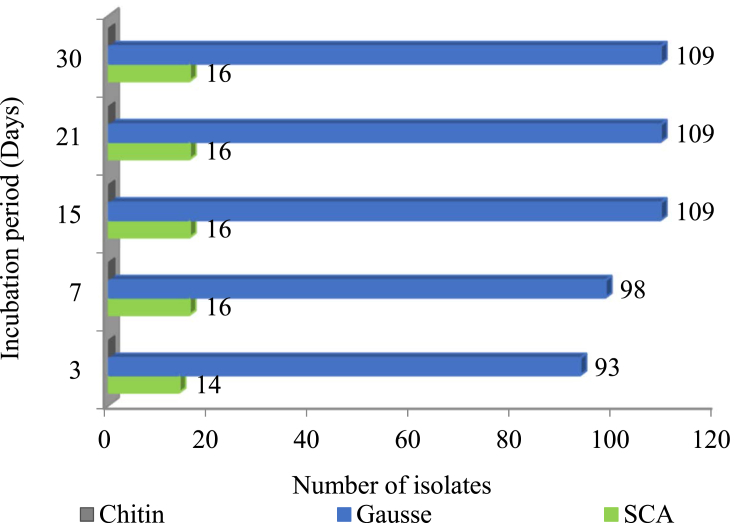
The Algerian Saharan desert is considered one of the largest hot desert in the world, including several landscapes and climate such as sand and stone deserts known for extreme aridity and heat. In the present study, Saharan saline soil samples were collected and studied to recover actinobacteria. 125 Isolates were obtained according to classical suspension dilution technique, using three synthetic culture media (Gausse, starch casein agar (SCA) and Chitin agar). The findings depicted the potency of Gausse agar medium in the recovery of actinobacteria from this niche giving the highest number of isolates (109) in comparison to chitin agar medium where no isolates were recovered. Colony numbers after 3, 7, 15, 21 and 30 days of the samples incubation under different culture media are given in Fig. 1.
Fig. 1.
Actinobacteria colony number per culture media (Chitin, Gausse and Starch Casein Agar) and incubation time obtained from Saharan soil samples.
The 125 isolates were distributed into series according to the color of mature sporulated aerial mycelia (Table 5). Their differences in aerial mycelia color as well as those of the produced pigments may be an indication of actinobacteria diversity in the investigated sites.
Table 5.
Distribution of actinobacteria isolates from Saharan saline soil based on aerial mycelium color and culture media.
| Source | Number of isolates |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Color of aerial mycelium | |||||||||
| White | Yellow | Cream | Pink | Brown | Grey | No aerial mycelium | Total | Total (%) | |
| Gausse medium | 47 | 3 | 18 | 7 | 7 | 25 | 2 | 109 | 87.2 |
| SCA medium | 6 | 1 | 4 | 2 | 0 | 3 | 0 | 16 | 12.8 |
| Total | 53 | 4 | 22 | 9 | 7 | 28 | 2 | 125 | 100 |
| Total (%) | 42.4 | 3.2 | 17.6 | 7.2 | 5.6 | 22.4 | 1.6 | 100 | 100 |
Members of the white series were found to represent 42.4% from the total number of isolates and the lowest occurrence was observed for the strains harboring no aerial mycelia series (1.6%). 51 produced soluble pigments, representing 32.68% (Table 6).
Table 6.
Distribution of actinobacteria isolates from Saharan saline soil based on soluble pigments production color.
| Source | Number of isolates |
|||||
|---|---|---|---|---|---|---|
| Color of soluble pigment | ||||||
| No pigment | Yellow | Pink | Brown | Violet | Total | |
| Soil sample | 74 | 12 | 23 | 14 | 2 | 125 |
| Total (%) | 59.2 | 9.6 | 18.4 | 11.2 | 1.6 | 100 |
77 isolates were found to produce metabolites active against both Gram positive and negative pathogenic germs while 48 isolates exhibited no antagonistic activity. The active actinobacterial strains were cultivated on Gausse and SCA media broths for an ulterior screening. The antibacterial activity of the filtrates was then determined by well diffusion method. Based on inhibition zones formation, only 45 strains confirmed their higher antagonistic potential and the strain GSBNT10 revealed a potent bioactivity against both Gram positive and negative bacteria but also against MRSA (Table 7) mainly attributed to the production of antibacterial compounds. Numerous investigations confirmed the presence of diverse culturable rare and novel actinobacteria able to produce new natural products (Lamari et al., 2002; Zitouni et al., 2004; Saker et al., 2015; Chaabane-Chaouch et al., 2016).
Table 7.
Antimicrobial activity, expressed as inhibition zone diameter in mm, of the actinobacteria strains isolated from Saharan saline soil.
| Strains | Activity (mm) |
||||
|---|---|---|---|---|---|
| E. coli | S. aureus | MRSA | B. subtilis | P. aeruginosa | |
| GSBNT5 | 11.47 ± 0.63 | 21.82 ± 2.61 | - | 16.29 ± 1.58 | - |
| GSBNT7 | - | - | - | 14.59 ± 1.39 | - |
| GSBNT9 | - | 23.71 ± 2.58 | - | 18.13 ± 1.34 | - |
| GSBNT10 | 18.36 ± 1.86 | 26.87 ± 1.19 | 22.71 ± 1.36 | 26.12 ± 2.40 | - |
| GSBNT11 | - | 8.23 ± 0.73 | - | - | - |
| GSBNT16 | - | 20.89 ± 1.42 | - | 15.86 ± 1.16 | - |
| GSBNT17 | 12.57 ± 1.14 | 20.49 ± 1.69 | 12.03 ± 1.37 | 13.92 ± 1.70 | - |
| GSBNT19 | - | - | - | 10.78 ± 0.84 | - |
| GSBNT20 | - | 22.63 ± 2.12 | - | - | - |
| GSBNT24 | - | - | - | 12.71 ± 1.26 | - |
| GSBNT27 | - | 10.90 ± 0.71 | - | - | - |
| GSBNT33 | - | 15.58 ± 1.32 | - | - | - |
| GSBNT35 | - | - | - | 11.94 ± 1.08 | - |
| GSBNT36 | - | 9.25 ± 0.68 | 12.47 ± 1.06 | - | - |
| GSBNT37 | 10.65 ± 1.13 | 24.84 ± 1.58 | - | 16.12 ± 1.68 | - |
| GSBNT40 | - | - | - | 12.47 ± 0.79 | - |
| GSBNT41 | - | - | - | 17.89 ± 1.88 | - |
| GSBNT42 | - | 12.25 ± 1.34 | - | - | - |
| GSBNT48 | - | - | - | 10.59 ± 1.36 | - |
| GSBNT49 | - | - | - | 12.43 ± 1.27 | - |
| GSBNT50 | 9.12 ± 0.80 | 18.72 ± 2.26 | 14.11 ± 2.06 | - | 9.25 ± 1.14 |
| GSBNT52 | - | 20.11 ± 2.19 | - | - | - |
| GSBNT55 | - | - | - | 15.02 ± 1.13 | - |
| GSBNT56 | - | 7.43 ± 0.86 | - | - | - |
| GSBNT57 | - | - | - | 10.88 ± 1.26 | - |
| GSBNT62 | - | 14.78 ± 0.95 | - | - | - |
| GSBNT63 | 12.78 ± 1.33 | 16.64 ± 1.48 | - | 14.27 ± 1.48 | - |
| GSBNT65 | - | 9.35 ± 1.08 | - | - | - |
| GSBNT66 | - | - | - | 10.93 ± 1.62 | - |
| GSBNT72 | 15.26 ± 2.48 | 23.10 ± 2.39 | - | 13.42 ± 1.71 | - |
| GSBNT74 | - | - | - | 9.23 ± 1.07 | - |
| GSBNT75 | - | - | - | 16.89 ± 1.77 | - |
| GSBNT78 | - | 11.25 ± 0.84 | - | - | - |
| GSBNT93 | - | - | - | 15.03 ± 1.96 | - |
| GSBNT95 | - | - | - | 11.72 ± 1.62 | - |
| GSBNT103 | 9.44 ± 0.68 | - | - | - | 9.12 ± 0.69 |
| GSBNT107 | - | - | - | 10.45 ± 0.63 | - |
| WSBNT111 | - | 9.05 ± 0.79 | - | 9.19 ± 0.72 | 11.10 ± 1.69 |
| WSBNT112 | - | 22.29 ± 1.88 | 19.61 ± 1.17 | 14.89 ± 1.45 | 12.06 ± 2.49 |
| WSBNT113 | - | - | - | 15.18 ± 1.79 | - |
| WSBNT114 | - | 25.04 ± 1.59 | 20.65 ± 1.55 | 21.89 ± 2.35 | 8.64 ± 0.38 |
| WSBNT115 | - | 22.04 ± 2.06 | 18.43 ± 1.87 | 16.78 ± 1.51 | 10.04 ± 2 |
| WSBNT117 | - | 23.59 ± 2.21 | 16.63 ± 1.29 | 18.53 ± 2.49 | - |
| WSBNT118 | 14.83 ± 1.23 | 12.34 ± 1.04 | 9.18 ± 0.98 | 10.16 ± 1.93 | - |
| WSBNT119 | - | 22.59 ± 1.48 | 14.47 ± 1.43 | 17.10 ± 2.16 | - |
3.2. Characterization of the selected strain GSBNT10
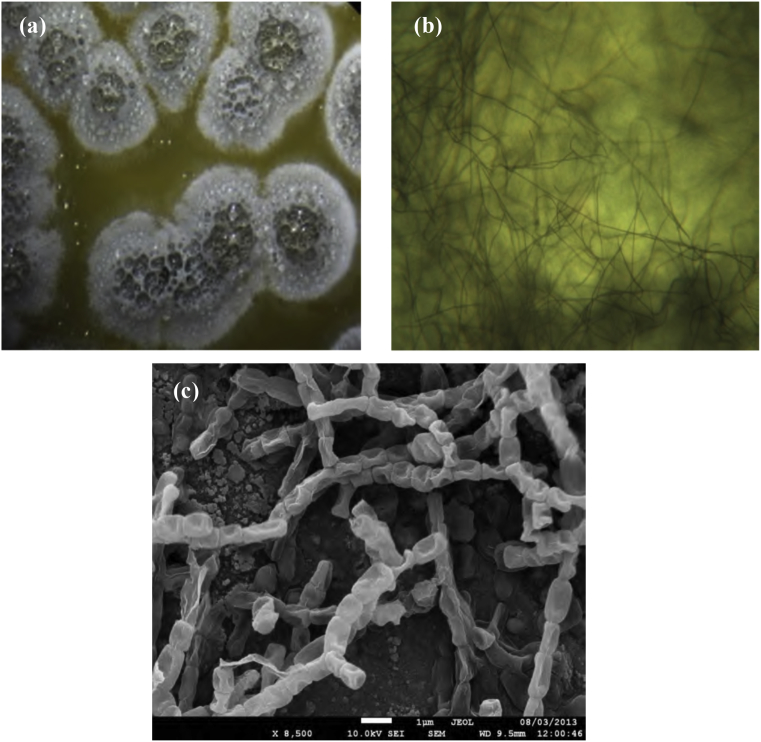
The morphological features of the selected strain GSBNT10 was undertaken on 7 culture media. The isolate is Gram positive filamentous bacterium, harboring pale white and grey aerial spore mass and grey to yellow-brown reverse side color. It exhibited a good growth on most of the used culture media showing gray aerial mycelium with different color of vegetative mycelium varying from grey to yellow with formation of yellow diffusible pigment. It showed morphological properties characteristics of Streptomyces genus. Scanning electron microscopy showed hyphae bearing short chains of spores as well as single spore cells (Fig. 2). The spores were round to oval with smooth surface. With regard to chemotaxonomic characteristics, the presence of LL-diaminopimelic acid (DAP) and glycin amino acids as well as glucose in the cell wall convincingly categorized the strain GSBNT10 to the cell wall type-IC (Lechevalier and Lechevalier, 1970). Phospholipid pattern consisted of phosphatidylethanolamine (62%), phosphatidylglycerol (30%), phosphatidylcholin (5%) and sphingomyelin (3%) as well as several uncharacterized polar lipid components.
Fig. 2.
Hyphae and spore chains of strain GSBNT10 grown on Gausse medium at 28 °C for 7 days; (a) A sporulated aerial mycelium, (b) Optic microscopy view (X40) and (c) Scanning electron microscopy, bar = 10 μm, (x 8500).
According to the obtained results, we can deduce that GSBNT10 strain is pertaining to PII type phospholipids group of Streptomyces genus (Lechevalier et al., 1977) characterized by the predominant presence of phosphatidyl-ethanolamine.
Physiological tests indicated that the isolate GSBNT10 is able to use a variety of organic compounds such as starch, D-galactose, D-glucose, glycerol, inositol, D-lactose, maltose, rhamnose, ribose, trehalose, D-xylose, tween 80, but not adenine, guanine, hypoxanthine, gelatin, casein and cellulose with however a weak growth observed for adonitol, arabinose, mannose, raffinose, sorbitol and sucrose. The isolate was found to use alanine, proline and threonine as nitrogen source whereas it is not able to utilize aspartic acid, cysteine, tyrosine, serine, glycine and tryptophan, and a weak growth was observed in presence of methionine, histidine, arginine, leucine and isoleucine amino acids.
Development of the strain GSBNT10 was good at pH 5, 7, 9 and 11, and from 28 to 45 °C. However, no growth was obtained at 4 °C. The organism was found to be susceptible to Erythromycin (15μg), Gentamycin (10μg) and Vancomycin (30μg) but resistant to Ampicillin (10μg) and Clavulanic acid/Aztreonam. The growth was good in the presence of sodium chloride at a concentration of 10% (w/v).
An almost complete 16S rRNA gene sequence analysis (1491 nucleotides) for strain GSBNT10 was determined and assigned in GenBank under accession number MF399050.1. A phylogenetic tree was constructed based on 16S rRNA gene sequences to show the comparative relationship between strain GSBNT10 and other related Streptopmyces species (Fig. 3). The comparative analysis of 16S rRNA gene sequence and phylogenetic relationship showed that it forms a clade with S. crystallinus NBRC 15401T with which it shares a 16S rRNA gene sequence similarity of 98.47% and supported by high bootstrap value (93%). It is generally accepted that microorganisms displaying 16S rRNA sequence similarity values of 98. 65% or less belong to different species (Kim et al., 2014). Indeed, our strain could be distinguished from S. crystallinus NBRC 15401T by some phenotypic properties such as the color of aerial mycelium on all tested ISP media; red for S. crystallinus and varied from gray to brown for GSBNT10 (Table 8) and the utilization of D-xylose, sucrose, L-rhamnose, lactose, maltose (as sole carbone sources). The strain GSBNT10 was able to grow in the presence of a wide range of NaCl (1–9%) and temperature (10–50 °C) (Table 9). Moreover, the phylogenetically closest related Streptomyces strain was not known to produce act-D, thus this strain is a new source of this antibiotic. These features separate the strain GSBNT10 from the type strain S. crystallinus NBRC 15401T suggesting that the strain GSBNT10 may represent a novel species of Streptomyces. This finding is in accordance with previous reports about the abundance and the large distribution of Streptomyces genus in nature. They have long been considered as a significant source of secondary metabolites in particular antibiotics with diverse structures and properties but also for their ability to produce hydrolytic enzymes of major biotechnological interest (Lewin et al., 2016).
Fig. 3.
Phylogenetic tree calculated from almost-complete 16S rRNA gene sequences (1491 nt) showing relationships between the strain GSBNT10 and the most related type species of the genus Streptomyces. The tree was constructed using Jukes and Cantor (1969) evolutionary distance methods and the neighbour-joining method of Saitou and Nei (1987). Bootstrap values above 50% (percentages of 1000 replications) are indicated at nodes. Bar, 0.02 substitutions per nucleotide position. Actinopolyspora mortivallis DSM 44261Twas used as the out group.
Table 8.
Culture properties of strain Streptomyces sp. GSBNT10 and S. crystallinus NBRC 15401 type strain.
| Characteristic on ISP | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| Strain GSBNT10 | |||||||
| Growth | + | ± | + | + | + | ± | ± |
| Aerial spore mass | Light grey | Light brown | Light grey | Dark grey | White grey | Light grey | Beige |
| Substrate mycelium | Light brown | Light brown | Light brown | Light brown | Light brown | Dark grey | Light brown |
| Diffusible pigments | + | + | + | + | + | Yellow | Yellow |
| S. crystallinusNBRC 15401 | |||||||
| Growth | None | ± | + | ± | ± | ± | ± |
| Aerial spore mass | None | Red | White | Red | Red | Red | Red |
| Substrate mycelium | None | Light brown | Dark brown | Light brown | Light yellow | Light brown | Light brown |
| Diffusible pigments | None | None | + | None | None | Dark brown | Dark brown |
+ good growth, ± weakly positive.
Table 9.
Physiological properties of strain Streptomyces sp. GSBNT10 and S. crystallinus NBRC 15401 type strain.
| Characteristic | Strain GSBNT10 | S. crystallinus NBRC 15401 |
|---|---|---|
| Carbon source utilization | ||
| D-xylose | ++ | - |
| D-mannose | ± | - |
| D-glucose | +++ | + |
| D-galactose | +++ | + |
| Sucrose | + | - |
| L-rhamnose | +++ | - |
| Raffinose | ± | ++ |
| Mannitol | / | - |
| L-arabinose | ± | - |
| Meso-inositol | ++ | - |
| Lactose | +++ | + |
| Maltose | +++ | - |
| Trehalose | +++ | ++ |
| D-ribose | +++ | - |
| D-fructose | +++ | - |
| Nitrogen source utilization | ||
| L-glycine | - | + |
| L-valine-L-leucine | ± | - |
| L-histidine | ± | - |
| L-phenylalanine | / | - |
| L-asparagine | / | + |
| L-methionine | ± | - |
| Maximum NaCl tolerance (%, w/v) | 9 | 5 |
| Range of Temperature (°C) | 10–45 | 30–45 |
| Growth in presence of (%, w/v) | ||
| Phenol (0.1) | ± | + |
| Sodium azide (0.01) | - | - |
| Hydrolysis of starch | + | + |
| Hydrolysis of Tween 80 | + | - |
| Hydrolysis of proteins | - | + |
3.3. Selection of basal medium
It has been already established that culture conditions including abiotic and nutritional factors influence the production of bioactive molecules by actinobacteria. This was verified ones again for GSBNT10 which showed a significant difference in its ability to produce bioactive compounds according to the culture medium composition, with the highest bioactivity observed on Czapeck culture medium (18.89 mm, 17.17 mm and 16.17 mm) against E. coli, S. aureus and MRSA respectively, containing per liter: 10g of soluble starch, 3g of NaNO3, 1g of K2HPO4, 0.5g of MgSO4·7H2O, 0.5g of KCl, 10mg of FeSO4·7H2O. Therefore, Czapeck was selected for further optimization study using statistical approaches.
3.4. Purification and characterization of active metabolites
3.4.1. Isolation and identification of act-D
Ethyl acetate crude extract (300 mg) was dissolved in 3 mL of methanol, then subjected to flash column chromatography on silica gel using dichloromethane-methanol (90:10), and 100% methanol collecting two fractions.
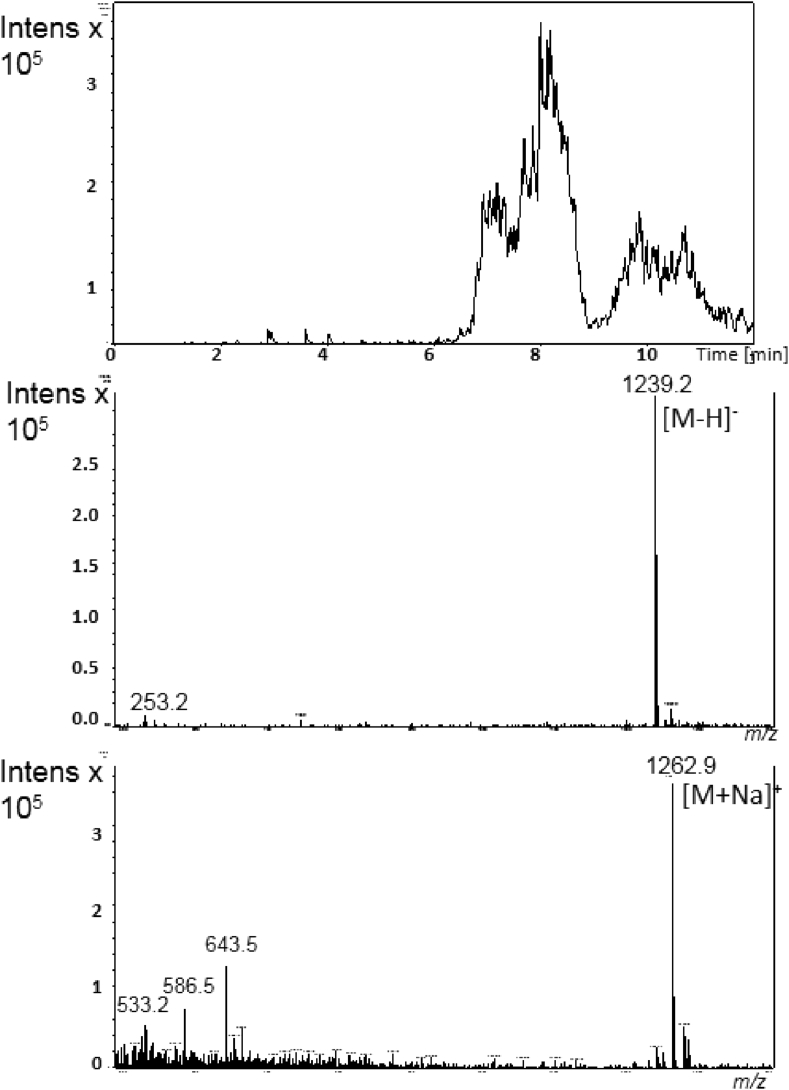
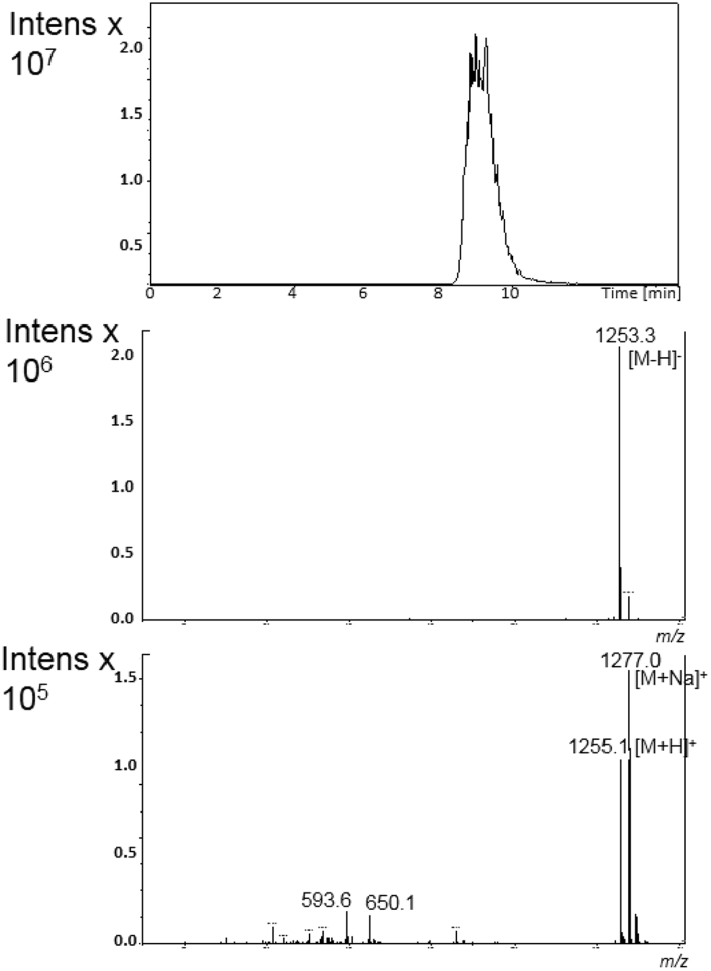
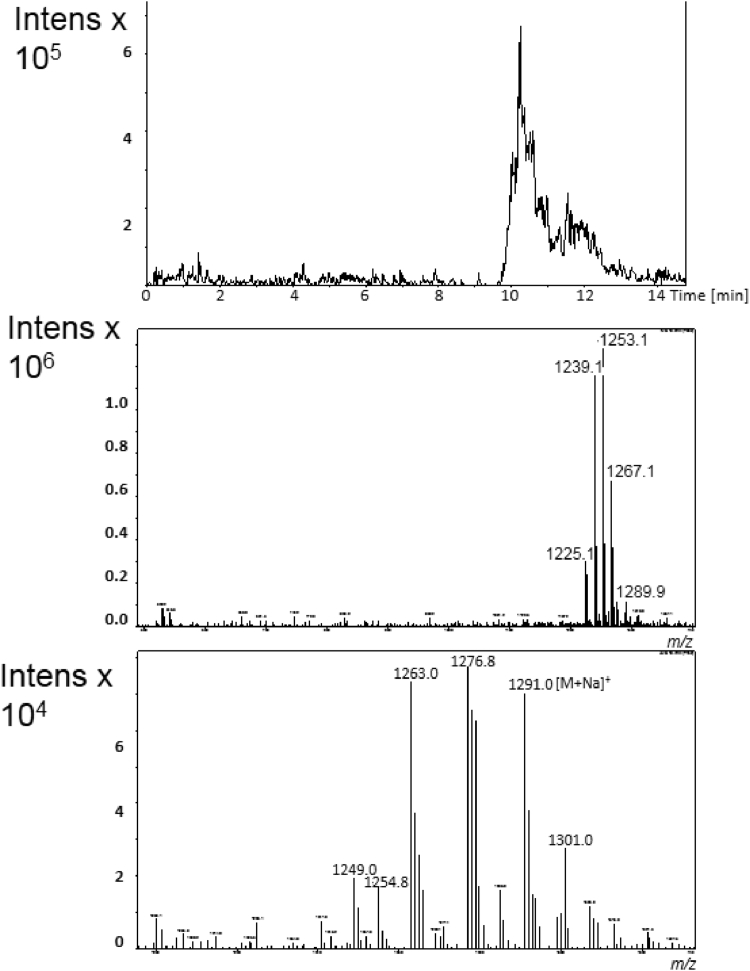
The chromatographic profile of the crude extract, performed by DAD-HPLC analysis at 245 nm, exhibited a major signal at retention time tR = 9.3 min. The LC-MS spectrum in positive ion mode showed three signals with very close retention times harboring different mass values. In detail, i) by analysis in negative ion mode, the peak at tR = 8.0 min was associated the MS signal at m/z 1239 corresponding to [M-H]- ion, in agreement with the signal at m/z 1263 attributed to [M + Na]+ ion in the spectrum recorded in positive mode condition (Fig. 4); ii) the peak at tR = 9.3 min was associated to the intense signal at m/z 1253 for [M-H]- ion in negative mode, and to m/z 1255 and 1277 corresponding to [M + H]+ and [M + Na]+ ions respectively under positive mode detection (Fig. 5); iii) the peak at tR = 10.4 min was associated to the signal at m/z 1291 corresponding to [M + Na]+ ion, as deduced by the corresponding signal at m/z 1267 for the [M-H]- ion in negative mode (Fig. 6).
Fig. 4.
Online LC-MS analysis of ethyl acetate crude extract displaying actinomycin D analogue at tR = 8.0 min from Streptomyces sp. GSBNT10: extracted–ion chromatogram (top), and corresponding MS spectra in negative (middle) and in positive ion mode (bottom).
Fig. 5.
Online LC-MS analysis of ethyl acetate crude extract displaying the major metabolite actinomycin D at tR = 9.3 min from GSBNT10: extracted–ion chromatogram (top), and corresponding MS spectra in negative (middle) and in positive ion mode (bottom).
Fig. 6.
Online LC-MS analysis of ethyl acetate crude extract displaying actinomycin X2 at tR = 10.4 min from GSBNT10 strain: extracted –ion chromatogram (top), and corresponding MS spectra in negative (middle) and in positive ion mode (bottom).
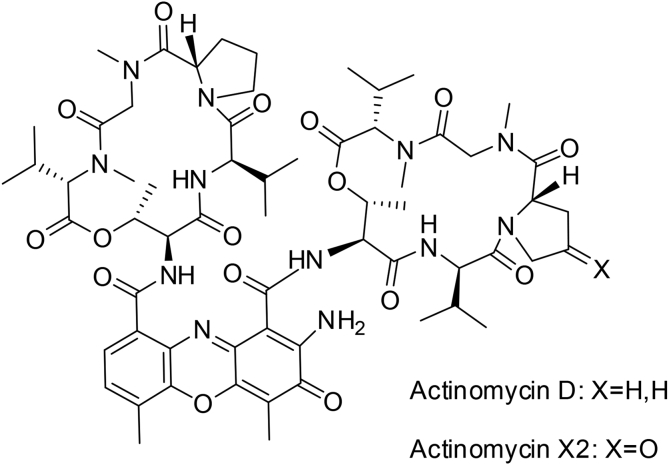
Further analytic and preparative RP18 HPLC purification of this fraction led to the identification of act-D (Fig. 7) and two minor products: Actinomycin X2 and an act-D analogue. It was further confirmed by comparing the NMR and MS spectra with data reported (Lackner, 1975; Mahmoud, 2004). The structural assignment of act-D analogue was prevented due to its small amount.
Fig. 7.
Molecular structures of metabolites produced by Streptomyces sp. GSBNT10 strain: the major actinomycin D (act-D) and the minor actinomycin X2.
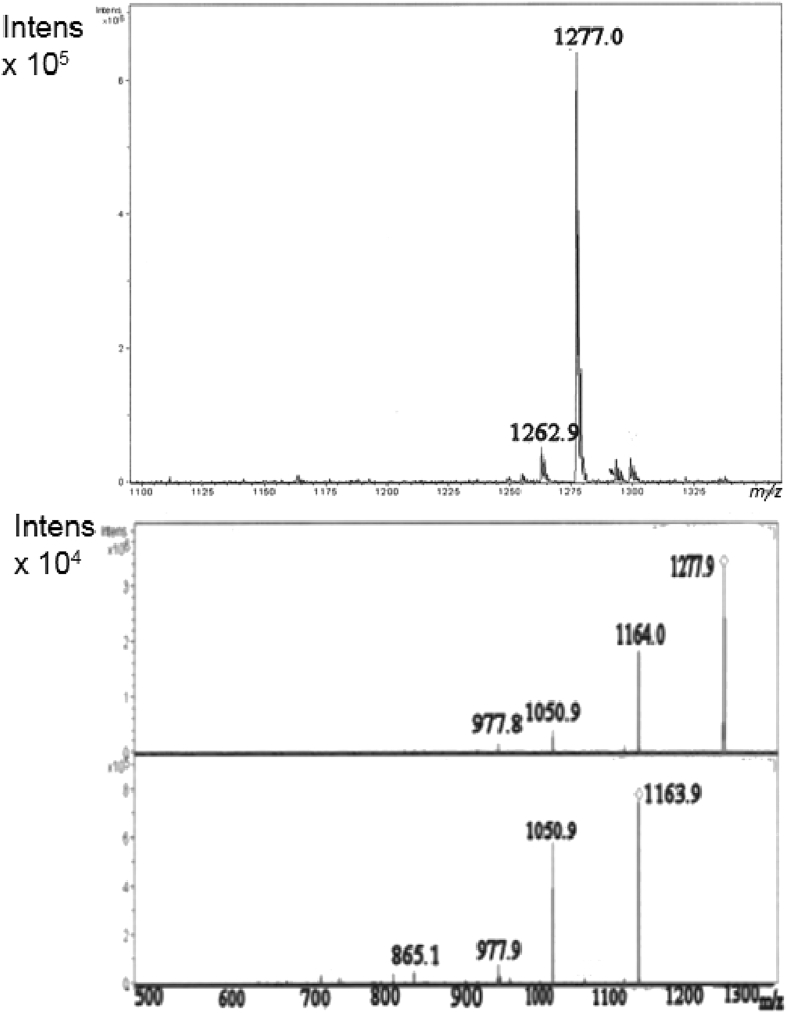
Act-D (C62H86N12O16, MM = 1255.4) was isolated as a reddish-orange powder (10 mg) from fraction 1 (40 mg) by preparative HPLC. On TLC, it turned to red by anisaldehyde/sulphuric acid Rf = 0.4 (CH2Cl2/MeOH 90:10). (+) ESI MS: m/z 1277 ([M + Na]+, 1255 ([M + H]+; (+) ESI MS2 (1277): m/z1164; (+) ESI MS3 (1164): m/z 1051, 977, 865 (Fig. 8). (-)-ESI MS: m/z 1253 ([M-H]-).
Fig. 8.
ESI -MS of act-D in positive ion mode: m/z 1277 corresponding to [M + Na]+ ion (top); MS/MS (middle) and MS3 (bottom) spectra by fragmentation experiment on m/z 1277.
3.5. Screening of physical and nutritional parameters for act-D production by PBD method
Czapeck medium was adopted for the selection of significant components and parameters influencing the production of act-D by Streptomyces sp. GSBNT10. The production of the metabolite was correlated with the growth curve of GSBNT10 strain where the maximum bioactivity was observed after 6 days of incubation.
Six different culture parameters including organic, inorganic salts and physical parameters factors were evaluated for their suitability to sustain increased act-D production by GSBNT10. Plackett-Burman design was used for initial screening of medium components (Tables 1 and 2).
The first order polynomial model obtained using PB design which represented the act-D production as a function of six independent variables screened without interaction terms is as follow (equation 1):
| (1) |
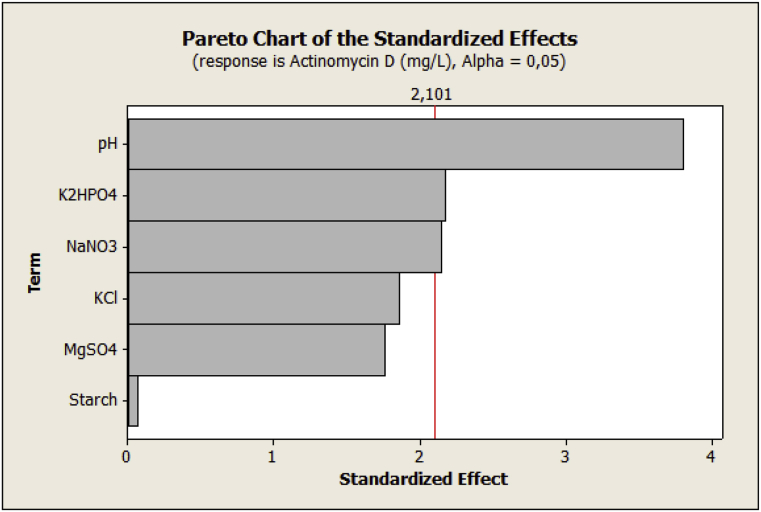
The significance of each coefficient was determinde by P-values (Table 10). Probability values less than 0.05 (p < 0.05) indicated significance of the model term whereas P-value greater than 0.05 (p > 0.05) indicated model terms were not signficant. Therefore, the results reported in Table 10 show that the variables, NaNO3 (x3) (t = 2.15, p = 0.045), K2HPO4 (x4) (t = 2.17, p = 0.044) and pH (x6) (t = 3.81, p = 0.001) were considered as significant factors for act-D production. The remaining variables, including starch (x1), KCl (x2) and MgSO4 (x5), had P- value more than 0.05 and were considered not significant.
Table 10.
Estimated regression coefficients of the model developed by Plackett-Burman design (PBD).
| Term | Effect | Coef | SE Coeff | T | P-value |
|---|---|---|---|---|---|
| Constant | 206.63 | 14.13 | 14.62 | 0.000 | |
| Starch (x1) | -1.84 | -0.92 | 14.13 | -0.07 | 0.949 |
| KCl (x2) | -52.39 | -26.19 | 14.13 | -1.85 | 0.080 |
| NaNO3 (x3) | 60.73 | 30.37 | 14.13 | 2.15 | 0.045 |
| K2HPO4 (x4) | 61.34 | 30.67 | 14.13 | 2.17 | 0.044 |
| MgSO4 (x5) | -49.57 | -24.79 | 14.13 | -1.75 | 0.096 |
| pH (x6) | 107.61 | 53.80 | 14.13 | 3.81 | 0.001 |
From Eq. (1), it was predicted that increasing concentrations of NaNO3 (x3), K2HPO4 (x4) and medium pH (x6) in their explored intervals should enhance the act-D production because of their positive effects. pH has the strongest effect on the response since coefficient of (b6 = +53.8) is larger than the coefficients of the other investigated factors.
The results could be also presented through the Pareto chart of effects (Fig. 9). The figure showed each of the estimated effect. Thereafter, the exact optimal values for the individual factors were estimated using the Central Composite Design (CCD).
Fig. 9.
Pareto chart showing the effect of 6 variables on act-D production.
3.6. Optimization of selected medium components
CCD was applied not only to study the interactions between the three significant variables (NaNO3, K2HPO4, pH) but also to determine their optimal levels. All trials in the CCD design supported the production of act-D, moreover its concentration varied from 74.07 to 635.86 mgL-1 (Table 4). act-D was observed to be improved by optimizing pH, K2HPO4 and NaNO3 to 8.2 value, 0.2 g and 3 g respectively. Similar findings have been obtained by Praveen et al. (2008a) showing that the incorporation of NaNO3 in the production medium improved the yield of act-D.
The second order polynomial model for act-D production obtained after performing 17 experiments (Table 4) is as follows:
| (2) |
The results of ANOVA are shown in Table 11. The model had a low P-value (P = 0.035), indicating that the model fitted the experimental data very significantly. The fitness of the model was examined by the determination coefficient (R2 = 0.82), which indicated that 82 % of data variability could be explained by the model (Table 11). The F value (9.8) and P-value (0.125) of lack-of-fit was not significantly relative to the pure error. All the statistical results of the model showed a high accuracy and general applicability of the second order model, and was adequate to describe responses observed in the experiments.
Table 11.
Variance analysis (ANOVA) of the second order model.
| Source | DF | Seq SS | Adj SS | Adj MS | F | P |
|---|---|---|---|---|---|---|
| Regression | 9 | 338355 | 338355 | 37595 | 3,54 | 0,035 |
| Linear | 3 | 184515 29439 54975 | 184515 29439 54975 | 61505 | 5,79 | 0,026 |
| 1 | 100100 | 100100 | 29439 54975 | 2,77 | 0,140 0,057 | |
| 1 | 100100 | 5,17 | 0,018 | |||
| 1 | 9,42 | |||||
| Square | 3 | 139644 | 139644 | 46548 | 4,38 | 0,049 |
| 1 | 46131 | 60205 | 60205 | 5,66 | 0,049 | |
| 1 | 22863 | 4347 | 4347 | 0,41 | 0,543 | |
| 1 | 70651 | 70651 | 70651 | 6,65 | 0,037 | |
| Interaction | 3 | 14195 | 14195 | 4732 | 0,45 | 0,728 |
| 1 | 6600 | 6600 | 6600 | 0,62 | 0,457 | |
| . | 1 | 6216 | 6216 | 6216 | 0,58 | 0,469 |
| 1 | 1379 | 1379 | 1379 | 0,13 | 0,729 | |
| Lack-of-Fit | 5 | 71487 | 71487 | 14297 | 9,8 | 0,125 |
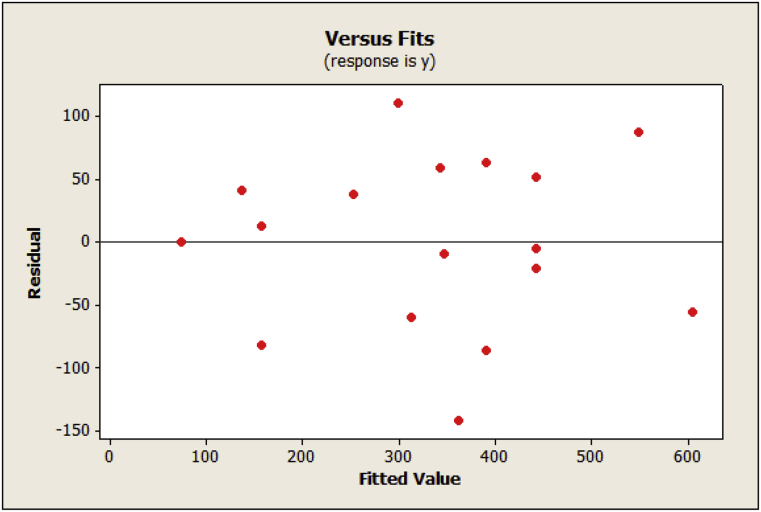
It is usually necessary to check the fitted model to ensure that it provides an adequate approximation to the real system. Fig. 10 presents a plot of residuals versus the predicted response. The general impression is that the residuals displayed randomness in scattering and suggested that the variance of the original observation was constant.
Fig. 10.
Plot of residuals versus fitted values for act-D production.
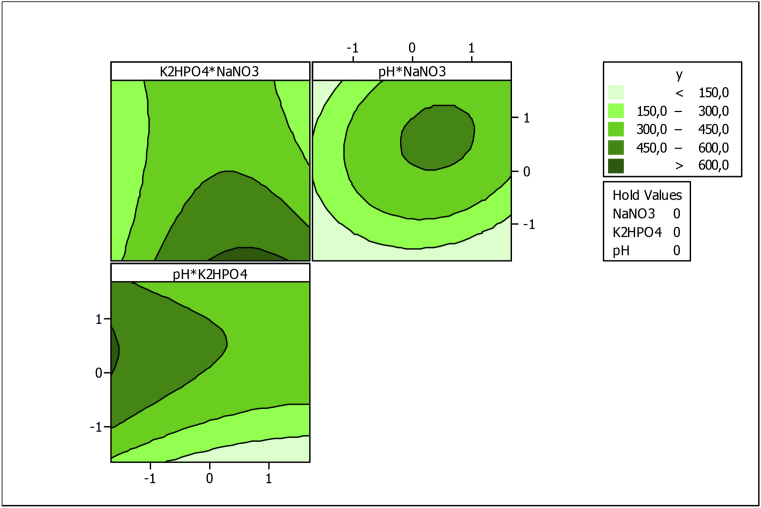
The regression equation is expressed graphically in contour and surface plots which depicted the interactions among the independent variables and their influence on act-D yield. The model was validated by comparing the observed and predicted values at the optimal conditions. A good correlation verified the accuracy of the model.
Fig. 11 shows contour plots for the act-D concentration generated by the predicted model using minitab 16.0 software. Each 2D plots presented the effect of two variables while the third one was fixed at middle level (level 0). According to the contour plots drawn, the absence of mutual interactions is clearly observed between K2HPO4 and NaNO3 as well as pH and NaNO3, and between pH and K2HPO4. The third plots show that an optimal act-D concentration upper than 600 mg/L could be determined for NaNO3 concentration from 3 gL-1 to 4 gL-1 (coded value, 0 to +1), K2HPO4 concentration lower than 0.6 gL-1 (coded value, -1) and medium pH from 7.2 et 9.2 (coded value, 0 to +1).
Fig. 11.
Contours plots of the predicted act-D concentration.
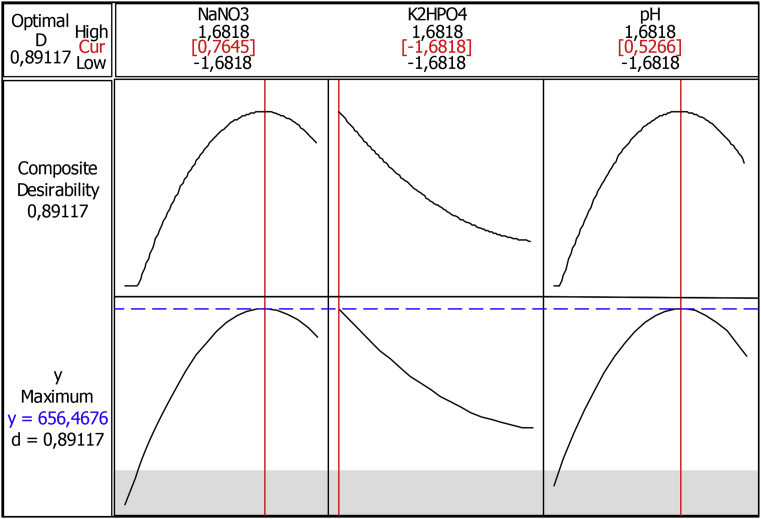
Fig. 12 is the result of the response optimizer using desirability function to show which levels of independent variables produce the most desirable predicted response. The profiles for predicted response and the desirability level for factors indicate that NaNO3 3.76 gL-1 (x3 = 0.7645), K2HPO4 0.2 gL-1 (x4 = -1.6818) and pH 8.25 (x6 = 0.5266) allow to reach 656.46 mgL-1 of act-D production.
Fig. 12.
Predicted and actual values of media composition and act-D production.
3.7. Model validation and confirmation
In order to determine the model adequacy, the optimized predicted levels of pH value, K2HPO4, and NaNO3 were considered for experimental production of act-D for comparison with the predictive data.
The experiment was run in triplicate. The measured act-D concentration resulted 632.53 mg/L, close to the predicted one 656.46 mgL-1 and so revealing a high degree of accuracy. Thus, this statistical approach allowed the determination of optimum levels of culture parameters favoring 58.56% increase of act-D yield. Starting from an initial 414 mgL-1 yield, it reached 656.46 mgL-1 after optimization with 1.58 fold increase. This validation proves that the developed model has resulted in an appreciable enhancement of act-D production.
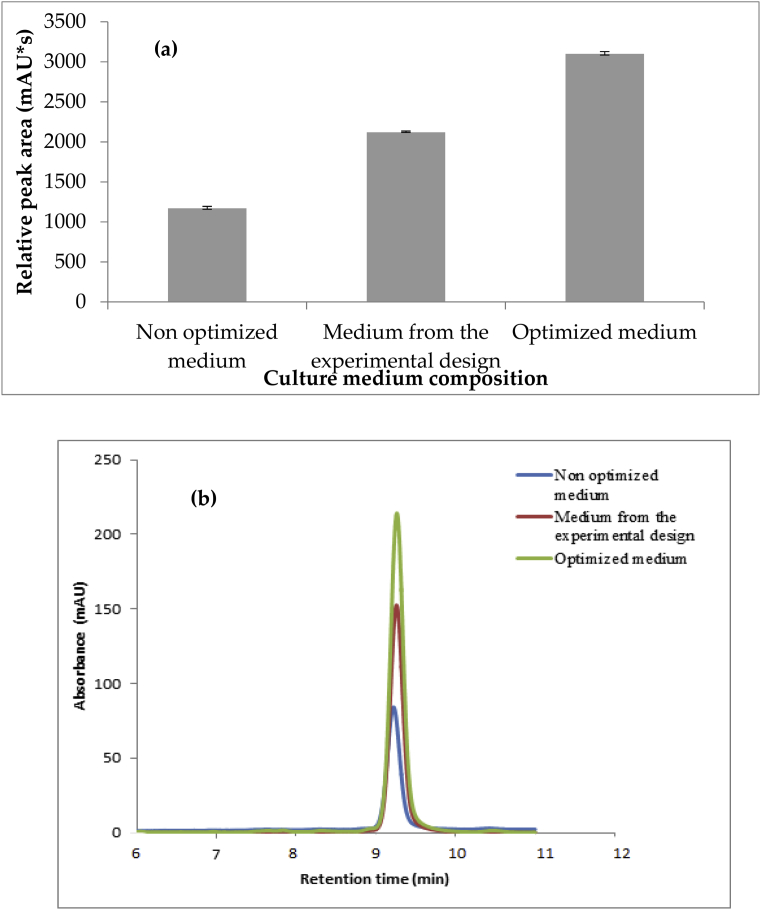
DAD-HPLC/ESI-MS analysis of crude extracts obtained from both unoptimized and optimized levels culture media revealed a significant improve in the production of act-D, as deduced by the relative areas of chromatographic peaks (Fig. 13).
Fig. 13.
(a) Relative peak areas of act-D eluted at tR = 9.3 min in the chromatographic profile of the strain Streptomyces sp. GSBNT10, (b) Relative intensity of chromatographic peaks of act-D eluted tR = 9.3 min by UV-DAD HPLC analysis (reversed phase C18 column, methanol-water 75:25, flow 1mLmin-1, detection at 440 nm).
Some studies have been reported on the improvement of act-D yield by different optimization methods. Williams and Katz (1977) developed a culture medium, using the classical one-factor-at-a-time approach, yielding 500 to 600 mgL-1 of act-D by S. parvulus ATCC 12434 cultured in shaken flasks. Dalili and Chau (1988) achieved a maximum act-D yield of 80 mgL-1 under immobilized condition of the strain. Furthermore, Praveen et al. (2008a) achieved 365 mgL-1 act-D yield obtained from S. sindenensis cultivated in fermenter combining statistical approaches (PBD and RSM by CCD), while Sousa et al. (2002) reached 635 mgL-1 using one factor-at-a-time experiments from culture of S. parvulus and Hamza et al. (2013) optimized act-D production (306 mgL-1) by the strain Streptomyces sp. AH 11.4E3, applying the same approach. Moreover, few actinobacteria strains have been reported to produce considerable amount of act-D, including S. avermitilis (1770 mgL-1) (Chen et al., 2012), S. flavogriseus (960 mgL-1) (Wei et al., 2017) and S. sindenensis mutant strain (850 mgL-1) (Praveen et al., 2008b).
It is noteworthy that HPLC-DAD/ESI-MS, known as a technique widely used in metabolites analysis due to its high sensitivity, specificity and molecular structure information (Shi et al., 2011), emerged also in this work as a powerful tool for the fast identification and quatitative detection of metabolites in extracts of cultures under different conditions.
4. Conclusion
A new act-D producing strain GSBNT10 was isolated from Saharan soil samples and identified as a novel species of Streptomyces genus. act-D yield was successfully enhanced using Plackett-Burman and Central Composite designs of RSM and the mathematical model was perfectly confirmed by an efficient LC-MS analysis of the crude extracts. Maximum act-D production was obtained on Czapeck medium at initial pH value of 8.2, K2HPO4 0.2 gL-1 and NaNO3 3gL-1 showing a remarkable increase (58.56%), from 414 mgL-1 under unoptimzed conditions to 656.46 mgL-1 by response surface methodology.
The agreement and the close concordance of predicted value with the experimental one confirmed the validation of the model. Furthermore, the considerable amount of act-D produced by this new strain on solid state fermentation and under the optimized conditions established in this study, suggests a potential scale-up of the process for the access of this biologically relevant metabolite.
Declarations
Author contribution statement
Ibtissem Djinni: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Andrea Defant, Warda Djoudi: Conceived and designed the experiments; Analyzed and interpreted the data.
Faouzia Chaabane Chaouch: Analyzed and interpreted the data.
Samiha Souagui: Conceived and designed the experiments.
Mouloud Kecha: Contributed reagents, materials, analysis tools or data.
Ines Mancini: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by the Algerian Ministry of Higher Education and Scientific Research in the frame of CNEPRU research project N° F00620140028.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
Authors thank Adriano Sterni and Mario Rossi, University of Trento- Italy for recording mass spectra and IR spectra, respectively.
References
- Ahsan T., Chen J., Wu Y., Irfan M. Application of response surface methodology for optimization of medium components for the production of secondary metabolites by Streptomyces diastatochromogenes KX852460. Amb. Express. 2017;7:96. doi: 10.1186/s13568-017-0388-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avendaño C., Menéndez J.C. Anticancer drugs acting via radical species, photosensitizers and photodynamic therapy of cancer. In: Avendaño C., Menéndez J.C., editors. Medicinal Chemistry of Anticancer Drugs. first ed. Elsevier Science; Linacre House, Jordan Hill, Oxford, OX2 8DP, UK: 2008. pp. 93–138. [Google Scholar]
- Chaabane Chaouch F., Bouras N., Mokrane S., Zitouni A., Schumann P., Spröer C., Sabaou N., Klenk H.P. Streptosporangium becharense sp. nov., an actinobacterium isolated from desert soil. Int. J. Syst. Bacteriol. 2016;66:2484–2490. doi: 10.1099/ijsem.0.001077. [DOI] [PubMed] [Google Scholar]
- Chen C., Wang Q., Song F., Abdel-Mageed W.M., Guo H., Fu C., Hou W., Dai H., Liu X., Yang N., Xie F., Yu K., Chen R., Zhang L. A marine-derived Streptomyces sp. MS449 produces high yield of actinomycin X2 and actinomycin D with potent anti-tuberculosis activity. Appl. Microbiol. Biotechnol. 2012;95:919–927. doi: 10.1007/s00253-012-4079-z. [DOI] [PubMed] [Google Scholar]
- Dalili M., Chau P.C. Production of Actinomycin-D with immobilized Streptomyces parvulus under nitrogen and carbon starvation conditions. Biotechnol. Lett. 1988;10:331–336. [Google Scholar]
- Djinni I., Djoudi W., Souagui S., Rabia F., Rahmouni S., Mancini I., Kecha M. Streptomyces thermoviolaceus SRC3 strain as a novel source of the antibiotic adjuvant streptazolin: a statistical approach toward the optimized production. J. Microbiol. Methods. 2018;148:161–168. doi: 10.1016/j.mimet.2018.04.008. [DOI] [PubMed] [Google Scholar]
- Eun H.E. DNA polymerases. In: Eun H.E., editor. Enzymology Primer for Recombinant DNA Technology. first ed. Academic Press: 525 B Street, Suite 1900; San Diego, California 92101-4495, USA: 1996. pp. 345–489. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fontes G.C., Finotellib P.V., Rossic A.M., Rocha-Leão M.H.M. Optimization of penicillin G microencapsulation with OSA starch by factorial design. Chem. Eng. Trans. 2012;27:85–90. [Google Scholar]
- Gordon R.E., Barnett D.A., Handarhan J.E., Hor-Nay-Pang C. Nocardia coeliaca, Nocardia autotrophica and the nocardia strains. Int. J. Syst. Bacteriol. 1974;24:54–63. [Google Scholar]
- Goupy J. 1999. Plans d’expériences pour surfaces de réponses, Ed. Dunod, Paris, France. [Google Scholar]
- Gürtler V., Stanisich V.A. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. An. Microbiol. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- Hamza A.A., Ali H.A., Clark B.R., Murphy C.D., Elobied E.A. Optimization of fermentation conditions for actinomycin D production by a newly isolated Streptomyces sp. AH 11.4. E3 J. Biotechnol. Pharm. Res. 2013;4:29–34. [Google Scholar]
- Jukes T.H., Cantor C.R. Evolution of protein molecules. In: Munro H.N., editor. Mammalian Protein Metabolism. first ed. Academic Press; New York, USA: 1969. pp. 21–132. [Google Scholar]
- Kieser T., Bibb M.J., Butner M.J., Charter K.F., Hopwood D.A. Preparation and analysis of the genomic and plasmid DNA. In: Kieser T., editor. Practical Streptomyces Genetics. The John Innes Foundation; Norwich, England: 2000. pp. 162–170. [Google Scholar]
- Kim Y.H., Park B.S., Bhatia S.K., Seo H.M., Jeon J.M., Kim H.J., Yi D.H., Lee J.H., Choi K.Y., Park H.Y., Kim Y.G., Yang Y.H. Production of rapamycin in Streptomyces hygroscopicus from glycerol-based media optimized by systemic methodology. J. Microbiol. Biotechnol. 2014;24:1319–1326. doi: 10.4014/jmb.1403.03024. [DOI] [PubMed] [Google Scholar]
- Kleeff J., Kornmann M., Sawhney H., Korc M. Actinomycin D induces apoptosis and inhibits growth of pancreatic cancer cells. Int. J. Cancer. 2000;86:399–407. doi: 10.1002/(sici)1097-0215(20000501)86:3<399::aid-ijc15>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Krassilnikov N.A. Intra-strain and intra-sp. antagonism among microorganisms. Dokl. Akad. Nauk SSSR. 1960;77 117-119, 725-728. [Google Scholar]
- Kumar V., Bharti A., Gusain O., Singh B.G. Scanning electron microscopy of Streptomyces without use of any chemical fixatives. Scanning. 2011;33:446–449. doi: 10.1002/sca.20261. [DOI] [PubMed] [Google Scholar]
- Kurosawa K., Bui V.P., VanEssendelft J.L., Willis L.B., Lessard P.A., Ghiviriga I., Sambandan T.G., Rha C.K., Sinskey A.J. Characterization of Streptomyces MITKK-103, a newly isolated actinomycin X2-producer. Appl. Microbiol. Biotechnol. 2006;72:145–154. doi: 10.1007/s00253-005-0240-2. [DOI] [PubMed] [Google Scholar]
- Lackner H. Three-dimensional structure of the actinomycins. Angew Chem. Int. Ed. Engl. 1975;14:375–386. doi: 10.1002/anie.197503751. [DOI] [PubMed] [Google Scholar]
- Lamari L., Zitouni A., Boudjella H., Badji B., Sabaou N., Lebrihi A., Lefebvre G., Seguin E., Tillequin F. New dithiolopyrrolone antibiotics from Saccharothrix sp. SA 233. I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. 2002;55:696–701. doi: 10.7164/antibiotics.55.696. [DOI] [PubMed] [Google Scholar]
- Lechevalier M.P., Lechevalier H. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int. J. Syst. Bacteriol. 1970;20:435–443. [Google Scholar]
- Lechevalier M.P., De Bievre C., Lechevalier H.A. Chemotaxonomy of aerobic actinomycetes: phospholipid composition. Biochem. Syst. Ecol. 1977;5:249–260. [Google Scholar]
- Lewin G.R., Carlos C., Chevrette M.G., Horn H.A., McDonald B.R., Stankey R.J., Fox B.G., Currie C.R. Evolution and ecology of Actinobacteria and their bioenergy applications. Annu. Rev. Microbiol. 2016;70:235–254. doi: 10.1146/annurev-micro-102215-095748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohani N., Singh N.H., Moganty R.R. Structural aspects of the interaction of anticancer drug Actinomycin-D to the GC rich region of hmgb1 gene. Int. J. Biol. Macromol. 2016;87:433–442. doi: 10.1016/j.ijbiomac.2016.02.060. [DOI] [PubMed] [Google Scholar]
- Mahalaxmi Y., Sathish T., Prakasham R.S. Development of balanced medium composition for improved rifamycin B production by isolated Amycolatopsis sp. RSP-3. Lett. Appl. Microbiol. 2009;49:533–538. doi: 10.1111/j.1472-765X.2009.02701.x. [DOI] [PubMed] [Google Scholar]
- Mahmoud M.A.S. University of Göttingen; Germany: 2004. Bioactive Secondary Metabolites from marine and Terrestrial Bacteria: Isoquinolinequinones, Bacterial Compounds with a Novel Pharmacophor. PhD Thesis. [Google Scholar]
- Pasciak M., Holst O., Lindner B., Mordarska H., Gamian A. Novel bacterial polar lipids containing ether-linked alkyl chains, the structures and biological properties of the four major glycolipids from Propionibacterium propionicum PCM 2431 (ATCC 14157 T) J. Biol. Chem. 2003;278:3948–3956. doi: 10.1074/jbc.M206013200. [DOI] [PubMed] [Google Scholar]
- Praveen V., Tripathi D., Tripathi C.K.M., Bihari V. Nutritional regulation of actinomycin-D production by a new isolate of Streptomyces sindenensis using statistical methods. Indian J. Exp. Biol. 2008;46:138–144. [PubMed] [Google Scholar]
- Praveen V., Tripathi C.K.M., Bihari V., Srivastava S.C. Production of actinomycin-D by the mutant of a new isolate of Streptomyces sindenensis. Braz. J. Microbiol. 2008;39:689–692. doi: 10.1590/S1517-838220080004000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Saker R., Bouras N., Meklat A., Zitouni A., Schumann P., Sproer C., Sabaou N., Klenk H.P. Prauserella isguenensis sp. nov., a halophilic actinomycete isolated from desert soil. Int. J. Syst. Bacteriol. 2015;65:1598–1603. doi: 10.1099/ijs.0.000145. [DOI] [PubMed] [Google Scholar]
- Schmidt C., Shubert N.A., Brabetz S., Mack N., Schwalm B., Chan J.A., Selt F., Herold-Mend C., Witt O., Milde T., Pfister S.M., Korshunov A., Kool M. Preclinical drug screen reveals topotecan, actinomycin D, and volasertib as potential new therapeutic candidates for ETMR brain tumor patients. Neuro Oncol. 2017;19:1607–1617. doi: 10.1093/neuonc/nox093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P., Zhang Y., Qu H., Fan X. Systematic characterisation of secondary metabolites from Ixeris sonchifolia by the combined use of HPLC-TOFMS and HPLC-ITMS. Phytochem. Anal. 2011;22:66–673. doi: 10.1002/pca.1253. [DOI] [PubMed] [Google Scholar]
- Singh S.B., Guenilloud O., Pelaez F. Terrestrial microorganisms–Filamentous bacteria. In: Liu H.W., Mander L., editors. Comprehensive Natural Products II, Chemistry and Biology. first ed. Elsevier Science., The Boulevard Longford Lane, Kidlington OX5 1GB; UK: 2010. pp. 109–140. [Google Scholar]
- Singh V., Haque S., Niwas R., Srivastava A., Pasupuleti M., Tripathi C.K.M. Strategies for fermentation medium optimization: an in-depth review. Front. Microbiol. 2017;7:2087. doi: 10.3389/fmicb.2016.02087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa M.F.V.Q., Lopes C.E., Pereira N. Development of a bioprocess for the production of actinomycin-D. Braz. J. Chem. Eng. 2002;19:277–285. [Google Scholar]
- Srinivasulu B., Kim Y., Chang Y.K., Shang G., Yu T.W., Floss H.G. Construction of asm2 deletion mutant of Actinosynnema pretiosum and medium optimization for ansamitocin P-3 production using statistical approach. J. Microbiol. Biotechnol. 2006;16:1338–1346. [Google Scholar]
- Staneck J.L., Roberts G.D. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl. Microbiol. 1974;28:226–231. doi: 10.1128/am.28.2.226-231.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayabharathi R., Brunthadevi P., Sathyabama S., Bruheim P., Priyadarisini V.B. Optimization of resistomycin production purified from Streptomyces aurantiacus AAA5 using response surface methodology. J. Biochem. Technol. 2012;3:402–408. [Google Scholar]
- Wei Z., Xu C., Wang J., Lu F., Bie X., Lu Z. Identification and characterization of Streptomyces flavogriseus NJ-4 as a novel producer of actinomycin D and holomycin. PeerJ. 2017;5 doi: 10.7717/peerj.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams W.K., Katz E. Development of a chemically defined medium for the synthesis of actinomycin D by Streptomyces parvulus. Antimicrob. Agents Chemother. 1977;11:281–290. doi: 10.1128/aac.11.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S.H., Ha S.M., Kwon S., Lim J., Kim Y., Seo H., Chun J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitouni A., Boudjella H., Mathieu F., Sabaou N., Lebrihi A. Mutactimycin PR, a new anthracycline antibiotic from Saccharothrix sp. SA 103. I. Taxonomy, fermentation, isolation and biological activities. J. Antbiot. 2004;57:367–372. doi: 10.7164/antibiotics.57.367. [DOI] [PubMed] [Google Scholar]