Abstract
In the event of a bioterror attack, a prompt, sensitive and definite identification of the agents involved is of major concern for confirmation of the event and for mitigation of countermeasures. Whether the information from intelligence forces is limited concerning the biothreat identity or one suspects the presence of a novel or engineered agent, the genetic identification of microorganisms in an unknown sample is challenging. High-throughput sequencing (HTS) technologies can sequence a heterogeneous mixture of genetic materials with high sensitivity and speed; nevertheless, despite the enormous advantages of HTS, all previous reports have analyzed unknown samples in a timeframe of a few days to a few weeks. This timeframe might not be relevant to an emergency scenario. Here, we present an HTS-based approach for deciphering the genetic composition of unknown samples within a working day. This outcome is accomplished by a rapid library preparation procedure, short-length sequencing and a prompt bioinformatics comparison against all available microbial genomic sequences. Using this approach, as a proof of concept, we were able to detect two spiked-in biothreat agents, B. anthracis and Y. pestis, in a variety of environmental samples at relevant concentrations and within a short timeframe of eight hours.
Keywords: Bioinformatics, Genetics, Microbiology, Molecular biology
1. Introduction
The ever-growing concern about the abuse of biothreat agents to create terror has emerged in the past few decades as a primary international security challenge, owing to the simple and inexpensive production, easy dissemination, and complicated detection of these agents; the expense of protection against these agents; and the psychological, economic and social impact of bioterror acts. Early detection and identification of biothreat agents during an intentional release is essential to initiate emergency responses and mitigate such an incident. Efforts are being made across the globe to develop state-of-the-art technologies and systems for the detection and identification of biothreat agents. However, until now, there is no single system that can detect all biothreat agents.
In the relatively simple scenario where there is a solid suspicion of a specific biothreat agent, molecular biology techniques, which are mostly PCR-based and involve the amplification of a defined target, are employed. For instance, CDC-defined Tier 1 biothreat agents such as B. anthracis and Y. pestis [1] are well-defined and have multiple genetic tests for detection [2, 3, 4, 5, 6, 7]. These methods allow a rapid and straightforward identification compared to conventional microbiological methods, which are time-consuming. These techniques are highly sensitive and specific, and they can yield results within one or two hours from only a few copies of the target nucleic acid in the sample. However, PCR-based methods demand a level of prior knowledge that is not always present. Furthermore, newly emerged biothreat agents or genetically modified agents might not be detected at all. When there is no previous information about the content of a sample (i.e., an unknown sample), the ultimate nucleic acid-based detection method should be based on DNA sequencing. Sequencing of the entire content of a sample might not only provide an answer for the 'yes or no question' of whether a specific biothreat agent is present in the sample but also answer the 'what question', i.e., the question of what the sample contains, including the characterization of different genomic traits. Such an approach is not dependent on the growth of organisms; only the presence of their nucleic acid in the sample is needed for identification.
Until a decade ago, the use of DNA sequencing to decipher the content of an unknown sample was not feasible. Traditional Sanger-based sequencing was not suitable for analyzing mixtures and was expensive, time-consuming and laborious. The emergence of high-throughput sequencing (HTS) technologies paved the way for the comprehensive detection of pathogens without any prior knowledge. These massively parallel sequencing platforms can sequence a heterogeneous mixture of genetic materials with high sensitivity and speed and with a lower cost per base compared to the traditional Sanger sequencing method [8, 9]. HTS has other benefits apart from the improved detection of known and unknown pathogens in different samples; among these benefits are the ability to detect nonculturable organisms and the ability to detect coinfections, drug resistance and antibiotic resistance [10]. The use of HTS for the characterization of biothreats in unknown environmental samples is therefore potent and promising. In a study by Be1 et al., HTS was used for the detection of B. anthracis in soil and air samples, and a detection level of as few as 10 genomic equivalents of B. anthracis DNA per nanogram of background nucleic acid was demonstrated [11]. More recently, Plaire et al. performed a comparative analysis of the sensitivity of metagenomic sequencing and PCR in detecting a simulant of B. anthracis (Bacillus atrophaeus) in soil samples and found a similar detection level [12].
Despite the enormous advantages of HTS in comparison with Sanger sequencing, all the previous reports that used HTS in this context analyzed the genetic content of a sample in a timeframe of a few days to a few weeks. This timeframe might not be relevant to an emergency scenario such as a major biological attack. Here, we present, for the first time, the use of HTS for deciphering the genetic content of unknown samples within a working day. This result is accomplished by an approach we developed that comprises a combination of a rapid library preparation procedure utilizing Nextera XT technology, short (60 nt) HTS with the MiSeq sequencer and prompt bioinformatics comparison against all available bacterial, viral and fungal genomic sequences. Using this approach, as a proof of concept, we were able to detect two CDC Tier 1 agents, B. anthracis and Y. pestis, in a variety of environmental samples at relevant concentrations and within a relatively short timeframe.
2. Methods
2.1. Environmental sample collection
Environmental samples were collected in the Ness Ziona town area, Shfela District, Israel. The samples were taken from places that represent a typical town, e.g., asphalt-paved roads, sidewalks, orchards and parking lots (see details in Table 1). No specific permissions were required for the acquisition of the environmental samples, as all collections were performed on unprotected land. These environmental studies did not involve any endangered or protected species.
Table 1.
Environmental sample collection.
| Sample # | Source | Spike-in (concentration) | DNA Con (ng/μl) |
|---|---|---|---|
| 1–104 | Sidewalk mixed with soil | BA (104/ml) | ND |
| 1–105 | Sidewalk mixed with soil | BA (105/ml) | ND |
| 1–106 | Sidewalk mixed with soil | BA (106/ml) | ND |
| 1–107 | Sidewalk mixed with soil | BA (107/ml) | ND |
| 2 | Public stairway | BA (105/ml) | ND |
| 3 | Sidewalk | BA (105/ml) | ND |
| 4 | Parking lot | BA (105/ml) | ND |
| 5 | Asphalt road | BA (105/ml) | ND |
| 6 | Flowerbed soil | BA (105/ml) | 0.13 |
| 7 | Orchard soil | BA (105/ml) | 0.15 |
| 8 | Grass | BA (105/ml) | 0.10 |
| 9 | Rock, lichen | BA (105/ml) | ND |
| 10 | Rock, lichen | BA (105/ml) | ND |
| 11–104 | Asphalt road | YP (104/ml) | ND |
| 11–105 | Asphalt road | YP (105/ml) | ND |
| 11–106 | Asphalt road | YP (106/ml) | ND |
| 11–107 | Asphalt road | YP (107/ml) | ND |
| 11–108 | Asphalt road | YP (108/ml) | 0.90 |
| 11–109 | Asphalt road | YP (109/ml) | 12.1 |
*BA = B. anthracis strain Vollum (ATCC 14578), YP = Y. pestis strain EV76, ND = not determined, <0.1 ng/μl, measured with the Qubit high-sensitivity DNA kit.
The samples were collected from a 20 cm2 area using two swabs soaked in 1× PBS and one dry swab. The swabs were then vortexed vigorously in 5 ml PBS. At this stage, the samples were inoculated with serial dilutions of B. anthracis strain Vollum (ATCC 14578) spores or Y. pestis strain EV76 [13] bacteria corresponding to 104–109 CFU/ml. The samples were aliquoted to 1 ml and then centrifuged at 14,000 rpm for 5 min. Nine hundred microliters of the supernatant was removed, and the remaining 100 μl was resuspended in 100 μl 20% Triton X-100 (Sigma). The samples were then microwaved at maximum power for 7 min for sterilization and were used for DNA extraction.
2.2. DNA extraction, quantification and carrier DNA supplementation
DNA was extracted from the samples using the DNA QIAamp DNA Blood Mini Kit (Qiagen) with a protocol for blood and body fluids in a QIAcube robot and was recovered in a 100 μl elution volume in ddH2O. Analysis of DNA in the extracts using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific) yielded values below the limit of accurate detection (<5 ng/μl). Consequently, the DNA was quantified using a Qubit fluorometer and the Qubit DNA HS Assay Kit (Invitrogen), which can measure DNA at subnanogram concentrations. The DNA was diluted to 0.2 ng/μl, as required by the Nextera XT sample preparation protocol. If the concentration of the extracted DNA was below 0.2 ng/μl, carrier DNA (B. subtilis strain 168) was added to a total amount of 1 ng in 5 μl, according to the Nextera XT sample preparation protocol.
2.3. Library preparation and HTS
1 ng of DNA was transferred to the library preparation step utilizing the Nextera XT DNA Sample Preparation Kit (Illumina Inc., San Diego, CA, USA). The standard manufacturer protocol was modified to optimize library preparation from environmental samples. For each sample, the 20 μl tagmentation reaction contained 5 μl of amplicon tagment mix (ATM), which includes the enzyme used for tagmentation, 10 μl of TD buffer, 1 ng of input DNA (4 μl combined, with or without carrier DNA) and 1 μl of 20 mg/ml bovine serum albumin (BSA). The tagmentation reactions were incubated in a thermal cycler at 50 °C for 5 min. Subsequently, the tagmented DNA was amplified via limited-cycle PCR. The quality and quantity of the purified libraries were assessed using the high-sensitivity (HS) DNA kit on an Agilent 2100 Bioanalyzer. The libraries were normalized to 3 nM, denatured with 0.2 N NaOH for 5 min and diluted 1:100 in HT1 buffer (Illumina) to a final concentration of 15 pM. The diluted libraries were sequenced with an Illumina MiSeq with 50 bp v2 run chemistry. The sequencing length was 60 nt in single-read mode. Each sample was spiked with 0.2 pM phiX174 library (Illumina).
2.4. Bioinformatic analyses
All reads generated by HTS were taxonomically profiled with Pathoscope 2.0 [14] using a constructed target genome library containing all complete bacterial (5414), viral (5777) and fungal (3451) genomes downloaded from NCBI. The reads were aligned to these databases using the Bowtie2 algorithm [15] with the default Pathoscope parameters. The phiX174 sequence and similar sequences were manually curated from the results.
2.5. Real-time PCR assays for B. anthracis detection
Real time PCR assays were performed in a 25 μl reaction volume using the SensiFAST™ Probe Lo-ROX kit (BIOLINE) on a 7500 real-time PCR system (Applied Biosystems). The PCR was carried out under the following conditions: 2 min at 95 °C followed by 40 cycles at 30 s 95 °C and 30 s 60 °C. The primer and probes for B. anthracis quantification were adapted from Wielinga et al. 2011 [16] and were purchased from Integrated DNA Technologies, Inc. The primers and probe sequences were as follows:
PL3_f:AAAGCTACAAACTCTGAAATTTGTAAATTG;
PL3_r:CAACGATGATTGGAGATAGAGTATTCTTT.
Tqpro_PL3: FAM-AACAGTACGTTTCACTGGAGCAAAATCAA-BHQ1.
3. Results
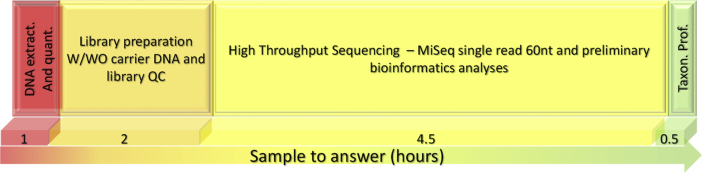
The approach for the rapid identification of pathogens in unknown environmental samples using HTS is summarized in Fig. 1. We established a rapid and comprehensive procedure that starts with an unknown environmental sample and ends with a detailed metagenomic profile of the identified agents in a timeframe of eight hours.
Fig. 1.
Schematic presentation of the sample profiling steps. Each step is represented by a box of a different color, with the time for each step (in hours) noted below each box. W/WO = with/without; Taxon. Prof. = taxonomic profiling.
In our approach, the DNA is first extracted and quantified utilizing the sensitive Qubit apparatus, which can detect subnanogram quantities of DNA. The concentration of most of the samples was below 0.2 ng/μl (1 ng total in 5 μl), which is the recommended starting concentration for library preparation using the Nextera technology. This technology simplifies the library preparation procedure, reduces the preparation time to less than two hours and requires the smallest amount of DNA as a template compared with the requirements of other HTS library preparation procedures. The issue of insufficient amounts of DNA in our samples was resolved by adding carrier DNA to a total of 1 ng. Another modification of the Nextera protocol was the addition of BSA in the tagmentation step for the enhancement of the enzymatic reactions. The resulting libraries were sequenced with the short sequencing length of 60 nt and then taxonomically profiled against all the available bacterial, viral and fungal genome sequences using the Pathoscope framework for rapid and accurate metagenomic profiling.
Since we wanted to implement our approach with environmental samples collected from locations that represent a typical town, all samples in this work were collected from zones such as asphalt-paved roads, sidewalks, orchards and parking lots (see details in Table 1).
3.1. Identification of B. anthracis spores serially diluted in an environmental sample
We first sought to examine the feasibility of our approach by analyzing an environmental sample to which varying concentrations of B. anthracis spores (104 to 107/ml) were added. The environmental sample was collected from a sidewalk that was partially covered with sand and dust. The DNA extracted from the sample did not reach the minimal concentration needed for library preparation; therefore, 1 ng of carrier DNA was added. The sequencing and analysis results for the samples containing B. anthracis spores are summarized in Table 2. The reads were mapped to B. anthracis for all the spike-in concentrations (104–107 CFU/ml). The lower concentrations, namely, 104-105 spiked-in spores/ml, were run in uniplex mode to maximize the yield, and an average of 14,662,024 ± 730,159 reads were generated, resulting in the detection of B. anthracis with 4,404 and 1,125 reads in the 105/ml and 104/ml spore spike-in conditions, respectively. The higher concentrations (106 and 107 spiked-in spores/ml) were then run in a multiplex mode and, therefore, yielded a somewhat lower number of total reads (below 2 million each). Furthermore, as expected, B. anthracis was clearly detected in these samples, with over 4000 mapped reads for these higher concentrations. The average fraction of mapped reads, which represents the total number of mapped reads divided by the total number of sequenced reads, was 0.35 ± 0.05.
Table 2.
Sequencing reads from the environmental samples spiked with a serial dilution of B. anthracis spores.
| Mapped reads | Spiked-in B. anthracis spores (CFU/ml). see Table 1 for details |
|||
|---|---|---|---|---|
| 104 | 105 | 106 | 107 | |
| Bacillus anthracis | 1,125 | 4,404 | 5,205 | 4,103 |
| Bacillus subtilis | 3,486,962 | 5,697,983 | 648,733 | 513,889 |
| Total # of reads | 13,931,864 | 15,392,183 | 1,900,913 | 1,429,718 |
| Mapped reads | 3,858,630 | 6,210,179 | 675,269 | 533,545 |
| Fraction mapped | 0.28 | 0.40 | 0.36 | 0.37 |
3.2. Identification of different environmental samples spiked with 105B. anthracis spores/ml
Following the results obtained in the serial dilution experiments, we extended our environmental samples to represent a variety of town/urban areas, e.g., asphalt-paved roads, sidewalks, orchards and parking lots (see details in Table 1). We chose to spike these samples with 105/ml B. anthracis spores, a concentration that was clearly detected in the previous step. A total of nine samples were collected. In six of the samples (2, 3, 4, 5, 9 and 10; Table 1) the extracted DNA did not reach the detection level of the Qubit HS DNA kit; therefore, 1 ng of carrier DNA was added for the library preparation procedure. The other samples (6–8) had a DNA concentration of 0.1–0.15 ng/μl, and carrier DNA was added to a total amount of 1 ng.
Table 3 summarizes the results for the different environmental samples spiked with 105/ml B. anthracis spores. The average total number of reads for the nine samples was 13,196,803 ± 3,780,546, and B. anthracis was clearly identified (based on 899–6201 mapped reads, average 2893 ± 1717) in all of the samples. The fraction of total mapped reads decreased from an average of 0.27 ± 0.03 for samples 2, 3, 4, 5, 9 and 10 to an average of 0.1 ± 0.01 for samples 6–8, probably due to the higher levels of environmental DNA.
Table 3.
Sequencing reads from the environmental samples spiked with 105/ml B. anthracis spores.
|
B. anthracis environmental sample # (see Table 1 for details) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Mapped reads | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| Bacillus anthracis | 4,349 | 899 | 1,538 | 1,503 | 2,490 | 1,431 | 3,013 | 6,201 | 5,241 |
| Bacillus subtilis | 3,948,977 | 2,933,010 | 840,997 | 4,030,754 | 1,249,576 | 694,450 | 1,415,279 | 3,035,291 | 2,457,851 |
| Total # of reads | 17,713,294 | 11,502,509 | 4,273,582 | 16,400,867 | 12,182,845 | 15,383,943 | 16,174,778 | 12,510,448 | 12,628,962 |
| Mapped reads | 4,610,922 | 3,462,640 | 905,393 | 4,614,614 | 1,321,132 | 1,510,390 | 1,492,309 | 3,841,713 | 2,969,562 |
| Fraction mapped | 0.26 | 0.30 | 0.21 | 0.28 | 0.11 | 0.10 | 0.09 | 0.31 | 0.24 |
3.3. Identification of Y. pestis bacteria serially diluted in an environmental sample
Following the identification of B. anthracis in environmental samples, we wanted to test our approach using another biothreat model. To this end, Y. pestis bacteria were spiked into an environmental sample swabbed from an asphalt road (sample 11, see details in Table 1). The DNA extracted from the sample did not reach the minimal concentration needed for library preparation for the spike-in concentrations of 104/ml-107/ml; therefore, 1 ng of carrier DNA was added. For the higher spike-in concentrations, namely, 108/ml-109/ml, no carrier DNA was added.
Table 4 summarizes the results for the serial dilution of Y. pestis bacteria in an environmental sample. These samples were sequenced in duplex mode with two samples in each sequencing run. Nevertheless, Y. pestis was identified in all the spike-in dilutions tested, with the number of mapped reads ranging from 2,690 for the 104/ml Y. pestis bacteria spike-in concentration to 4,248,594 for the 109/ml spike-in concentration. In addition, the fraction of mapped reads was higher than that observed in our previous experiments, with an average of 0.89 ± 0.02; this increase was probably due to the lower amounts of environmental DNA in the sample (asphalt road).
Table 4.
Sequencing reads from the environmental samples spiked with a serial dilution of Y. pestis bacteria.
|
Y. pestis bacteria spike-in (CFU/ml). see Table 1 for details | ||||||
|---|---|---|---|---|---|---|
| Mapped reads | 104 | 105 | 106 | 107 | 108 | 109 |
| Y. pestis | 2,690 | 8,411 | 25,743 | 970,236 | 4,087,184 | 4,248,594 |
| Bacillus subtilis | 4,771,579 | 6,216,021 | 4,665,644 | 3,527,201 | - | - |
| Total # of reads | 5,423,633 | 7,114,036 | 5,407,919 | 5,400,774 | 4,905,519 | 4,961,625 |
| Mapped reads | 4,793,585 | 6,237,920 | 4,705,463 | 4,649,273 | 4,461,149 | 4,572,890 |
| Fraction mapped | 0.88 | 0.88 | 0.87 | 0.86 | 0.91 | 0.92 |
4. Discussion
In the event of a bioterror attack, a prompt, sensitive and definite identification of the biothreat agents involved is of major concern for confirmation of the event and for mitigation by direct and indirect countermeasures, such as evacuation, regional curfew or administration of the appropriate drugs and vaccines, if available. Genetic methods, which are mainly PCR based, are highly sensitive but are limited to the identification of focused biothreats. Whether the information from intelligence forces is limited concerning the biothreat identity or one suspects the presence of a novel or engineered agent, genetic identification is challenging.
The remarkable progress, growing accessibility and falling cost of HTS-based technologies provide an opportunity to perform whole-genome analyses of environmental samples, which yield a breadth of information not available from other genetic-based assays. Previous studies demonstrated that HTS-based approaches can identify the presence of biothreat agents in environmental samples even when these agents are present in low copy numbers (for example, see references [11, 12, 17]). However, those studies were conducted with a timeframe of a few days to weeks from sample to answer. In the present study, we aimed to establish a comprehensive, HTS-based approach for deciphering the genetic content of a sample without any prior knowledge of its content (i.e., an unknown sample) that could be completed in a relevant timeframe for a bioterror event – within a working day.
In our approach, the DNA is extracted and quantified, and libraries are prepared using the Nextera technology, which shortens the library preparation step to less than two hours and utilizes an in vitro transposition method in which DNA is tagmented (fragmented and tagged simultaneously) [18, 19]. The resulting tagged fragments undergo a short-cycle PCR reaction to add sequencing adaptors, and the generated libraries are then sequenced for a relatively short single-end read of 60 nt with the MiSeq sequencer and subjected to a prompt bioinformatics analysis using Pathoscope, which enables taxonomic profiling of the sample against all available bacterial, viral and fungal genome sequences. In our hands, using the short 60 nt reads did not hamper the detection ability, as compared to using longer reads (150nt) in a subset of environmental samples (data not shown).
It is acknowledged that swab extracts that are collected from environmental samples do not always contain enough genetic material for an appropriate library preparation procedure [12]. Indeed, using our environmental sampling procedure, we did not reach the amount of 1 ng needed for library preparation with the Nextera method, which requires the lowest concentration of starting material compared with the requirements of other methods. The 1 ng of DNA needed for the Nextera library preparation procedure is equivalent to 1.85×108/ml copies of a bacteria with a genome size of 5 million bp. Thus, our previous attempts with amounts less than 1 ng yielded low-quality libraries or negative results [data not shown]. We chose to tackle this problem by adding carrier DNA to reach the needed amount. Theoretically, any DNA could be utilized for this purpose, but we elected to use DNA from B. subtilis, a bacterium from the Bacillus genus, as a further demonstration that our approach can distinguish between relatively closely related bacteria such as B. subtilis and B. anthracis, which have only an ∼32% divergence in their DNA sequences [20].
During the establishment of our approach, we encountered certain environmental samples for which the yield of the libraries was low or the library preparation procedure completely failed. These outcomes possibly occurred due to contaminants and/or inhibitors such as humic or flavic acid in the samples. The addition of BSA, which has been used successfully as an additive in several enzymatic reactions [21, 22], eliminated the problem and resulted in a library preparation that was close to optimum.
In the current study, we chose to validate/examine our approach with two of the main CDC Tier 1 biothreat agents, B. anthracis and Y. pestis [1], as simulants for a possible intentional biothreat spread. We initially tested our approach using a serial dilution (104-107/ml) of spiked-in spores of B. anthracis, which represents a typical gram-positive sporulating bacterium.
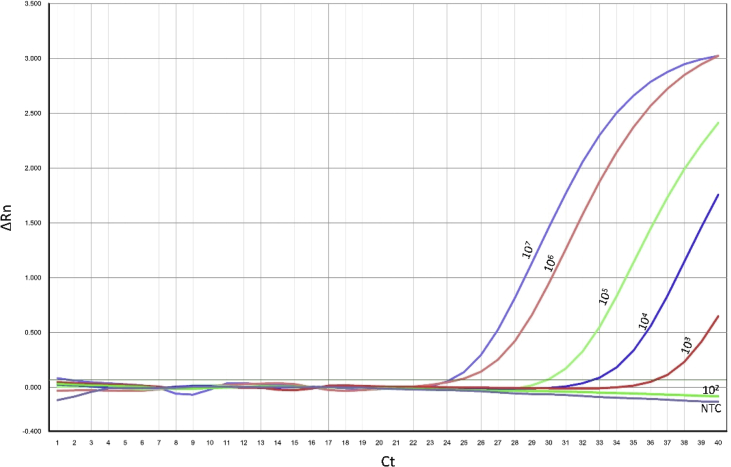
The B. anthracis spores were also analyzed using real-time PCR [16] showing a lower limit of detection of 103/ml (Fig. 2) in accordance to previous reported results in different environmental samples [2]. Based on experiments in animal models and environmental investigations of industrial human outbreaks, the human LD50 is estimated as 1000 to 4100 and the LD10 50–100 spores [23].
Fig. 2.
Real time PCR of B. anthracis environmental samples. Spiked B. anthracis environmental samples were analyzed by real-time PCR as previously reported [16]. The B. anthracis concentrations (spores/ml) are noted in the figure. NTC, no template control; Ct, is the intersection between an amplification curve and a threshold line; delta Rn is the ratio of the fluorescence of FAM™ Dye divided by the background fluorescence of ROX dye.
In all the spike-in dilutions analyzed, B. anthracis was identified based on over 1,000 mapped reads, demonstrating our ability to clearly detect the biothreat agent. We then extended our environmental sample repertoire by analyzing nine additional different environmental samples which were spiked with 105/ml B. anthracis spores. In all samples analyzed, B. anthracis was clearly detected, with a range of 899–6201 mapped reads. Subsequently, we demonstrated our approach with another biothreat agent, Y. pestis, which represents a typical gram-negative bacterium [24]. The limit of detection for this bacterium was previously reported to be 105 CFU/ml using real-time PCR in different environmental samples [3]. Based on epidemiological investigations and animal experiments, the human LD50 for Y. pestis is estimated as 500 to 15,000 CFU/HID50 [25].
In the current study, Y. pestis bacteria were spiked in (104-109/ml) and were clearly detected, with over 6700 mapped reads per sample. In this set of experiments, the number of mapped reads was significantly higher than that for the same concentrations of spiked-in B. anthracis spores (for instance, 6700 vs. 1125 mapped reads for Y. pestis and B. anthracis, respectively, in the 104/ml spike-in samples and 16180 vs. 4404 mapped reads for Y. pestis and B. anthracis, respectively, in the 105/ml spike-in samples). The lower number of mapped reads could be attributed to the somewhat less-efficient DNA extraction from B. anthracis spores compared with DNA extraction from vegetative bacteria such as Y. pestis.
The results presented in this paper demonstrate that our HTS-based approach is an efficient procedure for coping with the challenge of deciphering the composition of samples containing biothreat agents without any prior knowledge about their nature. Using our approach, within a very applicable timeframe of eight hours, we succeeded in correctly identifying two biothreat agent simulants in relevant concentrations (104/ml) in more than 10 environmental samples. Using our approach we detected bacteria which are relatively resistant in the environment. The use of this approach for the detection of sensitive microorganisms, such as DNA or RNA viruses might be a lot more challenging due to the low stability of those "environmentally weaker" genomes.
In conclusion, our approach as a whole is well suited for rapidly and unambiguously detecting any bacterial or viral agent with a DNA genome in a sample without any prior knowledge. At this point in time, RNA viruses have not been detected using our approach. Our next goal is to further generalize the approach and to develop a holistic procedure that will detect any agent in the sample regardless of its genetic material.
Declarations
Author contribution statement
Ofir Israeli: Conceived and designed the experiments; Performed the experiments; Wrote the paper.
Inbar Cohen-Gihon, Anat Zvia: Performed the experiments; Wrote the paper.
Shirley Lazar: Performed the experiments.
Ohad Shifman: Conceived and designed the experiments.
Haim Levy, Avital Tidhar: Performed the experiments; Contributed reagents, materials, analysis tools or data.
Adi Beth-Din: Conceived and designed the experiments; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
The raw data is available on demand from the authors.
References
- 1.Anonymous . 2018. Select Agents and Toxins List, on CDC.https://www.selectagents.gov/selectagentsandtoxinslist.html [Google Scholar]
- 2.Dauphin L.A., Bowen M.D. A simple method for the rapid removal of Bacillus anthracis spores from DNA preparations. J. Microbiol. Methods. 2009;76:212–214. doi: 10.1016/j.mimet.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Dauphin L.A., Stephens K.W., Eufinger S.C., Bowen M.D. Comparison of five commercial DNA extraction kits for the recovery of Yersinia pestis DNA from bacterial suspensions and spiked environmental samples. J. Appl. Microbiol. 2010;108:163–172. doi: 10.1111/j.1365-2672.2009.04404.x. [DOI] [PubMed] [Google Scholar]
- 4.Kim J., Gedi V., Lee S.C., Cho J.H., Moon J.Y., Yoon M.Y. Advances in anthrax detection: overview of bioprobes and biosensors. Appl. Biochem. Biotechnol. 2015;176:957–977. doi: 10.1007/s12010-015-1625-z. [DOI] [PubMed] [Google Scholar]
- 5.Mirski T., Bartoszcze M., Bielawska-Drozd A., Cieslik P., Michalski A.J., Niemcewicz M., Kocik J., Chomiczewski K. Review of methods used for identification of biothreat agents in environmental protection and human health aspects. Ann. Agric. Environ. Med. 2014;21:224–234. doi: 10.5604/1232-1966.1108581. [DOI] [PubMed] [Google Scholar]
- 6.Peruski L.F., Jr., Peruski A.H. Rapid diagnostic assays in the genomic biology era: detection and identification of infectious disease and biological weapon agents. Biotechniques. 2003;35:840–846. doi: 10.2144/03354ss01. [DOI] [PubMed] [Google Scholar]
- 7.Seiner D.R., Colburn H.A., Baird C., Bartholomew R.A., Straub T., Victry K., Hutchison J.R., Valentine N., Bruckner-Lea C.J. Evaluation of the FilmArray(R) system for detection of Bacillus anthracis, Francisella tularensis and Yersinia pestis. J. Appl. Microbiol. 2013;114:992–1000. doi: 10.1111/jam.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buermans H.P., den Dunnen J.T. Next generation sequencing technology: advances and applications. Biochim. Biophys. Acta. 2014;1842:1932–1941. doi: 10.1016/j.bbadis.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 9.van Dijk E.L., Auger H., Jaszczyszyn Y., Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30:418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Radford A.D., Chapman D., Dixon L., Chantrey J., Darby A.C., Hall N. Application of next-generation sequencing technologies in virology. J. Gen. Virol. 2012;93:1853–1868. doi: 10.1099/vir.0.043182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Be N.A., Thissen J.B., Gardner S.N., McLoughlin K.S., Fofanov V.Y., Koshinsky H., Ellingson S.R., Brettin T.S., Jackson P.J., Jaing C.J. Detection of Bacillus anthracis DNA in complex soil and air samples using next-generation sequencing. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plaire D., Puaud S., Marsolier-Kergoat M.C., Elalouf J.M. Comparative analysis of the sensitivity of metagenomic sequencing and PCR to detect a biowarfare simulant (Bacillus atrophaeus) in soil samples. PLoS One. 2017;12 doi: 10.1371/journal.pone.0177112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flashner Y., Mamroud E., Tidhar A., Ber R., Aftalion M., Gur D., Lazar S., Zvi A., Bino T., Ariel N., Velan B., Shafferman A., Cohen S. Generation of Yersinia pestis attenuated strains by signature-tagged mutagenesis in search of novel vaccine candidates. Infect. Immun. 2004;72:908–915. doi: 10.1128/IAI.72.2.908-915.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong C., Manimaran S., Shen Y., Perez-Rogers J.F., Byrd A.L., Castro-Nallar E., Crandall K.A., Johnson W.E. PathoScope 2.0: a complete computational framework for strain identification in environmental or clinical sequencing samples. Microbiome. 2014;2:33. doi: 10.1186/2049-2618-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wielinga P.R., Hamidjaja R.A., Agren J., Knutsson R., Segerman B., Fricker M., Ehling-Schulz M., de Groot A., Burton J., Brooks T., Janse I., van Rotterdam B. A multiplex real-time PCR for identifying and differentiating B. anthracis virulent types. Int. J. Food Microbiol. 2011;145(Suppl 1):S137–S144. doi: 10.1016/j.ijfoodmicro.2010.07.039. [DOI] [PubMed] [Google Scholar]
- 17.Gardner S.N., Frey K.G., Redden C.L., Thissen J.B., Allen J.E., Allred A.F., Dyer M.D., Mokashi V.P., Slezak T.R. Targeted amplification for enhanced detection of biothreat agents by next-generation sequencing. BMC Res. Notes. 2015;8:682. doi: 10.1186/s13104-015-1530-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adey A., Morrison H.G., Asan Xun X., Kitzman J.O., Turner E.H., Stackhouse B., MacKenzie A.P., Caruccio N.C., Zhang X., Shendure J. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caruccio N. Preparation of next-generation sequencing libraries using Nextera technology: simultaneous DNA fragmentation and adaptor tagging by in vitro transposition. Methods Mol. Biol. 2011;733:241–255. doi: 10.1007/978-1-61779-089-8_17. [DOI] [PubMed] [Google Scholar]
- 20.Helgason E., Tourasse N.J., Meisal R., Caugant D.A., Kolsto A.B. Multilocus sequence typing scheme for bacteria of the Bacillus cereus group. Appl. Environ. Microbiol. 2004;70:191–201. doi: 10.1128/AEM.70.1.191-201.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrell P.B., Foote R.H., McArdle M.M., Trouern-Trend V.L., Tardif A.L. Media and dilution procedures tested to minimize handling effects on human, rabbit, and bull sperm for computer-assisted sperm analysis (CASA) J. Androl. 1996;17:293–300. [PubMed] [Google Scholar]
- 22.Kreader C.A. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 1996;62:1102–1106. doi: 10.1128/aem.62.3.1102-1106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters C.J., Hartley D.M. Anthrax inhalation and lethal human infection. Lancet. 2002;359:710–711. doi: 10.1016/s0140-6736(02)07792-9. [DOI] [PubMed] [Google Scholar]
- 24.Stenseth N.C., Atshabar B.B., Begon M., Belmain S.R., Bertherat E., Carniel E., Gage K.L., Leirs H., Rahalison L. Plague: past, present, and future. PLoS Med. 2008;5:e3. doi: 10.1371/journal.pmed.0050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woods C. US Army Medical Research; Frederick, Maryland: 2005. USAMRIID's Medical Management of Biological Casualties Handbook. [Google Scholar]