Abstract
Previously, we have reported that the active vitamin D metabolite, calcitriol and vitamin D3 (cholecalciferol), both remarkably inhibit hepatitis C virus production. The mechanism by which vitamin D3 exerts its effect is puzzling due to the low levels of calcitriol produced in vitamin D3-treated Huh7.5 cells. In this study, we aimed to explore the mechanism of vitamin D3 anti-hepatitis C virus effect. We show that vitamin D3 activity is not mediated by its metabolic conversion to calcitriol, but may be due to its primary metabolic product 25(OH)D3. This is inferred from the findings that 25(OH)D3 could inhibit hepatitis C virus production in our system, and that adequate concentrations needed to exert this effect are produced in Huh7.5 cells treated with vitamin D3. Using the CRISPR-Cas9 editing technology to knockout the vitamin D receptor, we found that the antiviral activity of vitamin D3 and 25(OH)D3 was not impaired in the vitamin D receptor knockout cells. This result indicates that 25(OH)D3 anti-hepatitis C virus effect is exerted by a vitamin D receptor-independent mode of action. The possibility that vitamin D3 and 25(OH)D3, being 3β-hydroxysteroids, affect hepatitis C virus production by direct inhibition of the Hedgehog pathway in a vitamin D receptor-independent manner was ruled out. Taken together, this study proposes a novel mode of action for the anti-hepatitis C virus activity of vitamin D3 that is mediated by 25(OH)D3 in a vitamin D receptor-independent mechanism.
Keywords: vitamin D3, 25-hydroxyvitamin D3, vitamin D receptor, hepatitis C virus
1. Introduction
Vitamin D is increasingly recognized as an important physiological regulator with pleiotropic functions. Cholecalciferol (vitamin D3) the 3β-hydroxyl-secosteroid, which is mainly produced in the skin and partly supplied through diet, is an inert compound, which acquires its biological activity through two successive hydroxylations to provide 1α, 25-dihydroxyvitamin D3 (calcitriol) [1,2]. The first hydroxylation occurs in the liver, the main source for the major circulating form of vitamin D, 25-hydroxyvitamin D3 (25(OH)D3). It harbors the physiologically relevant vitamin D 25-hydroxylase (25(OH)ase): CYP2R1, but also three other 25(OH)ases: CYP27A1, CYP2J2, and CYP3A4 [3]. The second hydroxylation occurs in the kidney where 25(OH)D3 is metabolized to calcitriol by 25-hydroxyvitamin D 1α-hydroxylase encoded by Cyp27B1. Although the kidney was initially thought to be the sole organ expressing CYP27B1, it is now appreciated that its expression in tissues other than the kidney is widespread including in the hepatocarcinoma cell (HCC) line Huh7.5 [4]. Catabolism of calcitriol and 25(OH)D3 is governed by 25-hydroxyvitamin-D 24-hydroxylase (24-hydroxylase), encoded by Cyp24A1, which is a target to calcitriol action. Calcitriol exerts its biological activities by binding to the vitamin D receptor (VDR), a member of the nuclear receptor family, which forms a heterodimer with RXR and binds to vitamin D responsive elements (VDREs) in the promotor of vitamin D-target genes and mediate their transcription.
However, more than a decade ago the dogma that cholecalciferol is biologically inert was challenged when Biljsma et al. reported that 3β-hydroxysteroid in general, and cholecalciferol in particular, can directly inhibit the Hedgehog (Hh) pathway [5,6,7,8]. Hh signaling is mediated by the membrane protein, patched (Ptch1) which binds and inhibits smoothened (Smo) [9]. Activation of this pathway is initiated by binding of Hh to Ptch1, releasing Smo, and the downstream transcription factor the glioma-associated (Gli) from inhibition. Cholecalciferol was shown to directly bind Smo, thereby preventing the alleviation of Smo and Gli inhibition.
It is also argued that 25(OH)D3, which was long regarded as an inactive prohormone, is an agonistic vitamin D receptor ligand at high concentrations. It was shown to have gene regulatory activity with target gene profiles largely matching those of calcitriol [1,2,10].
The vitamin D system is known as essential to the skeletal system; however, during the last 50 years, a multitude of extraskeletal effects have been documented. Recently, a strong association between vitamin D deficiency and the clinical outcome and disease progression of hepatitis C virus (HCV) infections was demonstrated [11]. It was reported that supplementation of vitamin D to pegylated interferon and ribavirin therapy significantly improved sustained virologic response (SVR) rates in patients with chronic HCV infection [12,13,14]. We have recently shown that both calcitriol and vitamin D3 remarkably inhibited HCV production in an HCC line [15]. In our study, we found that nanomolar concentrations of calcitriol were required to attain substantial inhibition of HCV production in the Huh7.5 hepatoma cell line, while only picomolar concentrations of calcitriol were produced in these cultures when supplemented with HCV-inhibiting concentrations of vitamin D3. In view of this discrepancy, we challenge the presumption that calcitriol is the main and only mediator of the anti-HCV activity of vitamin D3 and examine the role of 25(OH)D3 as a VDR agonist and of cholecalciferol itself in this activity. Herein, we present evidence that the antiviral activity of vitamin D3 is most probably mediated by 25(OH)D3 in a VDR-independent mechanism.
2. Results
2.1. Involvement of Calcitriol in the Anti-HCV Activity of Vitamin D3
In our previous study, we found that the inhibition of HCV production by vitamin D3 is accompanied by calcitriol generation which results in the induction of the vitamin D target gene Cyp24A1. This led us to conclude that the newly produced calcitriol mediates the anti-HCV activity of the vitamin. However, we were troubled by the more than two orders of magnitude difference between calcitriol concentrations required to attain substantial inhibition of HCV production (nanomolar range) and the concentrations of the hormone produced in these cells (picomolar range). It might be argued that the intracellular calcitriol concentrations are higher than those secreted to the medium due to its local production by CYP27B1 expressed in these cells (Figure 2 and Reference [15]). To test this possibility, we aimed to inhibit calcitriol production and examine its intracrine activity. We used the antifungal drug ketoconazole, which is a mixed-function cytochrome P450 inhibitor, that has been shown to inhibit CYP27B1-dependent production of 1,25-dihydroxyvitamin D [16,17]. Cells were pretreated with ketoconazole in the presence or absence of vitamin D3 for 3 h and then were infected with the HCV intergenotypic HJ3-5 chimeric virus. As can be seen in Figure 1A, treatment with ketoconazole had no effect on HCV inhibition by vitamin D3 as determined by focus-forming unit (FFU) infectious virus assay. To ascertain that ketoconazole inhibited the production of calcitriol, we monitored its effect on the induction of the calcitriol target gene, Cyp24A1 [8]. Treatment of Huh7.5 cells with ketoconazole abolished Cyp24A1 mRNA induction (Figure 1B), indicating a markedly decreased production of calcitriol. These results suggest that the anti-HCV effect of vitamin D3 is not due to high local concentrations of in situ-produced calcitriol.
Figure 1.
Effect of ketoconazole on the anti-hepatitis C virus (HCV) activity of vitamin D3. Huh7.5 cells were treated with vitamin D3 (VD) (5 µM), ketoconazole (Keto) (1 µM), or both 3 h prior to infection. Nonsignificant is denoted by ns. (A) Inhibition of HCV HJ3-5 virus production, as determined by focus-forming unit (FFU) assay of virus released into cell culture media 24 h post-infection. (B) Real-time polymerase chain reaction (PCR) analysis of Cyp24A1 expression level in Huh7.5 treated cells. A representative of two experiments was performed in triplicates. The results are shown as the relative quantity (RQ) normalized to Gapdh mRNA values; the control cells were assigned a value of 1. Statistical significance was calculated by two-tailed Student’s t test ** p < 0.002, ns—nonsignificant.
2.2. The Role of 25(OH)D3 as a Direct Mediator of the Antiviral Activity of Vitamin D3
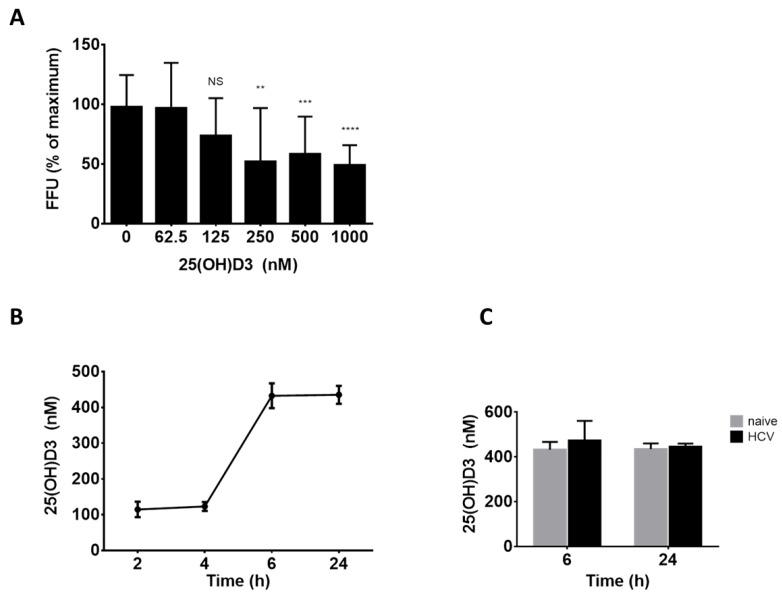
Hepatocytes are highly efficient in metabolizing vitamin D3 to 25(OH)D3 which, at high concentrations (>400 nM), is capable of binding to and activating VDR [12]. Excluding calcitriol in situ production as the mechanism of vitamin D3 antiviral activity, we thus examined the possible role of 25(OH)D3 generation by the hepatocarcinoma cells in this activity of vitamin D3. To this end, cells were treated with increasing concentrations of 25(OH)D3 and then infected with HCV. As shown in Figure 2A treatment with 25(OH)D3 at concentrations of 250–1000 nM efficiently inhibited HCV production (up to 50%). The inhibition was not due to a cytotoxic effect since treatment with 25(OH)D3 did not affect Huh7.5 cell viability (Figure S1A).
Figure 2.
Involvement of 25(OH)D3 in mediating vitamin D3 anti-HCV effect. (A) Inhibition of HCV HJ3-5 virus production as determined by FFU assay of virus released into media following infection and treatment with 25(OH)D3 (62.5–1000 nM). Percent of FFU was calculated by comparing with virus released in nontreated cell cultures (0). Mean values ± SD of three different experiments are presented. Statistical significance was calculated by two-tailed Student’s t test and is indicated as follows: ** p < 0.05, *** p < 0.01, **** p < 0.0001; ns—nonsignificant. (B) ELISA analysis of 25(OH)D3 levels produced by noninfected Huh7.5 cells 2–24 h post-treatment with vitamin D (5 μM) and (C) HCV infected and noninfected cells 6 and 24 h post-treatment with vitamin D (5 μM). A representative experiment out of two was performed in triplicates.
We then asked whether the 25(OH)D3 concentrations needed to inhibit HCV can be attained in Huh7.5 cell cultures treated with vitamin D3. Vitamin D3 can potentially be hydroxylated in our cell system by four known human liver vitamin D 25(OH)ases: CYP2R1, CYP27A1, and CYP2J2 and CYP3A4 to produce 25(OH)D3 [3]. To evaluate the potential of Huh7.5 cells to produce 25(OH)D3, we tested the expression level of the genes encoding for these enzymes. Interestingly, although CYP3A4 is the most abundant CYP450 in human liver [18], it was not detected in the Huh7.5 cells. However, CYP2R1, CYP2J2, and CYP27A1 hydroxylases were highly expressed in Huh7.5 cells (Figure S1B). As expected, and as previously reported, CYP27B1 was expressed at low levels.
These results show that Huh7.5 cells contain the metabolic machinery needed to produce 25(OH)D3. To evaluate the concentrations of 25(OH)D3 produced in Huh7.5 culture, cells (infected and naïve) were treated with vitamin D (5 µM) and 25(OH)D3 in the culture medium, measured by ELISA (Figure 2B). The 6 h post-vitamin D3-addition 25(OH)D3 concentration reached ~450 nM and leveled off thereafter up to 24 h. The extent of 25(OH)D3 production was not influenced by HCV infection (Figure 2C). Since these levels are sufficient to inhibit HCV (Figure 2A), it could be argued that 25(OH)D3 may be the active metabolite mediating the anti-HCV activity of vitamin D3.
2.3. The Role of VDR in the Anti-HCV Activity of Vitamin D3
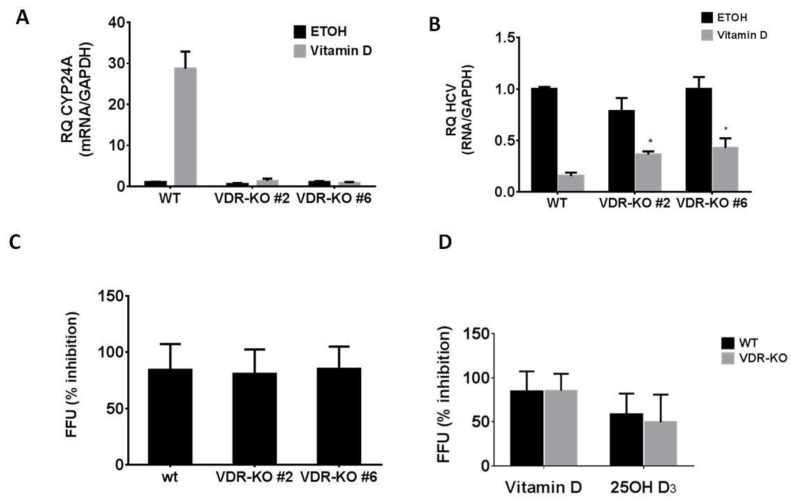
Similar to calcitriol, 25(OH)D3 was reported to mediate its effects through VDR engagement. Therefore, we further asked whether the anti-HCV effect of 25(OH)D3 and vitamin D3 is VDR-dependent. We used CRISPR-CAS9 to produce VDR knockout (VDR-KO) in Huh7.5 cells. To that end, a single guided RNA (sgRNA) targeting the coding sequence of the first translated exon of the VDR gene was cloned into the pSpCas9 plasmid, transfected into Huh7.5 cells and analyzed by targeted next-generation sequencing (NGS) (Figure S2). Using limiting dilution, we isolated cell clones with mutated VDR. Two clones (clone #6 and clone #2) were subjected to sequence analysis. Clone #2 apparently appeared to contain more than one isolated cell type as indicated by the spectrum of sequences obtained in the analysis (Figure S2Cb) and was considered as a mixed cell population. Sequence analysis of clone #6 revealed a nucleotide insertion which resulted in the putative disruption of the VDR open reading frame. VDR–KO was confirmed by the inability of these cells to upregulate Cyp24A1 expression in response to vitamin D3 (Figure 3A). Unexpectedly, vitamin D3 and 25(OH)D3 were still able to inhibit HCV infection as determined by HCV RNA and by FFU assay (Figure 3B–D), showing that inhibition of HCV production is not mediated by VDR activation.
Figure 3.
Vitamin D3 and 25(OH)D3 inhibit HCV production in a VDR-independent manner. (A) Real-time PCR analysis of Cyp24A1 mRNA expression levels in Huh7.5 wild-type (WT) and VDR knockout (VDR-KO) cells after 24 h treatment with vitamin D3 (5 µM) compared with nontreated cells. Results are presented as relative quantity (RQ) of the target gene mRNA normalized to Gapdh mRNA values; control cells mRNA levels were assigned a value of 1. (B) Real-time PCR analysis of HCV RNA expression levels in Huh7.5 WT and VDR-KO cells 24 h post-infection with HCV and treatment with vitamin D3 (5 µM) compared with nontreated cells. The results shown are the average of three independent experiments and presented as the relative quantity of the HCV noncoding region normalized to Gapdh mRNA values; the control cells were assigned a value of 1. (C) Inhibition of HCV HJ3-5 virus production by vitamin D3 determined by FFU assay of virus released into media following infection of Huh7.5 and VDR-KO cells. (D) Inhibition of HCV HJ3-5 virus production by 25(OH)D3 determined by FFU assay of virus released into media following infection of Huh7.5 and VDR-KO #6 cells. The results shown are the average of three independent experiments. Statistical significance was calculated by two-tailed Student’s t test (* p < 0.0005).
2.4. Involvement of the Hh Pathway in the Anti-HCV Activity of Vitamin D3
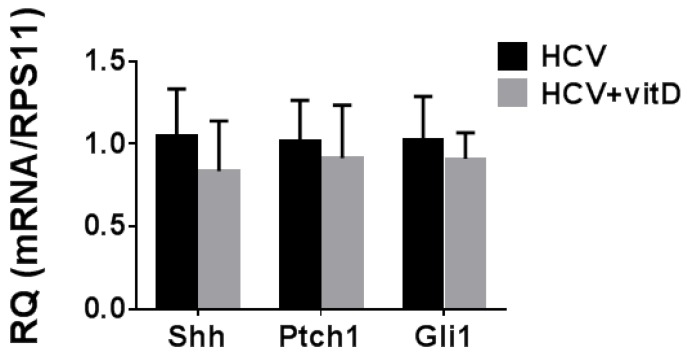
The finding of a VDR-independent mode of action of vitamin D3 raised the possibility that the anti-HCV effect is mediated by a direct action of the vitamin. The only direct biological activity reported to date for cholecalciferol is its inhibitory effect on the Hh pathway [9]. An association between HCV replication and Hh pathway activity was reported [19], suggesting that inhibition of the Hh pathway would inhibit HCV replication. The Hh pathway is frequently activated in HCCs and the expression of Hh target genes serves as an indication for pathway activation [20]. Therefore, we measured mRNA levels of the Hh pathway genes and downstream targets Shh, Ptch1, and Gli1 in the VDR-KO Huh7.5 cells. We found that the Hh pathway is active in the wild-type (WT) cells (data not shown) as well as in the VDR-KO Huh7.5 cells (Figure 4) in accordance with previous reports [19]. However, as shown in Figure 4, there was no effect of vitamin D3 on mRNA expression of these genes in these cells, implying that its antiviral effect in our experimental system is probably not mediated through the Hh pathway.
Figure 4.
Effect of vitamin D3 on mRNA levels of Hedgehog (Hh) pathway target genes in VDR-KO Huh7.5 cells. VDR-KO Huh7.5 cells were treated with vitamin D3 (5 µM) three hours prior to infection. Hh target genes expression levels were determined by real-time PCR analysis. Results are presented as relative quantity of target gene mRNA normalized to RPS11 mRNA values. HCV-infected cells were assigned a value of 1. Results are shown as means RQ ± SD of three experiments.
3. Discussion
The physiological activity of vitamin D3 is commonly attributed to direct binding of its metabolite 1α,25(OH)2D3, calcitriol, to the VDR. Calcitriol precursor, 25(OH)D3 is regarded as a nonactive prohormone. However, it is now established that 25(OH)D3 at supraphysiological concentrations can be a VDR agonist by itself [1]. Not long ago, the dogma that vitamin D3 itself is an inert compound was challenged. Vitamin D3 as a 3β-hydroxysteroid was shown to inhibit the Hh signaling pathway by directly binding to Smo, one of the elements regulating this pathway [9].
This study aimed to shed light on our puzzling finding that vitamin D3, similarly to calcitriol, remarkably inhibited HCV production in HCC cells [15]. We have previously proposed that the antiviral activity of vitamin D3 may be mediated by calcitriol produced in Huh7.5 cells by sequential hydroxylation of its 25 and 1 positions. However, examination of the amount of calcitriol generated by the HCV-infected HCC cells revealed that it was too low to account for the antiviral effect of vitamin D3. We argued that this antiviral activity may nevertheless be due to the presumed higher local concentrations of the intracellularly produced calcitriol, but could not ignore the possibility that it is mediated through a different mechanism. To distinguish between these possibilities, we inhibited the local conversion of vitamin D3 to calcitriol with ketoconazole, a known inhibitor of CYP27B1 [16,17]. We found that ketoconazole did not impair the anti-HCV activity of vitamin D3, while inhibiting the intracrine genomic activity of calcitriol as manifested by the lack of induction of its most sensitive target gene, Cyp24A1. It should be noted that the use of ketoconazole does not provide a direct evidence for the exclusion of calcitriol as the mediator of vitamin D3 antiviral activity. However, the fact that this activity is VDR-independent, strengthens the notion that the antiviral activity of vitamin D3 is not mediated by its metabolic conversion to calcitriol.
Another possible mediator of vitamin D3 anti-HCV activity is its primary metabolite 25(OH)D3. It is well-known that hepatocytes are capable of producing copious amounts of 25(OH)D3 that can activate the VDR, making this supposition plausible [1,2]. To explore the validity of this notion, the concentration of 25(OH)D3 required to reduce infectious virus production was determined and found to range between 250 nM and 1 μM (Figure 2). These results match perfectly with those of Matsumura et al. [21]. The finding, that such high concentrations of 25(OH)D3 can be attained in vitamin D3-supplemented Huh7.5 cell cultures, lend further support for this supposition.
Documented direct effects of 25(OH)D3 are generally attributed to its VDR-agonistic action. To ascertain that the anti-HCV activity of 25(OH)D3 in this study is VDR mediated, its inhibitory effect was examined in VDR-KO cells. We found that knocking out the VDR did not affect the antiviral activity of 25(OH)D3 and also of vitamin D3 while preventing the induction of Cyp24A1 by vitamin D3. These results indicate that the anti-HCV effects of 25(OH)D3 and vitamin D3 are exerted by a VDR-independent mode of action.
A VDR-independent mechanism that may account for the anti-HCV effect of vitamin D3 is its action on the Hh signaling pathway [9]. This supposition stems from the report showing the involvement of the Hh pathway in HCV replication in vitro [19]. While we found that the Hh pathway is constitutively activated in our cell system as demonstrated by the expression of its downstream target genes, vitamin D3 treatment did not affect Hh pathway activity. The resistance of the Hh pathway to vitamin D3 treatment may be due to the presence of point mutations in Smo that were shown to prevent the binding of 3β-hydroxysteroids and small molecule inhibitors of the Hh pathway in basal cell carcinoma (BCC) [9]. These results rule out inhibition of the Hh signaling pathway as a mechanism for HCV inhibition by vitamin D3.
The current and previous studies provide evidence for the occurrence of several mechanisms of action for the anti-HCV effect of the vitamin D system. We and others have shown that calcitriol inhibited HCV production [15,22,23]. We have found that knocking out the VDR in Huh7.5 cells abolished the anti-HCV activity of calcitriol (data not shown) indicating that this activity is mediated by the VDR. This conclusion is in accord with the report of Yu-Min Lin et al. [23]. It should be noted that two studies failed to show anti-HCV effect of calcitriol at physiological concentrations [21,22]. This discrepancy may stem from variation in VDR expression levels in the specific cell lines used in these studies.
In addition to the antiviral effect of calcitriol, we herein show that 25(OH)D3 is capable of inhibiting HCV production in a VDR-independent mechanism. The fact that 25(OH)D3 can inhibit HCV was reported previously by Matsumura et al. [21]. Although the authors attribute 25(OH)D3 activity to a VDR dependent mode of action, we suppose that, in this system too, 25(OH)D3 action may be VDR-independent, since calcitriol was inactive in this system.
We and others [15,22] have shown that treatment with vitamin D3 inhibits HCV production. This effect is most probably mediated by its conversion to 25(OH)D3. However, it cannot be ruled out that in addition, vitamin D3 has an independent direct anti-HCV activity. For example, a liponomic effect may underlie such an anti-HCV activity perturbing the structure of cellular and viral particle membranes which take part in all phases of the HCV life cycle [24]. Knocking out the various 25 vitamin D hydroxylases would provide evidence for a direct effect of cholecalciferol.
Recently, a VDR-independent effect of 25(OH)D3 on lipid metabolism was reported [25]. It was shown to impair the activation of the transcription factor sterol regulatory element-binding protein-2 (SREBP2), a master regulator of lipogenesis. This effect was specific to 25(OH)D3 and was not shared with vitamin D3. SREBP2 is an important transcription factor regulating the synthesis and uptake of lipids including cholesterol [26]. As every step of the virus life cycle is intimately associated with lipid metabolism and cholesterol homeostasis, we are now testing the possibility that vitamin D exerts its effect through regulation of the SREBP pathway.
4. Materials and Methods
4.1. Reagents
Cholecalciferol, 25-hydroxyvitamin D3, and ketoconazole were purchased from Sigma Chemical Co. (St. Louis, MO, USA) and dissolved in absolute ethanol.
4.2. Cells
Huh7.5 cells were grown in Dulbecco’s modified Eagle’s medium (Biological Industries, Kibbutz Beit-Haemek, Israel) supplemented with 10% fetal calf serum, penicillin, and streptomycin as described [15].
4.3. Inhibition of Infectious Virus Production
Virus assays were carried out with the intergenotypic HJ3-5 chimeric HCV virus. Huh7.5 cells were used for the production of virus stocks and for all assays.
The inhibitory action of vitamin D metabolites on HCV production was assessed essentially as described [15]. Huh7.5 cells were pretreated with vitamin D3, 25(OH)D3, or the vehicle ethanol for 3 h before infection with the HJ3-5 virus at a multiplicity of infection (moi) of 0.1–0.01. For determination of virus titer, the medium was replaced after 24 h with fresh medium not containing vitamin D3 or 25(OH)D3, and left for an extra 24 h incubation, in order to eliminate the reagents carry-over. Supernatant fluids were collected from the cell cultures and the titer of infectious virus was determined by the FFU assay, essentially as described [27].
4.4. Inhibition of 1,25-Dihydroxyvitamin D Production
Ketoconazole was used for the inhibition of vitamin D3 metabolism. Ketoconazole (1 μM) dissolved in ethanol was added concomitantly with vitamin D3 to Huh7.5 cells for 3 h before infection with the HJ3-5 virus as above. After 24 h, cells were collected for RNA extraction and analyzed for gene expression.
4.5. RNA Isolation and cDNA Synthesis
Total RNA was extracted from cells using EZ-10 DNAaway RNA Miniprep Kit (Bio Basic Inc., Markham, ON, Canada). Total RNA (1 µg) was subjected to reverse transcription (RT) using the qScript cDNA Synthesis Kit (Quantabio, Beverly, MA, USA).
4.6. Quantitative Real-time RT-PCR
Real-time RT-PCR assays were performed in the StepOnePlus Real-Time PCR Systems (Applied Biosystems, Foster City, CA, USA), by qScript 1-Step SYBR Green qRT-PCR Kit (Applied Quantabio, Beverly, MA, USA) using gene-specific primer pairs (Table 1).
Table 1.
Primer sets used for RT-PCR and PCR.
| Accession Number | Primer Name | Sequence (5′→3′) | Product Size (bp) |
|---|---|---|---|
| NM_001128915.1 | Cyp24A2-S | ACCCAAAGGAATTGTCCGCA | 111 |
| Cyp24A1-AS | CAAAACGCGATGGGGAGTTC | ||
| NM_024514.4 | Cyp2r1-S | TGGAGGCATATCAACTGTGGT | 133 |
| Cyp2r1-AS | GAGTAAGCCTCCCATTTTTGTCA | ||
| NM_000775.4 | Cyp2J2-S | TGGACCCCACCAAACTCTCT | 153 |
| Cyp2J2-AS | GGATTGCCTGTGTGCTTT | ||
| NM_000784.4 | Cyp27A1-S | GTTCACCACGGAAGGACACC | 163 |
| Cyp27A1-AS | GTTCCCCGAAGCACTCTCTG | ||
| NM_017460.6 | Cyp3A4-S | TGTGGGGCTTTTATGATGGT | 117 |
| Cyp3A4-AS | GACCAAAAGGCCTCCGGTTT | ||
| NM_000785.4 | Cyp27B1-S | GTGCTAAGACTGTACCCTGTGG | 150 |
| Cyp27B1-AS | ATTTGGCTCTGGGAACTGG | ||
| ENSG00000111424 | VDR-S | AGGGCGAATCATGTATGAGG | 396 |
| VDR-AS | TGCTTCTTCTCCCTCCCTTT | ||
| NM_000193.3 | SHH-S | GAAACTCCGAGCGATTTAAGGA | 228 |
| SHH-AS | GGCCCTCGTAGTGCAGAGA | ||
| NM_001083603.2 | PTCH1-S | TCTTGGTGTTGGTGTGGATG | 145 |
| PTCH1-AS | ATTGCTGATGGACGTGAGG | ||
| NM_005269.2 | Gli1-S | CATCAGGGAGGAAAGCAGAC | 146 |
| Gli1-AS | CATTGCCAGTCATTTCCACAC | ||
| NM_001015.5 | RPS11-S | GCCCTCAATAGCCTCCTTGG | 149 |
| RPS11-AS | TTCAGACTGAGCGTGCCTAC | ||
| NM_002046.7 | GAPDH-S | GAAGGTGAAGGTCGGAGTC | 226 |
| GAPDH-AS | GAAGATGGTGATGGGATTTC |
4.7. (OH)D3 Determination
The level of 25(OH)D3 in cell culture medium was determined by the 25-Hydroxyvitamin DS EIA Assay kit (Immunodiagnostic Systems Ltd., Boldon, UK) according to the manufacturer instructions.
4.8. Cell Viability Assay
Cell viability was determined by AlamarBlue Cell Viability Reagent (Invitrogen, Carlsbad, CA, USA) measuring fluorescence intensity in culture supernatants.
4.9. VDR Knockout Cells
A guided RNA (gRNA) sequence that targets the genomic sequence in the coding region of the VDR gene in position #63987 (ENSG00000111424) was designed using the in silico prediction tool (http://crispr.mit.edu). The gRNA was designed to be located on an XhoII restriction site sequence enabling an easy detection of genome modification (Figure S2). The gRNA sequence: 5’-CGGAACGTGCCCCGGATCTG-3’ was cloned into the pSpCas9(BB)-2A-GFP (PX458) (Addgene, Watertown, MA, USA) plasmid and transfected into the Huh7.5 cells using TransIT Transfection Kit (Mirus Bio LLC, USA). Next-generation sequencing and mutation analysis were performed by Hy Laboratories Ltd. (Israel). In brief, target loci PCR-amplification from the genomic DNA of the cells pool involved using the following primers:
CS1_VDRs-5’-ACACTGACGACATGGTTCTACAAGGGCGAATCATGTATGAGG-3′ and
CS2_VDR as 5’-TACGGTAGCAGAGACTTGGTCTTGCTTCTTCTCCCTCCCTTT-3′
A second PCR was performed on the obtained amplicons using the Access Array index primers for Illumina (Fluidigm) to add the adaptor and index sequences to the sample. The PCR product was purified using AMPure XP beads (Beckman-Coulter), the concentration was measured by Qubit (Invitrogen, USA), and the size determined by Tapestation analysis (Agilent Technologies, USA). The sample was then loaded on the Illumina Miseq and sequenced using a V2-500 cycle kit to generate 250 × 2 paired-end reads. Reads were demultiplexed to generate two FASTQ files and trimmed for quality and adaptor sequences, merged and mapped to the template provided to generate BA/BAI files for the mapping using CLC-Bio software (QIAGEN). More than 90% of the reads were mapped to the template. To measure the frequencies of indels in the target regions, we used the Cas-Analyzer algorithm (http://www.rgenome.net/cas-analyzer/#) [28].
Limiting dilution was performed to select for specific clones with VDR-KO gene. Successful transfection was assessed through the detection of green fluorescent protein (GFP)-derived fluorescence in cells. Selection of positive clones was performed by target PCR spanning the gRNA target site in the VDR gene (Table 1), digestion with MflI restriction enzyme (Takara) was used for identification of positive clones (Figure S2). Sequence analysis was performed to ascertain VDR-KO clone.
4.10. Statistical Analysis
Results are expressed as mean ± SD for replicate cultures. Statistical significance of differences between two experimental groups was determined by the unpaired Student’s t test. A value of p < 0.05 was considered statistically significant
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/9/2367/s1.
Author Contributions
Conceptualization, R.Z., A.R., A.I., R.T.-K.; Methodology, R.Z., A.R., N.R., A.E., L.B.; Investigation, R.Z., A.R., N.R., A.E., L.B., R.T.-K.; Writing—Original Draft Preparation, R.Z., A.R.; Writing—Review & Editing, R.Z., A.R., N.R., L.B., A.I., R.T.-K.; Supervision, R.Z., A.R.
Funding
This research received no external funding
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Lou Y.R., Molnár F., Peräkylä M., Qiao S., Kalueff A.V., St-Arnaud R., Carlberg C., Tuohimaa P. 25-Hydroxyvitamin D3is an agonistic vitamin D receptor ligand. J. Steroid Biochem. Mol. Biol. 2010;118:162–170. doi: 10.1016/j.jsbmb.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Lou Y.R., Laaksi I., Syvälä H., Bläuer M., Tammela T.L.J., Ylikomi T., Tuohimaa P. 25-hydroxyvitamin D3 is an active hormone in human primary prostatic stromal cells. FASEB J. 2004;18:332–334. doi: 10.1096/fj.03-0140fje. [DOI] [PubMed] [Google Scholar]
- 3.Jones G., Prosser D.E., Kaufmann M. Cytochrome P450-mediated metabolism of vitamin D. J. Lipid Res. 2014;55:13–31. doi: 10.1194/jlr.R031534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bikle D.D., Patzek S., Yang W. Physiologic and pathophysiologic roles of extra renal CYP27b1: Case report and review. Bone Rep. 2018;8:255–267. doi: 10.1016/j.bonr.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robin N.C., Agoston Z., Biechele T.L., James R.G., Berndt J.D., Moon R.T. Accumulation of the Vitamin D Precursor Cholecalciferol Antagonizes Hedgehog Signaling to Impair Hemogenic Endothelium Formation Mauricio. Stem Cell Rep. 2014;2:9–17. doi: 10.1016/j.stemcr.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bijlsma M.F., Spek C.A., Zivkovic D., van de Water S., Rezaee F., Peppelenbosch M.P. Repression of smoothened by patched-dependent (pro-)vitamin D3 secretion. PLoS Biol. 2006;4:1397–1410. doi: 10.1371/journal.pbio.0040232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang J.Y., Xiao T.Z., Oda Y., Chang K.S., Shpall E., Wu A., So P.L., Hebert J., Bikle D., Epstein E.H. Vitamin D3 inhibits hedgehog signaling and proliferation in murine basal cell carcinomas. Cancer Prev. Res. 2011;4:744–751. doi: 10.1158/1940-6207.CAPR-10-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerjee U., Ghosh M., Kyle Hadden M. Evaluation of vitamin D3 A-ring analogues as Hedgehog pathway inhibitors. Bioorganic Med. Chem. Lett. 2012;22:1330–1334. doi: 10.1016/j.bmcl.2011.12.081. [DOI] [PubMed] [Google Scholar]
- 9.Hadden M.K. Hedgehog and Vitamin D Signaling Pathways in Development and Disease. 1st ed. Vol. 100. Elsevier Inc.; Amsterdam, The Netherlands: 2016. [DOI] [PubMed] [Google Scholar]
- 10.Susa T., Iizuka M., Okinaga H., Tamamori-Adachi M., Okazaki T. Without 1α-hydroxylation, the gene expression profile of 25(OH)D3treatment overlaps deeply with that of 1,25(OH)2D3in prostate cancer cells. Sci. Rep. 2018;8:2–11. doi: 10.1038/s41598-018-27441-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beloso C., Souto J., Fábregat M., Romanelli G., Javiel G.M.A. Vitamin D deficiency and hepatitis viruses-associated liver diseases: A literature review. World J. Diabetes. 2018;9:157–164. doi: 10.4239/wjd.v9.i9.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nimer A., Mouch A. Vitamin D improves viral response in hepatitis C genotype 2-3 naïve patients. World J. Gastroenterol. 2012;18:800–805. doi: 10.3748/wjg.v18.i8.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bitetto D., Fabris C., Fornasiere E., Pipan C., Fumolo E., Cussigh A., Bignulin S., Cmet S., Fontanini E., Falleti E., et al. Vitamin D supplementation improves response to antiviral treatment for recurrent hepatitis C. Transpl. Int. 2011;24:43–50. doi: 10.1111/j.1432-2277.2010.01141.x. [DOI] [PubMed] [Google Scholar]
- 14.Villar L.M., Del Campo J.A., Ranchal I., Lampe E., Romero-Gomez M. Association between vitamin D and hepatitis C virus infection: A meta-analysis. World J. Gastroenterol. 2013;19:5917–5924. doi: 10.3748/wjg.v19.i35.5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gal-Tanamy M., Bachmetov L., Ravid A., Koren R., Erman A., Tur-Kaspa R., Zemel R. Vitamin D: An innate antiviral agent suppressing hepatitis C virus in human hepatocytes. Hepatology. 2011;54:1570–1579. doi: 10.1002/hep.24575. [DOI] [PubMed] [Google Scholar]
- 16.Bland R., Walker E.A., Hughes S.V., Stewart P.M., Hewison M. Constitutive expression. of 25-hydroxyvitamin D3-1alpha-hydroxylase in a transformed human proximal tubule cell line: Evidence for direct regulation of vitamin D metabolism by calcium. Endocrinology. 1999;140:2027–2034. doi: 10.1210/endo.140.5.6683. [DOI] [PubMed] [Google Scholar]
- 17.Kongsbak M., Von Essen M.R., Boding L., Levring T.B., Schjerling P., Lauritsen J.P.H., Woetmann A., Ødum N., Bonefeld C.M., Geisler C. Vitamin D up-regulates the vitamin D receptor by protecting it from proteasomal degradation in human CD4+T cells. PLoS ONE. 2014;9:e96695. doi: 10.1371/journal.pone.0096695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta R.P., Hollis B.W., Patel S.B., Patrick K.S., Bell N.H. CYP3A4 is a Human Microsomal Vitamin D 25-Hydroxylase. J. Bone Miner. Res. 2003;19:680–688. doi: 10.1359/JBMR.0301257. [DOI] [PubMed] [Google Scholar]
- 19.Mason A. Upregulation of Hedgehog Pathway is Associated with Cellular Permissiveness for Hepatitis C Virus Replication. Hepatology. 2009;19:389–399. doi: 10.1002/hep.24576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang S., He J., Zhang X., Bian Y., Yang L., Xie G., Zhang K., Tang W., Stelter A.A., Wang Q., et al. Activation of the hedgehog pathway in human hepatocellular carcinomas. Carcinogenesis. 2006;27:1334–1340. doi: 10.1093/carcin/bgi378. [DOI] [PubMed] [Google Scholar]
- 21.Matsumura T., Kato T., Sugiyama N., Tasaka-Fujita M., Murayama A., Masaki T., Wakita T., Imawari M. 25-hydroxyvitamin D 3 suppresses hepatitis C virus production. Hepatology. 2012;56:1231–1239. doi: 10.1002/hep.25763. [DOI] [PubMed] [Google Scholar]
- 22.Julio A., Gutierrez K.A., Jones R.F., Singhania A., Christopher H., Woelk R.T.S., AWyles D.L. Vitamin D Metabolites Inhibit Hepatitis C Virus and Modulate Cellular Gene Expression. J. Virol. Antiviral Res. 2014;3:3. doi: 10.4172/2324-8955.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y.M., Sun H.Y., Chiu W.T., Su H.C., Chien Y.C., Chong L.W., Chang H.C., Bai C.H., Young K.C., Tsao C.W. Calcitriol inhibits HCV infection via blockade of activation of PPAR and interference with endoplasmic reticulum-associated degradation. Viruses. 2018;10:E57. doi: 10.3390/v10020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alaei M., Negro F. Hepatitis C virus and glucose and lipid metabolism. Diabetes Metab. 2008;34:692–700. doi: 10.1016/S1262-3636(08)74606-8. [DOI] [PubMed] [Google Scholar]
- 25.Asano L., Watanabe M., Ryoden Y., Usuda K., Yamaguchi T., Khambu B., Takashima M., Sato S.-I., Sakai J., Nagasawa K., et al. Vitamin D Metabolite, 25-Hydroxyvitamin D, Regulates Lipid Metabolism by Inducing Degradation of SREBP/SCAP. Cell Chem. Biol. 2017;24:207–217. doi: 10.1016/j.chembiol.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 26.Walker A.K., Näär A.M. SREBPs: Regulators of cholesterol/lipids as therapeutic targets in metabolic disorders, cancers and viral diseases. Clin. Lipidol. 2012;7:27–36. doi: 10.2217/clp.11.67. [DOI] [Google Scholar]
- 27.Yi M., Ma Y., Yates J., Lemon S.M. Compensatory Mutations in E1, p7, NS2, and NS3 Enhance Yields of Cell Culture-Infectious Intergenotypic Chimeric Hepatitis C Virus. J. Virol. 2007;81:629–638. doi: 10.1128/JVI.01890-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J., Lim K., Kim J.S., Bae S. Cas-analyzer: An online tool for assessing genome editing results using NGS data. Bioinformatics. 2017;33:286–288. doi: 10.1093/bioinformatics/btw561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.