Abstract
Intratumoral injection of oncolytic viruses provides a direct means of tumor cell destruction for inoperable tumors. Unfortunately, oncolytic vectors based on human adenovirus (HAdV) typically do not spread efficiently throughout the tumor mass, reducing the efficacy of treatment. In this study, we explore the efficacy of a conditionally replicating HAdV vector expressing the p14 Fusion-Associated Small Transmembrane (FAST) protein (CRAdFAST) in both immunocompetent and immunodeficient mouse models of cancer. The p14 FAST protein mediates cell-cell fusion, which may enhance spread of the virus-mediated, tumor cell-killing effect. In the murine 4T1 model of cancer, treatment with CRAdFAST resulted in enhanced cell death compared to vector lacking the p14 FAST gene, but it did not reduce the tumor growth rate in vivo. In the human A549 lung adenocarcinoma model of cancer, CRAdFAST showed significantly improved oncolytic efficacy in vitro and in vivo. In an A549 xenograft tumor model in vivo, CRAdFAST induced tumor cell fusion, which led to the formation of large acellular regions within the tumor and significantly reduced the tumor growth rate compared to control vector. Our results indicate that expression of p14 FAST from an oncolytic HAdV can improve vector efficacy for the treatment of cancer.
Keywords: adenovirus, cancer, FAST, cell fusion, oncolytic
Introduction
Despite continuing efforts to curb the immense burden of cancer through treatment, research, and prevention, approximately 40% of the human population will be diagnosed with some form of cancer during their lifetime.1 Approximately half of these patients will succumb to the disease, underscoring the need for new therapeutic options. Oncolytic viruses, which are designed to specifically replicate in and kill cancer cells, have shown tremendous efficacy in preclinical studies.2, 3, 4 Currently, there are a large number of oncolytic viruses in phase I, II, and III clinical trials, based on a diverse array of viruses, including adenovirus, reovirus, and vaccinia, among others, while one, the herpes simplex virus type 1 (HSV-1)-based T-Vec, is approved for use in patients by the United States Food and Drug Administration, the European Medical Agency, and the Australian Therapeutic Goods Administration.5, 6, 7, 8, 9 The development and improvement of oncolytic viruses continues to be an important field of study.
Human adenovirus (HAdV) has been explored extensively as an anti-cancer agent, using two main strategies. Replication-deficient HAdV vectors can be used to deliver anti-cancer genes (e.g., p53), immunomodulators (e.g., interleukin [IL]-12), or other potentially therapeutic genes to tumor cells.10, 11 Indeed, a non-replicating HAdV encoding p53, termed Gendicine, is widely available to patients in China.12 Alternatively, oncolytic HAdV can be engineered to replicate specifically in cancer cells while leaving normal tissue relatively unaffected,13 and Oncorine, a conditionally replicating HAdV, is commercially available to patients in China.14 Unfortunately, when injected intratumorally, HAdV typically does not disperse far from the injection tract, leaving much of the tumor unaffected.15, 16 Indeed, although HAdV-5 (serotype 5) encodes proteins within the early region 3 (E3) specifically dedicated to enhancing release of virus from the infected cell at very late times post-infection,17 much of the virus still remains cell associated.
For this reason, a number of approaches have been explored to improve HAdV dispersion in a tumor, including the expression of extracellular matrix-degrading enzymes, junction-opening peptides, and proteins intended to improve cytolysis.18, 19, 20, 21 Alternatively, several research groups have evaluated the ability of fusogenic proteins, such as those encoded by the gibbon ape leukemia virus (GALV) or measles virus, to enhance spread of virus-associated cell death through the tumor mass through direct cell-cell fusion.22, 23, 24, 25 However, one issue with the use of many of these fusion proteins is their size: inclusion of the coding sequences for these fusogenic proteins, such as the GALV fusion protein (2,004 bp), in an oncolytic HAdV approaches or exceeds the upper limits for DNA packaging in the viral capsid.26, 27 This DNA-packaging limit may preclude the use of other DNA elements designed to enhance vector efficacy, such as tumor-specific promoters to confer selective viral replication or inclusion of other therapeutic genes designed to arm the oncolytic HAdV vector. Although inclusion of the E3 can increase oncolytic efficacy, its inclusion further limits the ability of such viruses to harbor ancillary regulatory elements or genes.28, 29
The p14 Fusion-Associated Small Transmembrane (p14 FAST) protein is a small (14-kDa, 375-bp) fusogenic protein from a reptilian reovirus.30 Unlike most fusogenic proteins, p14 FAST is non-structural (i.e., not a component of an infecting virion), but it likely enhances lateral virus spread between host cells through its ability to promote cell-cell fusion. The exact mechanism of p14 FAST protein-associated fusion has yet to be elucidated; however, due to its small size and relatively minute ectodomain, p14 FAST protein likely does not act through a mechanism similar to typical structural viral fusion proteins.31, 32 Early studies showing that plasmid-mediated expression of p14 FAST protein induced syncytium formation and, subsequently, apoptosis in the fused cells suggested that p14 FAST protein may show efficacy as a sole therapeutic or could be used to enhance the efficacy of oncolytic viruses.33 Indeed, expression of p14 FAST protein from an oncolytic vesicular stomatitis virus (VSV) (VSVΔ51FAST) enhanced the therapeutic efficacy of the virus, and co-delivery of VSV-FAST with an oncolytic vaccinia virus also provided a synergistic effect in cell culture and ex vivo models of cancer.34, 35
In previous studies, our lab explored the efficacy of an early region 1 (E1)-deleted, replication-deficient HAdV expressing the p14 FAST protein (AdFAST) in both immunodeficient human A549 lung adenocarcinoma xenograft-bearing CD-1 nude mice and immunocompetent BALB/c 4T1 tumor models.36, 37 In cell culture, AdFAST induced significant cell fusion and cell death relative to control vector. Higher levels of virus were needed to achieve this effect in the mouse 4T1 cell line, in part due to lower infectivity and p14 FAST protein expression relative to the human A549 cell line. Unfortunately, these very promising in vitro results did not translate in vivo, where no effects on tumor growth or mouse survival were observed. Interestingly, during the course of our in vitro studies, we showed that, under conditions in which the E1-deleted AdFAST virus could replicate (i.e., in the E1-complementing 293 cell line), p14 FAST protein caused significantly more efficient cell fusion and killing with greatly reduced quantity of vector. This observation suggests that p14 FAST protein may provide the greatest efficacy when expressed from a replicating, oncolytic HAdV. In this context, expression of the p14 FAST protein would amplify as the virus replicates, potentially providing greater cell fusion and enhanced induction of apoptosis, in addition to facilitating vector spread through the tumor.
In this study, we have investigated the efficacy of an oncolytic, conditionally replicating HAdV (CRAd) vector encoding the p14 FAST protein in tissue culture and animal models of cancer.
Results
CRAdFAST Can Replicate and Express Viral Genes but Does Not Produce Progeny Virions in Mouse 4T1 Cells
Previously, we showed that vectors based on HAdV-5 infected 4T1 mouse mammary carcinoma cells at an efficiency approximately one-sixth that of A549 cells, a cell line that infects well with HAdV and is used commonly in many HAdV studies.37 HAdV can replicate its DNA and express both early and late genes in many mouse cell lines, but typically infection is not productive (i.e., progeny virions are not produced). We therefore examined whether HAdV infection was productive in 4T1 cells.
4T1 cells were infected at an MOI of 3 with AdFAST (E1 deleted, replication defective) or CRAdFAST (E1AΔ24, replication competent), and total DNA was harvested for qPCR analysis of viral genome content 4–72 h later. The HAdV vector structures are shown in Figure 1. As shown in Figure 2A, as expected, infection of 4T1 cells with the replication-defective AdFAST vector did not result in an increase in viral genome copy number over the course of the experiment. In contrast, we observed an approximate 1,000-fold increase in viral genome copy number in CRAdFAST-treated 4T1 cells between 4 and 72 h post-infection (hpi), clearly showing that the virus can replicate its DNA in this cell line.
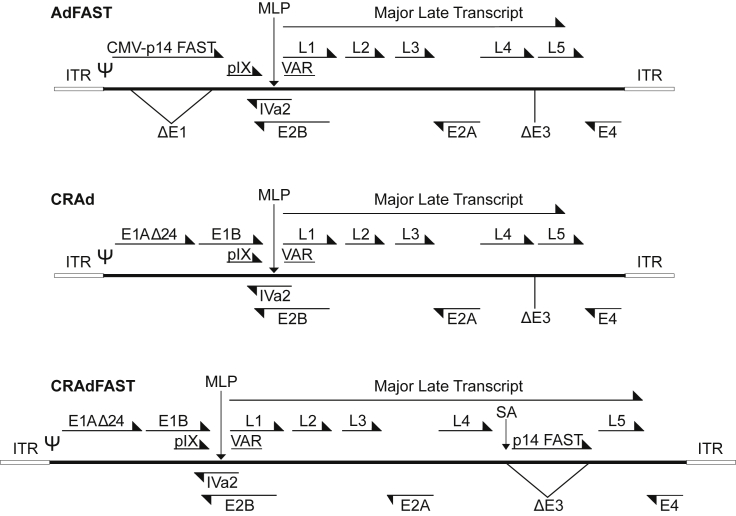
Figure 1.
Adenovirus Constructs
Schematic diagrams of HAdV vectors used in this study: AdFAST, an E1- and E3-deleted non-replicating HAdV vector expressing p14 FAST from the cytomegalovirus immediate early enhancer-promoter in the E1; CRAd, an E1Δ24, E3-deleted conditionally replicating vector; and CRAdFAST, an E1Δ24, E3-deleted conditionally replicating vector expressing p14 FAST from within the E3 region through the inclusion of a major late promoter splice acceptor (SA).
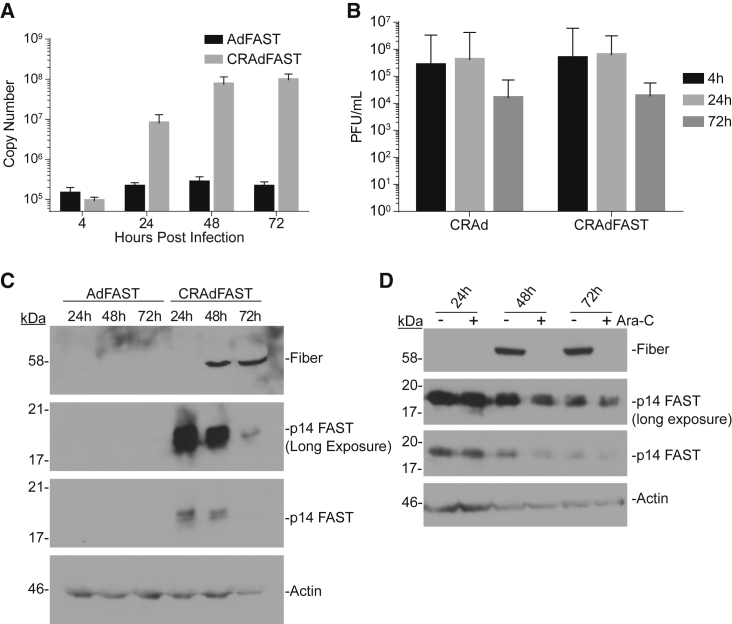
Figure 2.
Replication of HAdV-5 Oncolytic Vectors in 4T1 Cells Is Non-productive
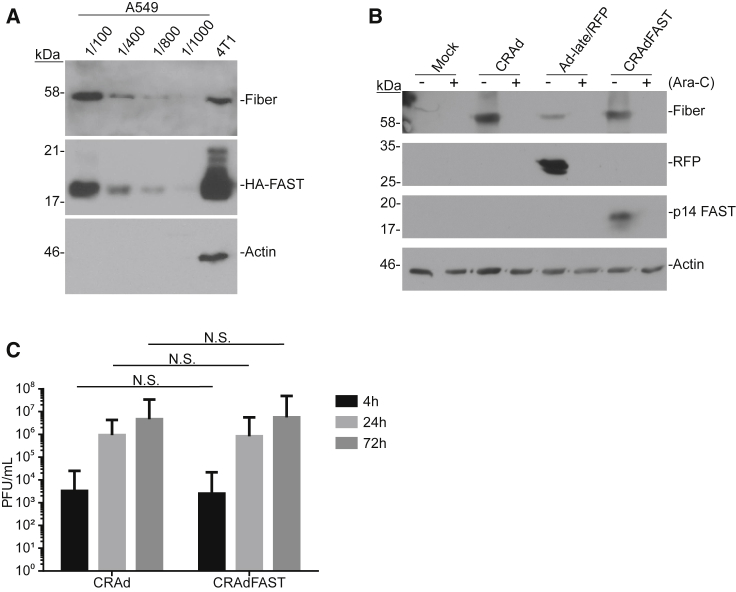
(A) 4T1 cells were infected with either AdFAST or CRAdFAST at an MOI of 10. At 4, 24, 48, and 72 hpi, media were removed and the cells were harvested in SDS-Proteinase K buffer for DNA extraction. qPCR was performed on 200 ng total isolated DNA for the Ad hexon genome region. Error bars represent mean ± SD. (B) 4T1 cells were infected with either CRAd or CRAdFAST at an MOI of 10. Cells and medium were collected at 4, 24, and 72 hpi. Viral titers were determined by plaque assay on 293 cells. Error bars represent mean ± SD. (C) 4T1 cells were infected with either AdFAST or CRAdFAST at an MOI of 3 and harvested at 24, 48, and 72 hpi. Immunoblot analysis was performed to examine HAdV fiber and HA (p14 FAST) protein levels, and actin was used as a loading control. (D) 4T1 cells were infected at an MOI of 50 with CRAdFAST for 1 h, and they were overlaid with medium lacking (−) or containing (+) 20 μg/mL cytosine arabinoside (Ara-C) to inhibit DNA replication. Crude protein samples were harvested at 24, 48, and 72 hpi, and they were assayed by immunoblot for the expressions of fiber protein, p14 FAST protein, and actin (loading control).
To determine if infection of 4T1 cells was productive, cells were infected at an MOI of 10 with CRAd or CRAdFAST, and the cells were harvested into the medium at various times post-infection and titered for virus. As shown in Figure 2B, we did not observe an increase in the amount of recovered virus between 4 and 72 hpi, suggesting that CRAd and CRAdFAST did not give rise to progeny virions during this time period. However, the increase in genome copy number as the virus replicates its DNA should effectively amplify viral gene expression, leading to the accumulation of high levels of virus-encoded proteins, including the p14 FAST protein, within the infected cells.
We next evaluated the expression of virus-encoded proteins, specifically the native viral fiber gene and p14 FAST. CRAdFAST contains the p14 FAST cDNA preceded by a splice acceptor (SA) site replacing the E3 deletion, which should allow for high-level expression of p14 FAST only when the virus replicates, a strategy that we and others have employed previously.38, 39, 40 4T1 cells were infected with AdFAST (E1 deleted, replication defective) or CRAdFAST, and protein expression was examined at 24, 48, and 72 hpi. As expected, treatment of cells with AdFAST did not result in a detectable expression of fiber protein (Figure 2C). At this low MOI of 3, p14 FAST protein expression was also not detectable in AdFAST-treated cells, consistent with our previous observation that a high MOI (>100) is required to visualize FAST expression in this cell line.37 In contrast, likely due to the increase in template copy number within the cells infected with CRAdFAST (Figure 2A), expressions of both p14 FAST protein and the late fiber protein were easily detectable for this virus (Figure 2C).
Interestingly, we detected the expression of p14 FAST protein at a much earlier time point compared to fiber protein. If the onset of fiber protein expression correlates with active virus DNA replication, it suggests that p14 FAST protein expression may precede replication (i.e., is under early regulation) in 4T1 cells. To assess this possibility, we examined the expression of fiber and p14 FAST protein in the presence or absence of cytosine arabinoside (cytarabine, Ara-C), a compound that blocks viral DNA replication. Since the major late promoter is not activated until viral genome replication begins,41 treatment with Ara-C should prevent the expression of fiber and p14 FAST protein if they are indeed under late regulation. 4T1 cells were infected with CRAdFAST at an MOI of 50 in the absence or presence of 20 μg/mL Ara-C, and they were harvested at 24, 48, and 72 hpi. As shown in Figure 2D, while fiber expression was indeed inhibited by treatment with Ara-C, p14 FAST expression was unaffected by this treatment. Thus, although CRAdFAST can provide very high-level expression of p14 FAST protein in the 4T1 mouse cell line, unlike endogenous viral genes (i.e., fiber), its expression does not appear to be conditionally tied to active virus replication.
Expression of p14 FAST Protein from CRAd Enhances 4T1 Cell Killing In Vitro
We next examined whether the elevated expression of p14 FAST protein from CRAdFAST enhanced the ability of the oncolytic vector to mediate cell fusion and cause cancer cell death in 4T1 cells in culture. 4T1 cells were infected with AdFAST, CRAd, or CRAdFAST at an MOI of 100 or mock infected with PBS, and the cells were analyzed visually for cell fusion by phase-contrast microscopy at varying times post-infection. We did not observe evidence of fusion in mock- or AdFAST-treated cells at any of the time points (Figure 3A), consistent with our previous work showing that a very high MOI of AdFAST was required to achieve visible fusion in 4T1 cells.37 Cells treated with CRAd showed evidence of cytopathic effect (CPE), but only at the 72-h time point, and no cell fusion occurred. However, CRAdFAST-treated 4T1 cells showed clear evidence of cell fusion as early as 24 hpi, which increased over the 72-h time course. By 72 hpi, the entire monolayer of CRAdFAST-treated 4T1 cells had fused and exhibited extensive CPE. Thus, treatment of 4T1 cells with the replication-competent CRAdFAST results in greatly enhanced fusion relative to the replication-defective AdFAST.
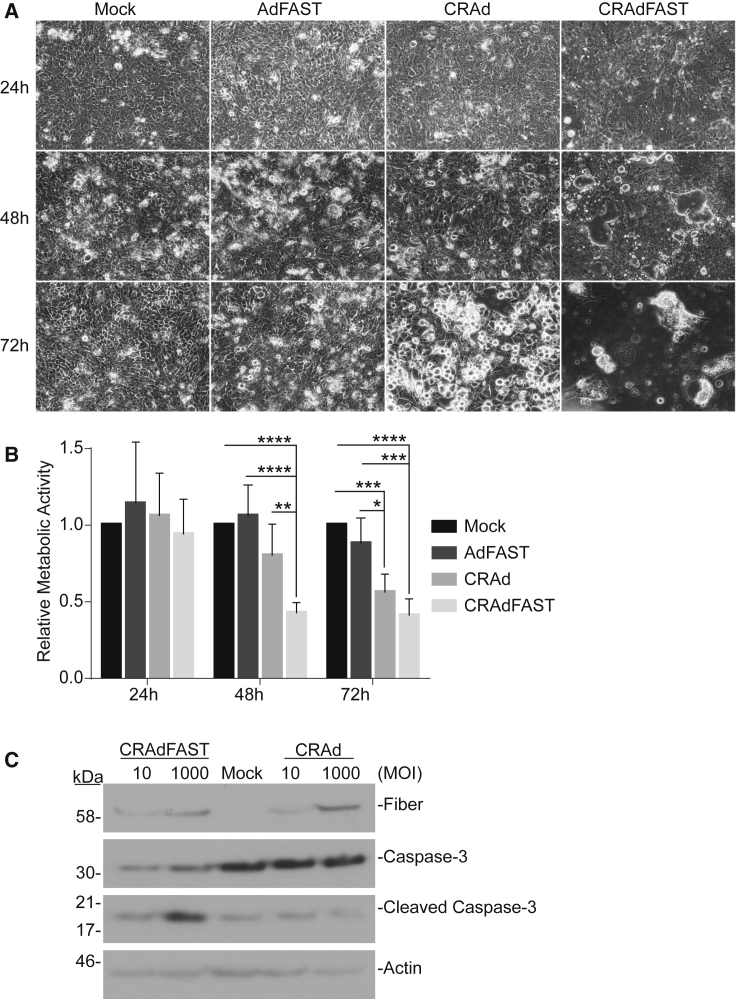
Figure 3.
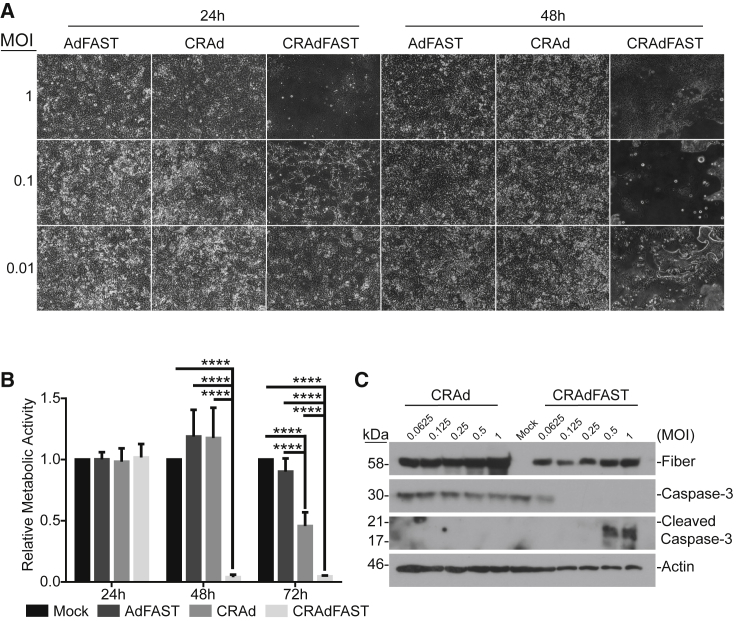
CRAdFAST Induces Cellular Fusion and Cell Death in 4T1 Cells More Efficiently Than Non-replicating AdFAST
(A) 4T1 cells were mock infected or infected with AdFAST, CRAd, or CRAdFAST at an MOI of 100. Phase-contrast images were obtained at 24, 48, and 72 hpi. (B) 4T1 cells were mock infected or infected with AdFAST, CRAd, or CRAdFAST at an MOI of 1,000 in triplicate wells (n = 6 technical replicates, n = 2 independent experimental replicates). At 48 hpi, metabolic activity was assessed by MTS assay, and the values are reported relative to metabolic activity in mock-infected cells. Mean values reported ± SD (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). (C) 4T1 cells were mock infected or infected with CRAd or CRAdFAST at MOIs of 10 and 1,000, and protein samples were harvested at 48 hpi. Protein lysates from infected cells were subjected to immunoblot for HAdV fiber, full-length caspase-3, and cleaved caspase-3, with actin used as a loading control.
To determine whether enhanced cell fusion mediated by CRAdFAST was associated with 4T1 cell death, we first examined metabolic activity of the treated cells over time. 4T1 cells were infected at an MOI of 1,000 with AdFAST, CRAd, or CRAdFAST or mock infected with PBS, and metabolic activity was examined at 24-h intervals by MTS assay. Treatment of cells with AdFAST did not result in a decrease in metabolic activity at any of the time points examined (Figure 3B). In contrast, CRAdFAST significantly reduced metabolic activity of the cells as early as 48 hpi. By 72 hpi, treatment with CRAd also induced a decrease in metabolic activity compared to AdFAST- and mock-treated cells.
We also examined the cells for evidence of apoptotic cell death by immunoblot analysis for cleaved caspase-3 (Figure 3C). Treatment of 4T1 cells with CRAd did not result in enhanced levels of cleaved caspase-3 compared to untreated cells. In contrast, treatment of cells with CRAdFAST did result in enhanced cleavage of caspase-3, but only at a high MOI. Taken together, these results indicate that treatment of 4T1 cells with CRAdFAST results in high-level expression of p14 FAST protein and significant cell fusion, resulting in a reduction in cellular metabolic activity and the onset of apoptosis-like cell death. Importantly, all of these effects are enhanced relative to 4T1 cells treated with AdFAST or CRAd, suggesting that arming an oncolytic HAdV vector with p14 FAST protein enhances the anti-neoplastic potential of the vector.
Expression of p14 FAST Protein from CRAd Does Not Improve Therapeutic Efficacy in the 4T1 Murine Model of Cancer in Immunocompetent Mice
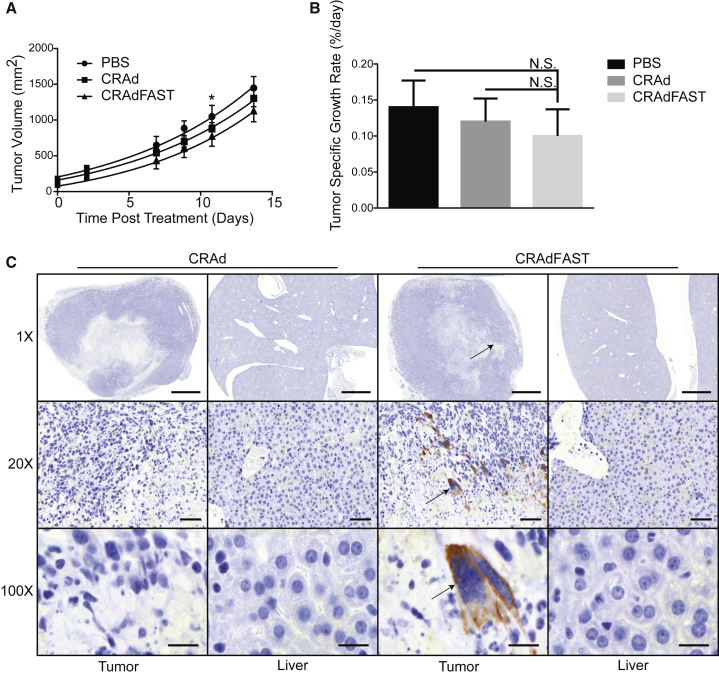
We next examined whether p14 FAST protein expression could improve the efficacy of CRAd in the immunocompetent 4T1 mouse model of cancer. BALB/c mice bearing 4T1 subcutaneous tumors were injected intratumorally with PBS or 7 × 109 plaque-forming units (PFU) of CRAd or CRAdFAST, and tumor size was measured by caliper three times weekly. As shown in Figures 4A and 4B, treatment of tumors with either CRAd or CRAdFAST resulted in a trend toward a reduced growth rate, but it did not reach statistical significance. Tumor-specific growth rates of PBS-treated tumors (0.159%/day) were not significantly different from those of CRAd- (0.143%/day, p = 0.74) or CRAdFAST-treated tumors (0.110%/day, p = 0.097) (Figure 4B). We did not observe tumor regression or stasis, and all animals reached endpoint and were euthanized at day 14. Tumor tissues removed at day 5 post-treatment were stained for hemagglutinin (HA) to detect p14 FAST protein and examined visually for evidence of syncytia. As expected, CRAd-treated tumors showed no evidence of p14 FAST protein or syncytial formation. p14 FAST protein was detected in tumors from CRAdFAST-treated mice, and syncytia were observed in isolated regions within the treated tumors (Figure 4C). Livers from mice injected with the CRAd or CRAdFAST showed no obvious pathology. Thus, CRAdFAST can infect 4T1 tumors, giving rise to the expression of p14 FAST protein, resulting in syncytium formation, but, ultimately, it does not reduce the tumor growth rate or enhance animal survival.
Figure 4.
Expression of p14 FAST Protein Does Not Improve Oncolytic Efficacy of CRAd in the Murine 4T1 Model of Cancer
Tumor-bearing mice were injected intratumorally with PBS or 7 × 109 PFU CRAd or CRAdFAST, and tumor volumes were measured for eight mice per group three times weekly. (A) Symbols represent mean calculated tumor volumes over time. Lines represent non-linear regression model of tumor growth rates. At day 11, PBS-treated tumors were significantly larger than CRAdFAST-treated tumors (*p = 0.03). Error bars represent mean ± SEM; * p = 0.033 for CRAdFAST compared to PBS. (B) Tumor-specific growth rates were calculated for each tumor and averaged; error bars represent SD. (C) Sections of tumor and liver from mice sacrificed at day 5 post-injection were probed with anti-HA antibody (p14 FAST) and counterstained with hematoxylin to assess p14 FAST expression. At 1×, 20×, and 100× magnifications, scale bars represent 2,000, 100, and 20 μm, respectively.
CRAdFAST Replicates Efficiently in Human A549 Cells
In general, HAdV has an enhanced ability to infect human cell lines compared to mouse cell lines. To examine this in our system, A549 and 4T1 cells were infected at an MOI of 10 with CRAdFAST; crude protein samples were collected 48 hpi, and they were subjected to immunoblot for the relative levels of HAdV fiber and p14 FAST protein (Figure 5A). We compared the undiluted 4T1-infected cell sample to a dilution series of the A549-infected cell sample, to get a more accurate estimation of the relative viral protein expression. As shown in Figure 5A, the quantity of HAdV fiber protein in the 4T1 sample was between 1/100 and 1/400 that present in A549 cells, which would reflect the cumulative effect of differences in infection efficiency, genome replication, and gene expression between the two cell lines. Interestingly, the ratio of p14 FAST protein present in the CRAdFAST-infected 4T1 cells compared to A549 cells appeared proportionately higher than that of fiber. Nevertheless, CRAdFAST can achieve higher levels of infection and gene expression in A549 cells compared to 4T1 cells.
Figure 5.
Analysis of Protein Expression and Virus Yield of CRAdFAST in A549 Cells In Vitro
(A) A549 and 4T1 cells were infected at an MOI of 10 and harvested at 48 hpi. A549 samples were diluted to 1/100, 1/400, 1/800, and 1/1,000 to compare protein levels to those obtained in 4T1 cells. (B) A549 cells were infected at an MOI of 3 with CRAd, AdRFP, or CRAdFAST or mock infected, and they were overlaid with medium lacking (−) or containing (+) 20 μg/mL Ara-C to inhibit viral DNA replication. Cells were harvested at 24 hpi, and immunoblot analysis was performed to examine HAdV fiber, RFP, and HA (p14 FAST) protein levels, with actin used as a loading control. (C) A549 cells were infected at an MOI of 10 with either CRAd or CRAdFAST. Cells and media were harvested at 4, 24, and 72 hpi. Viral titers were determined by plaque assay. Error bars represent mean ± SD.
To confirm that p14 FAST protein expression was tied to virus replication in human A549 cells, we again examined gene expression in the presence or absence of the DNA replication inhibitor Ara-C. A549 cells were infected at an MOI of 10 with CRAd, Ad-late/red fluorescent protein (RFP), or CRAdFAST, in the presence or absence of 20 μg/mL Ara-C, and protein expression was examined 24 h later. Ad-late/RFP is similar in structure to CRAdFAST, but it contains a wild-type E1 and a SA-RFP expression cassette instead of SA-p14 FAST within the E3.39 As shown in Figure 5B, treatment of cells with Ara-C abrogated the expression of the late fiber protein for all of the viruses, and it also inhibited the expression of RFP and p14 FAST from Ad-late/RFP and CRAdFAST, respectively. Thus, inclusion of an expression cassette consisting of a SA and transgene replacing the E3 of an HAdV-5 vector allows for replication-dependent expression of the transgene in human A549 cells, but not the murine 4T1 cell line.
We also examined whether expression of p14 FAST protein altered the replication kinetics of the virus by examining virus yield at varying times post-infection. A549 cells were infected at an MOI of 10, and the cells were harvested into the medium at varying times post-infection and, subsequently, assayed for virus titer. As shown in Figure 5C, infection of A549 cells with both CRAd and CRAdFAST resulted in an ∼300-fold increase in viral titer between 4 and 24 hpi, which further increased by 72 hpi. Importantly, there was no difference in virus recovery between CRAd containing or lacking the p14 FAST gene, suggesting that the expression of p14 FAST protein did not adversely affect virus growth and recovery.
CRAdFAST Shows Enhanced Cancer Cell Killing In Vitro in Human A549 Cells
We next examined whether expression of p14 FAST protein from CRAdFAST led to fusion and cell death of A549 cells in culture. A549 cells were infected at MOIs 1, 0.1, and 0.01 with AdFAST, CRAd, and CRAdFAST, and phase-contrast images were obtained at 24 and 48 hpi. No fusion was observed in AdFAST-infected cells (Figure 6A), which is consistent with our previous studies showing that an MOI of ∼50 of this replication-defective virus is required to achieve fusion in this cell line.36 Only minor evidence of CPE was observed in CRAd-infected cells at the highest MOI (MOI = 1) and at the latest time point (48 hpi) examined. In contrast, syncytia were observed in cells treated with CRAdFAST at an MOI as low as 0.1 and as early as 24 hpi. By 48 hpi, even an MOI as low as 0.01 yielded complete monolayer fusion, suggesting that CRAdFAST is very efficient at inducing fusion in A549 cells.
Figure 6.
CRAdFAST Mediates Enhanced Cell Fusion and Cell Death in A549 Cells In Vitro
(A) Phase-contrast images taken at 24 and 48 hpi of A549 cells infected at MOIs of 0.01, 0.1, and 1 with AdFAST, CRAd, or CRAdFAST. (B) A549 cells were mock infected or infected with AdFAST, CRAd, or CRAdFAST at an MOI of 1 in triplicate (n = 6 technical replicates, n = 2 independent experimental replicates). At 24, 48, and 72 hpi, metabolic activity was assessed by MTS assay; values reported are relative to mock-infected cells and represent mean ± SD (****p < 0.0001). (C) A549 cells were mock infected or infected at MOIs of 0.0625, 0.125, 0.25, 0.5, and 1 with CRAd or CRAdFAST, and they were harvested for immunoblot analysis at 48 hpi. Blots were probed for HAdV fiber, full-length caspase-3, or cleaved caspase-3, and actin was used as a loading control.
To determine if CRAdFAST-mediated cell fusion impacted A549 cell viability, A549 cells were infected with AdFAST, CRAd, or CRAdFAST or mock infected with PBS, and metabolic activity was assayed at varying times post-infection. Treatment of A549 cells with CRAdFAST caused a dramatic decrease in metabolic activity relative to cells treated with AdFAST or CRAd (Figure 6B). At 48 hpi, cells treated with CRAdFAST showed a metabolic activity of only 4% of untreated cells, which was also significantly lower than that observed for CRAd and AdFAST (117% and 118%, respectively, compared to 4% for CRAdFAST, p < 0.0001). By 72 hpi, metabolic activity of CRAdFAST-infected cells remained significantly lower than that of cells treated with the control viruses.
To determine whether this decrease in metabolic activity was associated with cell death, we examined treated cells for the presence of cleaved caspase-3 by immunoblot (Figure 6C). A549 cells were either mock infected or infected with varying amounts of virus (MOI 0.0625–1), and crude protein extracts were prepared 48 h later. Infection of cells with CRAd did not lead to a decline in the cellular quantity of full-length caspase-3 or formation of the active cleavage product. In contrast, in cells treated with CRAdFAST, full-length caspase-3 was only detectable at the lowest MOI tested, suggesting processing of the protein to its active form, and cleaved caspase-3 was readily detectable at MOIs of 0.5 and higher. Taken together, these data indicate that expression of p14 FAST protein from a CRAd enhances the oncolytic potential of the virus, resulting in significant cell fusion and a decline in cellular metabolic activity due to the induction of apoptotic-like cell death in A549 cells.
Expression of p14 FAST Protein Improves CRAd Efficacy in the Human A549 Xenograft Tumor Model In Vivo
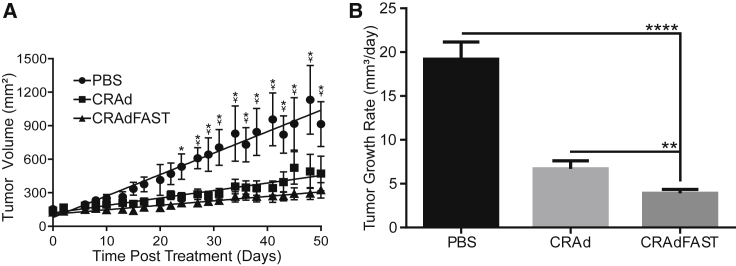
As CRAdFAST showed significant efficacy as an oncolytic virus in A549 cells in culture, we tested its potency in vivo using an A549 xenograft model of cancer. CD-1 nude mice bearing A549 tumors were injected with a single dose of vehicle (PBS) or 1 × 109 PFU of CRAd or CRAdFAST, and treated tumors were measured by caliper three times weekly. A statistically significant decrease in tumor size was achieved with both CRAd (beginning at day 27) and CRAdFAST (beginning at day 24) relative to animals treated with vehicle alone (Figure 7A). Although CRAdFAST-treated tumors trended toward a smaller size than those treated with CRAd, there was no statistically significant difference between these treatments by day 50 when mice were euthanized. However, tumors treated with CRAdFAST showed a statistically significant reduction in growth rate (CRAdFAST = 3.9 ± 0.45 mm3/day) compared to tumors treated with either PBS (19.2 ± 1.95 mm3/day, p < 0.0001) or CRAd (6.7 ± 0.92 mm3/day, p = 0.0013) (Figure 7B). Thus, arming an oncolytic HAdV with p14 FAST enhanced oncolytic efficacy of the vector in vivo in A549 tumors.
Figure 7.
Expression of p14 FAST Protein Significantly Improves Oncolytic Efficacy of CRAdFAST following Intratumoral Injection in a Human A549 Xenograft Model of Cancer
Mice bearing A549 tumors were injected intratumorally with PBS, CRAd, or CRAdFAST. Tumor volumes were measured for five mice per group, three times weekly. (A) Symbols represent calculated tumor volumes over time. Lines represent linear regression model of tumor growth rates. Beyond day 29, PBS-treated tumors are significantly larger than both CRAd- and CRAdFAST-treated tumors. Error bars represent mean ± SEM; *p <0.05 for CRAdFAST compared to PBS; Y represents p < 0.05 for CRAd compared to PBS. (B) Tumor growth rates were calculated for each tumor and averaged; error bars represent SD (**p = 0.0013, ****p < 0.0001).
We next examined tumor sections histologically. Since vectors based on HAdV tend to accumulate in the liver, even following intratumoral injection,42 we also examined the livers from treated animals for signs of syncytia or other pathology. Tumor and liver tissue were harvested at 5 and 50 days post-treatment, processed, and stained with anti-HAdV antibody to detect viral gene expression and anti-HA antibody to detect the expression of FAST protein. Anti-HAdV antibody primarily detects structural (late) proteins, which are only expressed after DNA replication has initiated, and it is thus a surrogate marker for active virus replication.43
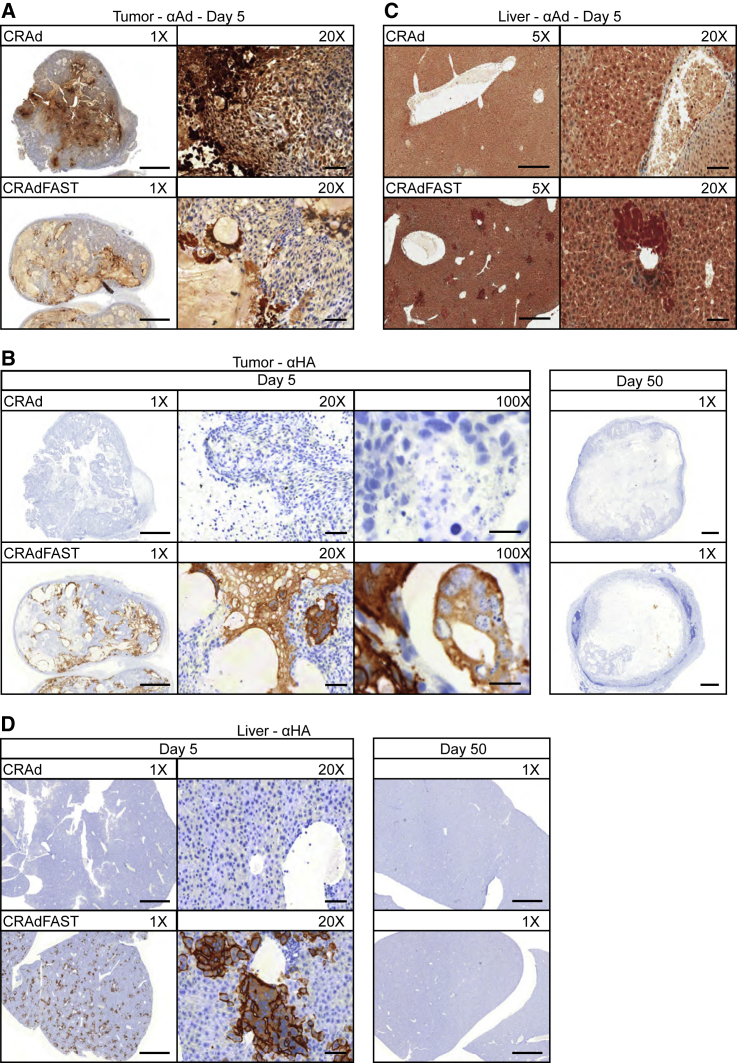
At 5 days post-injection, both CRAd and CRAdFAST appeared to have replicated efficiently in A549 tumors, as evidenced by the presence of viral late proteins; however, the two viruses showed strikingly different patterns of spatial spread (Figure 8A). While CRAd-treated tumors appeared to have intense HAdV staining localized to pockets in the interior of the tumor, sections from CRAdFAST-treated tumors showed more diffuse staining spread throughout the tumor mass. As expected, immunohistochemistry for the HA-tagged p14 FAST protein was negative in CRAd-treated tumors, and there was no evidence of multinucleated syncytia at either day 5 or day 50 (Figure 8B). In contrast, in CRAdFAST-treated animals, expression of the HA-tagged p14 FAST protein was readily detected in tumors at day 5 and, to a much lesser extent, also at day 50 (Figure 8B). Importantly, HA-positive areas of the tumor showed clear evidence of syncytium formation. Interestingly, although livers from mice injected with CRAdFAST stained positive with anti-HAdV at day 5 post-vector injection, no anti-HAdV staining was observed for CRAd (Figure 8C). Staining of liver sections for the HA-tagged p14 FAST protein showed small regions of positive staining around blood vessels at day 5 (Figure 8D), accompanied by apparent syncytium formation, which was resolved by day 50, suggesting this transient effect resolved without intervention and produced no obvious long-term effect on liver health.
Figure 8.
Treatment with CRAdFAST Induces Syncytium Formation in A549 Tumors In Vivo
(A) Sections of A549 xenograft tumors from mice treated with CRAd or CRAdFAST and sacrificed at day 5 post-injection were probed with anti-HAdV-5 antibody and counterstained with hematoxylin to assess HAdV replication. Images were taken at 1× and 20× magnifications. (B) Tumor sections from mice treated with CRAd or CRAdFAST and sacrificed at day 5 and day 50 post-injection were probed with anti-HA (p14 FAST) antibody and counterstained with hematoxylin to assess p14 FAST expression. Images were taken at 1×, 20×, and 100× magnifications. (C) Liver sections from mice treated with CRAd or CRAdFAST and sacrificed at day 5 and day 50 post-injection were probed with anti-HA (p14 FAST) and counterstained with hematoxylin to assess p14 FAST expression. Images were taken at 1× and 20× magnifications. (D) Liver sections from mice treated with CRAd or CRAdFAST and sacrificed at day 5 post-injection were probed with anti-HAdV-5 and counterstained with hematoxylin to assess HAdV replication. Images were taken at 5× and 20× magnifications. At 1×, 5×, 20×, and 100×, scale bars represent 2,000, 500, 100, and 20 μm, respectively.
Taken together, our results indicate that treatment of A549 cells with a CRAd vector expressing p14 FAST protein induces dramatic cell fusion, with a concomitant decline in cellular metabolic activity and induction of apoptosis, resulting in a statistically significant reduction in tumor growth rate in vivo, relative to CRAd alone.
Discussion
Oncolytic viruses have shown tremendous potential in preclinical studies, which, in many cases, has not translated to significant efficacy in clinical trials. Like many oncolytic viruses, HAdV has a poor ability to home to the tumor mass following systemic delivery, and, instead, it is primarily sequestered in the liver.44, 45 Although the amount of virus delivered to the tumor can be increased through direct injection of the vector into the tumor, HAdV typically does not transit far from the injection tract, leaving much of the tumor mass unaffected by the virus.15, 16, 46 One approach to increase the fraction of the tumor impacted by the gene therapy vector is to engineer the virus to express fusion proteins.25 In this manner, the few tumor cells that are initially infected with the virus will fuse with adjacent cells, producing multinucleated syncytia, thus directly spreading the area of the tumor impacted by the virus. This approach can be applied to both non-replicating vectors, allowing a co-expressed therapeutic transgene (e.g., p53) to spread between cells, and replicating oncolytic vectors. For oncolytic vectors, the virus can not only be spread through classical release of progeny virions and uptake by adjacent cells but also through fusion of the infected cell with adjacent naive cells.
In this study, we armed an oncolytic HAdV vector with the p14 FAST protein, and we evaluated the efficacy of the vector in tissue culture and animal models of cancer. Our oncolytic HAdV vector contained the E1AΔ24 mutation, which confers selective replication in cancer cells that are deficient in the Rb pathway.47 To restrict expression of the p14 FAST protein only to cancer cells, we placed it within the late transcription unit, under control by the HAdV major late promoter (MLP) (Figure 1). Transcripts under the control of the MLP are expressed poorly at early times of infection, but they increase to a great extent following the initiation of HAdV DNA replication.48 Previously, we used this approach to achieve high-level expression of proteins for retargeting HAdV infection40 and to design a reporter virus for high-throughput screening of small molecules affecting HAdV replication.39
Under conditions in which CRAdFAST could replicate, very high-level expression of the p14 FAST protein was achieved (Figures 2 and 5). Interestingly, in 4T1 cells in culture, we observed different temporal expression patterns between p14 FAST and fiber protein (Figure 2B). The p14 FAST protein was detected at an earlier time point (24 hpi) and declined at later time points (72 hpi), whereas HAdV fiber was first detected at 48 hpi and remained constant at 72 hpi. This led us to perform experiments with Ara-C, a DNA replication inhibitor that allows the differentiation between early and late gene expression. In the 4T1 cell line, p14 FAST expression from CRAdFAST appears to be under early, rather than late, transcriptional control (Figure 2). Although the majority of the E3 region is deleted in this viral construct, the early E3 promoter is retained, and it may be influencing the expression of p14 FAST. In human A549 cells, this same vector showed DNA replication-dependent expression for both the late fiber and p14 FAST proteins (Figure 5). Thus, there appear to be fundamental differences in promoter utilization and regulation in the murine 4T1 and human A549 cell lines. It is likely that these differences contribute to species-specific replication of HAdV.
The efficacy of oncolytic vectors is attributed in part to their ability to directly replicate in and kill cancer cells and also through their ability to act as an adjuvant to induce antitumor immunity.3, 49 Thus, we initially tested CRAdFAST in the 4T1 immunocompetent mouse model of cancer. However, in general, HAdV-5 has a relatively poor ability to infect mouse cells, including 4T1 cells,37 and infection is typically not productive (Figure 2). Although treatment of 4T1 cells in culture with CRAdFAST led to cell fusion, a reduction in cellular metabolic activity, and induction of apoptosis (Figure 3), no beneficial effect was achieved in vivo (Figure 4). However, we did observe regions of syncytium formation in CRAdFAST-treated 4T1 tumors, which was not observed in CRAd-treated animals, suggesting that p14 FAST protein could mediate cell fusion in vivo (Figure 4).
While inclusion of retargeting ligands can increase infection efficiency,40, 50, 51, 52, 53, 54 the inability of HAdV to productively replicate in mouse cells is a distinct limitation to testing HAdV in mouse models of cancer. Researchers have identified other animal models that are more permissive to HAdV replication, such as the Syrian hamster and cotton rat,55, 56 which may be more amenable to studying CRAdFAST in the context of an intact innate immune system. The mechanism for species-specific restriction of HAdV replication is unclear, although all aspects of the virus life cycle appear affected (i.e., early gene expression, DNA replication, and late gene expression).57, 58 HAdV is reported to productively replicate in mouse CMT-64 cells,28, 59, 60, 61 suggesting that this cell line may be a more appropriate model in which to assess efficacy of oncolytic HAdV in immunocompetent animals.
We next evaluated CRAdFAST in a human A549 xenograft model of cancer in immunodeficient mice. In this model, CRAd and CRAdFAST would be expected to replicate and spread to adjacent cells, but the effect of these vectors on antitumor immunity cannot be evaluated. In tissue culture, a very low quantity of CRAdFAST was required to achieve complete fusion of the cell monolayer, which was accompanied by a dramatic decline in cellular metabolic activity and induction of apoptotic-like cell death, as indicated by cleavage of caspase-3 (Figure 6). In vivo, treatment with CRAdFAST caused a statistically significant decline in tumor growth rate (Figure 7).
ICOVIR16, a conditionally replicating HAdV expressing the GALV fusion protein, induced a similar reduction in SkMel-28 tumor growth rates following a single intratumoral injection.22 Indeed, inclusion of the GALV fusion protein in ICOVIR16 appeared to have a greater therapeutic effect than we observed for p14 FAST. It is possible that the efficiency with which HAdV can replicate and spread within A549 cells may mask some of the benefits induced by intratumoral syncytium formation in our experiments. Immunohistological examination of tumors treated with CRAd showed intense staining primarily in the center regions of the tumor, whereas CRAdFAST showed less intense but more diffuse staining (Figure 8A). In addition, CRAdFAST-treated tumors had large acellular pockets bordered by regions of significant anti-HA (p14 FAST) staining (Figure 8B). Multinucleated syncytia were also noted in CRAdFAST-treated A549 tumors. Nuclear fragmentation, which is a hallmark of apoptotic cell death, was evident in both CRAd- and CRAdFAST-treated tumors (Figure 8A, 100×). Interestingly, not all fusogenic proteins induce apoptosis,62, 63 suggesting that different fusogenic proteins may induce alternate forms of cell death.
Since a large proportion of the initial injectate can escape the tumor and infect the liver,42, 44 we also examined pathology of the liver in treated animals. Interestingly, although the livers of all animals did not show any gross abnormal pathology at 5 days post-treatment, we did observe significant anti-HAdV staining primarily adjacent to blood vessels in the liver of CRAdFAST-treated animals, which was not present in animals treated with CRAd (Figure 8C). Similarly, anti-HA (p14 FAST) staining revealed the formation of small syncytia in regions adjacent to blood vessels in the liver of CRAdFAST-treated animals (Figure 8D).
The lack of anti-HAdV staining in the liver of mice treated with CRAd suggests that p14 FAST conferred some property that allowed for the accumulation of viral proteins in the liver of CRAdFAST-treated animals. p14 FAST protein may allow for enhanced replication of CRAd in mouse hepatocytes, although the mechanism by which this would occur is unclear. Cell fusion or syncytium formation within the CRAdFAST-infected tumor may allow for enhanced release of progeny virus into circulation, resulting in enhanced uptake of virus in the liver, which would not occur in CRAd-treated animals. In this scenario, the presence of HAdV proteins in the liver would be due to de novo replication within the liver, which we find unlikely given the cancer-specific replication conferred by the E1AΔ24 mutation.
Alternatively, we and others have shown that expression of fusion proteins, including p14 FAST protein,37 can lead to the enhanced release of exosome-like particles termed syncytiosomes.64, 65 Syncytiosomes are hypothesized to have the capacity to transport tumor-associated antigen outside of the tumor micro-environment. Thus, the HAdV and p14 FAST proteins present in the liver in CRAdFAST-treated animals may have been released from infected tumor cells in syncytiosomes, which were subsequently taken up by the liver. Consistent with this scenario, we have observed enhanced release of exosome-like particles from CRAdFAST-infected cells (data not shown). Syncytiosomes are also extremely efficient at delivering their contents to dendritic cells, thereby enhancing tumor-associated antigen cross-presentation and activation of T cells.64, 65 This latter point would obviously be extremely beneficial for the formation of anti-tumor immunity to help mediate tumor regression, thus improving CRAd efficacy in permissive and immunocompetent animal models of cancer and, ultimately, human patients.
Materials and Methods
Cell Culture
Human A549 (ATCC CRM-CCL-185)66 and 293 cells (ATCC CRL-1573)67 were maintained in minimal essential medium (MEM) (Sigma-Aldrich, Oakville, ON, Canada) supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich), 2 mM GlutaMAX (Invitrogen, Waltham, MA, USA), and 1× antibiotic-antimycotic (Invitrogen). Mouse 4T1 (ATCC CRL-2539)68 cells were maintained in DMEM-high glucose (Sigma-Aldrich) with identical supplements.
Adenoviral Constructs
Schematic representations of all HAdV vectors are shown in Figure 1. All HAdV vectors are based on serotype 5 (HAdV-5), and they were generated by a combination of traditional and RecA-mediated cloning.69 AdFAST-HA is an E1- and E3-deleted HAdV vector, it contains the p14 FAST gene under regulation of the human cytomegalovirus (CMV) immediate early enhancer-promoter and bovine growth hormone polyadenylation sequence (BGHpA) replacing the E1 region, and it has been described previously.36 For simplicity, this vector was referred to as AdFAST. HAdV vectors CRAd and CRAdFAST used in this study contain the E1AΔ24 mutation and are also deleted of the E3 region. E1AΔ24 prevents the E1A protein from binding the retinoblastoma (Rb) protein, which is required for efficient replication in normal tissues, thus conferring selective replication in cancer cells defective in the Rb pathway.47
To link p14 FAST protein expression to virus replication, we placed an expression cassette comprising an SA site-p14 FAST coding sequence replacing the E3 deletion, which places p14 FAST expression under regulation of the viral MLP.38, 39, 40 Since the MLP is only active at appreciable levels following active virus DNA replication,48 p14 FAST protein is only expressed in cells in which the virus can replicate. The p14 FAST proteins expressed from AdFAST and CRAdFAST contain a C-terminal HA epitope tag, as previously described.36 Ad-late/RFP is similar in structure to CRAdFAST, but it contains a wild-type E1 and encodes an SA-monomeric RFP expression cassette within the E3, rather than SA-p14 FAST, and it has been described previously.39 HAdV vector stocks were amplified in 293N3S cells, which were a kind gift of Dr. Frank Graham (McMaster University),70 with the exception that CRAdFAST was amplified in A549 cells in suspension culture. Viruses were purified and titered as described previously.71
Analysis of CRAdFAST Growth Kinetics
To determine whether expression of p14 FAST altered the growth kinetics of the vector, 35-mm dishes of A549 and 4T1 cells were infected at an MOI of 10 PFU/cell with either CRAd or CRAdFAST. At 2, 24, and 72 hpi, the cells were collected into the medium and subjected to two rounds of freeze-thaw to release progeny virions. Viral titers in the initial inoculum and at the various time points were determined by plaque assay.71
To quantify HAdV genome replication in 4T1 cells, 35-mm dishes of 4T1 cells were infected at an MOI of 3 in 200 μL PBS with either AdFAST or CRAdFAST for 1 h, washed with PBS, and medium was replaced. At 4, 24, 48, and 72 hpi, medium was removed and the cells were incubated with 500 μL SDS-Proteinase K buffer (10 mM Tris-HCl [pH 7.4], 10 mM EDTA, 1% SDS [w/v], and 1 mg/mL proteinase K) overnight at 37°C. DNA extraction was performed as described previously,71 and qPCR was performed using 200 ng isolated DNA and the following primer sets: Hexon (5′-CTTACCCCCAACGAGTTTGA-3′ and 5′-GGAGTACATGCGGTCCTTGT-3′) and actin (5′-CCGTCAGGCAGCTCATAGCTCTTC-3′ and 5′-CTGAACCCTAAGGCCAACCGT-3′). qPCR for the actin gene was performed to verify that equal quantities of DNA were present in the samples (data not shown). In parallel, a standard curve of known viral genome quantities was prepared, and it was used to calculate genome concentrations in each sample. Copy numbers were calculated by the method previously described by Whelan et al.72
Immunoblot Analysis
Cells were seeded in 35-mm culture dishes at a density of 6.0 × 105 cells/well. The next day, the cells were infected with virus at varying MOIs for 1 h and the medium was replaced. For experiments including Ara-C, medium containing 20 μg/mL drug was placed onto the cells after the 1-h infection. At varying times post-infection, the medium was removed, the cells were harvested in 2× Laemmli buffer (62.5 mM Tris HCl [pH 6.8], 25% glycerol, 2% SDS, 0.01% Bromophenol Blue, and 5% 2-mercaptoethanol), and the samples were stored at −20°C. Harvested samples were boiled for 5 min at 100°C, separated by SDS-PAGE, and transferred using a semi-dry transfer protocol onto polyvinylidene fluoride (PVDF) membrane (Millipore, Etobicoke, ON, Canada). Membranes were probed for β-actin (1:10,000, Clone AC-15 A1978, Sigma-Aldrich), HAdV-5 fiber (1/10,000 dilution, MS-1027-P0, Neomarkers [Fremont, CA, USA]), caspase-3 (1:10,000, 9662, Cell Signaling Technology [Beverly, MA, USA]), cleaved caspase-3 (1:1,000, 9664, Cell Signaling Technology), or HA epitope (1:3,000, 2367, Cell Signaling Technology). Immunoblots were developed using Luminata Classico horseradish peroxidase (HRP) substrate (Millipore) and visualized by autoradiography.
Microscopy
Phase-contrast images were acquired using a Zeiss Axio Observer Microscope (Carl Zeiss Canada, North York, ON, Canada) and analyzed using Zen 2 (Blue Edition) software (Carl Zeiss Canada). Images were processed using GNU Image Manipulation Program (GIMP)73 and compiled using Inkscape software.74
Metabolic Activity Assay
Metabolic activity was quantified using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA). 4T1 and A549 cells were seeded at 2 × 104 cells/well in flat-bottom 96-well cell culture dishes (Thermo Scientific, Waltham, MA, USA). The next day, the cells were infected with varying MOIs of AdFAST, CRAd, or CRAdFAST in 25 μL PBS for 1 h, and medium was replaced. Metabolic activity was evaluated at 24, 48, and 72 hpi. 20 μL MTS substrate was added to each well and incubated in the dark for 1 h. Color development was analyzed in the 96-well plate using a SpectraMax 190 plate spectrophotometer (Molecular Devices, Sunnyvale, CA, USA) at 490 nm.
4T1 Syngeneic Cancer Model
All mouse experiments were approved by the Animal Care Committee at the University of Ottawa. For the 4T1 mouse mammary carcinoma tumor model, 30 BALB/c (Charles River Laboratories, Wilmington, MA, USA) mice were injected subcutaneously with 1 × 105 cells in the right hind flank. When tumors reached approximately 5 × 5 mm (5 days post-injection), the mice received an intratumoral injection of 50 μL PBS or 7 × 109 PFU CRAd or CRAdFAST diluted in PBS to a total volume of 50 μL. Then 3 days after vector delivery, two mice from each group were euthanized for histological analysis of tumor and liver samples. For the remainder of the animals, tumor size was measured three times weekly via electronic calipers: two measurements were taken, one at the largest diameter and another perpendicular to the first, and volumes were calculated as (length × width2)/2. The experiment achieved endpoint when any tumor reached 15 × 15 mm, had a volume of approximately 1,687.5 mm3, or the mice showed signs of ulceration.
A549 Xenograft Tumor Model
For the A549 human lung adenocarcinoma tumor model, 21 CD-1 nude mice (Charles River Laboratories) were injected subcutaneously with 1 × 106 A549 cells in the right hind flank. Then 15 days later, when the tumors reached approximately 5 × 5 mm, the mice received an intratumoral injection of 50 μL PBS or 1.0 × 109 PFU CRAd or CRAdFAST diluted to a volume of 50 μL in PBS. Mice were monitored for tumor growth and survival as described above.
Tumor Histology and Immunohistochemistry
Immediately upon removal, tumors and livers were placed in 4% paraformaldehyde (PFA) solution. Then 48 h later, the samples were transferred to 70% ethanol, and they were further processed, embedded, sectioned, and stained with H&E at the University of Ottawa Histology Core Facility. Formalin-fixed, paraffin-embedded slides were deparaffinized in xylene and rehydrated through a 100%–70% ethanol gradient. Heat-induced epitope retrieval was performed at 110°C for 12 min with citrate buffer (pH 6.0), and 3% H202 was used to block endogenous peroxidases. Sections were further blocked with Background Sniper blocking reagent (BioCare Medical, CA, USA) and incubated with rabbit anti-HA antibody (Cell Signaling Technology, C29F4; 1:1,000 liver, 1:2,000 tumor) and/or rabbit anti-HAdV-5 (Abcam, ab6982; 1:2,000) for 1 h at room temperature. Antibody binding was visualized using the MAHC4TM + DAB detection system according to the manufacturer’s instructions (BioCare Medical). Slides were then counterstained with hematoxylin, and images were obtained using the Zeiss Mirax Midi whole-slide digital scanner. Images were analyzed using Pannoramic Viewer software (3DHISTECH, build 1.15.4.43061, Budapest, Hungary), and they were compiled using Inkscape software.74
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 6 (version 6.01; GraphPad, San Diego, CA, USA). All bar graphs represent mean, while error bars represent SD. MTS data presented in bar graphs were compared by two-way ANOVA, followed by Tukey’s honestly significant difference (HSD) post hoc analysis. 4T1 tumor growth rates presented in bar graphs were modeled using tumor-specific growth rate, as described by Mehrara et al.,75 and compared by one-way ANOVA. For the A549 xenograft tumor model, a linear growth model was chosen to analyze growth, as the r2 values overall were higher for linear regression (0.53 for PBS, 0.33 for CRAd, and 0.42 for CRAdFAST) than for non-linear regression using an exponential model (0.38 for PBS, 0.27 for CRAd, and 0.34 for CRAdFAST). There is significant evidence that tumors often take on a linear growth pattern.76
Author Contributions
The project was conceived by J.C.B. and R.J.P. and designed by J.D.P. and R.J.P. Data were collected and analyzed by J.D.P. J.P. and J.C.B. contributed to the design and implementation of animal trials. The manuscript was written by J.D.P. and R.J.P. and edited by all authors.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank Kathy Poulin for excellent technical assistance and advice and Dr. Christina Addison (Ottawa Hospital Research Institute) for providing cytosine arabinoside. Funding was provided by grants to R.J.P. from the Cancer Research Society, Canada (grant number 19363), Canadian Institutes of Health Research, Canada (grant numbers MOP-142316 and MOP-136898), and the Natural Sciences and Engineering Research Council of Canada, Canada (grant number RGPIN-2014-04810). J.D.P. is a recipient of a Queen Elizabeth II Graduate Scholarship in Science and Technology, Ontario, Canada and an Ontario Graduate Scholarship, Ontario, Canada.
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Stojdl D.F., Lichty B., Knowles S., Marius R., Atkins H., Sonenberg N., Bell J.C. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat. Med. 2000;6:821–825. doi: 10.1038/77558. [DOI] [PubMed] [Google Scholar]
- 3.Parato K.A., Senger D., Forsyth P.A., Bell J.C. Recent progress in the battle between oncolytic viruses and tumours. Nat. Rev. Cancer. 2005;5:965–976. doi: 10.1038/nrc1750. [DOI] [PubMed] [Google Scholar]
- 4.Lawler S.E., Speranza M.-C., Cho C.-F., Chiocca E.A. Oncolytic viruses in cancer treatment: a review. JAMA Oncol. 2017;3:841–849. doi: 10.1001/jamaoncol.2016.2064. [DOI] [PubMed] [Google Scholar]
- 5.DeWeese T.L., van der Poel H., Li S., Mikhak B., Drew R., Goemann M., Hamper U., DeJong R., Detorie N., Rodriguez R. A phase I trial of CV706, a replication-competent, PSA selective oncolytic adenovirus, for the treatment of locally recurrent prostate cancer following radiation therapy. Cancer Res. 2001;61:7464–7472. [PubMed] [Google Scholar]
- 6.Vidal L., Pandha H.S., Yap T.A., White C.L., Twigger K., Vile R.G., Melcher A., Coffey M., Harrington K.J., DeBono J.S. A phase I study of intravenous oncolytic reovirus type 3 Dearing in patients with advanced cancer. Clin. Cancer Res. 2008;14:7127–7137. doi: 10.1158/1078-0432.CCR-08-0524. [DOI] [PubMed] [Google Scholar]
- 7.Eder J.P., Kantoff P.W., Roper K., Xu G.X., Bubley G.J., Boyden J., Gritz L., Mazzara G., Oh W.K., Arlen P. A phase I trial of a recombinant vaccinia virus expressing prostate-specific antigen in advanced prostate cancer. Clin. Cancer Res. 2000;6:1632–1638. [PubMed] [Google Scholar]
- 8.Andtbacka R.H., Kaufman H.L., Collichio F., Amatruda T., Senzer N., Chesney J., Delman K.A., Spitler L.E., Puzanov I., Agarwala S.S. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J. Clin. Oncol. 2015;33:2780–2788. doi: 10.1200/JCO.2014.58.3377. [DOI] [PubMed] [Google Scholar]
- 9.Rehman H., Silk A.W., Kane M.P., Kaufman H.L. Into the clinic: Talimogene laherparepvec (T-VEC), a first-in-class intratumoral oncolytic viral therapy. J. Immunother. Cancer. 2016;4:53. doi: 10.1186/s40425-016-0158-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang F.F., Bruner J.M., Fuller G.N., Aldape K., Prados M.D., Chang S., Berger M.S., McDermott M.W., Kunwar S.M., Junck L.R. Phase I trial of adenovirus-mediated p53 gene therapy for recurrent glioma: biological and clinical results. J. Clin. Oncol. 2003;21:2508–2518. doi: 10.1200/JCO.2003.21.13.2508. [DOI] [PubMed] [Google Scholar]
- 11.Narvaiza I., Mazzolini G., Barajas M., Duarte M., Zaratiegui M., Qian C., Melero I., Prieto J. Intratumoral coinjection of two adenoviruses, one encoding the chemokine IFN-γ-inducible protein-10 and another encoding IL-12, results in marked antitumoral synergy. J. Immunol. 2000;164:3112–3122. doi: 10.4049/jimmunol.164.6.3112. [DOI] [PubMed] [Google Scholar]
- 12.Peng Z. Current status of gendicine in China: recombinant human Ad-p53 agent for treatment of cancers. Hum. Gene Ther. 2005;16:1016–1027. doi: 10.1089/hum.2005.16.1016. [DOI] [PubMed] [Google Scholar]
- 13.Khuri F.R., Nemunaitis J., Ganly I., Arseneau J., Tannock I.F., Romel L., Gore M., Ironside J., MacDougall R.H., Heise C. a controlled trial of intratumoral ONYX-015, a selectively-replicating adenovirus, in combination with cisplatin and 5-fluorouracil in patients with recurrent head and neck cancer. Nat. Med. 2000;6:879–885. doi: 10.1038/78638. [DOI] [PubMed] [Google Scholar]
- 14.Russell S.J., Peng K.-W., Bell J.C. Oncolytic virotherapy. Nat. Biotechnol. 2012;30:658–670. doi: 10.1038/nbt.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sauthoff H., Hu J., Maca C., Goldman M., Heitner S., Yee H., Pipiya T., Rom W.N., Hay J.G. Intratumoral spread of wild-type adenovirus is limited after local injection of human xenograft tumors: virus persists and spreads systemically at late time points. Hum. Gene Ther. 2003;14:425–433. doi: 10.1089/104303403321467199. [DOI] [PubMed] [Google Scholar]
- 16.Heise C.C., Williams A., Olesch J., Kirn D.H. Efficacy of a replication-competent adenovirus (ONYX-015) following intratumoral injection: intratumoral spread and distribution effects. Cancer Gene Ther. 1999;6:499–504. doi: 10.1038/sj.cgt.7700071. [DOI] [PubMed] [Google Scholar]
- 17.Tollefson A.E., Scaria A., Hermiston T.W., Ryerse J.S., Wold L.J., Wold W.S. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J. Virol. 1996;70:2296–2306. doi: 10.1128/jvi.70.4.2296-2306.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barton K.N., Paielli D., Zhang Y., Koul S., Brown S.L., Lu M., Seely J., Kim J.H., Freytag S.O. Second-generation replication-competent oncolytic adenovirus armed with improved suicide genes and ADP gene demonstrates greater efficacy without increased toxicity. Mol. Ther. 2006;13:347–356. doi: 10.1016/j.ymthe.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Al-Zaher A.A., Moreno R., Fajardo C.A., Arias-Badia M., Farrera M., de Sostoa J., Rojas L.A., Alemany R. Evidence of anti-tumoral efficacy in an immune competent setting with an iRGD-modified hyaluronidase-armed oncolytic adenovirus. Mol. Ther. Oncolytics. 2018;8:62–70. doi: 10.1016/j.omto.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guedan S., Rojas J.J., Gros A., Mercade E., Cascallo M., Alemany R. Hyaluronidase expression by an oncolytic adenovirus enhances its intratumoral spread and suppresses tumor growth. Mol. Ther. 2010;18:1275–1283. doi: 10.1038/mt.2010.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yumul R., Richter M., Lu Z.Z., Saydaminova K., Wang H., Wang C.H., Carter D., Lieber A. Epithelial Junction Opener Improves Oncolytic Adenovirus Therapy in Mouse Tumor Models. Hum. Gene Ther. 2016;27:325–337. doi: 10.1089/hum.2016.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guedan S., Grases D., Rojas J.J., Gros A., Vilardell F., Vile R., Mercade E., Cascallo M., Alemany R. GALV expression enhances the therapeutic efficacy of an oncolytic adenovirus by inducing cell fusion and enhancing virus distribution. Gene Ther. 2012;19:1048–1057. doi: 10.1038/gt.2011.184. [DOI] [PubMed] [Google Scholar]
- 23.Guedan S., Gros A., Cascallo M., Vile R., Mercade E., Alemany R. Syncytia formation affects the yield and cytotoxicity of an adenovirus expressing a fusogenic glycoprotein at a late stage of replication. Gene Ther. 2008;15:1240–1245. doi: 10.1038/gt.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Haviv Y.S., Derdeyn C.A., Lam J., Coolidge C., Hunter E., Curiel D.T., Blackwell J.L. Human immunodeficiency virus type 1-mediated syncytium formation is compatible with adenovirus replication and facilitates efficient dispersion of viral gene products and de novo-synthesized virus particles. Hum. Gene Ther. 2001;12:2155–2165. doi: 10.1089/10430340152710504. [DOI] [PubMed] [Google Scholar]
- 25.Del Papa J., Parks R.J. Adenoviral Vectors Armed with Cell Fusion-Inducing Proteins as Anti-Cancer Agents. Viruses. 2017;9:13. doi: 10.3390/v9010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saha B., Wong C.M., Parks R.J. The adenovirus genome contributes to the structural stability of the virion. Viruses. 2014;6:3563–3583. doi: 10.3390/v6093563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bett A.J., Prevec L., Graham F.L. Packaging capacity and stability of human adenovirus type 5 vectors. J. Virol. 1993;67:5911–5921. doi: 10.1128/jvi.67.10.5911-5921.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y., Hallden G., Hill R., Anand A., Liu T.-C., Francis J., Brooks G., Lemoine N., Kirn D. E3 gene manipulations affect oncolytic adenovirus activity in immunocompetent tumor models. Nat. Biotechnol. 2003;21:1328–1335. doi: 10.1038/nbt887. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki K., Alemany R., Yamamoto M., Curiel D.T. The presence of the adenovirus E3 region improves the oncolytic potency of conditionally replicative adenoviruses. Clin. Cancer Res. 2002;8:3348–3359. [PubMed] [Google Scholar]
- 30.Shmulevitz M., Duncan R. A new class of fusion-associated small transmembrane (FAST) proteins encoded by the non-enveloped fusogenic reoviruses. EMBO J. 2000;19:902–912. doi: 10.1093/emboj/19.5.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corcoran J.A., Salsman J., de Antueno R., Touhami A., Jericho M.H., Clancy E.K., Duncan R. The p14 fusion-associated small transmembrane (FAST) protein effects membrane fusion from a subset of membrane microdomains. J. Biol. Chem. 2006;281:31778–31789. doi: 10.1074/jbc.M602566200. [DOI] [PubMed] [Google Scholar]
- 32.Ciechonska M., Duncan R. Reovirus FAST proteins: virus-encoded cellular fusogens. Trends Microbiol. 2014;22:715–724. doi: 10.1016/j.tim.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Salsman J., Top D., Boutilier J., Duncan R. Extensive syncytium formation mediated by the reovirus FAST proteins triggers apoptosis-induced membrane instability. J. Virol. 2005;79:8090–8100. doi: 10.1128/JVI.79.13.8090-8100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Boeuf F., Gebremeskel S., McMullen N., He H., Greenshields A.L., Hoskin D.W., Bell J.C., Johnston B., Pan C., Duncan R. Reovirus FAST Protein Enhances Vesicular Stomatitis Virus Oncolytic Virotherapy in Primary and Metastatic Tumor Models. Mol. Ther. Oncolytics. 2017;6:80–89. doi: 10.1016/j.omto.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Boeuf F., Diallo J.-S., McCart J.A., Thorne S., Falls T., Stanford M., Kanji F., Auer R., Brown C.W., Lichty B.D. Synergistic interaction between oncolytic viruses augments tumor killing. Mol. Ther. 2010;18:888–895. doi: 10.1038/mt.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong C.M., Poulin K.L., Tong G., Christou C., Kennedy M.A., Falls T., Bell J.C., Parks R.J. Adenovirus-Mediated Expression of the p14 Fusion-Associated Small Transmembrane Protein Promotes Cancer Cell Fusion and Apoptosis In Vitro but Does Not Provide Therapeutic Efficacy in a Xenograft Mouse Model of Cancer. PLoS ONE. 2016;11:e0151516. doi: 10.1371/journal.pone.0151516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong C.M., Nash L.A., Del Papa J., Poulin K.L., Falls T., Bell J.C., Parks R.J. Expression of the fusogenic p14 FAST protein from a replication-defective adenovirus vector does not provide a therapeutic benefit in an immunocompetent mouse model of cancer. Cancer Gene Ther. 2016;23:355–364. doi: 10.1038/cgt.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carette J.E., Graat H.C., Schagen F.H., Abou El Hassan M.A., Gerritsen W.R., van Beusechem V.W. Replication-dependent transgene expression from a conditionally replicating adenovirus via alternative splicing to a heterologous splice-acceptor site. J. Gene Med. 2005;7:1053–1062. doi: 10.1002/jgm.754. [DOI] [PubMed] [Google Scholar]
- 39.Saha B., Parks R.J. Histone deacetylase inhibitor SAHA suppresses human adenovirus gene expression and replication. J. Virol. 2019 doi: 10.1128/JVI.00088-19. Published online April 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poulin K.L., Lanthier R.M., Smith A.C., Christou C., Risco Quiroz M., Powell K.L., O’Meara R.W., Kothary R., Lorimer I.A., Parks R.J. Retargeting of adenovirus vectors through genetic fusion of a single-chain or single-domain antibody to capsid protein IX. J. Virol. 2010;84:10074–10086. doi: 10.1128/JVI.02665-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaynor R.B., Tsukamoto A., Montell C., Berk A.J. Enhanced expression of adenovirus transforming proteins. J. Virol. 1982;44:276–285. doi: 10.1128/jvi.44.1.276-285.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bramson J.L., Hitt M., Gauldie J., Graham F.L. Pre-existing immunity to adenovirus does not prevent tumor regression following intratumoral administration of a vector expressing IL-12 but inhibits virus dissemination. Gene Ther. 1997;4:1069–1076. doi: 10.1038/sj.gt.3300508. [DOI] [PubMed] [Google Scholar]
- 43.Saha B., Parks R.J. Human adenovirus type 5 vectors deleted of early region 1 (E1) undergo limited expression of early replicative E2 proteins and DNA replication in non-permissive cells. PLoS ONE. 2017;12:e0181012. doi: 10.1371/journal.pone.0181012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernt K.M., Ni S., Gaggar A., Li Z.Y., Shayakhmetov D.M., Lieber A. The effect of sequestration by nontarget tissues on anti-tumor efficacy of systemically applied, conditionally replicating adenovirus vectors. Mol. Ther. 2003;8:746–755. doi: 10.1016/j.ymthe.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 45.Ross P.J., Parks R.J. Oncolytic adenovirus: getting there is half the battle. Mol. Ther. 2003;8:705–706. doi: 10.1016/j.ymthe.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y., Hu J.K., Krol A., Li Y.-P., Li C.-Y., Yuan F. Systemic dissemination of viral vectors during intratumoral injection. Mol. Cancer Ther. 2003;2:1233–1242. [PubMed] [Google Scholar]
- 47.Fueyo J., Gomez-Manzano C., Alemany R., Lee P.S., McDonnell T.J., Mitlianga P., Shi Y.X., Levin V.A., Yung W.K., Kyritsis A.P. A mutant oncolytic adenovirus targeting the Rb pathway produces anti-glioma effect in vivo. Oncogene. 2000;19:2–12. doi: 10.1038/sj.onc.1203251. [DOI] [PubMed] [Google Scholar]
- 48.Thomas G.P., Mathews M.B. DNA replication and the early to late transition in adenovirus infection. Cell. 1980;22:523–533. doi: 10.1016/0092-8674(80)90362-1. [DOI] [PubMed] [Google Scholar]
- 49.Lindenmann J., Klein P.A. Springer Science & Business Media; 2012. Immunological Aspects of Viral Oncolysis. [Google Scholar]
- 50.Smith T.A., Idamakanti N., Marshall-Neff J., Rollence M.L., Wright P., Kaloss M., King L., Mech C., Dinges L., Iverson W.O. Receptor interactions involved in adenoviral-mediated gene delivery after systemic administration in non-human primates. Hum. Gene Ther. 2003;14:1595–1604. doi: 10.1089/104303403322542248. [DOI] [PubMed] [Google Scholar]
- 51.Tao N., Gao G.-P., Parr M., Johnston J., Baradet T., Wilson J.M., Barsoum J., Fawell S.E. Sequestration of adenoviral vector by Kupffer cells leads to a nonlinear dose response of transduction in liver. Mol. Ther. 2001;3:28–35. doi: 10.1006/mthe.2000.0227. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Z., Krimmel J., Zhang Z., Hu Z., Seth P. Systemic delivery of a novel liver-detargeted oncolytic adenovirus causes reduced liver toxicity but maintains the antitumor response in a breast cancer bone metastasis model. Hum. Gene Ther. 2011;22:1137–1142. doi: 10.1089/hum.2011.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coughlan L., Vallath S., Gros A., Giménez-Alejandre M., Van Rooijen N., Thomas G.J., Baker A.H., Cascalló M., Alemany R., Hart I.R. Combined fiber modifications both to target α(v)β(6) and detarget the coxsackievirus-adenovirus receptor improve virus toxicity profiles in vivo but fail to improve antitumoral efficacy relative to adenovirus serotype 5. Hum. Gene Ther. 2012;23:960–979. doi: 10.1089/hum.2011.218. [DOI] [PubMed] [Google Scholar]
- 54.Poulin K.L., Tong G., Vorobyova O., Pool M., Kothary R., Parks R.J. Use of Cre/loxP recombination to swap cell binding motifs on the adenoviral capsid protein IX. Virology. 2011;420:146–155. doi: 10.1016/j.virol.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 55.Li X., Wang P., Li H., Du X., Liu M., Huang Q., Wang Y., Wang S. The efficacy of oncolytic adenovirus is mediated by T-cell responses against virus and tumor in Syrian hamster model. Clin. Cancer Res. 2017;23:239–249. doi: 10.1158/1078-0432.CCR-16-0477. [DOI] [PubMed] [Google Scholar]
- 56.Steel J.C., Morrison B.J., Mannan P., Abu-Asab M.S., Wildner O., Miles B.K., Yim K.C., Ramanan V., Prince G.A., Morris J.C. Immunocompetent syngeneic cotton rat tumor models for the assessment of replication-competent oncolytic adenovirus. Virology. 2007;369:131–142. doi: 10.1016/j.virol.2007.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silverstein G., Strohl W.A. Restricted replication of adenovirus type 2 in mouse Balb/3T3 cells. Arch. Virol. 1986;87:241–264. doi: 10.1007/BF01315303. [DOI] [PubMed] [Google Scholar]
- 58.Blair G.E., Dixon S.C., Griffiths S.A., Zajdel M.E.B. Restricted replication of human adenovirus type 5 in mouse cell lines. Virus Res. 1989;14:339–346. doi: 10.1016/0168-1702(89)90026-9. [DOI] [PubMed] [Google Scholar]
- 59.Halldén G., Hill R., Wang Y., Anand A., Liu T.-C., Lemoine N.R., Francis J., Hawkins L., Kirn D. Novel immunocompetent murine tumor models for the assessment of replication-competent oncolytic adenovirus efficacy. Mol. Ther. 2003;8:412–424. doi: 10.1016/s1525-0016(03)00199-0. [DOI] [PubMed] [Google Scholar]
- 60.Cheong S.C., Wang Y., Meng J.H., Hill R., Sweeney K., Kirn D., Lemoine N.R., Halldén G. E1A-expressing adenoviral E3B mutants act synergistically with chemotherapeutics in immunocompetent tumor models. Cancer Gene Ther. 2008;15:40–50. doi: 10.1038/sj.cgt.7701099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu T.-C., Hallden G., Wang Y., Brooks G., Francis J., Lemoine N., Kirn D. An E1B-19 kDa gene deletion mutant adenovirus demonstrates tumor necrosis factor-enhanced cancer selectivity and enhanced oncolytic potency. Mol. Ther. 2004;9:786–803. doi: 10.1016/j.ymthe.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 62.Bateman A., Bullough F., Murphy S., Emiliusen L., Lavillette D., Cosset F.L., Cattaneo R., Russell S.J., Vile R.G. Fusogenic membrane glycoproteins as a novel class of genes for the local and immune-mediated control of tumor growth. Cancer Res. 2000;60:1492–1497. [PubMed] [Google Scholar]
- 63.Tan L., Jia H., Liu R., Wu J., Han H., Zuo Y., Yang S., Huang W. Inhibition of NF-kappaB in fusogenic membrane glycoprotein causing HL-60 cell death: implications for acute myeloid leukemia. Cancer Lett. 2009;273:114–121. doi: 10.1016/j.canlet.2008.07.035. [DOI] [PubMed] [Google Scholar]
- 64.Errington F., Jones J., Merrick A., Bateman A., Harrington K., Gough M., O’Donnell D., Selby P., Vile R., Melcher A. Fusogenic membrane glycoprotein-mediated tumour cell fusion activates human dendritic cells for enhanced IL-12 production and T-cell priming. Gene Ther. 2006;13:138–149. doi: 10.1038/sj.gt.3302609. [DOI] [PubMed] [Google Scholar]
- 65.Linardakis E., Bateman A., Phan V., Ahmed A., Gough M., Olivier K., Kennedy R., Errington F., Harrington K.J., Melcher A., Vile R. Enhancing the efficacy of a weak allogeneic melanoma vaccine by viral fusogenic membrane glycoprotein-mediated tumor cell-tumor cell fusion. Cancer Res. 2002;62:5495–5504. [PubMed] [Google Scholar]
- 66.Giard D.J., Aaronson S.A., Todaro G.J., Arnstein P., Kersey J.H., Dosik H., Parks W.P. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 67.Graham F.L., Smiley J., Russell W.C., Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 68.Dexter D.L., Kowalski H.M., Blazar B.A., Fligiel Z., Vogel R., Heppner G.H. Heterogeneity of tumor cells from a single mouse mammary tumor. Cancer Res. 1978;38:3174–3181. [PubMed] [Google Scholar]
- 69.Chartier C., Degryse E., Gantzer M., Dieterle A., Pavirani A., Mehtali M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 1996;70:4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Graham F.L. Growth of 293 cells in suspension culture. J. Gen. Virol. 1987;68:937–940. doi: 10.1099/0022-1317-68-3-937. [DOI] [PubMed] [Google Scholar]
- 71.Ross P.J., Parks R.J. Construction and characterization of adenovirus vectors. Cold Spring Harb. Protoc. 2009;2009 doi: 10.1101/pdb.prot5011. pdb.prot5011. [DOI] [PubMed] [Google Scholar]
- 72.Whelan J.A., Russell N.B., Whelan M.A. A method for the absolute quantification of cDNA using real-time PCR. J. Immunol. Methods. 2003;278:261–269. doi: 10.1016/s0022-1759(03)00223-0. [DOI] [PubMed] [Google Scholar]
- 73.Kimball, S., and Mattis, P. (1995). GNU Image Manipulation Program, version 2.8.22. https://www.gimp.org/.
- 74.Harrington, B. (2005). Inkscape. https://inkscape.org/.
- 75.Mehrara E., Forssell-Aronsson E., Ahlman H., Bernhardt P. Specific growth rate versus doubling time for quantitative characterization of tumor growth rate. Cancer Res. 2007;67:3970–3975. doi: 10.1158/0008-5472.CAN-06-3822. [DOI] [PubMed] [Google Scholar]
- 76.Brú A., Albertos S., Luis Subiza J., García-Asenjo J.L., Brú I. The universal dynamics of tumor growth. Biophys. J. 2003;85:2948–2961. doi: 10.1016/S0006-3495(03)74715-8. [DOI] [PMC free article] [PubMed] [Google Scholar]