Abstract
Rhabdomyosarcoma (RMS) is one of the most common types of soft tissue sarcoma in children; however, the pathogenesis of RMS is unclear. MicroRNAs (miRs) are involved in the development and progression of RMS. The role of miR-410-3p in RMS cell invasion, migration, proliferation and apoptosis, and its possible mechanism were investigated in the current study. Reverse transcription-quantitative polymerase chain reaction and western blot analysis were performed to detect the expression of miR-410-3p in RMS tissues and cells. In addition, the present study investigated the expression levels of molecules associated with the epithelial-mesenchymal transition (EMT), including E-cadherin, N-cadherin, Slug and Snail, and apoptotic factors, including Bcl-2-associated X protein (bax), cleaved-caspase 3, cleaved poly (ADP-ribose) polymerase (PARP), p53 and Bcl-2. Cell Counting Kit-8, terminal deoxynucleotidyl transferase dUTP nick end labeling and Transwell assays were conducted to determine the functional roles of miR-410-3p. Exogenous expression of miR-410-3p inhibited RMS cell invasion, migration and proliferation, induced apoptosis, suppressed the expression of Snail, Slug, N-cadherin and Bcl-2, and increased the expression of E-cadherin, bax, cleaved-caspase 3, cleaved PARP and p53. In summary, it was proposed that miR-410-3p overexpression suppressed invasion, migration and proliferation, downregulated the expression of EMT-associated molecules, and promoted apoptosis and the expression of apoptotic factors in RMS cells. Therefore, miR-410-3p may serve as a novel tumor suppressor gene in RMS, and could possess diagnostic and therapeutic potentials for the treatment of RMS.
Keywords: microRNA-410-3p, epithelial-mesenchymal transition, rhabdomyosarcoma, invasion, migration
Introduction
Rhabdomyosarcoma (RMS) is one of the most common types of soft tissue sarcoma (STS) in children, and typically manifests in the head and neck region, genitourinary system, and the extremities (1,2). The subtypes of RMS include embryonal RMS (ERMS), which is the main histological subtype accounting for ~60% of all pediatric RMS, and alveolar RMS (ARMS), which accounts for 20–25% (3,4). The 5-year overall survival rate of RMS ranges from 25–65%, and the main clinicopathological parameters that affect the 5-year survival rate are age and tumor characteristics (5,6). Specifically, alveolar histology and metastasis are key prognosis variables that predict poor outcome (5,7).
MicroRNAs (miRNAs/miRs) are associated with the mechanisms of RMS tumorigenesis (8). miRNAs are non-coding RNAs that serve a pivotal role in post-transcriptional control (9,10), and are involved in the development and progression of RMS (11). This includes miR-21, miR-183, miR-372 and miR-373, which serve as oncogenes (12–14), and let-7, miR-124 and miR-200a, which serve as tumor suppressor genes (15–17). miR-410-3p is a tumor suppressor that suppresses cancer cell growth and invasion in osteosarcoma (18), pancreatic cancer (19) and breast cancer (20) when minimally expressed, whereas miR-410-3p acts as an oncogene in liver, colorectal (21,22) and non-small-cell lung cancer (23). Therefore, miR-410-3p serves different roles in various types of tumor. An important step in the invasion and metastasis process is the epithelial-mesenchymal transition (EMT), which is regulated by miR-135b (24) and miR-130a (25) in osteosarcoma, and by miR-410-3p in breast (20) and gastric cancer cells (26). The Snail gene is the direct target gene of miR-410-3p in breast cancer cells (20); however, the role of miR-410-3p in RMS development and progression remains unclear.
In the present study, compared with normal controls, reduced miR-410-3p expression levels were detected in RMS tissues and three RMS cells, including RD, PLA802 and RH30. Upregulated miR-410-3p expression was identified to suppress cell invasion, migration and proliferation, the EMT process, and induce apoptosis and the expression of apoptotic factors in RMS. These results highlighted the function of miR-410-3p in RMS and proposed a novel mechanism of miR-410-3p in RMS.
Materials and methods
Patients and tissue samples
All formalin-fixed paraffin-embedded (FFPE) tissue samples were selected from a collection of the Department of Pathology of The First Affiliated Hospital of Shihezi University School of Medicine (Shihezi, China) between January 2010 and December 2017. This comprised 22 RMS samples obtained from 10 female and 12 male patients aged between 2.5 and 68 years (mean age, 21 years) and nine normal skeletal muscle tissue samples from nine female patients aged between 25 and 52 years (mean age, 37 years). All patients involved in the study provided written informed consent. The study was approved by the institutional Ethics Committee of The First Affiliated Hospital of Shihezi University School of Medicine (Shihezi, China) and conducted in accordance with the ethical guidelines of the Declaration of Helsinki. Two senior pathology professors performed the histomorphological evaluation of all samples to select the qualifying samples.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the paraffin-embedded tissues and cell lines using the miRNeasy FFPE kit (cat. no. 217504; Qiagen GmbH, Hilden, Germany) or the miRNeasy Mini kit (cat. no. 217004; Qiagen GmbH) according to the manufacturer's protocol. Complementary DNA of miRNA or mRNA was generated using the mi Script II RT kit (Qiagen, No. 218075). Total miRNA or RNA (1 µg) was reverse-transcribed in 20 µl reaction mix using the miScript II RT kit according to the manufacturer's recommendations; reverse transcription was performed in 37°C for 60 min and 95°C for 5 min. qPCR was performed using the mi Script SYBR Green PCR kit (Qiagen, No. 218075) or SYBR Green PCR Master mix (Qiagen GmbH) on a 7500 Fast Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and the parameters were set in accordance with the manufacturer's protocol. The thermocycling conditions were as follows: The initial denaturation step (95°C for 15 min), followed by 40 cycles of denaturation (94°C for 15 sec), annealing (55°C for 30 sec) and extension (70°C for 30 sec). U6 small RNA (U6) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA were used as internal controls. All reactions were performed in triplicate. The primers for miR-410-3p (cat. no. 218300-MS00004144; Qiagen GmbH) and U6 (cat. no. 218300-MS00033740; Qiagen GmbH) were purchased.
The following EMT-associated molecule primer sequences were used: Snail forward, 5′-GGTTCTTCTGCGCTACTGCT-3′ and reverse, 5′-TAGGGCTGCTGGAAGGTAAA-3′; Slug forward, 5′-GGGGAGAAGCCTTTTTCTTG-3′ and reverse, 5′-TAATCATGTTTGTGCAGGAG-3′; zinc finger E-box binding homeobox 1 (ZEB1) forward, 5′-GCACAGAAGAGCCACAAGTA-3′ and reverse, 5′-GCAAGACAAGTTCAAGGGTTC-3′; vimentin forward, 5′-CCAGGCAAAGCAGGAGTC-3′ and reverse, 5′-GGGTATCAACCAGAGGGAGT-3′; E-cadherin forward, 5′-TGCCCAGAAAATGAAAAAGG-3′ and reverse, 5′-GTGTATGTGGCAATGCGTTC-3′; N-cadherin forward, 5′-TGGACCATCACTCGGCTTA-3′ and reverse, 5′-ACACTGGCAAACCTTCACG-3′; and GAPDH forward and reverse, 5′-ACCCAGAAGACTGTGGATGG-3′; and reverse, 5′-TCTAGACGGCAGGTCAGGTC-3′. The triplicate comparative quantification cycle (Cq) values were averaged and the relative expression levels of the tissues were determined as 2−ΔCq, where ΔCq=Cq of miR-410-3p-Cq of U6, and the relative miR-410-3p or EMT-associated molecule expression levels were quantified by the 2−ΔΔCq method (27).
Cell culture and transfection
The human ARMS cell line PLA802 was obtained from the Cell Bank of Chinese Academy of Science (Shanghai, China). The human ERMS cell line RD, human ARMS cell line RH30 and the human skeletal muscle cells (HSkMC, ATCC® PCS-950-010™) were obtained from the American Type Culture Collection (Manassas, VA, USA) and maintained according to the manufacturer's protocol. PLA802 cells were cultured in RPMI 1640 basic medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin. RH30, RD and HSkMC cells were maintained in Dulbecco's Modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10%, 100 U/ml penicillin and 100 mg/ml streptomycin. All cells were cultured in a humidified 5% CO2 atmosphere culture at 37°C.
miR-410-3p and control miRNA (miR-control) plasmids were designed and synthesized by Shanghai GeneChem Co., Ltd. (Shanghai, China) by adding pri-mir-410 to a pcDNA3.1 vector. The sequence of pri-mir-410 was TCTCACCTTTGATGTCCCATCCGTCCTCAGGACTGCTTCCCGGGGGCAGCGCTGGCACCACGGGACGCGGCAGCCACGTTCTTGAGCCGATGGCACTCTGGGTACCTGAGAAGAGGTTGTCTGTGATGAGTTCGCTTTTATTAATGACGAATATAACACAGATGGCCTGTTTTCAGTACCGCTACCGCCCGGTGGTGTGCGGGCGCCACGCCTGAGGCGGGACTTTCCAGGGTACGTGAATGCATGGGCTGGTGTGACATTTGTCATCC. The recombinant pGV268/miR-410-3p plasmid was transformed into DH5α-competent cells (cat. no. CB140401; Tiangen Biotech Co., Ltd., Beijing, China), and a single clone was selected and cultured in lysogeny broth media with 1:1,000 ampicillin (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) overnight at 37°C. Plasmids were extracted using an Endo-free Plasmid Mini kit purchased from Omega Bio-Tek, Inc. (Norcross, GA, USA). The concentration and purity were measured using a Nanodrop Instrument (NanoDrop Technologies; Thermo Fisher Scientific, Inc., Pittsburgh, PA, USA). The plasmids were stored at −20°C prior to subsequent use. The RMS cell lines were transiently transfected with 1 µg plasmids using 2 µl Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h, according to the manufacturer's protocol.
Cell migration and invasion assays
A 24-well plate containing 8-µm pore size chamber inserts (Costar; Corning Inc., Corning, NY, USA) was used to evaluate tumor cell migration and invasion, according to the manufacturer's protocol. For the migration assay, 5×104 cells transfected for 24 h were seeded in the upper chamber. For the invasion assay, the membrane was coated with Matrigel to form a matrix barrier and 2×105 cells transfected for 48 h were placed in the upper chamber. Cells in 100 µl serum-free DMEM were carefully loaded onto each filter insert in the upper chamber and 600 µl DMEM with 10% FBS was added to each lower chamber. The cells were incubated at 37°C for 24 and 48 h for the migration and invasion assays, respectively. Subsequently, the filter inserts were removed from the chambers. Migrated or invaded cells were fixed and stained with 0.1% crystal violet in 4°C for 15 min, and 5 photographs were obtained and analyzed in each group using an inverted light microscope at ×400 magnification.
Cell proliferation assays
A Cell Counting Kit-8 assay (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was used to assess cell proliferation, according to the manufacturer's protocol. Following 0, 24, 48 and 72 h of transfection, the absorbance of the miR-410-3p-transfected RH30 cells at 450 nm was evaluated using an XMARK Microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). All experiments were repeated three times independently.
Apoptosis assay
Apoptosis was investigated by the one-step terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) apoptosis assay kit (cat. no. C1089; Beyotime Institute of Biotechnology, Haimen, China). Following 48 h of RH30 cell transfection, the cells were treated with 4% paraformaldehyde solution at room temperature for 1 h, washed once with phosphate buffered saline (PBS), then permeabilized by PBS containing 0.1% Triton X-100 for 5 min in an ice bath and incubated in test solution at 37°C for 1 h, according to the manufacturer's protocol. Subsequently, the positive cells were detected by fluorescent microscopy (Olympus BX51 microscope, Tokyo, Japan) at ×400 magnification using 488 nm excitation and 530 nm emission wavelengths. The cells with red fluorescence were defined as positive apoptotic cells. The percentage of apoptotic cells was calculated as the percentage of cells with positive TUNEL staining of five randomly selected fields in each slide; ≥100 cells were assessed in each field. The apoptotic rate was calculated as follows: Apoptotic rate=number of apoptotic cells/total number of cells ×100%.
The Cancer Genome Atlas (TCGA) data analysis
TCGA contains multimodal genomics, epigenomics and proteomics data for thousands of tumor samples across >20 types of cancer, including sarcoma (28). TCGA data analyses were performed using Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/), a web-based tool that delivers fast and customizable functionalities based on TCGA and Genotype-Tissue Expression data, developed by Tang et al (29). GEPIA provides key interactive and customizable functions, including patient survival analysis. Among the sarcoma samples that were not provided by the website, 262 samples possessed disease-free survival (DFS) data. The GEPIA used the log-rank test for the Kaplan-Meier survival analysis of the DFS data. The patients were divided equally into two groups by Snail expression level (high and low Snail expression groups) by GEPIA.
The cBioPortal for Cancer Genomics (30) (http://cbioportal.org) was used to evaluate the survival analysis of miR-410-3p copy number alterations (CNAs) in the sarcoma data from TCGA, and was employed for exploring, visualizing and analyzing multidimensional cancer genomics data from TCGA (30). The miR-410-3p expression levels in RMS cells were further investigated using the National Cancer Institute Development Therapeutics Program (DTP) website (31) (https://sarcoma.cancer.gov/sarcoma/webpages/searchCriteria.xhtml).
Snail expression in datasets
Snail expression was compared between RMS tissue samples and normal samples by analyzing mRNA profiling datasets. The high-throughput sequencing dataset GSE108022 was downloaded from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) database (32). This dataset included RNA-seq processed data for 106 samples, including 5 normal muscle samples and 101 RMS samples. In addition, the DESeq2 package was used to handle the GSE108022 dataset (33).
Western blot analysis
Transfected RD and RH30 cells were harvested and lysed using radioimmunoprecipitation assay buffer (Beijing Solarbio Science & Technology Co., Ltd.). Protein concentrations were determined using a Nanodrop 2000 spectrophotometer (Thermo Scientific, USA) by measuring optical density at 280 nm. Proteins (50 µg/lane) were separated on 10% SDS-PAGE gels, electrotransferred onto polyvinylidene fluoride membranes and immersed in a blocking solution containing 5% non-fat milk and 0.1% Tween-20 at room temperature for 1.5 h. Following blocking, the membranes were incubated with antibodies targeting E-cadherin (rabbit-derived; 1:1,000; 135 kDa; cat. no. 3195; Cell Signaling Technology, Inc., Danvers, MA, USA), N-cadherin (rabbit-derived; 1:1,000; 140 kDa; cat. no. 13116; Cell Signaling Technology, Inc.), Snail (rabbit-derived; 1:1,000; 29 kDa; cat. no. 3879; Cell Signaling Technology, Inc.), Slug (rabbit-derived; 1:1,000; 30 kDa; cat. no. 9585; Cell Signaling Technology, Inc.), Bcl-2-associated X protein (bax; rabbit-derived; 1:1,000; 21 kDa; cat. no. ab32503; Abcam, Cambridge, UK), p53 (mouse-derived; 1:1,000; 53 kDa; cat. no. ab16465; Abcam), cleaved-caspase 3 (rabbit-derived; 1:1,000; 17 kDa; cat. no. 9664; Cell Signaling Technology, Inc.), cleaved-poly (ADP-ribose) polymerase (PARP; rabbit-derived; 1:1,000; 25 kDa; cat. no. ab32064; Abcam), Bcl-2 (rabbit-derived; 1:1,000; 25 kDa; cat. no. AB112; Beyotime Institute of Biotechnology) and β-actin (mouse-derived; 1:1,000; 40 kDa; cat. no. TA-09; OriGene Technologies, Inc., Beijing, China) at 4°C overnight. Subsequently, the secondary antibodies horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G (IgG; 1:1,000; cat. no. TA130003; OriGene Technologies, Inc.) or HRP-conjugated goat anti-rabbit IgG (1:5,000; cat. no. TA140003; OriGene Technologies, Inc.) were added for 2 h at room temperature. Finally, detection was performed using an enhanced chemiluminescence kit (Thermo Fisher Scientific, Inc.).
Statistical analysis
SPSS 19.0 (IBM Corp., Armonk, NY, USA) was used for all statistical analysis. Data are presented as the mean ± standard deviation. One-way analysis of variance followed by the least significant difference post hoc test was used to determine statistical significance. P<0.05 was considered to indicate a statistically significant difference. All figures were created with GraphPad Prism 7.0 (GraphPad Software, Inc., La Jolla, CA, USA).
Results
miR-410-3p is endogenously expressed at low levels in RMS tissues and cell lines
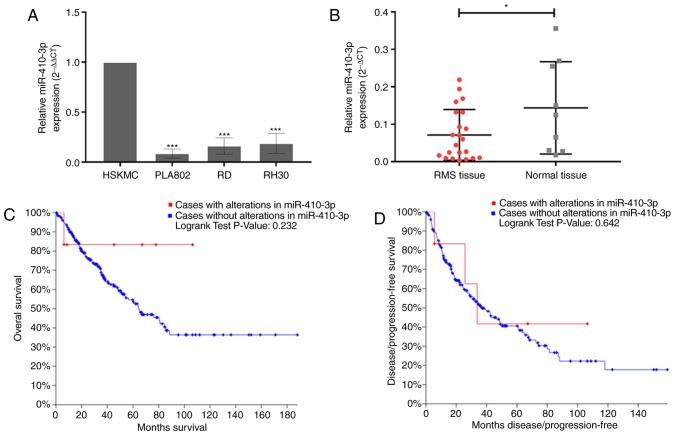
RT-qPCR was performed to determine the expression levels of miR-410-3p in three RMS cell lines. miR-410-3p was revealed to exhibit significantly low expression levels in the three RMS cell lines compared with HSkMCs (Fig. 1A). The miR-410-3p expression levels in RMS cells were further investigated using the DTP website. The miR-410-3p expression levels in RH30, RD and normal human skeletal muscle cells was 5.64, 5.94 and 6.59, respectively, which supported the aforementioned results. In addition, the expression of miR-410-3p in tissues were consistent with that in cells (Fig. 1B). Subsequently, the cBioPortal for Cancer Genomics (30) was used to investigate the association between overall survival, DFS and miR-410 CNAs in sarcoma using data obtained from TCGA. No significant associations were identified between miR-410-3p CNAs and survival (Fig. 1C and D). However, according to the survival curve, patients with altered miR-410-3p expression (red line) exhibited an improved overall survival rate. Therefore, changes of miR-410-3p may serve as tumor regulatory factors in patients.
Figure 1.
RMS cells and tissues exhibit low miR-410-3p expression levels. (A) The expression levels of miR-410-3p were significantly lower in the three RMS cells, PLA802, RD and RH30, compared with HSkMCs, as revealed by RT-qPCR analysis. U6 expression was used as a control for normalization. Three independent experiments were performed. ***P<0.001 vs. HSkMC. (B) The expression levels of miR-410-3p were significantly lower in 22 RMS tissue samples compared with nine normal skeletal muscle tissue samples, as revealed by RT-qPCR analysis. U6 expression was used as a control for normalization. Three independent experiments were performed. *P<0.05. (C and D) The associations between copy number alterations of miR-410 and overall or disease-free survival of patients with sarcoma were obtained using the cBioPortal database. Red lines represent cases with miR-410 alterations and the blue lines represent cases without miR-410 alterations. RMS, rhabdomyosarcoma; miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; HSkMC, human skeletal muscle cell.
Upregulated miR-410-3p inhibits the invasion and migration of RMS cells
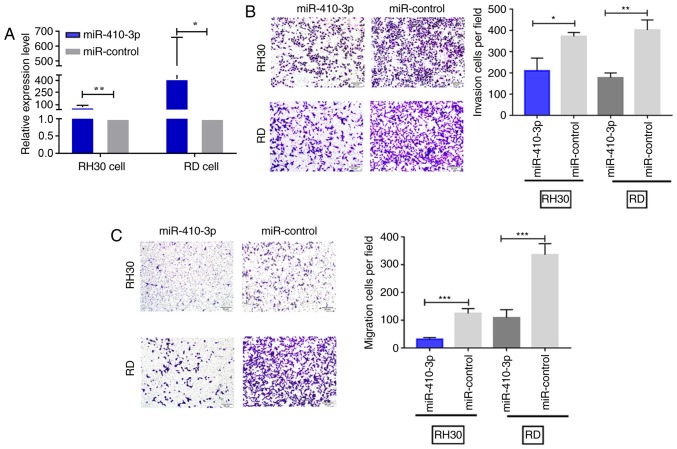
When transfected with the miR-410-3p plasmid, RD and RH30 cells exhibited significantly higher expression levels of miR-410-3p compared with cells transfected with miR-control (Fig. 2A). A Transwell assay was then used to investigate the cell invasion and migration abilities in vitro. The number of invasive and migratory cells was significantly lower in the miR-410-3p-transfected group compared with the control group (P<0.05; Fig. 2B and C).
Figure 2.
miR-410-3p inhibits RMS cell invasion and migration in vitro. (A) The miR-410-3p expression levels were measured in RMS cells transiently transfected with miR-410-3p plasmid. U6 was used as an internal reference. (B and C) The invasion and migration potentials were significantly lower for RH30 and RD cells in the transfected group compared with the control group. All experiments were performed three times. Representative images are presented. Scale bar, 50 µm; magnification, ×400. *P<0.05, **P<0.01 and ***P<0.001. miR, microRNA; RMS, rhabdomyosarcoma.
Upregulated miR-410-3p inhibits the expression of molecules associated with EMT
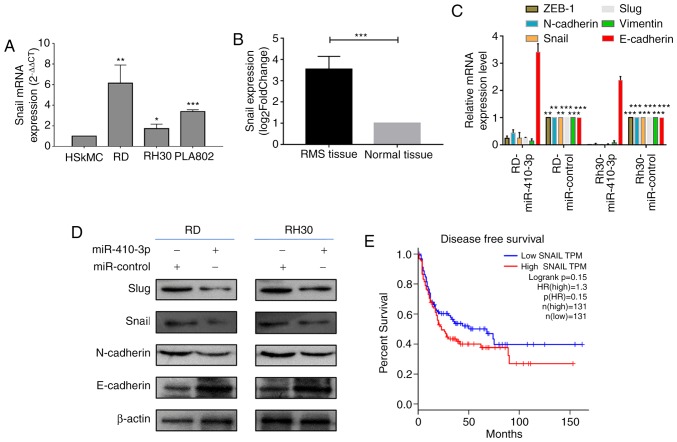
In a previous study, miR-410-3p inhibited EMT in breast cancer cells by targeting Snail (20). Therefore, the current study analyzed the expression of Snail in RMS cells. The expression levels of Snail mRNA were significantly higher in the three RMS cells, RD, RH30 and PLA802, compared with HSkMCs (Fig. 3A). Subsequently, the high-throughput sequencing dataset GSE108022 was downloaded and Snail expression in this dataset was analyzed. The expression levels of Snail were identified to be significantly higher in RMS tissues compared with normal tissues (Fig. 3B). As demonstrated in Fig. 3C and D, the mRNA and protein expression levels of Snail notably decreased following the overexpression of miR-410-3p in RD and RH30 cells. To identify the association of Snail expression with patient survival, the present study analyzed the mRNA expression profiles of patients with sarcoma whose DFS data were available in the sarcoma dataset of TCGA (Fig. 3E). Patients with low Snail expression exhibited increased survival; however, this was not significantly higher compared with patients exhibiting upregulated Snail expression (P=0.15). It may be suggested that miR-410-3p inhibits RMS cell invasion and migration by targeting Snail.
Figure 3.
Upregulated miR-410-3p inhibits the expression of EMT-associated molecules. (A) RT-qPCR was used to investigate Snail mRNA expression levels in HSkMCs and the three RMS cell lines. GADPH was used as an internal reference. *P<0.05, **P<0.01 and ***P<0.001 vs. HSkMC. (B) miR-410-3p expression levels in RMS (n=101) and normal skeletal muscle tissues (n=5) were obtained from the GSE108022 dataset. ***P<0.001 (C) RT-qPCR was used to determine mRNA expression levels of EMT-associated molecules, including Snail, Slug, zinc finger E-box binding homeobox 1 (ZEB1), vimentin, E-cadherin and N-cadherin, in RMS cells following 48 h of transfection with miR-410-3p. GADPH was used as an internal reference. **P<0.01 and ***P<0.001 vs. corresponding control. (D) Western blot analysis was used to detect the expression of EMT-associated molecules in RMS cells following 48 h of transfection with miR-410-3p. β-actin was used an internal control. (E) Kaplan-Meier survival analysis of the DFS of patients with sarcoma whose DFS information was available in The Cancer Genome Atlas sarcoma dataset. The patients were divided into high and low Snail expression groups. No significant difference was observed in the DFS rate between the two groups. All experiments were performed three times. miR, microRNA; EMT, epithelial-mesenchymal transition; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; HSkMC, human skeletal muscle cells; DFS, disease free survival; RMS, rhabdomyosarcoma; HR, hazard ratio; ZEB-1, zinc finger E-box binding homeobox 1.
RT-qPCR analysis revealed the mRNA expression levels of molecules associated with EMT (Fig. 3C). Compared with cells transfected with miR-control, RD and RH30 cells transfected with miR-410-3p exhibited significantly lower mRNA expression levels of Slug, ZEB-1, Vimentin and N-cadherin (P<0.05), and significantly higher expression levels of E-cadherin (P<0.05; Fig. 3C). Western blot analysis revealed that miR-410-3p overexpression enhanced the expression of the epithelial marker E-cadherin, but decreased that of mesenchymal markers Slug and N-cadherin in RD and RH30 cells (Fig. 3D). Therefore, it can be suggested that miR-410-3p inhibits tumor invasion and migration by regulating EMT-associated proteins in RMS cells.
Upregulated miR-410-3p suppresses proliferation and promotes RMS apoptosis
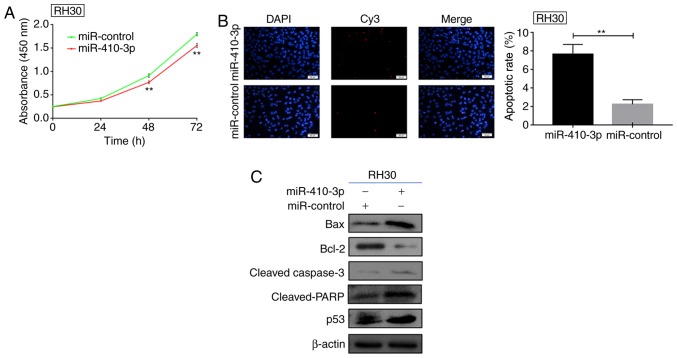
Following the transfection of RH30 cells with miR-410-3p or control plasmids, the effect of miR-410-3p on proliferative ability was assessed by a CCK-8 assay. Exogenous expression of miR-410-3p significantly inhibited RH30 cell proliferation 48 and 72 h following transfection compared with the control (Fig. 4A). Subsequently, a TUNEL assay was employed for the detection of apoptotic RH30 cells (Fig. 4B). The cells with overexpressed miR-410-3p demonstrated a significantly increased apoptotic rate. Western blot analysis was used to detect the expression of apoptotic proteins following 48 h of transfection with miR-410-3p. Exogenous miR-410-3p expression promoted the expression of apoptotic proteins, including bax, cleaved-caspase 3, cleaved PARP and p53, and inhibited the expression of Bcl-2 protein (Fig. 4C). This suggests RH30 cells may have underwent apoptosis as reflected by alterations in the expression of the aforementioned proteins. In summary, miR-410-3p was identified to be downregulated in RMS, promote apoptosis and suppress the proliferation of RMS cells in vitro, which indicated that miR-410-3p may serve as a tumor suppressor in RMS.
Figure 4.
Upregulated miR-410-3p suppresses proliferation and enhances apoptosis. (A) A Cell Counting Kit-8 assay demonstrated the proliferative ability of RH30 cells transfected with miR-410-3p. **P<0.01 vs. miR-control. (B) Terminal deoxynucleotidyl transferase dUTP nick end labeling assay indicated the apoptotic rate of RH30 cells 48 h post-transfection. Representative images are presented and the histogram represents the quantified apoptotic rates. Scale bars, 50 µm. **P<0.01. (C) Apoptotic proteins were detected by western blot analysis in the control and transfected RH30 cells. The expression levels of bax, cleaved-caspase 3, cleaved PARP and p53 were upregulated following transfection with miR-410-3p; however, the expression of Bcl-2 decreased. β-actin was used as an internal control. All experiments were repeated three times. miR, microRNA; bax, Bcl-2-associated X protein; PARP, poly(ADP-ribose) polymerase.
Discussion
RMS is one of the most common types of STS in children and exhibits a poor prognosis (34); however, to the best of our knowledge, the invasion and metastasis mechanisms of RMS remain to be clarified. Numerous miRNAs suppress various cancer-associated genes by acting as oncogenes or tumor suppressors (35). Endogenously low expression of miR-410-3p has been demonstrated in various types of tumor, including osteosarcoma (18), cholangiocarcinoma (36), pancreatic cancer (19) and breast cancer (20), which is consistent with the current results. In addition, analysis using the DTP website also confirmed low expression of miR-410-3p in RMS cells. The cBioPortal for Cancer Genomics revealed that the miR-410 alterations may act as regulatory factors of sarcoma in patients. miR-410-3p serves different roles in various types of tumor; however, the role of miR-410-3p in RMS remains unclear.
miR-410-3p inhibits cell invasion and migration in numerous cancer types (18–20,36,37), including pancreatic cancer (19), breast cancer (20) and cholangiocarcinoma (36). The present results demonstrated that upregulated miR-410-3p suppressed the proliferation and promoted RMS cell apoptosis by regulating the expression of apoptotic proteins, as is the case in cholangiocarcinoma cells (36) and human colorectal cancer cells (21). Furthermore, miR-410-3p was revealed to inhibit the invasion and migration abilities of RMS cells.
EMT and its reverse process, mesenchymal-epithelial transition, serve central roles during early embryonic development (38). However, EMT also serves a critical role in tumor invasion and metastasis (39,40). A number of studies have reported that sarcoma aggressiveness is associated with key factors of the phenotypic plasticity of tumors (41), including Snail, of which high expression was associated with reduced DFS of patients with sarcoma (42), and ZEB1, which is upregulated in patients with metastasized osteosarcoma compared with those without metastasis (43). The current results indicated that upregulated miR-410-3p inhibits the expression of EMT-associated molecules; Snail, Slug and N-cadherin were downregulated, and E-cadherin was upregulated. Epithelial-derived tumors can activate associated embryonic pathways, while mesenchymal tumors transform into increased epithelial phenotypes (44). As of the role of EMT at the onset of the metastatic process, regulating EMT in tumors is considered a promising strategy for inhibiting metastasis and improving the survival of patients with cancer (45). miR-410-3p inhibits EMT in breast cancer cells by targeting Snail (20). The expression profiling datasets revealed that Snail was upregulated in RMS tissues and Snail mRNA was overexpressed in RMS cells. When miR-410-3p was upregulated in RMS cells, the mRNA and protein expression levels of Snail were notably downregulated. Therefore, it can be suggested that miR-410-3p may inhibit the invasion and migration of RMS cells by targeting Snail, which may be beneficial for the development of drugs.
miR-27a overexpression has been demonstrated to promote RMS cell proliferation by directly targeting retinoic acid α receptor and retinoic X receptor α (46). miR-206 can mediate RMS differentiation by directly regulating SET and MYND domain-containing protein 1 and glucose 6-phosphate dehydrogenase (47). In addition, by targeting transcription factors of early growth response-1, miR-183 promotes the migration of RMS cells (48). Therefore, growing evidence suggests that the aberrant expression of miRNAs serves important roles in diverse biological processes, including development, differentiation, growth and metabolism (10,49). The current results demonstrated that miR-410-3p overexpression can inhibit RMS cell proliferation and Bcl-2 protein expression, induce apoptosis and promote the expression of bax, cleaved-caspase 3, cleaved PARP and p53.
It was hypothesized that miR-410-3p inhibits the invasion and migration of RMS by inhibiting EMT. Therefore, the current study first detected the expression of miR-410-3p in RMS tissues and cells, and identified that miR-410-3p expression was reduced in RMS tissues and cells compared with the healthy controls. Therefore, the low expression of miR-410-3p may exhibit a potential role in RMS development. Furthermore, exogenous miR-410-3p expression was revealed to induce apoptosis and inhibit proliferation by enhancing the expression of pro-apoptotic proteins, and suppress RMS cell invasion and migration by regulating the EMT process.
In conclusion, miR-410-3p overexpression was proposed to suppress invasion, migration, proliferation and the EMT process, and promote apoptosis and apoptotic factor expression in RMS cells. These findings may provide a novel insight for the identification of potential therapeutic targets for the treatment of RMS.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant nos. 81660441 and 81460404).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
LZ and YP performed the experiments and wrote the manuscript. CL and FL designed the study and edited the manuscript. XC, WJ, WC and YL collected and analyzed the data.
Ethics approval and consent to participate
The study was approved by the institutional Ethics Committee of the First Affiliated Hospital of Shihezi University School of Medicine (Shihezi, China) and conducted in accordance with the ethical guidelines of the Declaration of Helsinki. All patients involved in the study provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jo VY, Doyle LA. Refinements in sarcoma classification in the current 2013 World Health Organization classification of tumours of soft tissue and bone. Surg Oncol Clin N Am. 2016;25:621–643. doi: 10.1016/j.soc.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Toro JR, Travis LB, Wu HJ, Zhu K, Fletcher CD, Devesa SS. Incidence patterns of soft tissue sarcomas, regardless of primary site, in the surveillance, epidemiology and end results program, 1978–2001: An analysis of 26,758 cases. Int J Cancer. 2006;119:2922–2930. doi: 10.1002/ijc.22239. [DOI] [PubMed] [Google Scholar]
- 3.Pastore G, Peris-Bonet R, Carli M, Martínez-García C, Sánchez de Toledo J, Steliarova-Foucher E. Childhood soft tissue sarcomas incidence and survival in European children (1978–1997): Report from the automated childhood cancer information system project. Eur J Cancer. 2006;42:2136–2149. doi: 10.1016/j.ejca.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 4.Weihkopf T, Blettner M, Dantonello T, Jung I, Klingebiel T, Koscielniak E, Lückel M, Spix C, Kaatsch P. Incidence and time trends of soft tissue sarcomas in German children 1985–2004-A report from the population-based German Childhood Cancer Registry. Eur J Cancer. 2008;44:432–440. doi: 10.1016/j.ejca.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 5.Punyko JA, Mertens AC, Baker KS, Ness KK, Robison LL, Gurney JG. Long-term survival probabilities for childhood rhabdomyosarcoma. A population-based. Cancer. 2005;103:1475–1483. doi: 10.1002/cncr.20929. [DOI] [PubMed] [Google Scholar]
- 6.Sultan I, Qaddoumi I, Yaser S, Rodriguez-Galindo C, Ferrari A. Comparing adult and pediatric rhabdomyosarcoma in the surveillance, epidemiology and end results program, 1973 to 2005: An analysis of 2,600 patients. J Clin Oncol. 2009;27:3391–3397. doi: 10.1200/JCO.2008.19.7483. [DOI] [PubMed] [Google Scholar]
- 7.Kelly KM, Womer RB, Sorensen PH, Xiong QB, Barr FG. Common and variant gene fusions predict distinct clinical phenotypes in rhabdomyosarcoma. J Clin Oncol. 1997;15:1831–1836. doi: 10.1200/JCO.1997.15.5.1831. [DOI] [PubMed] [Google Scholar]
- 8.Hron AJ, Asakura A. An examination of the role of transcriptional and posttranscriptional regulation in rhabdomyosarcoma. Stem Cells Int. 2017;2017:2480375. doi: 10.1155/2017/2480375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Rota R, Ciarapica R, Giordano A, Miele L, Locatelli F. MicroRNAs in rhabdomyosarcoma: Pathogenetic implications and translational potentiality. Mol Cancer. 2011;10:120. doi: 10.1186/1476-4598-10-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Xiao W, Chen W, Luo L, Ye S, Liu Y. The epigenetic modifier trichostatin A, a histone deacetylase inhibitor, suppresses proliferation and epithelial-mesenchymal transition of lens epithelial cells. Cell Death Dis. 2013;4:e884. doi: 10.1038/cddis.2013.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 14.Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 2010;70:9570–9580. doi: 10.1158/0008-5472.CAN-10-2074. [DOI] [PubMed] [Google Scholar]
- 15.Oh JS, Kim JJ, Byun JY, Kim IA. Lin28-let7 modulates radiosensitivity of human cancer cells with activation of K-Ras. Int J Radiat Oncol Biol Phys. 2010;76:5–8. doi: 10.1016/j.ijrobp.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 16.Liang YJ, Wang QY, Zhou CX, Yin QQ, He M, Yu XT, Cao DX, Chen GQ, He JR, Zhao Q. MiR-124 targets Slug to regulate epithelial-mesenchymal transition and metastasis of breast cancer. Carcinogenesis. 2013;34:713–722. doi: 10.1093/carcin/bgs383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eades G, Yao Y, Yang M, Zhang Y, Chumsri S, Zhou Q. miR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. J Biol Chem. 2011;286:25992–26002. doi: 10.1074/jbc.M111.229401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao D, Jia P, Wang W, Zhang G. VEGF-mediated suppression of cell proliferation and invasion by miR-410 in osteosarcoma. Mol Cell Biochem. 2015;400:87–95. doi: 10.1007/s11010-014-2265-2. [DOI] [PubMed] [Google Scholar]
- 19.Guo R, Gu J, Zhang Z, Wang Y, Gu C. MicroRNA-410 functions as a tumor suppressor by targeting angiotensin II type 1 receptor in pancreatic cancer. IUBMB Life. 2015;67:42–53. doi: 10.1002/iub.1342. [DOI] [PubMed] [Google Scholar]
- 20.Zhang YF, Yu Y, Song WZ, Zhang RM, Jin S, Bai JW, Kang HB, Wang X, Cao XC. miR-410-3p suppresses breast cancer progression by targeting Snail. Oncol Rep. 2016;36:480–486. doi: 10.3892/or.2016.4828. [DOI] [PubMed] [Google Scholar]
- 21.Liu C, Zhang A, Cheng L, Gao Y. miR410 regulates apoptosis by targeting Bak1 in human colorectal cancer cells. Mol Med Rep. 2016;14:467–473. doi: 10.3892/mmr.2016.5271. [DOI] [PubMed] [Google Scholar]
- 22.Ye X, Jiang F, Li Y, Mu J, Si L, Wang X, Ning S, Li Z. Retracted: Glabridin attenuates the migratory and invasive capacity of breast cancer cells by activating microRNA-200c. Cancer Sci. 2014;105:875–882. doi: 10.1111/cas.12426. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Li D, Yang Y, Zhu G, Liu X, Zhao M, Li X, Yang Q. MicroRNA-410 promotes cell proliferation by targeting BRD7 in non-small cell lung cancer. FEBS Lett. 2015;589:2218–2223. doi: 10.1016/j.febslet.2015.06.031. [DOI] [PubMed] [Google Scholar]
- 24.Shen S, Huang K, Wu Y, Ma Y, Wang J, Qin F, Ma J. A miR-135b-TAZ positive feedback loop promotes epithelial-mesenchymal transition (EMT) and tumorigenesis in osteosarcoma. Cancer Lett. 2017;407:32–44. doi: 10.1016/j.canlet.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, Yan D, Wu W, Zhu J, Ye W, Shu Q. MicroRNA-130a promotes the metastasis and epithelial-mesenchymal transition of osteosarcoma by targeting PTEN. Oncol Rep. 2016;35:3285–3292. doi: 10.3892/or.2016.4719. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Zheng L, Sun Y, Wang T, Wang B. Tangeretin enhances radiosensitivity and inhibits the radiation-induced epithelial-mesenchymal transition of gastric cancer cells. Oncol Rep. 2015;34:302–310. doi: 10.3892/or.2015.3982. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Y, Qiu P, Ji Y. TCGA-assembler: Open-source software for retrieving and processing TCGA data. Nat Methods. 2014;11:599–600. doi: 10.1038/nmeth.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grever MR, Schepartz SA, Chabner BA. The National Cancer Institute: Cancer drug discovery and development program. Semin Oncol 622–638. 1992 [PubMed] [Google Scholar]
- 32.Hayes MN, McCarthy K, Jin A, Oliveira ML, Iyer S, Garcia SP, Sindiri S, Gryder B, Motala Z, Nielsen GP, et al. Vangl2/RhoA signaling pathway regulates stem cell self-renewal programs and growth in rhabdomyosarcoma. Cell Stem Cell. 2018;22:414–427. doi: 10.1016/j.stem.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breneman JC, Lyden E, Pappo AS, Link MP, Anderson JR, Parham DM, Qualman SJ, Wharam MD, Donaldson SS, Maurer HM, et al. Prognostic factors and clinical outcomes in children and adolescents with metastatic rhabdomyosarcoma-a report from the Intergroup Rhabdomyosarcoma Study IV. J Clin Oncol. 2003;21:78–84. doi: 10.1200/JCO.2003.06.129. [DOI] [PubMed] [Google Scholar]
- 35.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 36.Palumbo T, Poultsides GA, Kouraklis G, Liakakos T, Drakaki A, Peros G, Hatziapostolou M, Iliopoulos D. A functional microRNA library screen reveals miR-410 as a novel anti-apoptotic regulator of cholangiocarcinoma. BMC Cancer. 2016;16:353. doi: 10.1186/s12885-016-2384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L, Zhang J, Feng Y, Li R, Sun X, Du W, Piao X, Wang H, Yang D, Sun Y, et al. MiR-410 regulates MET to influence the proliferation and invasion of glioma. Int J Biochem Cell Biol. 2012;44:1711–1717. doi: 10.1016/j.biocel.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 38.Yang J, Weinberg RA. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 40.Gavert N, Ben-Ze'ev A. Epithelial-mesenchymal transition and the invasive potential of tumors. Trends Mol Med. 2008;14:199–209. doi: 10.1016/j.molmed.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 41.Somarelli JA, Shetler S, Jolly MK, Wang X, Bartholf Dewitt S, Hish AJ, Gilja S, Eward WC, Ware KE, Levine H, et al. Mesenchymal-Epithelial transition in sarcomas is controlled by the combinatorial expression of MicroRNA 200s and GRHL2. Mol Cell Biol. 2016;36:2503–2513. doi: 10.1128/MCB.00373-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alba-Castellón L, Batlle R, Francí C, Fernández-Aceñero MJ, Mazzolini R, Peña R, Loubat J, Alameda F, Rodríguez R, Curto J, et al. Snail1 expression is required for sarcomagenesis. Neoplasia. 2014;16:413–421. doi: 10.1016/j.neo.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen A, Zhang Y, Yang H, Xu R, Huang G. Overexpression of ZEB1 relates to metastasis and invasion in osteosarcoma. J Surg Oncol. 2012;105:830–834. doi: 10.1002/jso.23012. [DOI] [PubMed] [Google Scholar]
- 44.Baum B, Settleman J, Quinlan MP. Transitions between epithelial and mesenchymal states in development and disease. Semin Cell Dev Biol. 2008;19:294–308. doi: 10.1016/j.semcdb.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida T, Ozawa Y, Kimura T, Sato Y, Kuznetsov G, Xu S, Uesugi M, Agoulnik S, Taylor N, Funahashi Y, Matsui J. Eribulin mesilate suppresses experimental metastasis of breast cancer cells by reversing phenotype from epithelial-mesenchymal transition (EMT) to mesenchymal-epithelial transition (MET) states. Br J Cancer. 2014;110:1497–1505. doi: 10.1038/bjc.2014.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tombolan L, Zampini M, Casara S, Boldrin E, Zin A, Bisogno G, Rosolen A, De Pittà C, Lanfranchi G. MicroRNA-27a contributes to rhabdomyosarcoma cell proliferation by suppressing RARA and RXRA. PLoS One. 2015;10:e0125171. doi: 10.1371/journal.pone.0125171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coda DM, Lingua MF, Morena D, Foglizzo V, Bersani F, Ala U, Ponzetto C, Taulli R. SMYD1 and G6PD modulation are critical events for miR-206-mediated differentiation of rhabdomyosarcoma. Cell Cycle. 2015;14:1389–1402. doi: 10.1080/15384101.2015.1005993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 2010;70:9570–9580. doi: 10.1158/0008-5472.CAN-10-2074. [DOI] [PubMed] [Google Scholar]
- 49.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.