Abstract
Chromosomal translocation is a key process in the oncogenic transformation of somatic cells. Previously, artificial induction of chromosomal translocation was performed using homologous recombination-mediated loxP labeling of target regions followed by Cre-mediated recombination. Recent progress in genome editing techniques has facilitated the easier induction of artificial translocation by cutting two targeted genome sequences from different chromosomes. The present study established a system to induce t(11;14)(q13;q32), which is observed primarily in multiple myeloma (MM) and involves the repositioning of the cyclin D1 (CCND1) gene downstream of the immunoglobulin heavy chain (IgH) constant region enhancers by translocation. The placing of tandem gRNAs designed to cut both the IgH Eµ and CCND1 15-kb upstream regions in lentiCRISPRv2 enabled the induction of chromosomal translocation in 293T cells, with confirmation by translocation-specific PCR and fluorescence in situ hybridization probing with IgH and CCND1. At the translocation junctions, small deletions and the addition of DNA sequences (indels) were observed in several clones. Cloned cells with t(11;14) exhibited slower growth and lower CCND1 mRNA expression compared to the parent cells, presenting the opposite phenomena induced by t(11;14) in MM cells, indicating that the silent IgH gene juxtaposed to CCND1 may negatively affect CCND1 gene expression and cell proliferation in the non-B lymphocyte lineage. Therefore, the present study achieved the induction of silent promoter/enhancer translocation in t(11;14)(q13;q32) as a preparatory experiment to study the role of IgH constant region enhancer-driven CCND1 overexpression in oncogenic transformation processes in B lymphocytes.
Keywords: multiple myeloma, CRISPR/CRISPR-associated protein 9, t(11;14)(q13;q32), immunoglobulin H enhancer, cyclin D1
Introduction
Chromosomal translocations have been detected in a variety of cancers. Certain types of translocation are known to cause specific cancers. Translocations include several types of fusion genes, for example, fusion genes in which a chimeric open reading frame produces a chimeric protein after inter-intronic recombination between two different genes, and promoter/enhancer translocation upstream of a different gene, with subsequent control of expression by the translocated promoter/enhancer (1,2).
Among fusion genes involved in hematopoietic malignancies, the first to be cloned was the BCR/ABL fusion gene from the Philadelphia chromosome, t(9;22), associated with chronic myelogenous leukemia, although dozens of translocated genes were already identified (3). Multiple myeloma (MM) results from malignantly transformed plasma cells and is reportedly associated with specific translocations, such as t(11;14), t(4;14), and t(14;16) (4–6). As a common feature, the enhancer region of the immunoglobulin heavy chain (IgH) gene is rearranged to the target gene; transcriptional activation of the gene in a B lymphocyte-specific manner results in its functioning as an oncogene. The most frequently detected translocation in MM is rearrangement between the constant region of the IgH gene on the long arm of chromosome 14 and the 5′-upstream region of the cyclin D1 (CCND1) gene on the long arm of chromosome 11. Similar translocations were initially identified in the locus of B-cell CLL/lymphoma 1 (Bcl1) in t(11;14)(q13;q32) (7,8). CCND1 is a key molecule of the cell cycle control machinery, involved in regulating the G1/S transition process (9). Cyclin activity is controlled by transcription level. In the middle of the G1 phase, transcriptional activation of the cyclin D1 gene results in an increase in protein production, which then complexes with and induces the kinase activity of cdk4 and cdk6, in turn stimulating cell cycle progression. CCND1 gene juxtaposed to the downstream of IgH constant region shows accelerated expression of CCND1 proteins by the effect of IgH super-enhancer in B lymphocyte-specific manner, therefore, t(11;14)-harboring cells indicate increased cell proliferation (10).
Sequencing of the junction point of t(11; 14)(q13; q32) in B-cell malignancies revealed a difference between B-cell lymphoma and MM (4,11,12). Two types of gene rearrangement mechanisms are involved in chromosomal translocations associated with B-cell malignancies: one that involves VDJ recombination mediated by recombination activating gene (RAG)1/2, and another that involves activation-induced cytidine deaminase (AID) in immunoglobulin class switching processes (3). Translocation in MM is induced by AID (13,14). The resulting differences in the origin of translocation-containing malignant cells in various B-cell differentiation stages cause differences in clinical condition (4,15).
Genome editing systems are now widely utilized in many research fields (16,17). Among these systems, the Streptococcus pyogenes type II clustered, regularly interspaced, short palindromic repeats (CRISPR)-Cas9 system enables efficient sequence-specific editing, enhancing the homologous recombination-mediated template replacement of target genes (18–20). Software that provides for effective visual target estimation of CRISPR/Cas9 in the genome database in combination with an evaluation score enhances the convenience of the system (21). Recent reports have described the induction of chromosomal translocations by simultaneous cutting of two specific DNA sequences using CRISPR-Cas9 (22–27) to engineer translocations that produce chimeric proteins from the fusion genes. Translocation of promotor/enhancer regions such as the B-cell lymphoma t(11;14) has not been reported, however.
In this study, we developed a CRISPR/Cas9-mediated genome editing system to induce the MM-specific chromosomal translocation t(11;14)(q13;q32) in cultured cells and confirmed the induction of a translocation identical to the experimental design. In addition, we analyzed the DNA sequence of the translocation junction, the gene expression and growth characteristics of t(11;14)-positive cells.
Materials and methods
Materials
Cell culture reagents, Dulbecco's modified Eagle's medium (DMEM) and penicillin/streptomycin, were obtained from Nakalai Tesque (Kyoto, Japan). Plasmids used in this study, lentiCRISPRv2 (#98290), pCAG-EGxxFP (#50716) (28), pX330 Cetn1/1 (#50718), and pCAG-EGxxFP-Cetn1 (#50717), were obtained from Addgene (www.addgene.org). lentiCRISPRv2 puro was a gift from Brett Stringer. Synthetic oligonucleotides were obtained from Eurofin Genomics (Tokyo, Japan). DNA iso, RNAiso, Guide-it mutation detection kit, and pMD20-T TA cloning vector were purchased from Takara bio (Kyoto, Japan). Thunderbird® SYBR qPCR Mix, KOD-PLUS, and ReverTraAce were obtained from Toyobo (Tokyo, Japan). PEImax 40000 was obtained from Polyscience Inc. (Warrington, PA, USA).
Cell culture, transfection, and virus packaging
293 and 293T (SV40 T antigen introduced 293, 293T) cells originated from Japanese Collection of Research Bioresources Cell Bank were cultured in Dulbecco's modified Eagle medium containing 10% fetal calf serum and incubated in a humidified chamber at 37°C with 95% air and 5% CO2. These cells were confirmed to be mycoplasma free. Plasmid transfection was carried out using PEImax 40000, following the manufacturer's protocol. For lentivirus packaging, plasmids were introduced into 293T cells, with subsequent recovery of the culture supernatant. Virus particles were recovered by centrifugation of the supernatant following addition of PEG6000 and NaCl (29).
CRISPR target localization
CRISPRscan (www.crisprscan.org/) (21) was used to localize the 20-bp target sequences of the IgH Eµ region and CCND1 gene 5′-upstream region on the UCSC genome browser (http://www.ucsc.genome.edu/). Candidate gRNAs without potential off-targets were selected using Cutting Frequency Determination score (30) indicated by CRISPRscan.
Plasmid construction
Double-stranded oligonucleotides for the guide RNA of IgH Eµ (5′-CACCGGACTGGCCTAGCGGAGGCTC-3′ and 5′-AAACGAGCCTCCGCTAGGCCAGTCC-3′ for candidate A, 5′-CACCGGAGAACATACCAAGCCCCAC-3′ and 5′-AAACGTGGGGCTTGGTATGTTCTCC-3′ for candidate B) and CCND1 (5′-CACCGGGGGTAGGAAGCCTCGGCTGTGG-3′ and 5′-AAACCCACAGCCGAGGCTTCCTACCCCC-3′ for candidate A, 5′-CACCGGTGGCGAGGTGGGACCGCGG-3′ and 5′-AAACCCGCGGTCCCACCTCGCCACC-3′ for candidate B) were ligated into lentiCRISPRv2 vector digested with BsmB1. For construction of pCAG-EGxxFP vectors to monitor gRNA activity, IgH or CCND1 gene target sequences were PCR-amplified from 293T cell genomic DNA using 5′-TTAGACAAGGGCGATGCCAG-3′ and 5′-TCAAGACCACTTTTCAACTACTCAC-3′ for IgH candidate A, 5′-TCATTACCACCCTCCACTACCT-3′ and 5′-CCACTAGAAGGGGAACTGGTC-3′ for IgH candidate B, 5′-CACATGCCCGAAGTCAAACC-3′ and 5′-ATCACCGAGATCAGAAGGCT-3′ for CCND1 candidate A, and 5′-CTTCTCACGAGCTGCCTTTG-3′ and 5′-GCTCATCACACAGCTTGACG-3′ for CCND1 candidate B with KOD-Plus DNA polymerase. After cloning into pMD20-T for DNA sequence confirmation, the target sequences were cloned into the pCAG-EGxxFP cleavage site. For the IgH-CCND1 tandem-lentiCRISPRv2 vector, the U6-IgH gRNA B expression cassette cloned into pMD20-T was PCR-amplified from IgH-B lentiCRISPRv2 using 5′-GCAGAGATCCAGTTTGGTTAAT-3′ and 5′-ACCTAGCTAGCGtATTCAAAAA-3′ with KOD-Plus DNA polymerase and then cloned into CCND1 B lentiCRISPRv2.
Homology-directed repair (HDR)-mediated EGxxFP repair monitoring of gRNA activity
IgH or CCND1 gRNA activity was monitored according to a previous report (28). Briefly, IgH (or CCND1, IgH-CCND1 tandem) lentiCRISPRv2 and pCAG-EGxxFP IgH (or CCND1) vectors were co-transfected into 293T cells. Two days later, EGFP fluorescence was observed using a fluorescence microscope (Axio Vert.A1, Zeiss, Oberkochen, Germany) and a flow cytometer (S3e cell sorter, Bio-Rad, Hercules, CA, USA). The fluorescence of co-transfected cells was compared to that of the positive control, pX330-Cetn1/1 and pCAG-EGxxFP Cetn1 combination, and the negative control, empty lentiCRISPRv2 and pCAG-EGxxFP. In flow cytometric analyses, mean fluorescence intensity of GFP-positive live cells was obtained using forward/side scatter-gating and FL1 window.
Confirmation of CRISPR genome editing activity
The genome editing activity of the IgH-CCND1 CRISPR vectors was confirmed by infecting 293T and 293 cells with lentiCRISPRv2 virus. These cells were selected as previous reports successfully established artificial translocation using CRISPR/Cas9 (22,25). Cells were infected with virus suspension and cultured for 14 days with puromycin. Colonies were picked, and DNA was recovered using a DNAiso kit according to the manufacturer's protocol. The genome regions of gRNA target sites were then PCR-amplified from puromycin-selected cells and parent cells using the primers used in EGxxFP vector construction. A Guide-IT mutation detection kit was used to detect mutations introduced into the genomic DNA of CRISPR target sites. Purified PCR products of selected and parental origin were mixed equivalently, denatured, reannealed, incubated with Guide-it nuclease, and electrophoresed to detect mismatch-directed cleavage. In addition, to confirm genome editing at DNA sequence level, PCR-amplified fragments were recombined into the pMD20-T TA cloning vector, and inserted DNA was sequenced by Fasmac Co. (Kanagawa, Japan).
Confirmation of translocation
PCR confirmation of translocation was performed using the IgH/CCND1 translocation specific primer pair (5′-AAGGGTGCGATGATGACCTAC-3′ for IgH side and 5′-AGCTGTTCTTGTAGTGGTGCC-3′ for CCND1 side) with Thunderbird® SYBR qPCR Mix and a Light Cycler Nano (Roche Diagnostics, Basel, Switzerland). For calibration of DNA content in quantitative PCR (Q-PCR), primers for ACTB gene (5′-AGAAAATCTGGCACCACACC-3′ and 5′-AACGGCAGAAGAGAGAACCA-3′) were used for reference. PCR condition was, denaturation of template: 94°C 120 sec, DNA amplification cycles: 50 cycles of (94°C 10 sec-62°C 10 sec-72°C 20 sec), melting temperature measurement: 72°C to 94°C at 0.1°C/sec. The presence of translocation-positive clones was assessed based on the melting temperature of PCR-amplified DNA compared with the positive control DNA. To confirm the efficiency of IgH/CCND1 CRISPR/Cas9-induced translocation, translocation-specific Q-PCR was performed using DNA of IgH/CCND1 lentiCRISPRv2-infected cells. Positive control DNA was amplified from genomic DNA 5′-TCATTACCACCCTCCACTAC-3′ and 5′-TTTGCTAGCCACTGGCATCGCCCT-3′ for the IgH side and 5′-GCTCATCACACAGCTTGACG-3′ and 5′-TTTGCTAGCGCGGTGGGGTCTTGTGTTG-3′ for the CCND1 side, and the two PCR-amplified fragments were recombined into a translocation-mimic PCR template (5′-IgH-CCND1−3′). PCR-amplified DNA of candidate positive cells was cloned into TA cloning vector, and inserted DNA was sequenced. DNA sequence alignment was performed using ApE (http://jorgensen.biology.utah.edu/wayned/ape/).
RT-qPCR
RNA was isolated from sub-confluent cultures of 293T and t(11;14) positive cells two days after inoculation using RNAiso reagent according to the manufacturer's instructions. cDNAs were synthesized from total RNA using random primers and RevTraAce reverse transcriptase. Quantitative PCR (Q-PCR) was performed using the primers 5′-GACCCCGCACGATTTCATTG-3′ and 5′-CTCTGGAGAGGAAGCGTGTG-3′ for CCND1 and 5′-ACACTTCTGCTCGTTGCCTT-3′ and 5′-ACACAAATGCTCCTCTCACC-3′ for MYEOV as the target and 5′-CACCAGGGCTGCTTTTAACTCT-3′ and 5′-TGGGATTTCCATTGATGACAAG-3′ for GAPDH as the reference with Thunderbird qPCR mix and a LightCycler Nano (n=3) using the same condition to DNA Q-PCR described above. Ratio of 2−Δ∆Cq values were calculated from the values obtained by LightCyclerNano Software (31).
Growth curve analysis
A total of 5×104 cells were inoculated into 35-mm dishes and cultured. The number of viable attached cells was determined using a dye-exclusion assay with trypan blue (n=3).
Statistical analysis
Quantitative data were presented as mean ± standard deviation obtained from three independent replicates. Statistical analyses were performed using Welch's t-test on Microsoft Excel version 14 on Windows 7. P<0.05 was considered to indicate a statistically significant difference.
Results
Construction of IgH/cyclinD1-specific CRISPR/Cas9
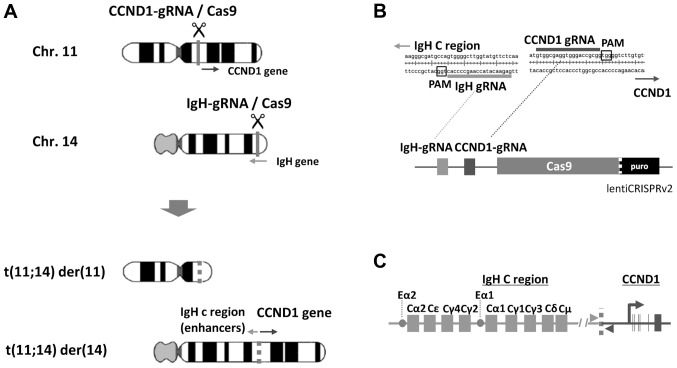
According to the scheme to obtain artificial induction of t(11;14) by cutting CCND1 upstream on chromosome (chr) 11 and IgH Eµ and Iµ regions on chr 14 (Fig. 1A), we designed two efficient gRNA sequences to express in one CRISPR/Cas9 vector (Fig. 1B) as described below. In order to mimic t(11;14) in MM (Fig. 1C), we searched target sequences of CRISPR/Cas9 around the previously reported junction sequence (32) using CRISPR Scan. As Ronchetti et al (32) were unable to identify clear clustering of junction sites of CCDN1, we searched the targets from 10 to 20 kb upstream of the protein-coding sequence. For IgH genes, Eµ and Iµ regions were searched. Candidate gRNA sequences predicted to exhibit high editing activity with minimal off-target activity were obtained. Four gRNA target sequences were selected for IgH and CCND1 genes, and synthetic oligonucleotides of these gRNA candidates were recombined into lentiCRISPRv2.
Figure 1.
Experimental design of t(11;14). (A) Schematic illustration of IgH-CCND1 CRISPR/Cas9-mediated induction of t(11;14). (B) gRNA target sequences and tandem-gRNA vector design. (C) Schematic illustration of the IgH-CCND1 fusion gene. CCND1 expression is driven by IgH C region enhancers in B lymphocytes. Two gray arrowheads at the junction (thick gray dotted line) indicate primer positions used for detection of fusion gene. IgH, immunoglobulin heavy chain; CCND1, cyclin D1; Chr, chromosome.
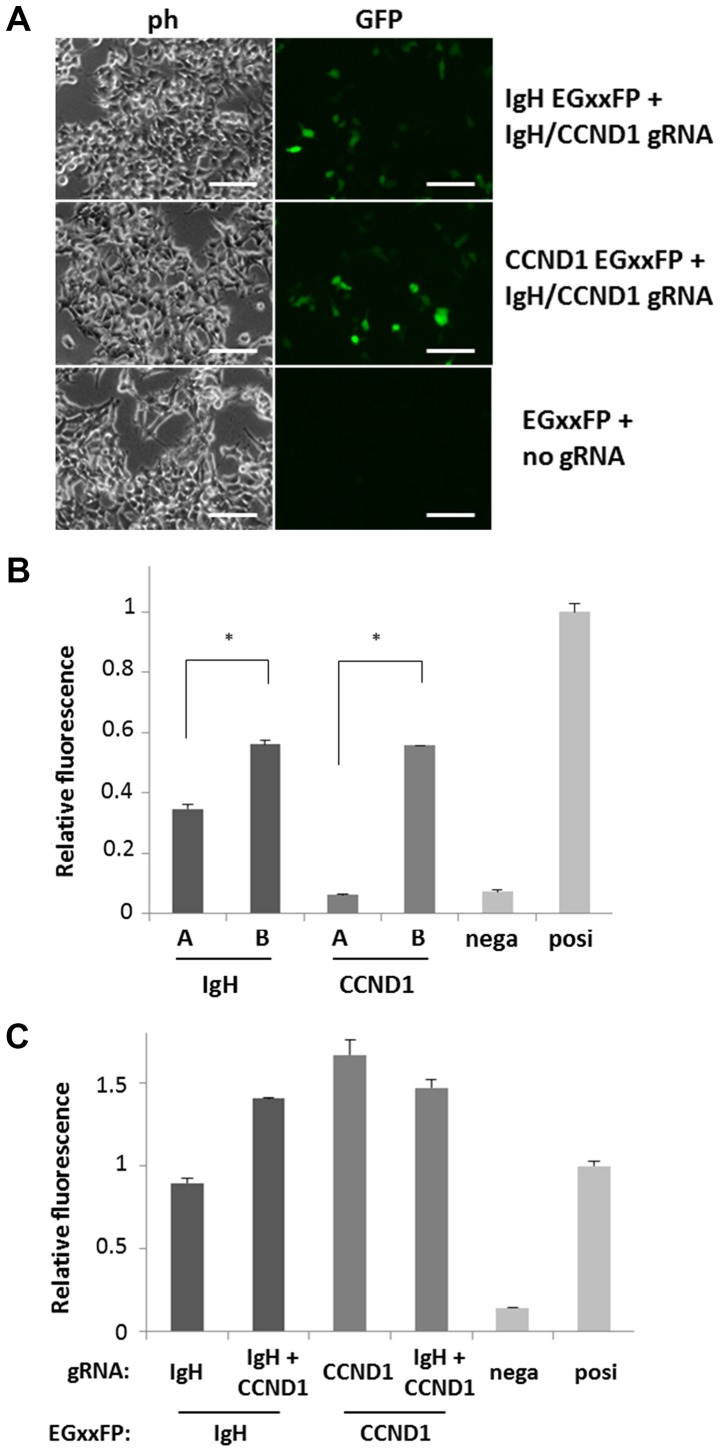
The potency of gRNA sequences was monitored using pCAG-EGxxFP. Both gRNA-coding lentiCRISPRv2 (IgH-A, -B, CCND1-A or -B) and corresponding gRNA target sequence-containing pCAG-EGxxFP (IgH-A, -B, CCND1-A or -B) were introduced into 293T cells, and editing activity was observed by recovery of EGFP fluorescence after HDR (Fig. 2A). EGFP fluorescence of cells into which the candidates were introduced was compared to that of positive control cells into which mouse Cetn1/1 pX330 and pCAG-EGxxFP Cetn1 were introduced, and candidates exhibiting fluorescence similar to that of Cetn1/1 cells were chosen (Fig. 2B). By comparing two gRNA candidates A and B for IgH or CCND1, selected gRNA sequences were 5′-GAGAACATACCAAGCCCCACTGG−3′ (IgH-B) for IgH, Chr14 genome position 105861042-105861064 in Genome Reference Consortium Human 38 (GRCh38), and 5′-GTGGCGAGGTGGGACCGCGGTGG−3′ (CCND1-B) for CCND1, Chr11 genome position 69627757-69627779 in GRCh38 (underlined sequences indicate protospacer adjacent motif, PAM).
Figure 2.
Confirmation of DNA cutting activity of the gRNA candidates. (A) 293T cells were introduced with target DNA (IgH, CCND1 or empty)-containing EGxxFP and gRNA (IgH and CCND1 or empty) expressing lentiCRISPRv2 vectors. Cells with high fluorescence indicate efficient gRNA activity to cut target DNA region. Scale bars, 50 µm. (B) gRNA activity are compared among IgH and CCND1 gRNA candidates-coding lentiCRISPRv2. Bars A and B indicate the negative control (empty lentiCRISPRv2) and positive control (pX330 Cetn1), respectively. Every gRNA vector was co-transfected with the gRNA target DNA-containing EGxxFP vector, and the intensity of cell fluorescence was analyzed by flow cytometry. Relative fluorescence compared to positive control is indicated (n=3). IgH-B and CCND1-B exhibited stronger fluorescence compared to IgH-A and CCND1-A, respectively. *P<0.001, as indicated. (C) Activity of single (IgH-B or CCND1-B) gRNA and dual (IgH-B and CCND1-B) gRNA expressing vectors were compared by co-transfecting with the gRNA target DNA-containing EGxxFP vectors (indicates as EGxxFP:). Negative and positive controls are the same as those in 2B. IgH, immunoglobulin heavy chain; CCND1, cyclin D1; ph, phase contrast micrograph; GFP, green fluorescent protein; nega, negative; posi, positive.
To induce chromosomal translocation with a single vector capable of editing both IgH and CCND1, the U6-CCND1 gRNA portion of CCND1-B lentiCRISPRv2 was PCR-amplified and cloned downstream of the U6-IgH gRNA of IgH-B lentiCRISPRv2 (Fig. 1B). The tandem gRNA (IgH-CCND1) lentiCRISPRv2 vector exhibited activity similar to that of EGxxFP fluorescence recovery compared with the IgH or CCND1 single-target vectors using IgH-B or CCND1-B EGxxFP vectors (Fig. 2C). There were some differences of activities between single and tandem gRNA vectors because of unknown reasons. However, we could detect similar activities to that of the positive control reproducibly using these vectors.
Induction of chromosomal translocation
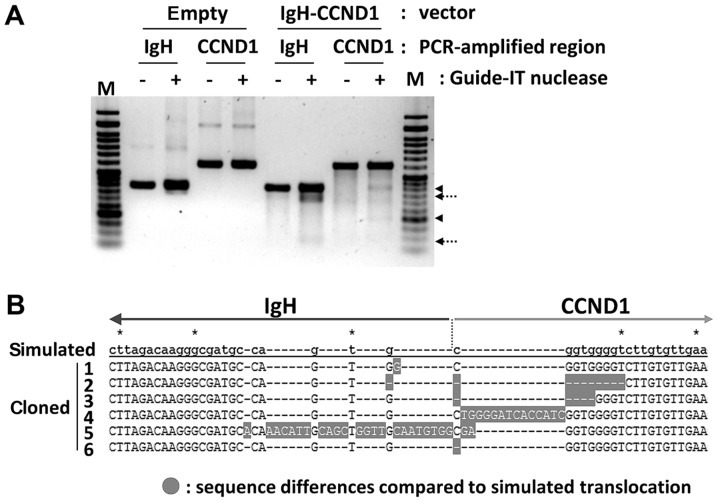
The dual gRNA lentiCRISPRv2 vector was packaged into a lentivirus to observe its translocation inducing activity, and then 293T cells were infected with the virus. Puromycin resistant cells were isolated after seven days. Genome editing activity was confirmed using a mismatch cleavage assay with PCR-amplified IgH and CCND1 genome regions from the DNA of puromycin-resistant cells. Mismatch cleavages by Guide-IT nuclease were indicated in vector-containing cells, demonstrating that Cas9-mediated double-strand breaks and repair by non-homologous end joining (NHEJ) had occurred (Fig. 3A). Translocation-specific PCR was then performed to confirm the presence of the designed translocation in puromycin-resistant cell DNA. Q-PCR analysis using translocation control DNA as the reference revealed a ratio of translocation-positive cells of 1/2,400 in 293T cells, whereas the ratio was 1/70 in 293 cells. Then, DNA bands of the expected length in translocation-positive cells were isolated and cloned into the TA vector, and the DNA sequence of several clones was determined. The observation of characteristic insertion and deletion of several base pairs in the DNA sequencing results for six clones showed that NHEJ had occurred (Fig. 3B). In addition, IgH and CCND1 gRNA target regions in the non-translocated allele were also PCR-amplified and cloned. DNA sequence of four clones each for IgH and CCND1 had small deletions showing that genome editing occurred even in the non-translocated alleles (data not shown).
Figure 3.
Confirmation of genome editing activity in IgH and CCND1 loci and induced translocation. (A) IgH and CCND1 target regions were PCR-amplified from infected 293T cells and forced using a mismatch cleavage assay. Fragments cleaved by Guide-IT nuclease are indicated by arrows with dotted lines (IgH), and arrowheads (CCND1). (B) t(11;14) translocation-specific PCR products from infected cells were cloned into the TA cloning vector, followed by DNA sequencing. Sequences of 6 clones are aligned against the simulated IgH/CCND1 sequence. The junction between IgH and CCND1 is indicated by a dotted line. Sequence differences relative to the simulated sequence are highlighted in gray boxes. IgH, immunoglobulin heavy chain; CCND1, cyclin D1; empty, empty lentiCRISPRv2-introduced; IgH-CCND1, IgH-CCND1 lentiCRISPRv2-introduced; IgH and CCND1 groups, PCR-amplified regions; - and +, Guide-IT nuclease addition; M, DNA marker.
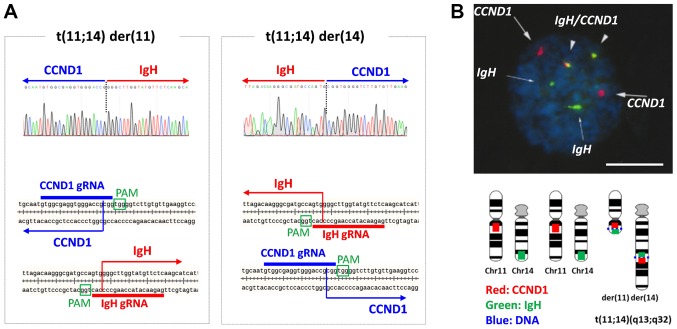
To clone translocation-positive 293T cells, Q-PCR was first used to identify wells containing positive cells in 48-well plates inoculated with 200 cells/well. Positive wells were then reseeded at 20 cells/well in 24-well plates, with subsequent identification of wells containing positive cells by Q-PCR as described above. Positive wells were inoculated into dishes at 100 cells/dish, and single colonies were then picked and expanded. We ultimately succeeded in isolating translocation-positive 293T cells using this serial dilution and PCR confirmation approach (data not shown). Direct sequencing of translocation-specific PCR products showed that the junction points of t(11;14) der(11) with the Chr11 centromere and der(14) with the Chr14 centromere were 3 bp from the PAM sequences shown in Fig. 3A. As reported, the Cas9 editing site is 3 bp from the PAM; direct rejoining occurred after editing in this cell clone. Reciprocal chromosomal translocation was also confirmed by fluorescence in situ hybridization (FISH) probing (IgH [green] and CCND1 [red] in Fig. 4B). In triploid karyotype of 293T cells, only one pair of chromosome 11 and 14 was engaged with t(11;14), whereas other two were not.
Figure 4.
Characterization of IgH-CCND1 CRISPR/Cas9-induced t(11;14). (A) t(11;14) translocation-specific PCR products from cloned cells were directly sequenced. Junctions in the sequence diagrams are indicated by dotted lines. CCND1 and IgH genome sequences included in der(11) and der(14) are indicated by blue (CCND1) and red (IgH) arrows with gRNA sequences. (B) t(11;14)-specific fluorescence in situ hybridization. One of the triploid Chr11 and Chr14 was reciprocally translocated. CCND1 and IgH probes are labeled with red and green boxes, respectively. Scale bar, 5 µm. PAM, protospacer adjacent motif; IgH, immunoglobulin heavy chain; CCND1, cyclin D1; Chr, chromosome.
Characteristics of t(11;14)-positive cells
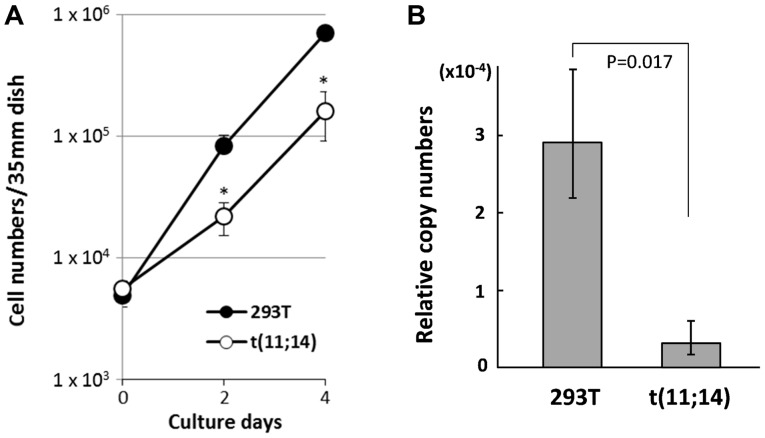
IgH enhancer-driven CCND1 overexpression by t(11;14) is one of the key factors in oncogenic cell proliferation in MM cells, because the immunoglobulin genes are highly active to produce antibodies only in the B lymphocyte lineage. Therefore, it is difficult to observe IgH enhancer-mediated activation of CCND1 gene unless the cells with t(11;14) differentiate to B lymphocytes. However, silent IgH gene in other cell species including 293, which is embryonal kidney origin, may negatively affect neighboring gene expression (33). And it is possible that such macroscopic changes induced by chromosomal translocation could alter the cellular characteristics through gene expression. Accordingly, we compared the cell proliferation rate and CCND1 gene expression in t(11;14)-positive cells to that of parental 293T cells. As shown in Fig. 5A, t(11;14)-positive cells exhibited slower proliferation than 293T cells. A comparison of gene expression of the CCND1 using GAPDH as control by RT-Q-PCR revealed lower CCND1 expression in the t(11;14)-positive cells compared with parental 293T cells (Fig. 5B). As a previous report suggested that the level of CCND1 expression is positively correlated with cell proliferation (34), it is possible that decreased CCND1 expression caused by the silent IgH enhancer repositioned in the upstream of CCND1 gene in t(11;14)-positive cells could result in slower growth compared with parental 293T cells in strong contrast to MM cells. In addition, MYEOV gene which locates in the upstream of CCND1 gene and is often overexpressed in MM cells by the translocated IgH variable region in t(11;14) der(11) (35) was also analyzed. But we couldn't detect MYEOV gene expression by RT-PCR both in t(11;14)-positive cells and parental 293T cells because of the restricted expression of this gene (gtexportal.org/home/gene/ENSG00000172927.3, The Genotype-Tissue Expression database).
Figure 5.
Characteristics of 293T cells harboring t(11;14). (A) Growth curves of t(11;14) 293T cells and parental cells. Viable cell numbers are indicated on days 0, 2, and 4 (n=3). Error bars indicate the standard deviation. *P<0.05 vs. 293T. (B) Reverse transcription-quantitative PCR analysis of CCND1. Relative copy numbers were calculated using GAPDH as a reference (n=3). CCND1, cyclin D1.
Discussion
In this study, we successfully induced artificial chromosomal translocation between the IgH constant region promoter/enhancer and the CCND1 protein-coding region using an IgH and CCND1-specific CRISPR/Cas9 genome editing system. In contrast to previous reports describing induction of growth-enhancing fusion genes using CRISPR/Cas9 (22–27), we encountered growth repression in the non-B lymphocyte target cells, which could have been associated with the transcriptionally silent IgH gene. These data suggest that selection pressure specific to translocation-positive cells is important to isolate the cells with such growth-suppressing translocations.
Recent reports have described efficient induction of targeted chromosomal translocation by introduction of a HDR template DNA which mimics translocation junction sequences (27,36). The template DNA carries a 5′-translocation donor gene fragment and 3′-recipient gene fragment separated by a selection marker, and is introduced into cells with routine translocation site-specific CRISPR/Cas9 vectors. After the two target sequences are specifically cut by Cas9s, breakpoints are repaired by HDR using the template DNA to form chromosomal translocation. Furthermore, the use of HSVtk for negative selection of off-target recombination of template DNA (37), knockout of p53 to suppress apoptotic cell death (38,39), and addition of NHEJ inhibitors to shift DNA double strand break repair system to HDR (40–42) also enhance HDR-template mediated genome editing. As application of these techniques can accelerate selective induction of difficult chromosomal translocations, we are planning to explore use of these techniques in combinations for future studies.
Although CCND1 expression in cells with translocations is thought to be associated with the transcriptional silence of IgH in cells other than B lymphocytes, we were able to obtain cells with t(11;14), suggesting that we should be able to induce this type of translocation in other cells. In addition, different types of immunoglobulin-associated translocations observed in B lymphocytic malignancies could be induced using a similar CRISPR/Cas9 system. For studying the process of myelomagenesis in the viewpoint of IgH enhancer-driven CCND1 overexpression, we plan to induce t(11;14) in B lymphocyte-derived iPS cells (BiPSCs) in which IgH genes are silent (43) using this technique. As we have already confirmed that our BiPSCs can be differentiated into hematopoietic stem cells (HSCs), we will check if t(11;14)-carrying BiPSCs are also capable of differentiating into HSCs and B lymphocyte lineage in vitro. Moreover, we will analyze the possible function of t(11;14) and CCND1 overexpression on B-cell differentiation and tumor development after transplantation into mouse bone marrow by comparing t(11;14)-BiPSC and parental BiPSC.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by grants-in-aid for Scientific Research (grant nos. 15K00543 and 16K18424) from the Japanese Ministry of Education, Culture, Sports, Science, and Technology.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
NT designed and performed the study. YA conducted the growth analysis and RT-qPCR. AY, YY and MS contributed to the cell culture and gene delivery experiments. AK and FK were involved in the design and construction of the CRISPR vectors. KK was involved in the experimental design. AS conceived and designed the study. All authors read and approved the manuscript, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Rabbitts TH. Chromosomal translocations in human cancer. Nature. 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 2.Nambiar M, Kari V, Raghavan SC. Chromosomal translocations in cancer. Biochim Biophys Acta. 2008;1786:139–152. doi: 10.1016/j.bbcan.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Lieber MR. Mechanisms of human lymphoid chromosomal translocations. Nat Rev Cancer. 2016;16:387–398. doi: 10.1038/nrc.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chesi M, Bergsagel PL, Brents LA, Smith CM, Gerhard DS, Kuehl WM. Dysregulation of cyclin D1 by translocation into an IgH gamma switch region in two multiple myeloma cell lines. Blood. 1996;88:674–681. [PubMed] [Google Scholar]
- 5.Chesi M, Nardini E, Brents LA, Schrock E, Ried T, Kuehl WM, Bergsagel PL. Frequent translocation t(4;14)(p16.3;q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3. Nat Genet. 1997;16:260–264. doi: 10.1038/ng0797-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chesi M, Bergsagel PL, Shonukan OO, Martelli ML, Brents LA, Chen T, Schrock E, Ried T, Kuehl WM. Frequent dysregulation of the c-maf proto-oncogene at 16q23 by translocation to an Ig locus in multiple myeloma. Blood. 1998;91:4457–4463. [PubMed] [Google Scholar]
- 7.Inaba T, Matsushime H, Valentine M, Roussel MF, Sherr CJ, Look AT. Genomic organization, chromosomal localization, and independent expression of human cyclin D genes. Genomics. 1992;13:565–574. doi: 10.1016/0888-7543(92)90126-D. [DOI] [PubMed] [Google Scholar]
- 8.Szepetowski P, Perucca-Lostanlen D, Gaudray P. Mapping genes according to their amplification status in tumor cells: Contribution to the map of 11q13. Genomics. 1993;16:745–750. doi: 10.1006/geno.1993.1257. [DOI] [PubMed] [Google Scholar]
- 9.Sherr CJ. Mammalian G1 cyclins. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- 10.Fonseca R, Blood EA, Oken MM, Kyle RA, Dewald GW, Bailey RJ, Van Wier SA, Henderson KJ, Hoyer JD, Harrington D, et al. Myeloma and the t(11;14)(q13;q32); evidence for a biologically defined unique subset of patients. Blood. 2002;99:3735–3741. doi: 10.1182/blood.V99.10.3735. [DOI] [PubMed] [Google Scholar]
- 11.Fenton JA, Pratt G, Rothwell DG, Rawstron AC, Morgan GJ. Translocation t(11;14) in multiple myeloma: Analysis of translocation breakpoints on der(11) and der(14) chromosomes suggests complex molecular mechanisms of recombination. Genes Chromosomes Cancer. 2004;39:151–155. doi: 10.1002/gcc.10304. [DOI] [PubMed] [Google Scholar]
- 12.Walker BA, Wardell CP, Johnson DC, Kaiser MF, Begum DB, Dahir NB, Ross FM, Davies FE, Gonzalez D, Morgan GJ. Characterization of IGH locus breakpoints in multiple myeloma indicates a subset of translocations appear to occur in pregerminal center B cells. Blood. 2013;121:3413–3419. doi: 10.1182/blood-2012-12-471888. [DOI] [PubMed] [Google Scholar]
- 13.Xu Z, Zan H, Pone EJ, Mai T, Casali P. Immunoglobulin class-switch DNA recombination: Induction, targeting and beyond. Nat Rev Immunol. 2012;12:517–531. doi: 10.1038/nri3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casellas R, Basu U, Yewdell WT, Chaudhuri J, Robbiani DF, Di Noia JM. Mutations, kataegis and translocations in B cells: Understanding AID promiscuous activity. Nat Rev Immunol. 2016;16:164–176. doi: 10.1038/nri.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nambiar M, Raghavan SC. How does DNA break during chromosomal translocations? Nucleic Acids Res. 2011;39:5813–5825. doi: 10.1093/nar/gkr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van der Oost J. Molecular biology. New tool for genome surgery. Science. 2013;339:768–770. doi: 10.1126/science.1234726. [DOI] [PubMed] [Google Scholar]
- 17.Gaj T, Gersbach CA, Barbas CF., III ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ. CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun. 2014;5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breese EH, Buechele C, Dawson C, Cleary ML, Porteus MH. Use of genome engineering to create patient specific MLL translocations in primary human hematopoietic stem and progenitor cells. PLoS One. 2015;10:e0136644. doi: 10.1371/journal.pone.0136644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang J, Zhang L, Zhou X, Chen X, Huang G, Li F, Wang R, Wu N, Yan Y, Tong C, et al. Induction of site-specific chromosomal translocations in embryonic stem cells by CRISPR/Cas9. Sci Rep. 2016;6:21918. doi: 10.1038/srep21918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lekomtsev S, Aligianni S, Lapao A, Bürckstummer T. Efficient generation and reversion of chromosomal translocations using CRISPR/Cas technology. BMC Genomics. 2016;17:739. doi: 10.1186/s12864-016-3084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reimer J, Knoss S, Labuhn M, Charpentier EM, Gohring G, Schlegelberger B, Klusmann JH, Heckl D. CRISPR-Cas9-induced t(11;19)/MLL-ENL translocations initiate leukemia in human hematopoietic progenitor cells in vivo. Haematologica. 2017;102:1558–1566. doi: 10.3324/haematol.2017.164046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanoli F, Tomishima M, Feng W, Lamribet K, Babin L, Brunet E, Jasin M. CRISPR-Cas9-guided oncogenic chromosomal translocations with conditional fusion protein expression in human mesenchymal cells. Proc Natl Acad Sci USA. 2017;114:3696–3701. doi: 10.1073/pnas.1700622114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mashiko D, Fujihara Y, Satouh Y, Miyata H, Isotani A, Ikawa M. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep. 2013;3:3355. doi: 10.1038/srep03355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cepko C. Large-scale preparation and concentration of retrovirus stocks. Curr Protoc Mol Biol. 1997 doi: 10.1002/0471142727.mb0912s37. Chapter 9: Unit 0.12. doi: 10.1002/0471142727.mb0912s37. [DOI] [PubMed] [Google Scholar]
- 30.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Ronchetti D, Finelli P, Richelda R, Baldini L, Rocchi M, Viggiano L, Cuneo A, Bogni S, Fabris S, Lombardi L, et al. Molecular analysis of 11q13 breakpoints in multiple myeloma. Blood. 1999;93:1330–1337. [PubMed] [Google Scholar]
- 33.Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJ, Grosveld FG, Knoch TA, Murre C. The 3D structure of the immunoglobulin heavy-chain locus: Implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zwijsen RM, Klompmaker R, Wientjens EB, Kristel PM, van der Burg B, Michalides RJ. cyclin D1 triggers autonomous growth of breast cancer cells by governing cell cycle exit. Mol Cell Biol. 1996;16:2554–2560. doi: 10.1128/MCB.16.6.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janssen JW, Vaandrager JW, Heuser T, Jauch A, Kluin PM, Geelen E, Bergsagel PL, Kuehl WM, Drexler HG, Otsuki T, et al. Concurrent activation of a novel putative transforming gene, myeov, and cyclin D1 in a subset of multiple myeloma cell lines with t(11;14)(q13;q32) Blood. 2000;95:2691–2698. [PubMed] [Google Scholar]
- 36.Spraggon L, Martelotto LG, Hmeljak J, Hitchman TD, Wang J, Wang L, Slotkin EK, Fan PD, Reis-Filho JS, Ladanyi M. Generation of conditional oncogenic chromosomal translocations using CRISPR-Cas9 genomic editing and homology-directed repair. J Pathol. 2017;242:102–112. doi: 10.1002/path.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gondo Y, Nakamura K, Nakao K, Sasaoka T, Ito K, Kimura M, Katsuki M. Gene replacement of the p53 gene with the lacZ gene in mouse embryonic stem cells and mice by using two steps of homologous recombination. Biochem Biophys Res Commun. 1994;202:830–837. doi: 10.1006/bbrc.1994.2005. [DOI] [PubMed] [Google Scholar]
- 38.Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med. 2018;24:927–930. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 39.Ihry RJ, Worringer KA, Salick MR, Frias E, Ho D, Theriault K, Kommineni S, Chen J, Sondey M, Ye C, et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med. 2018;24:939–946. doi: 10.1038/s41591-018-0050-6. [DOI] [PubMed] [Google Scholar]
- 40.Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, Liu H, La Russa M, Xie M, Ding S, Qi LS. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16:142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song J, Yang D, Xu J, Zhu T, Chen YE, Zhang J. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat Commun. 2016;7:10548. doi: 10.1038/ncomms10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li G, Zhang X, Zhong C, Mo J, Quan R, Yang J, Liu D, Li Z, Yang H, Wu Z. Small molecules enhance CRISPR/Cas9-mediated homology-directed genome editing in primary cells. Sci Rep. 2017;7:8943. doi: 10.1038/s41598-017-09306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawamura F, Inaki M, Katafuchi A, Abe Y, Tsuyama N, Kurosu Y, Yanagi A, Higuchi M, Muto S, Yamaura T, et al. Establishment of induced pluripotent stem cells from normal B cells and inducing AID expression in their differentiation into hematopoietic progenitor cells. Sci Rep. 2017;7:1659. doi: 10.1038/s41598-017-01627-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.