Abstract
Gastric cancer (GC) is one of the most common types of malignant tumor. Due to the lack of effective drugs and the emergence of chemotherapy resistance, patients with GC exhibit a poor prognosis and low survival rate. MicroRNAs (miRNAs/miRs) serve an important role in drug resistance of different types of cancer. They may be suitable for use as biomarkers in the diagnosis, treatment and prognosis of tumors. The present study aimed to investigate the molecular mechanism underlying the ability of miR-200c-3p to reverse drug resistance in a SGC7901/DDP GC cell line, particularly its effects on the ERCC excision repair 3, TFIIH core complex helicase subunit (ERCC3) and ERCC excision repair 4, endonuclease catalytic subunit (ERCC4) proteins in the nucleotide excision repair (NER) pathway. Reverse transcription-quantitative polymerase chain reaction demonstrated that miR-200c-3p expression in cisplatin-resistant SGC7901/DDP cells was lower than in parental SGC7901 cells, whereas the protein expression levels of ERCC3 and ERCC4 in these cells were higher by western blot analysis. In SGC7901/DDP-derived miR-200c-3p overexpressing cells, ERCC3 expression, ERCC4 expression and cisplatin resistance were decreased compared with in parental SGC7901/DDP cells and SGC7901/DDP-derived vector control cells. In SGC7901-derived miR-200c-3p knockdown cells, ERCC3 expression, ERCC4 expression and cisplatin resistance were increased compared with in parental SGC7901 cells and SGC7901-derived vector control cells. In conclusion, overexpression of miR-200c-3p may reverse drug resistance in the SGC7901/DDP GC cell line via downregulation of ERCC3 and ERCC4, which suggested this may be part of a mechanism involving the NER pathway.
Keywords: gastric cancer, microRNA-200c, cisplatin resistance, molecular mechanism
Introduction
Globally, gastric cancer (GC) is the third and fifth most common cause of cancer-associated mortality in men and women, respectively (1). Patients with GC usually exhibit no subjective symptoms in early stages, and the majority of cases are diagnosed only at advanced stages. Chemotherapy is the first-line treatment for patients with advanced GC, and platinum-based drugs are the most commonly used (2,3). However, primary and secondary drug resistance often lead to treatment failure and reduced survival rates. It has been reported that alterations in the DNA repair mechanism cause platinum resistance (4). The nucleotide excision repair (NER) pathway serves an important role in this process (5), and miRNAs can regulate this pathway (6,7). miRNAs are a class of non-coding, short, single-stranded regulatory RNAs. They do not encode proteins, but they can bind to the 3′-untranslated region (3′-UTR) of target mRNAs in a complementary manner and inhibit translation (8). Abnormal miRNAs expression is associated with drug resistance in numerous types of tumor. For example, in cisplatin-resistant SKOV3/DDP and A2780/DDP ovarian cancer cell lines, miR-152 and miR-185 can inhibit cell proliferation and promote apoptosis by inhibiting DNA methyltransferase 1, and thus, reverse platinum resistance (9). miR-508-5p can decrease ATP binding cassette subfamily B member 1 expression by inhibiting zinc ribbon domain-containing 1 to reverse multi-drug resistance of GC (10). In addition, miR-17-5p can render colon cancer cells insensitive to platinum-based treatment by inhibiting the expression of phosphate and tensin homolog (PTEN) (11).
It has previously been reported that miR-200c-3p-induced reversal of drug resistance and inhibition of proliferation in SGC7901/DDP GC cells is associated with the induction of E-cadherin, PTEN and B-cell lymphoma-2 (Bcl-2) associated X protein (BAX) protein expression, as well as the inhibition of protein kinase B (Akt) pathway activation and downregulation of Bcl-2 protein expression (12). Additionally, miRNAs are associated with DNA repair. Notably, miR-890 can inhibit the expression of mitotic arrest deficient 2 like 2, WEE1 G2 checkpoint kinase and XPC complex subunit, DNA damage recognition and repair factor, to delay DNA repair caused by ionizing radiation (IR), and as a result, prostate cancer cells exhibit increased sensitivity to IR (13). miR-145 can inhibit the activity of RAD18 E3 ubiquitin protein ligase to restrain DNA repair and reverse 5-flurouracil resistance of colon cancer cells (14). A previous study demonstrated that miR-192 in HepG2.2.15 liver cancer cells can inhibit the NER pathway through targeted regulation of ERCC excision repair 3, TFIIH core complex helicase subunit (ERCC3) and ERCC excision repair 4, endonuclease catalytic subunit (ERCC4) (7), but whether high miR-200c-3p expression in GC serves a role in the NER pathway is unknown. Therefore, the present study investigated the difference between SGC7901/DDP and SGC7901 cell lines with respect to NER ability, and measured miR-200c-3p expression. The present study revealed that upregulation or downregulation of miR-200c-3p expression can regulate the expression levels of key proteins in the NER pathway, including ERCC3 and ERCC4.
Materials and methods
Cell culture and lentiviral transduction
Human SGC7901/DDP and SGC7901 gastric adenocarcinoma cell lines were purchased from Nanjing KeyGen Biotech Co., Ltd. (Nanjing, China). Cells were grown in RPMI-1640 medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 U/ml streptomycin (all from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C in a humidified atmosphere with 5% CO2. To maintain the phenotype of cisplatin resistance, SGC7901/DDP cells were cultured in medium supplemented with 800 ng/ml cisplatin (Qilu Pharmaceutical Co., Ltd. Jinan, China). Lentiviral vectors carrying miR-200c-3p (pLV-miR-200c-3p), negative control (pLV-miR-200c-3p-NC) and the corresponding viruses [1×108 plaque forming units (PFU)] were all obtained from GeneCopoeia (Guangzhou, China). The lentiviruses were transfected into 293T cells for packaging, which was completed by GeneCopoeia prior to infecting GC cells. Lentiviral transduction to induce overexpression or knockdown of miR-200c-3p was performed according to the manufacturer's protocols. Specifically, cells were seeded in 12-well plates overnight to reach a density of 1×105 cells/well. Subsequently, cells were transduced at lentiviral multiplicity of infection 10 PFU/cell. After 72 h of transduction, SGC7901/DDP cells were cultured in medium supplemented with 2 µg/ml puromycin for 3 days. Similarly, lentiviral vector bearing a RNA interference sequence (5′-UCCAUCAUUACCCGGCAGUAUUA-3′) against miR-200c-3p was used to knockdown miR-200c-3p expression in SGC7901 cells. Following transduction, the cells were cultured in medium supplemented with 150 µg/ml puromycin for 5 days. Cell clones that survived the selection were used in subsequent experiments. The lentiviral vectors included green fluorescent protein or red fluorescent protein markers, which were used to monitor transduction efficiency under a fluorescence microscope. In summary, SGC7901/DDP-LV-NC-overexpression (NC-OE) cells and SGC7901/DDP-LV-miR-200c-3p-OE (miR-200c-3p-OE) cells were developed from SGC7901/DDP cells, whereas SGC7901-LV-miR-200c-3p-knockdown (miR-200c-3p-KD) cells and SGC7901-LV-NC-KD (NC-KD) cells were generated from SGC7901 cells.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from GC cell lines using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Total RNA concentration was evaluated at an absorbance of 260 nm using a NanoDrop-2000 spectrophotometer (NanoDrop; Thermo Fisher Scientific, Inc., Wilmington, DE, USA). RT-qPCR was performed on an Mx3000P qPCR system (Agilent Technologies, Inc., Santa Clara, CA, USA) with a SYBR Green single-step RT-qPCR kit (Biomics Biotechnologies Co., Ltd., Nantong, China) according to the manufacturer's protocols. U6 small nuclear RNA was selected as an endogenous control. The 25-µl RT-qPCR reaction system included 2 µl total RNA extract, 2X master mix, miR-200c-specific primers and U6-specific primers. PCR was performed under the following conditions: RT at 42°C for 30 min; 95°C denaturation for 10 min; 40 cycles of 95°C denaturation for 20 sec, 60°C annealing for 30 sec and 72°C extension for 30 sec; subsequently, fluorescent signals were collected at 72°C. The primer sequences were as follows: miR-200c, forward 5′-GGCGTAATACTGCCGGGTA-3′, reverse 5′-ATTGCGTGTCGTGGAGTCG-3′; and U6, forward 5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′. Data were analyzed using the following equation: ΔΔCq=Experimental (CqmiR-200-3p-Cqu6)-control (CqmiR-200-3p-Cqu6), and the relative expression levels of miR-200c-3p were calculated using the 2−ΔΔCq method (15). All experiments were performed in triplicate.
In vitro drug sensitivity test
SGC7901/DDP, SGC7901/DDP-LV-miR-200c-3p-OE, SGC7901/DDP-LV-NC-OE, SGC7901, SGC7901-LV-miR-200c-3p-KD and SGC7901-LV-NC-KD cells were suspended at a density of 1×105 cells/ml and seeded in 96-well plates, with each well containing 100 µl RPMI-1640 medium. After 24 h, freshly prepared cisplatin was added to SGC7901/DDP, SGC7901/DDP-LV-miR-200c-3p-OE and SGC7901/DDP-LV-NCscells at concentrations of 40, 20, 2, 0.2, 0.02 and 0.002 µg/ml; cisplatin was added to SGC7901, SGC7901-LV-miR-200c-3p-KD and SGC7901-LV-NC-KD at concentrations of 4, 2, 0.2, 0.02, 0.002 and 0.0002 µg/ml. A total of 48 h after the addition of cisplatin, original medium was replaced with 180 µl fresh medium, and 20 µl MTT (5 mg/ml) was added to each well. Cells were then cultured for 4 h at 37°C in a humidified atmosphere with 5% CO2, after which, the supernatant in the well was discarded. Subsequently, 150 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added to each well to dissolve crystals at 72°C for 10 min. Absorbance (490 nm) was measured using a spectrophotometer. The half maximal inhibitory concentration (IC50) of cisplatin was calculated according to relative cell survival. Each experiment was performed in triplicate.
Western blotting
Total protein was extracted from cells using radioimmunoprecipitation assay (Beyotime Institute of Biotechnology, Shanghai, China) lysis buffer containing protease inhibitor. Protein concentration was assessed using a Bicinchoninic Acid Protein Assay kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The primary antibodies against ERCC3 (1:2,000 dilution; cat. no. ab27317) and ERCC4 (1:1,000 dilution; cat. no. ab76948) (both from Abcam, Cambridge, UK) used were rabbit anti-human polyclonal antibodies. β-actin served as a loading control. Equal amounts of proteins (80 µg) per group were loaded and separated by 10 or 8% SDS-PAGE, transferred onto polyvinylidene fluoride membranes, and blocked with 5% non-fat milk for 1.5 h at room temperature. Membranes were incubated with the ERCC3, ERCC4 or β-actin (1:1,000 dilution; cat. no. ab8227; Abcam) primary antibodies overnight at 4°C. Membranes were washed three times with TBS-0.1% Tween (TBST; 10 min/wash) and then incubated with horseradish peroxidase-conjugated secondary antibodies (1:5,000 dilution; cat. no. 7074S; Shanghai Youningwei Biotechnology Co. Ltd., Shanghai, China) overnight at 4°C. Membranes were washed a further three times with TBST and signal detection was performed using a Super Enhanced Chemiluminescence Plus Detection reagent (Thermo Fisher Scientific, Inc.). Protein expression levels were normalized to β-actin, and the fold changes were calculated by densitometry (Tanon 2500 Fully Automatic Digital Gel Image Analysis system; Tanon Science and Technology Co., Ltd., Shanghai, China).
Statistical analysis
SPSS version 17.0 software (SPSS, Inc., Chicago, IL, USA) was used for data processing. All experiments were performed in triplicate, and all data are expressed as the means ± standard deviation. A Student's t-test was used to compare differences between two groups and one-way analysis of variance with a Student-Newman-Keuls post hoc test was used to compare differences among three or more groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-200c-3p, ERCC3 and ERCC4 in GC cell lines
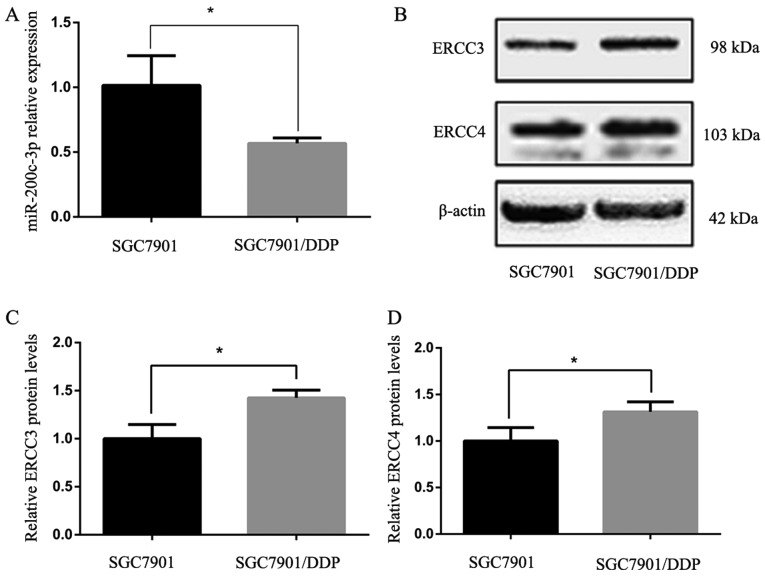
The expression levels of miR-200c-3p in drug-resistant SGC-7901/DDP cells were significantly lower than in the parent cell line SGC-7901 (P<0.05; Fig. 1A). Western blot analysis of ERCC3 and ERCC4 protein expression in SGC-7901 and SGC-7901/DDP cells indicated that ERCC3 and ERCC4 protein expression was significantly higher in SGC7901/DDP cells than in SGC7901 cells (P<0.05; Fig. 1B-D).
Figure 1.
Expression levels of miR-200c-3p, ERCC3 and ERCC4 in gastric cancer cell lines following lentiviral transduction. (A) Relative miR-200c-3p expression levels in SGC7901 cells and SGC7901/DDP cells, as measured by reverse transcription-quantitative polymerase chain reaction. The fold-change between SGC7901 cells and SGC7901/DDP cells was 1.788. (B) Western blot analysis detected higher ERCC3 and ERCC4 expression in SGC7901/DDP cells than in SGC7901 cells. (C and D) Semi-quantification of western blotting results shown in (B) via densitometry. *P<0.05. ERCC3, ERCC excision repair 3, TFIIH core complex helicase subunit; ERCC4, ERCC excision repair 4, endonuclease catalytic subunit; miR-200c-3p, microRNA-200c-3p.
Modifying miR-200c-3p expression levels with lentiviral vectors
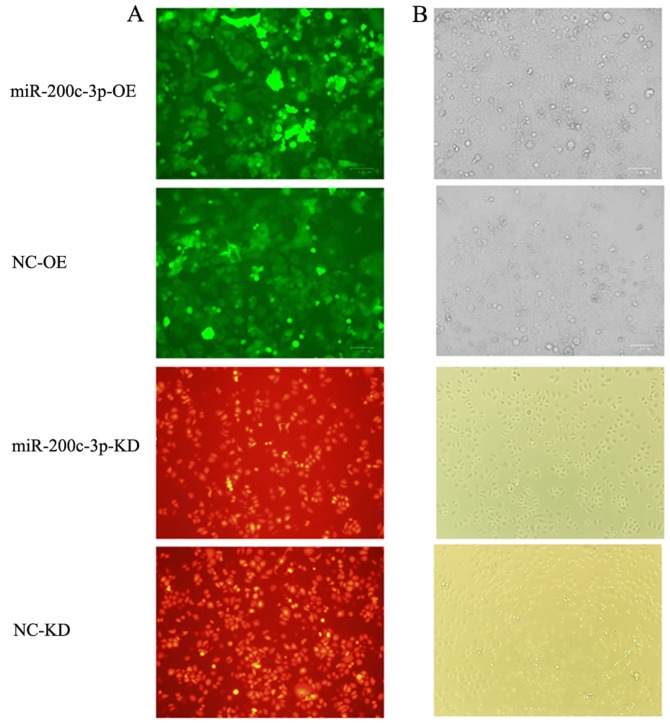
Fluorescence microscopy confirmed that SGC7901/DDP-LV-miR-200c-3p-OE cells with upregulated miR-200c-3p and SGC7901/DDP-LV-NC-OE cells transduced with an empty vector exhibited green fluorescence, whereas SGC7901-LV-miR-200c-3p-KD cells with downregulated miR-200c-3p and SGC7901-LV-NC-KD cells transduced with an empty vector exhibited red fluorescence (Fig. 2), suggesting that vectors carrying the target gene were successfully transduced.
Figure 2.
Transduction efficiency observed under a fluorescence microscope. Cells with miR-200c-3p OE and empty vector control cells fluoresced green. Cells with miR-200c-3p KD fluoresced red, indicating that vectors carrying the target gene were transfected into cells. (A) Fluorescence microscope images (magnification, ×100). (B) Optical microscope images (magnification, ×100). KD, knockdown; LV, lentivirus; miR-200c-3p, microRNA-200c-3p; NC, negative control; OE, overexpression.
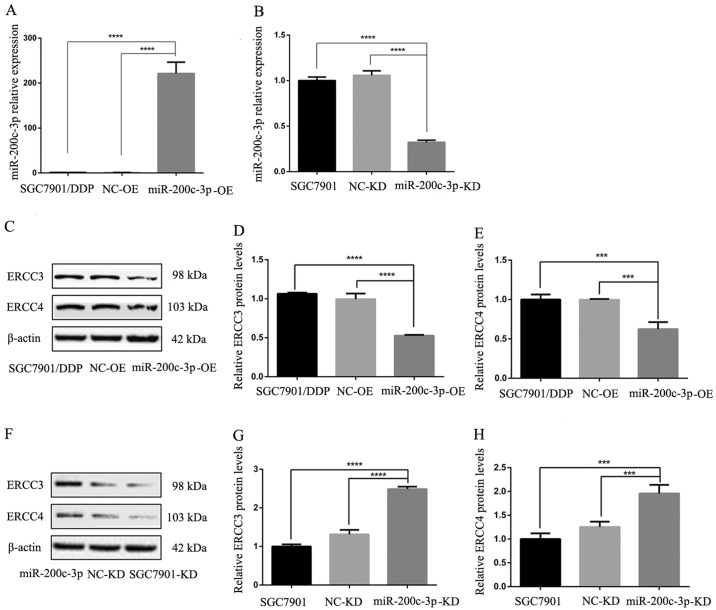
RT-qPCR confirmed that the expression levels of miR-200c-3p were significantly higher in SGC7901/DDP-LV-miR-200c-3p-OE cells compared with in SGC-7901/DDP and SGC7901/DDP-LV-NC-OE cells (P<0.0001; Fig. 3A). Following lentiviral transduction, miR-200c-3p was significantly downregulated in SGC7901/DDP-LV-miR-200c-3p-KD cells compared with in SGC7901 and SGC7901-LV-NC-KD cells (P<0.0001; Fig. 3B). Therefore, the expression levels of the target gene were altered in cells following lentiviral transduction.
Figure 3.
Reverse transcription-quantitative polymerase chain reaction revealed the expression levels of miR-200c-3p, ERCC3 and ERCC4 in cells following lentiviral transduction. ERCC3 and ERCC4 protein expression in gastric cancer cells was detected by western blot analysis. (A) Upregulation of miR-200c-3p in SGC7901/DDP-LV-miR-200c-3p-OE cells compared with in SGC7901/DDP and SGC7901/DDP-LV-NC-OE cells. (B) Downregulation of miR-200c-3p in SGC7901-LV-miR-200c-3p cells compared with in SGC7901 and SGC7901-LV-NC-KD cells. (C) Western blot analysis detected lower ERCC3 and ERCC4 expression in SGC7901/DDP-LV-miR-200c-3p-OE cells compared with in SGC7901/DDP and SGC7901/DDP-LV-NC-OE cells. (D) Relative expression levels of ERCC3 in each cell type. (E) Relative expression levels of ERCC4 in each cell type. (F) Western blot analysis detected higher ERCC3 and ERCC4 expression in SGC7901-LV-miR-200c-3p-KD cells compared with in SGC7901 and SGC7901-LV-NC-KD cells. (G) Relative expression levels of ERCC3 in each cell type. (H) Relative expression levels of ERCC4 in each cell type. ***P<0.001, ****P<0.0001. ERCC3, ERCC excision repair 3, TFIIH core complex helicase subunit; ERCC4, ERCC excision repair 4, endonuclease catalytic subunit; KD, knockdown; LV, lentivirus; miR-200c-3p, microRNA-200c-3p; NC, negative control; OE, overexpression.
ERCC3 and ERCC4 expression post-transduction
Following lentiviral infection of the cisplatin-resistant SGC7901/DDP GC cell line, ERCC3 and ERCC4 expression levels in SGC7901/DDP-LV-miR-200c-3p-OE cells were significantly lower than in SGC7901/DDP and SGC7901/DDP-LV-NC-OE cells (P<0.0001 and P<0.001, respectively; Fig. 3C-E). These findings indicated that ERCC3 and ERCC4 expression were decreased following upregulation of miR-200c-3p. Following lentiviral infection of SGC7901 GC cells, the expression levels of ERCC3 and ERCC4 were significantly increased in SGC7901-LV-miR-200c-3p-KD cells compared with in SGC7901 and SGC7901-LV-NC-KD cells (P<0.0001 and P<0.001, respectively; Fig. 3F-H). These results indicated that the expression levels of ERCC3 and ERCC4 were increased following miR-200c-3p knockdown; therefore, miR-200c-3p may inhibit ERCC3 and ERCC4 protein expression.
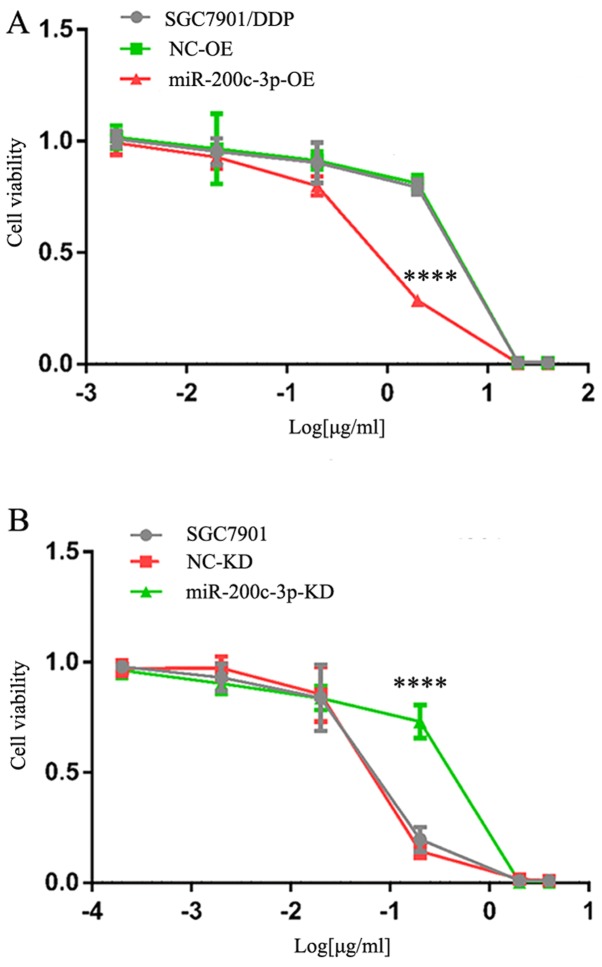
Alterations in cisplatin resistance
Cisplatin resistance was evaluated by calculating the IC50 value of DDP relative to untreated cells (Fig. 4). SGC7901/DDP-LV-miR-200c-3p-OE cells exhibited decreased cisplatin resistance compared with SGC7901/DDP and SGC7901/DDP-LV-NC-OE cells, and the differences were statistically significant (P<0.0001). These findings indicated that drug resistance was decreased following upregulation of miR-200c-3p. The IC50 for SGC7901-LV-miR-200c-3p-KD cells was greater than in SGC7901 and SGC7901-LV-NC-KD cells, and the differences were statistically significant (P<0.0001). These findings indicated that cisplatin resistance was increased following downregulation of miR-200c-3p. Therefore, upregulation of miR-200c-3p may reverse cisplatin resistance of SGC7901/DDP cells by inhibiting expression of the target proteins ERCC3 and ERCC4 in the NER pathway.
Figure 4.
Drug resistance of SGC7901/DDP cells following upregulation of miR-200c-3p and 48 h cisplatin treatment was determined using an MTT assay. (A) Cells were treated with 40, 20, 2, 0.2, 0.02 and 0.002 µg/ml cisplatin. IC50 of cisplatin was 7.63±0.25, 7.25±0.23 and 1.56±0.12 µg/ml in SGC7901/DDP, SGC7901/DDP-LV-NC-OE and SGC7901/DDP-LV-miR-200c-3p-OE cells. ****P<0.0001 vs. SGC7901/DDP and SGC7901/DDP-LV-NC-OE cells. (B) Cells were treated with 4, 2, 0.2, 0.02, 0.002 and 0.0002 µg/ml cisplatin. IC50 of cisplatin was 0.21±0.01, 0.20±0.003 and 0.59±0.04 µg/ml in SGC7901, SGC7901-LV-NC-KD and SGC7901-LV-miR-200c-3p-KD cells. ****P<0.0001 vs. SGC7901 and SGC7901-LV-NC-KD cells. IC50, half-maximal inhibitory concentration; KD, knockdown; LV, lentivirus; miR, microRNA; NC, negative control; OE, overexpression.
Discussion
miR-200c is a member of the miR-200 family, which is located on chromosome 12 (12p13.31). It has previously been reported that miR-200c is abnormally expressed in tumor tissues, and the majority of studies have suggested that low miR-200c is associated with poor prognosis (16–18). It is likely that miR-200c can inhibit the metastatic and invasive potential of tumors by inhibiting epithelial-mesenchymal transition (EMT) (19,20). Along with its important functions in EMT, tumor invasion and metastasis, its role in tumor drug resistance has been explored. Studies (21–23) describing multiple solid tumors have demonstrated that tubulin β3 (TUBB3) is involved in the regulation of sensitivity to microtubule-targeting chemotherapeutic drugs (e.g. paclitaxel and vincristine). miR-200c can directly target the 3′-UTR of TUBB3 mRNA to inhibit expression of TUBB3 at the post-transcriptional level (24), and thus increase the sensitivity of tumor cells to drugs, including paclitaxel and vincristine. Zhou et al (25) reported that miR-200c inhibited migration and invasion to reverses trastuzumab resistance by targeting zinc finger E-box binding homeobox (ZEB)1 and ZEB2. Several studies have indicated that miR-200c is associated with cisplatin resistance. The transcription factor AP-2α (AP-2) gene can increase sensitivity to cisplatin chemotherapy by promoting apoptosis. In the endometrium, miR-200b/200c/429 can bind to the 3′-UTR of the AP-2 gene, block protein expression and enhance cisplatin resistance (26). In esophageal cancer, miR-200c can inhibit expression of protein phosphatase 2 scaffold subunit Ab and promote activation of the downstream Akt pathway, which leads to cisplatin resistance (27). Chen et al (12) reported that drug resistance reversal and inhibition of proliferation by miR-200c in SGC7901/DDP cells is associated with induction of E-cadherin, PTEN and BAX protein expression, inhibition of Akt pathway activation, and downregulation of Bcl-2 protein expression. In addition, it has been reported that miR-200c can enhance the sensitivity of GC to cisplatin through direct targeted regulation of ZEB2 (28). Furthermore, Chang et al (29) demonstrated that miR-200c directly targets Rho family GTPase 3 (RhoE), and downgraded RhoE expression may enhance the sensitivity of SGC7901/DDP cells. Notably, these results suggested that miR-200c may serve a dual role in tumor chemosensitivity, which may be associated with the complex mechanisms of miR-200c. However, it has not been established whether miR-200c can regulate the NER pathway. Our previous study (30) demonstrated that miR-200c-3p is upregulated in tissues and plasma samples of patients with advanced GC, and is associated with improved efficacy of platinum-based chemotherapy and prognosis of patients. In order to further explore the mechanism by which miR-200c-3p promotes the sensitivity of platinum-containing chemotherapy in GC, the present study was conducted.
The present study revealed that miR-200c-3p was significantly downregulated in a drug-resistant SGC7901/DDP GC cell line compared with in the SGC7901 cell line, and that ERCC3 and ERCC4 in the NER pathway may be two targets of miR-200c-3p. Upregulation of miR-200c-3p decreased ERCC3 and ERCC4 expression in a SGC7901/DDP cell line and inhibited the NER pathway, decreasing cisplatin resistance. Downregulation of miR-200c-3p increased ERCC3 and ERCC4 expression in a SGC7901 GC cell line and may enhance the NER pathway, enhancing cisplatin resistance. These results suggested that upregulation of miR-200c-3p may inhibit ERCC3 and ERCC4 protein expression to interfere with the NER pathway in the SGC7901/DDP cell line, in order to reverse cisplatin resistance. ERCC3 and ERCC4 are important proteins in the NER pathway, and deletion or mutation of either can modify their activities (31–34). ERCC3 is located on human autosome 2q21 and encodes a protein with DNA helicase activity (35). Relying on the function of DNA ATPase and helicase, it can repair structure-distorting DNA damage and DNA around the RNAP II transcription promoter; therefore, it is an essential gene in the NER pathway, and in gene transcription generally. ERCC4 and ERCC excision repair 1, endonuclease non-catalytic subunit can form a heterodimer, which is a structure-specific endonuclease that can cut the DNA strand at the 5′-end of impaired DNA to facilitate removal of impaired DNA segments. However, the NER pathway involves multiple genes and each gene has a different function (36). In the present study, overexpression of miR-200c-3p only regulated two genes in the NER pathway, which would not completely inhibit the pathway. Our present study (37) indicated that ERCC1 might also be involved in the regulation of cisplatin resistance in gastric cancer. In addition, there are certain limitations of the present study. For example, only a single cell line (SGC7901/DDP) and a single drug (cisplatin) were investigated. The absence of direct binding luciferase assays to reveal the interaction between miR-200c-3p and ERCC3/4 was another limitation of the present study. In addition, no rescue experiments, such as upregulation or downregulation of ERCC3/4, were performed to confirm that miR-200c-3p reverses cisplatin resistance by regulating ERCC3/4. Therefore, further studies should be conducted to address these limitations and elucidate the detailed molecular mechanism.
In conclusion, to the best of our knowledge, the present study is the first to report that miR-200c-3p may reverse cisplatin resistance in a SGC7901/DDP GC cell line by inhibiting ERCC3 and ERCC4 protein expression, thus interfering with the NER pathway. This study provided novel insights into the mechanism underlying the regulatory effects of miR-200c-3p on cisplatin resistance.
Acknowledgements
Not applicable.
Funding
This study was supported by the Projects of Foreign Science and Technology Cooperation of Anhui Province (grant no. 1604b0602027).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
ML and KG conceived the hypothesis and study design. ML and MG wrote the manuscript and analyzed data. XX performed the experiments. YZ, JN and PL were involved in experimental design and gave important intellectual suggestions for the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Lordick F, Allum W, Carneiro F, Mitry E, Tabernero J, Tan P, Van Cutsem E, van de Velde C, Cervantes A. Unmet needs and challenges in gastric cancer: The way forward. Cancer Treat Rev. 2014;40:692–700. doi: 10.1016/j.ctrv.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Lordick F, Lorenzen S, Yamada Y, Ilson D. Optimal chemotherapy for advanced gastric cancer: Is there a global consensus? Gastric Cancer. 2014;17:213–225. doi: 10.1007/s10120-013-0297-z. [DOI] [PubMed] [Google Scholar]
- 4.Amable L. Cisplatin resistance and opportunities for precision medicine. Pharmacol Res. 2016;106:27–36. doi: 10.1016/j.phrs.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Basourakos SP, Li L, Aparicio AM, Corn PG, Kim J, Thompson TC. Combination platinum-based and DNA damage response-targeting cancer therapy: Evolution and future directions. Curr Med Chem. 2017;24:1586–1606. doi: 10.2174/0929867323666161214114948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69:1221–1229. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie QH, He XX, Chang Y, Sun SZ, Jiang X, Li PY, Lin JS. MiR-192 inhibits nucleotide excision repair by targeting ERCC3 and ERCC4 in HepG2.2.15 cells. Biochem Biophys Res Commun. 2011;410:440–445. doi: 10.1016/j.bbrc.2011.05.153. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Xiang Y, Ma N, Wang D, Zhang Y, Zhou J, Wu G, Zhao R, Huang H, Wang X, Qiao Y, et al. MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: A novel epigenetic therapy independent of decitabine. Oncogene. 2014;33:378–386. doi: 10.1038/onc.2012.575. [DOI] [PubMed] [Google Scholar]
- 10.Shang Y, Zhang Z, Liu Z, Feng B, Ren G, Li K, Zhou L, Sun Y, Li M, Zhou J, et al. miR-508-5p regulates multidrug resistance of gastric cancer by targeting ABCB1 and ZNRD1. Oncogene. 2014;33:3267–3276. doi: 10.1038/onc.2013.297. [DOI] [PubMed] [Google Scholar]
- 11.Fang L, Li H, Wang L, Hu J, Jin T, Wang J, Yang BB. MicroRNA-17-5p promotes chemotherapeutic drug resistance and tumor metastasis of colorectal cancer by repressing PTEN expression. Oncotarget. 2014;5:2974–2987. doi: 10.18632/oncotarget.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Zuo J, Liu Y, Gao H, Liu W. Inhibitory effects of miRNA-200c on chemotherapy-resistance and cell proliferation of gastric cancer SGC7901/DDP cells. Chin J Cancer. 2010;29:1006–1011. doi: 10.5732/cjc.010.10236. [DOI] [PubMed] [Google Scholar]
- 13.Hatano K, Kumar B, Zhang Y, Coulter JB, Hedayati M, Mears B, Ni X, Kudrolli TA, Chowdhury WH, Rodriguez R, et al. A functional screen identifies miRNAs that inhibit DNA repair and sensitize prostate cancer cells to ionizing radiation. Nucleic Acids Res. 2015;43:4075–4086. doi: 10.1093/nar/gkv273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu RL, Dong Y, Deng YZ, Wang WJ, Li WD. Tumor suppressor miR-145 reverses drug resistance by directly targeting DNA damage-related gene RAD18 in colorectal cancer. Tumour Biol. 2015;36:5011–5019. doi: 10.1007/s13277-015-3152-5. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Yu J, Ohuchida K, Mizumoto K, Sato N, Kayashima T, Fujita H, Nakata K, Tanaka M. MicroRNA, hsa-miR-200c, is an independent prognostic factor in pancreatic cancer and its upregulation inhibits pancreatic cancer invasion but increases cell proliferation. Mol Cancer. 2010;9:169. doi: 10.1186/1476-4598-9-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li RH, Chen M, Liu J, Shao CC, Guo CP, Wei XL, Li YC, Huang WH, Zhang GJ. Long noncoding RNA ATB promotes the epithelial-mesenchymal transition by upregulating the miR-200c/Twist1 axe and predicts poor prognosis in breast cancer. Cell Death Dis. 2018;9:1171. doi: 10.1038/s41419-018-1210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao W, Sun J. MicroRNA-200c promotes tumor cell proliferation and migration by directly targeting dachshund family transcription factor 1 by the Wnt/β-catenin signaling pathway in nasopharyngeal carcinoma. Anticancer Drugs. 2019;30:218–224. doi: 10.1097/CAD.0000000000000713. [DOI] [PubMed] [Google Scholar]
- 19.Chen ML, Liang LS, Wang XK. miR-200c inhibits invasion and migration in human colon cancer cells SW480/620 by targeting ZEB1. Clin Exp Metastasis. 2012;29:457–469. doi: 10.1007/s10585-012-9463-7. [DOI] [PubMed] [Google Scholar]
- 20.Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, Calin G, Wang H, Siefker-Radtke A, McConkey D, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15:5060–5072. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang HL, Ruan L, Zheng LM, Whyte D, Tzeng CM, Zhou XW. Association between class III β-tubulin expression and response to paclitaxel/vinorebine-based chemotherapy for non-small cell lung cancer: A meta-analysis. Lung Cancer. 2012;77:9–15. doi: 10.1016/j.lungcan.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 22.He W, Zhang D, Jiang J, Liu P, Wu C. The relationships between the chemosensitivity of human gastric cancer to paclitaxel and the expressions of class III β-tubulin, MAPT, and survivin. Med Oncol. 2014;31:950. doi: 10.1007/s12032-014-0950-3. [DOI] [PubMed] [Google Scholar]
- 23.Duran GE, Wang YC, Moisan F, Francisco EB, Sikic BI. Decreased levels of baseline and drug-induced tubulin polymerisation are hallmarks of resistance to taxanes in ovarian cancer cells and are associated with epithelial-to-mesenchymal transition. Br J Cancer. 2017;116:1318–1328. doi: 10.1038/bjc.2017.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cochrane DR, Spoelstra NS, Howe EN, Nordeen SK, Richer JK. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol Cancer Ther. 2009;8:1055–1066. doi: 10.1158/1535-7163.MCT-08-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X, Men X, Zhao R, Han J, Fan Z, Wang Y, Lv Y, Zuo J, Zhao L, Sang M, et al. miR-200c inhibits TGF-β-induced-EMT to restore trastuzumab sensitivity by targeting ZEB1 and ZEB2 in gastric cancer. Cancer Gene Ther. 2018;25:68–76. doi: 10.1038/s41417-017-0005-y. [DOI] [PubMed] [Google Scholar]
- 26.Wu Y, Xiao Y, Ding X, Zhuo Y, Ren P, Zhou C, Zhou J. A miR-200b/200c/429-binding site polymorphism in the 3′ untranslated region of the AP-2α gene is associated with cisplatin resistance. PLoS One. 2011;6:e29043. doi: 10.1371/journal.pone.0029043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamano R, Miyata H, Yamasaki M, Kurokawa Y, Hara J, Moon JH, Nakajima K, Takiguchi S, Fujiwara Y, Mori M, Doki Y. Overexpression of miR-200c induces chemoresistance in esophageal cancers mediated through activation of the Akt signaling pathway. Clin Cancer Res. 2011;17:3029–3038. doi: 10.1158/1078-0432.CCR-10-2532. [DOI] [PubMed] [Google Scholar]
- 28.Jiang T, Dong P, Li L, Ma X, Xu P, Zhu H, Wang Y, Yang B, Liu K, Liu J, et al. MicroRNA-200c regulates cisplatin resistance by targeting ZEB2 in human gastric cancer cells. Oncol Rep. 2017;38:151–158. doi: 10.3892/or.2017.5659. [DOI] [PubMed] [Google Scholar]
- 29.Chang L, Guo F, Wang Y, Lv Y, Huo B, Wang L, Liu W. MicroRNA-200c regulates the sensitivity of chemotherapy of gastric cancer SGC7901/DDP cells by directly targeting RhoE. Pathol Oncol Res. 2014;20:93–98. doi: 10.1007/s12253-013-9664-7. [DOI] [PubMed] [Google Scholar]
- 30.Li M, Gu K, Liu W, Xie X, Huang X. MicroRNA-200c as a prognostic and sensitivity marker for platinum chemotherapy in advanced gastric cancer. Oncotarget. 2017;8:51190–51199. doi: 10.18632/oncotarget.17087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang JR, Moncollin V, Vermeulen W, Seroz T, van Vuuren H, Hoeijmakers JH, Egly JM. A 3′->5′ XPB helicase defect in repair/transcription factor TFIIH of xeroderma pigmentosum group B affects both DNA repair and transcription. J Biol Chem. 1996;271:15898–15904. doi: 10.1074/jbc.271.27.15898. [DOI] [PubMed] [Google Scholar]
- 32.Lankoff A, Sochacki J, Spoof L, Meriluoto J, Wojcik A, Wegierek A, Verschaeve L. Nucleotide excision repair impairment by nodularin in CHO cell lines due to ERCC1/XPF inactivation. Toxicol Lett. 2008;179:101–107. doi: 10.1016/j.toxlet.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 33.McDaniel LD, Schultz RA. XPF/ERCC4 and ERCC1: Their products and biological roles. Adv Exp Med Biol. 2008;637:65–82. doi: 10.1007/978-0-387-09599-8_8. [DOI] [PubMed] [Google Scholar]
- 34.Andressoo JO, Weeda G, de Wit J, Mitchell JR, Beems RB, van Steeg H, van der Horst GT, Hoeijmakers JH. An Xpb mouse model for combined xeroderma pigmentosum and cockayne syndrome reveals progeroid features upon further attenuation of DNA repair. Mol Cell Biol. 2009;29:1276–1290. doi: 10.1128/MCB.01229-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poterszman A, Lamour V, Egly JM, Moras D, Thierry JC, Poch O. A eukaryotic XPB/ERCC3-like helicase in Mycobacterium leprae? Trends Biochem Sci. 1997;22:418–419. doi: 10.1016/S0968-0004(97)01124-9. [DOI] [PubMed] [Google Scholar]
- 36.Friedberg EC. How nucleotide excision repair protects against cancer. Nat Rev Cancer. 2001;1:22–33. doi: 10.1038/35094000. [DOI] [PubMed] [Google Scholar]
- 37.Ning J, Jiao Y, Xie X, Deng X, Zhang Y, Yang Y, Zhao C, Wang H, Gu K. miR-138-5p modulates the expression of excision repair cross-complementing proteins ERCC1 and ERCC4 and regulates the sensitivity of gastric cancer cells to cisplatin. Oncol Rep. 2019;41:1131–1139. doi: 10.3892/or.2018.6907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.